Abstract
Pseudomonas aeruginosa is an important pathogen commonly implicated in nosocomial infections. The occurrence of multidrug-resistant (MDR) P. aeruginosa strains is increasing worldwide and limiting our therapeutic options. The MDR phenotype can be mediated by a variety of resistance mechanisms, and the corresponding relative biofitness is not well established. We examined the prevalence, resistance mechanisms, and susceptibility of MDR P. aeruginosa isolates (resistant to ≥3 classes of antipseudomonal agents [penicillins/cephalosporins, carbapenems, quinolones, and aminoglycosides]) obtained from a large, university-affiliated hospital. Among 235 nonrepeat bloodstream isolates screened between 2005 and 2007, 33 isolates (from 20 unique patients) were found to be MDR (crude prevalence rate, 14%). All isolates were resistant to carbapenems and quinolones, 91% were resistant to penicillins/cephalosporins, and 21% were resistant to the aminoglycosides. By using the first available isolate for each bacteremia episode (n = 18), 13 distinct clones were revealed by repetitive-element-based PCR. Western blotting revealed eight isolates (44%) to have MexB overexpression. Production of a carbapenemase (VIM-2) was found in one isolate, and mutations in gyrA (T83I) and parC (S87L) were commonly found. Growth rates of most MDR isolates were similar to that of the wild type, and two isolates (11%) were found to be hypermutable. All available isolates were susceptible to polymyxin B, and only one isolate was nonsusceptible to colistin (MIC, 3 mg/liter), but all isolates were nonsusceptible to doripenem (MIC, >2 mg/liter). Understanding and continuous monitoring of the prevalence and resistance mechanisms of MDR P. aeruginosa would enable us to formulate rational treatment strategies to combat nosocomial infections.
Pseudomonas aeruginosa is an important pathogen commonly implicated in serious nosocomial infections such as pneumonia and sepsis. The occurrence of multidrug-resistant P. aeruginosa strains is increasing worldwide and limiting our therapeutic options. Resistance in P. aeruginosa may be mediated via several distinct mechanisms (3, 13). In multidrug-resistant isolates, the relative contributions of different molecular mechanisms toward phenotypic multidrug resistance are not well established. While there are multiple surveillance studies tracking the resistance of P. aeruginosa to various antimicrobial agents over the years (6, 7), studies reporting trends in concomitant resistance to multiple agents over time are scarce.
A hypermutable state is often observed in multidrug-resistant pathogens causing chronic infections in cystic fibrosis patients. It is often related to defects in genes involved in the mismatch repair system (e.g., mutS and mutL). It is also commonly believed that in multidrug-resistant P. aeruginosa isolates, reduced virulence may result due to decreased biofitness. However, recent data suggest otherwise, and multidrug-resistant P. aeruginosa may remain fully pathogenic (8).
Many first-line agents are ineffective if used alone against multidrug-resistant P. aeruginosa isolates. Consequently, unconventional agents or those that are less preferred (due to toxicity concerns) may have to be used. The objectives of this study were to examine (i) the prevalence of multidrug-resistant P. aeruginosa, (ii) the mechanism and biofitness cost of multidrug resistance, and (iii) the susceptibilities of multidrug-resistant P. aeruginosa isolates to polymyxin antibiotics and a newer carbapenem (doripenem). These investigations are expected to provide a rationale for effective treatment strategies to combat nosocomial infections.
(This study was presented in part at the 48th Interscience Conference on Antimicrobial Agents and Chemotherapy-Infectious Diseases Society of America 46th Annual Meeting, Washington, DC, 25 to 28 October 2008 [26].)
MATERIALS AND METHODS
Study site.
St. Luke's Episcopal Hospital is a 900-bed university-affiliated teaching hospital in Houston, TX. There are more than 120 intensive care unit beds, and more than 100 cardiothoracic surgeries are performed in the hospital each month. This study was approved by the institutional review boards of the hospital and the University of Houston.
