Abstract
Streptococcus uberis is an environmental pathogen commonly causing bovine mastitis, an infection that is generally treated with penicillin G. No field case of true penicillin-resistant S. uberis (MIC > 16 mg/liter) has been described yet, but isolates presenting decreased susceptibility (MIC of 0.25 to 0.5 mg/liter) to this drug are regularly reported to our laboratory. In this study, we demonstrated that S. uberis can readily develop penicillin resistance in laboratory-evolved mutants. The molecular mechanism of resistance (acquisition of mutations in penicillin-binding protein 1A [PBP1A], PBP2B, and PBP2X) was generally similar to that of all other penicillin-resistant streptococci described so far. In addition, it was also specific to S. uberis in that independent resistant mutants carried a unique set of seven consensus mutations, of which only one (Q554E in PBP2X) was commonly found in other streptococci. In parallel, independent isolates from bovine mastitis with different geographical origins (France, Holland, and Switzerland) and presenting a decreased susceptibility to penicillin were characterized. No mosaic PBPs were detected, but they all presented mutations identical to the one found in the laboratory-evolved mutants. This indicates that penicillin resistance development in S. uberis might follow a stringent pathway that would explain, in addition to the ecological niche of this pathogen, why naturally occurring resistances are still rare. In addition, this study shows that there is a reservoir of mutated PBPs in animals, which might be exchanged with other streptococci, such as Streptococcus agalactiae, that could potentially be transmitted to humans.
Penicillin resistance has been particularly well studied for the human pathogen Streptococcus pneumoniae (5, 15). In this organism, resistance occurs through modifications of penicillin-binding proteins (PBPs), leading to a decreased affinity for the drug. These modifications include mutations and/or mosaics in PBP2X and PBP2B, as well as in PBP1A for the highly resistant isolates (20). Several studies also indicated that full expression of resistance necessitates mutations both in the pbp genes and in other genes, of which only a few have been determined, namely, in the ciaRH, cpoA, and murMN loci (8, 9, 14). The same mechanism has also been found in viridians streptococci, which are occasionally responsible for human infections (18, 26). Very recent reports also demonstrated the implication of PBP mutations in S. agalactiae presenting a decreased susceptibility to penicillin (4, 17, 21). In contrast, the highly pathogenic Streptococcus pyogenes, which has been widely exposed to penicillin for decades, is apparently incapable of acquiring clinical resistance in vivo (22), even though laboratory mutants presenting decreased susceptibilities were reported (11). Thus, the capacity to develop penicillin resistance varies among different bacteria belonging to the same genus.
S. pneumoniae is primarily a human pathogen, while S. agalactiae infects humans and sometimes animals. In contrast, S. uberis and Streptococcus suis are primarily distributed among cattle and pigs, respectively, in which they may be responsible for serious and life-threatening diseases. For instance, S. uberis is commonly responsible for clinical and subclinical bovine mastitis, a type of infection that causes major economic loss in the dairy industry worldwide. Recent surveys in England and in France demonstrated that this bacterial species was implicated in about 20% of clinical and subclinical mastitis (2, 3). In addition, S. uberis can also be found in the environment surrounding the animal.
Currently penicillin remains one of the first-line antibiotics for prophylaxis and treatment of such pathologies. Moreover, despite several decades of widespread use of this molecule, it is still commonly believed that S. uberis, as well as other Streptococcus spp. implicated in animal intramammary infections, are susceptible to this drug. However, although it is true that no high-level penicillin resistance (MIC > 16 mg/liter) has yet been described for these bacteria, isolates with intermediate resistance (MICs ranging from 0.5 to 16 mg/liter) have been reported (10, 24).
The present study shows that decreased susceptibilities to penicillin can be achieved in S. uberis after repeated exposures to increasing drug concentrations in the laboratory, despite its phylogenetic link with S. pyogenes (27). Decreased susceptibility correlated with specific mutations in selected PBP genes. Moreover, similar PBP mutations were also detected in clinical isolates displaying decreased penicillin susceptibility that were collected in France, Holland, and Switzerland. These results highlight the reality of a reservoir of mutated PBPs in animals and suggest that resistance development in S. uberis might follow a quasiobligatory pathway.
(Part of this work was presented at the 1st ASM Conference on Antimicrobial Resistance in Zoonotic Bacteria and Food-borne Pathogens, Copenhagen, Denmark, 2008 [abstract C121].)
MATERIALS AND METHODS
Microorganisms and growth conditions.