Bacteria and multidrug resistance.
All nonrepeat bloodstream isolates of P. aeruginosa (collected >1 day apart) from January 2005 to December 2007 and their susceptibility profiles were obtained from the clinical microbiology laboratory. Multidrug-resistant isolates were defined as those resistant to three or more classes of antipseudomonal agents (i.e., penicillins/cephalosporins, carbapenems, fluoroquinolones, and aminoglycosides). If more than one agent within a class were tested (e.g., imipenem and meropenem were both tested), isolates must have been found to be resistant to all agents tested to be identified as resistant to that class. No isolate was obtained from patients with cystic fibrosis.
Genotyping analysis.
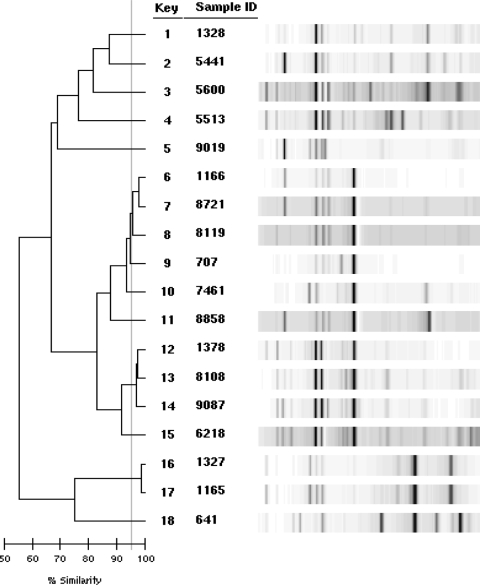
The clonal relatedness of the multidrug-resistant isolates was assessed using repetitive-element-based PCR (rep-PCR) (22, 24, 25). Genomic DNA was extracted using the UltraClean microbial DNA isolation kit according to the instructions of the manufacturer (Bacterial Barcodes, Inc., Athens, GA). Fifty nanograms of DNA was used as a template with the Pseudomonas fingerprinting kit (Bacterial Barcodes, Inc.). The DNA fragment patterns were separated by the Agilent 2100 bioanalyzer (Agilent Technologies, Inc., Santa Clara, CA), and comparisons were made both by visual examination and by analysis with the DiversiLab software using the Pearson correlation coefficient (Bacterial Barcodes, Inc.). The isolates were considered to be indistinguishable (having no difference in bands upon visual inspection, with similarity of the DNA fragment patterns of ≥99%), related (having one or two bands different, with pattern similarity of 95 to 98.9%), or distinct (having three or more bands different, with pattern similarity of <95%), as recommended by the software manufacturer.
Efflux pump overexpression.
Efflux pump (MexAB-OprM) overexpression was initially screened for by testing susceptibility to levofloxacin with and without an efflux pump inhibitor (MC-207110) (15); overexpression was subsequently confirmed by Western blotting of total membrane proteins (with total protein adjusted to 50 μg) using MexB-specific antibodies (16). Blots were developed using secondary rabbit immunoglobulins conjugated with horseradish peroxidase and the Immun-Star substrate kit (Bio-Rad, Hercules, CA). Strains PAO1 (the wild type) and PAO1 ΔmexR (a strain with known MexAB-OprM overexpression due to deletion of the MexR repressor gene regulating mexAB and oprM expression) were used as negative and positive controls, respectively (23). In view of the considerable proportion of isolates susceptible to aminoglycosides, the prevalence of MexXY-OprM overexpression was not thought to be high. Consequently, investigations to detect MexXY-OprM overexpression were not pursued.
Presence of metallo-beta-lactamase.
The presence of a carbapenemase was initially screened for by testing susceptibility to imipenem, with and without EDTA, using Etest according to the instructions of the manufacturer (AB Biodisk, Piscataway, NJ) (5). Subsequently, the presence of a carbapenemase was ascertained by a spectrophotometric (accelerated-hydrolysis) assay using 750 μl of imipenem (150 μM) in 100 μM phosphate buffer (pH 7.0) (12). The absorbance at 299 nm was measured at room temperature for 20 min after the addition of cell lysate (200 μl) to imipenem in buffer. The rate of hydrolysis was adjusted with respect to the total protein content. The inhibition of enzymatic activity was examined by measuring residual activity after preincubation of the cell lysate with EDTA (2 mM) for 20 min at 25°C. A standard wild-type strain, P. aeruginosa ATCC 27853 (American Type Culture Collection, Manassas, VA), and a clinical isolate harboring VIM-2 were used as negative and positive controls, respectively (14). Detection of the blaVIM gene by PCR was performed using forward (5′-ATCATGGCTATTGCGAGTCC) and reverse (5′-ACGACTGAGCGATTTGTGTG) primers, and comparison with the metallo-beta-lactamase (VIM-2) gene sequence with GenBank accession number AF191564 was made.
Point mutations in topoisomerases.
The quinolone resistance-determining regions (QRDR) of gyrA and parC were amplified by PCR and sequenced (2, 18). The corresponding sequences in PAO1 (GenBank accession numbers L29417 and AB003428) were used for comparison.
Hypermutable state.
To determine the mutation frequencies for the multidrug-resistant isolates, overnight cultures in cation-adjusted Mueller-Hinton II broth (BBL, Sparks, MD) were serially diluted 10-fold and plated quantitatively onto drug-free Mueller-Hinton II agar (MHA; BBL) and onto MHA supplemented with rifampin (300 mg/liter; Sigma-Aldrich, St. Louis, MO). Each isolate was tested at least twice on different days. A hypermutable state was defined by a mutation frequency >20 times that for PAO1 (17). PAO1 and PAO1 ΔmutS (a known hypermutable strain, a gift from Ryan Cirz and Floyd Romesberg) were used as negative and positive controls, respectively.
Growth rate determination.
To assess the in vitro growth rates of various multidrug-resistant isolates, a time-growth study was used. Briefly, a log-phase culture was diluted according to absorbance at 630 nm to approximately 105 CFU/ml in 1/4-strength Mueller-Hinton II broth (BBL). Samples (500 μl) were obtained in triplicate at the baseline and at 1, 2, 3, 4, 6, 8, and 24 h for quantitative culture. By fitting a growth model with a time lag term to the bacterial burden observed over time (29), the rate of growth was determined by the parameter estimate of the best-fit model.
Susceptibility testing.
Susceptibilities of multidrug-resistant isolates to polymyxin B, colistin, and doripenem were determined using Etest according to the instructions of the manufacturer (AB Biodisk, Piscataway, NJ) (5). P. aeruginosa ATCC 27853 was used as the control.
RESULTS
Bacteria and clonality.
Among 235 nonrepeat P. aeruginosa isolates screened, a total of 33 isolates (14.0%) from 20 patients were multidrug resistant. The crude multidrug resistance prevalence rates were 9 of 66 (13.6%) in 2005, 8 of 77 (10.4%) in 2006, and 16 of 92 (17.2%) in 2007. All the multidrug-resistant isolates were resistant to the carbapenems and quinolones tested, 91% were resistant to penicillins/cephalosporins, and 21% were resistant to the aminoglycosides. By using the first available isolate for each bacteremia episode (n = 18), 13 distinct clones were identified (Fig. 1 and Table 1).
FIG. 1.
DNA fingerprinting of the first multidrug-resistant P. aeruginosa isolates from 18 unique patients.
TABLE 1.
Point mutations detected, relative MexB expression, growth rates, and susceptibilities of multidrug-resistant isolatesa
| Isolate | Clone | MexB overexpression | Point mutation detected in: |
Growth rate (h−1) | MIC (μg/ml) of: |
|||
|---|---|---|---|---|---|---|---|---|
| gyrA | parC | Polymyxin B | Colistin | Doripenem | ||||
| PA27853 | 0.864 | 1.5 | 1.5 | 0.19 | ||||
| PA5441 | 2 | T83I | S87L | 0.935 | 1.5 | 1.5 | 4 | |
| PA5513 | 4 | T83I | S87L | 1.072 | 1.0 | 1.0 | >32 | |
| PA5600 | 3 | T83I | S87L | 0.944 | 1.5 | 1.0 | 4 | |
| PA6218 | 11 | + | D87N | 1.119 | 1.5 | 3.0 | >32 | |
| PA7461 | 8 | T83I | S87L | 1.146 | 2.0 | 2.0 | 12 | |
| PA8108 | 10 | + | T83I | 0.664 | 0.25 | 0.25 | >32 | |
| PA8119 | 6 | T83I | S87L | 0.723 | 1.5 | 1.5 | 16 | |
| PA8721 | 6 | T83I | S87L | 0.723 | 1.5 | 1.5 | 32 | |
| PA8858 | 9 | T83I | S87L | 0.844 | 2.0 | 1.5 | 8 | |
| PA9019 | 5 | D87G | S87L | 0.658 | 1.0 | 1.5 | >32 | |
| PA9087 | 10 | + | T83I | 0.469 | 0.5 | 0.38 | 6 | |
| PA641 | 13 | ND | P105T | 1.050 | 1.0 | 1.5 | >32 | |
| PA707 | 7 | + | T83I | S87L | 0.929 | 0.75 | 1.0 | >32 |
| PA1165 | 12 | + | T83I | S87L | 1.300 | 1.5 | 1.5 | 12 |
| PA1166 | 6 | T83I | S87L | 1.212 | 0.75 | 1.0 | 24 | |
| PA1327 | 12 | + | T83I | S87L | 1.423 | 1.0 | 2.0 | 8 |
| PA1328 | 1 | + | T83I | S87L | 1.029 | 1.5 | 1.5 | 3 |
| PA1378 | 10 | + | T83I | 0.908 | 1.5 | 2.0 | >32 | |
ND, not determined. Boldface type indicates nonsusceptibility based on CLSI (polymyxin)/FDA (doripenem) standards.
Mechanisms of resistance.
MexB overexpression was found in 8 of the 18 first-available isolates (Table 1). Significant reversal of imipenem susceptibility (≥8-fold reduction in the MIC) in the presence of EDTA was detected for one isolate (PA641). Rapid hydrolysis of imipenem and rate reduction in the presence of EDTA were also demonstrated. Subsequent PCR analysis revealed a sequence identical to that of VIM-2 (21). Point mutations resulting in amino acid changes were commonly found in the QRDR of gyrA (T83I, D87N, and D87G) and parC (S87L and P105T), as shown in Table 1.
Hypermutable state and growth rates.
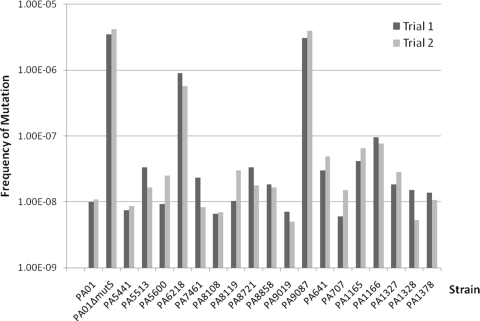
The mutation frequency for PAO1 was approximately 1 × 10−8, and that for PAO1 ΔmutS was considerably higher (approximately 5 × 10−5), as anticipated (17, 20). The mutation frequencies for all the multidrug-resistant isolates are shown in Fig. 2. Two multidrug-resistant isolates (PA6218 and PA9087) were found to be hypermutable. The in vitro growth rates of most of the multidrug-resistant isolates (15 of 18) were not found to be considerably different from that of the ATCC wild type (Table 1).
FIG. 2.
Mutation frequencies for various multidrug-resistant isolates. The mutation frequency for each isolate was determined in duplicate.
Susceptibilities to other agents.
Twenty-six multidrug-resistant isolates were available for susceptibility testing (selected isolates are listed in Table 1). All the isolates tested were susceptible to polymyxin B (MIC50, 1.5 mg/liter; MIC90, 2 mg/liter), and only one (PA6218) was nonsusceptible to colistin (MIC, 3 mg/liter). However, all the multidrug-resistant isolates examined were nonsusceptible (defined by a MIC of >2 mg/liter) to doripenem (MIC50, 12 mg/liter; MIC90, >32 mg/liter). Based on our previous investigations, the most common mechanism of imipenem/meropenem resistance in our hospital was reduced OprD expression (25). We suspected this would also be the mechanism of doripenem resistance.
DISCUSSION
Multidrug resistance in many nosocomial pathogens is on the rise, rendering many antimicrobial agents ineffective. A previous report indicated a rising trend of multidrug-resistant P. aeruginosa isolates being obtained from intensive care units in the United States (19); the prevalence rate was close to 15% in 2001 and 2002. The overall crude prevalence of multidrug-resistant bloodstream P. aeruginosa isolates in our institution from 2005 to 2007 was 14% (11.4% when calculated using the first isolate from each episode [≤30 days of index culture] only). Examining the prevalence for each year, we found a slight upward trend. However, a longer study would be necessary to ascertain this annual trend over time.
The multidrug-resistant phenotype may be mediated by more than one molecular mechanism. For example, in P. aeruginosa, multidrug resistance may be due to overexpression of a multidrug efflux pump(s) or be mediated via sequential accumulation of mutations each coding for resistance to one class of antimicrobial agents. In many surveillance studies, the specific mechanism of multidrug resistance is not thoroughly investigated. By using an efflux pump inhibitor, Mex pump overexpression was found previously in 56 to 86% of P. aeruginosa isolates resistant to both ciprofloxacin and a beta-lactam agent (9). These results were consistent with our observations. In this study, we found that 44% of the multidrug-resistant isolates had MexAB-OprM overexpression. The slightly lower prevalence may have been the result of a difference(s) in study design. Instead of using a nonspecific phenotypic method, we confirmed the overexpression of a specific (the most common) pump subtype. In addition, we also used more stringent criteria in defining the multidrug-resistant phenotype.
Metallo-beta-lactamases have been discovered previously in clinical isolates of P. aeruginosa in Texas. Isolates harboring blaVIM-2 and blaVIM-7 were reported by a neighboring cancer center within the Texas Medical Center (1, 28). These isolates were also multidrug resistant, but they were not available to ascertain if the isolate found in the present study to harbor a metallo-beta-lactamase (isolate PA641) was related to those from the neighboring cancer center. Of note, the patient from which PA641 was isolated did not have a medical history of cancer. To the best of our knowledge, this is the first report of a metallo-beta-lactamase in a clinical isolate of P. aeruginosa obtained in a facility other than a cancer center in Texas. The sequence of the blaVIM gene amplified using the internal primers listed above was identical to at least 94% of the published sequence of the blaVIM-2 gene. All the multidrug-resistant isolates were resistant to the fluoroquinolones. Mutations in the QRDR of gyrA and parC were common and consistent with previous findings (18). In addition, reduced biofitness (as assessed by in vitro growth rates) was observed only in a few isolates, as noted previously (10). The relationship between in vitro growth rates and in vivo virulence was not the focus of this study. While the data are compelling overall, it should be noted that the presence of various genetic mutations may or may not directly account for resistance patterns seen in a given isolate, unless the association is further confirmed with genetic complementation or knockout studies.
Slightly more than half of our multidrug-resistant isolates were not found to have efflux pump (MexAB-OprM) overexpression. Since hypermutation in P. aeruginosa is believed to be associated with resistance to multiple antimicrobial agents (20), we further explored the prevalence of a hypermutable state in these isolates. Among isolates obtained previously from cystic fibrosis patients, hypermutable isolates (54.4%) were commonly found (4); in contrast, only 2 of 18 multidrug-resistant isolates (11.1%) examined in the present study were found to be hypermutable. Furthermore, the in vitro growth rates of these hypermutable isolates were not consistently reduced, as reported previously (17). Taken as a whole, these results imply that additional environmental factors (e.g., oxidative stress) are likely to be involved in the pathogenesis of chronic infections in cystic fibrosis patients. Molecular investigations delineating the molecular mechanism(s) associated with hypermutation in our multidrug-resistant isolates are ongoing.
In view of a lack of new and effective agents against pathogens with the multidrug-resistant phenotype, alternative (even less preferred second-line) antimicrobial agents must be used clinically as a last resort. Based on the CLSI susceptibility interpretation criteria, almost all the multidrug-resistant isolates were susceptible to the polymyxins (polymyxin B and colistin; MICs, ≤2 mg/liter). However, all the isolates were nonsusceptible to doripenem according to the FDA susceptibility interpretation criteria (MIC, >2 mg/liter). Clinical isolates of P. aeruginosa with reduced susceptibility to the polymyxins have been reported previously (11, 27). As we are faced with an increasing demand to use these agents, it is important to monitor the susceptibilities of clinically important pathogens to these agents over time. The impact of multidrug resistance on clinical outcomes is currently under investigation.
In summary, the crude prevalence of multidrug-resistant P. aeruginosa bloodstream isolates was 14%; not all of these isolates had multidrug resistance due to the overexpression of an efflux pump. No significant biofitness cost was detected, and there was a low prevalence of hypermutation in these isolates. Understanding and continuous monitoring of the prevalence and resistance mechanisms of multidrug-resistant P. aeruginosa would enable us to formulate rational treatment strategies to combat nosocomial infections.
Acknowledgments
This study was conducted with support from the investigator-sponsored study program of AstraZeneca and the University of Houston College of Pharmacy summer student research program.
We thank Ryan Cirz and Floyd Romesberg for their control bacterial strains.
Footnotes
Published ahead of print on 19 January 2010.
REFERENCES
- 1.Aboufaycal, H., H. S. Sader, K. Rolston, L. M. Deshpande, M. Toleman, G. Bodey, I. Raad, and R. N. Jones. 2007. blaVIM-2 and blaVIM-7 carbapenemase-producing Pseudomonas aeruginosa isolates detected in a tertiary care medical center in the United States: report from the MYSTIC program. J. Clin. Microbiol. 45:614-615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akasaka, T., M. Tanaka, A. Yamaguchi, and K. Sato. 2001. Type II topoisomerase mutations in fluoroquinolone-resistant clinical strains of Pseudomonas aeruginosa isolated in 1998 and 1999: role of target enzyme in mechanism of fluoroquinolone resistance. Antimicrob. Agents Chemother. 45:2263-2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonomo, R. A., and D. Szabo. 2006. Mechanisms of multidrug resistance in Acinetobacter species and Pseudomonas aeruginosa. Clin. Infect. Dis. 43(Suppl. 2):S49-S56. [DOI] [PubMed] [Google Scholar]
- 4.Ciofu, O., B. Riis, T. Pressler, H. E. Poulsen, and N. Hoiby. 2005. Occurrence of hypermutable Pseudomonas aeruginosa in cystic fibrosis patients is associated with the oxidative stress caused by chronic lung inflammation. Antimicrob. Agents Chemother. 49:2276-2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clinical and Laboratory Standards Institute. 2007. Performance standards for antimicrobial susceptibility testing: seventeenth informational supplement. CLSI document M100-S17. Clinical and Laboratory Standards Institute, Wayne, PA.
- 6.Gales, A. C., H. H. Sader, and R. N. Jones. 2002. Respiratory tract pathogens isolated from patients hospitalized with suspected pneumonia in Latin America: frequency of occurrence and antimicrobial susceptibility profile: results from the SENTRY Antimicrobial Surveillance Program (1997-2000). Diagn. Microbiol. Infect. Dis. 44:301-311. [DOI] [PubMed] [Google Scholar]
- 7.Gaynes, R., and J. R. Edwards. 2005. Overview of nosocomial infections caused by gram-negative bacilli. Clin. Infect. Dis. 41:848-854. [DOI] [PubMed] [Google Scholar]
- 8.Hocquet, D., P. Berthelot, M. Roussel-Delvallez, R. Favre, K. Jeannot, O. Bajolet, N. Marty, F. Grattard, P. Mariani-Kurkdjian, E. Bingen, M. O. Husson, G. Couetdic, and P. Plesiat. 2007. Pseudomonas aeruginosa may accumulate drug resistance mechanisms without losing its ability to cause bloodstream infections. Antimicrob. Agents Chemother. 51:3531-3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kriengkauykiat, J., E. Porter, O. Lomovskaya, and A. Wong-Beringer. 2005. Use of an efflux pump inhibitor to determine the prevalence of efflux pump-mediated fluoroquinolone resistance and multidrug resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 49:565-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kugelberg, E., S. Lofmark, B. Wretlind, and D. I. Andersson. 2005. Reduction of the fitness burden of quinolone resistance in Pseudomonas aeruginosa. J. Antimicrob. Chemother. 55:22-30. [DOI] [PubMed] [Google Scholar]
- 11.Landman, D., S. Bratu, M. Alam, and J. Quale. 2005. Citywide emergence of Pseudomonas aeruginosa strains with reduced susceptibility to polymyxin B. J. Antimicrob. Chemother. 55:954-957. [DOI] [PubMed] [Google Scholar]
- 12.Lauretti, L., M. L. Riccio, A. Mazzariol, G. Cornaglia, G. Amicosante, R. Fontana, and G. M. Rossolini. 1999. Cloning and characterization of blaVIM, a new integron-borne metallo-β-lactamase gene from a Pseudomonas aeruginosa clinical isolate. Antimicrob. Agents Chemother. 43:1584-1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Livermore, D. M. 2002. Multiple mechanisms of antimicrobial resistance in Pseudomonas aeruginosa: our worst nightmare? Clin. Infect. Dis. 34:634-640. [DOI] [PubMed] [Google Scholar]
- 14.Lolans, K., A. M. Queenan, K. Bush, A. Sahud, and J. P. Quinn. 2005. First nosocomial outbreak of Pseudomonas aeruginosa producing an integron-borne metallo-β-lactamase (VIM-2) in the United States. Antimicrob. Agents Chemother. 49:3538-3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lomovskaya, O., M. S. Warren, A. Lee, J. Galazzo, R. Fronko, M. Lee, J. Blais, D. Cho, S. Chamberland, T. Renau, R. Leger, S. Hecker, W. Watkins, K. Hoshino, H. Ishida, and V. J. Lee. 2001. Identification and characterization of inhibitors of multidrug resistance efflux pumps in Pseudomonas aeruginosa: novel agents for combination therapy. Antimicrob. Agents Chemother. 45:105-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Middlemiss, J. K., and K. Poole. 2004. Differential impact of MexB mutations on substrate selectivity of the MexAB-OprM multidrug efflux pump of Pseudomonas aeruginosa. J. Bacteriol. 186:1258-1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Montanari, S., A. Oliver, P. Salerno, A. Mena, G. Bertoni, B. Tummler, L. Cariani, M. Conese, G. Doring, and A. Bragonzi. 2007. Biological cost of hypermutation in Pseudomonas aeruginosa strains from patients with cystic fibrosis. Microbiology 153:1445-1454. [DOI] [PubMed] [Google Scholar]
- 18.Mouneimne, H., J. Robert, V. Jarlier, and E. Cambau. 1999. Type II topoisomerase mutations in ciprofloxacin-resistant strains of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 43:62-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Obritsch, M. D., D. N. Fish, R. MacLaren, and R. Jung. 2004. National surveillance of antimicrobial resistance in Pseudomonas aeruginosa isolates obtained from intensive care unit patients from 1993 to 2002. Antimicrob. Agents Chemother. 48:4606-4610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oliver, A., B. R. Levin, C. Juan, F. Baquero, and J. Blazquez. 2004. Hypermutation and the preexistence of antibiotic-resistant Pseudomonas aeruginosa mutants: implications for susceptibility testing and treatment of chronic infections. Antimicrob. Agents Chemother. 48:4226-4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poirel, L., T. Naas, D. Nicolas, L. Collet, S. Bellais, J. D. Cavallo, and P. Nordmann. 2000. Characterization of VIM-2, a carbapenem-hydrolyzing metallo-β-lactamase and its plasmid- and integron-borne gene from a Pseudomonas aeruginosa clinical isolate in France. Antimicrob. Agents Chemother. 44:891-897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saeed, S., M. G. Fakih, K. Riederer, A. R. Shah, and R. Khatib. 2006. Interinstitutional and intrainstitutional transmission of a strain of Acinetobacter baumannii detected by molecular analysis: comparison of pulsed-field gel electrophoresis and repetitive sequence-based polymerase chain reaction. Infect. Control Hosp. Epidemiol. 27:981-983. [DOI] [PubMed] [Google Scholar]
- 23.Srikumar, R., C. J. Paul, and K. Poole. 2000. Influence of mutations in the mexR repressor gene on expression of the MexA-MexB-OprM multidrug efflux system of Pseudomonas aeruginosa. J. Bacteriol. 182:1410-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Syrmis, M. W., M. R. O'Carroll, T. P. Sloots, C. Coulter, C. E. Wainwright, S. C. Bell, and M. D. Nissen. 2004. Rapid genotyping of Pseudomonas aeruginosa isolates harboured by adult and paediatric patients with cystic fibrosis using repetitive-element-based PCR assays. J. Med. Microbiol. 53:1089-1096. [DOI] [PubMed] [Google Scholar]
- 25.Tam, V. H., K. T. Chang, M. T. LaRocco, A. N. Schilling, S. K. McCauley, K. Poole, and K. W. Garey. 2007. Prevalence, mechanisms, and risk factors of carbapenem resistance in bloodstream isolates of Pseudomonas aeruginosa. Diagn. Microbiol. Infect. Dis. 58:309-314. [DOI] [PubMed] [Google Scholar]
- 26.Tam, V. H., M. Ameka, K. T. Chang, L. A. McCaskey, J. S. Weston, J. P. Caeiro, and K. W. Garey. 2008. Abstr. 48th Annu. Intersci. Conf. Antimicrob. Agents Chemother. (ICAAC)-Infect. Dis. Soc. Am. (IDSA) 46th Annu. Meet., abstr. C2-204.
- 27.Tan, T. Y., and S. Y. Ng. 2006. The in-vitro activity of colistin in gram-negative bacteria. Singapore Med. J. 47:621-624. [PubMed] [Google Scholar]
- 28.Toleman, M. A., K. Rolston, R. N. Jones, and T. R. Walsh. 2004. blaVIM-7, an evolutionarily distinct metallo-β-lactamase gene in a Pseudomonas aeruginosa isolate from the United States. Antimicrob. Agents Chemother. 48:329-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Treyaprasert, W., S. Schmidt, K. H. Rand, U. Suvanakoot, and H. Derendorf. 2007. Pharmacokinetic/pharmacodynamic modeling of in vitro activity of azithromycin against four different bacterial strains. Int. J. Antimicrob. Agents 29:263-270. [DOI] [PubMed] [Google Scholar]