S. uberis ATCC 19436 and two penicillin-susceptible S. uberis strains isolated from cow mastitis cases were used as model organisms for the selection of laboratory-cycled penicillin-resistant mutants. These were compared to seven S. uberis isolates recovered from mastitis cases in France, Holland, and Switzerland and displaying decreased penicillin susceptibilities (MICs of 0.25 to 0.5 mg/liter). All strains were grown at 37°C either in brain heart infusion broth (BHI) (AES, Combourg, France) without aeration or on Columbia agar (Oxoid, Dardilly, France) supplemented with 3% of blood. Bacterial stocks were stored at −80°C in broth supplemented with 10% (vol/vol) of glycerol.
Antibiotics and chemicals.
Penicillin G was purchased from Sigma (Saint Quentin Fallavier, France). All other chemicals were reagent grade, commercially available products.
Antibiotic susceptibility.
The MICs of penicillin were determined by a previously described broth macrodilution method (1) with a final inoculum of ca. 106 CFU/ml. The MIC was defined as the lowest antibiotic concentration that inhibited visible bacterial growth after 24 h of incubation at 37°C.
Selection for penicillin resistance by successive cycling.
Selection for penicillin-resistant mutants was performed in broth cultures by exposing bacteria to stepwise-increasing concentrations of antibiotics (7). The whole cycling experiment was followed over a minimum of 50 cycles and repeated with 3 independent isolates (strains ATCC 19436, 8749, and 9529). Individual mutants were isolated by picking single colonies from agar plates, regrown in broth, and stored at −80°C for further study.
Identification of PBP genes in S. uberis.
The DNA sequences of genes encoding putative PBP1A, PBP1B, PBP2A, PBP2B, and PBP2X of S. uberis were determined by homology searches from its nonannotated sequence, available at http://www.sanger.ac.uk. BLAST searches were performed using the corresponding amino acid sequences from S. pyogenes (accession numbers NP_269695 [PBP1A], NP_268494 ′PBP1B[, NP_270001 [PBP2A], and NP_269705 [PBP2X]) and S. pneumoniae (NP_359110.1 [PBP2B]).
DNA preparation and genetic strategies.
Molecular techniques were carried out by using standard methods (25) or by following instructions provided with commercially available kits and reagents. Genomic DNA was extracted using the Qiagen DNeasy tissue kit (Qiagen, Courtaboeuf, France). Resistance mutations were sought in the transpeptidase domains of pbp genes and in the murMN operon. These genes, as well as fruA and pepC, which were used as negative-control genes, were amplified by PCR on a thermocycler (Bio-Rad, Richmond, CA). Primers were purchased from Sigma and are presented in Table 1. Cycling conditions consisted of 30 cycles at 94°C for 30 s, 52°C for 45 s, and 72°C for 1.5 min, followed by a 10-min delay period at 72°C after the last cycle. Amplicons were sequenced in both directions (Genome Express, Meylan, France). Sequences were analyzed using the LALIGN program on the Infobiogen website (www.fr.embnet.org).
TABLE 1.
Primers used for PCR amplificationa
| Gene | Forward sequence | Reverse sequence | Size (bp) |
|---|---|---|---|
| pbp1A | 5′-G572GCGACATCTGGATGAAAAT | 5′-G1289CCATTGTTCCAACATAATCA | 718 |
| pbp1B | 5′-C1167TTTGGCGGTTTGCTAGATG | 5′-G2056GATGGCGTTGGCTAGATTA | 890 |
| pbp2A | 5′-A1275GGGCTTGTTGGTCGTGTTA | 5′-C2008GGTCTTGTTAAAACCGATCC | 734 |
| pbp2B | 5′-C971TATGTCGGGCTTGTCTCGT | 5′-T1849GGCAACAGCTACTTCAGGA | 879 |
| pbp2X | 5′-T936GCAGATACTTTAGAAGGCTTGAA | 5′-G1739GAACCATTGCTACGACTGAG | 804 |
| murM | 5′-T−27GCATTACGTCAGTGGGTTT | 5′-T+55CAACTGATTCAGAAAAGGTTTCA | 1,306 |
| murN | 5′-C−25GCAAACAATTAAGGAGTAAACA | 5′-G+21CTGAAGCACTTGGTTTTTGA | 1,279 |
| fruA | 5′-C901CATTTGTTATTGGTGGTGGT | 5′-G1790CACCAGTTAAGGCAGAACC | 890 |
| pepC | 5′-G136CAACCGCTGATCAAGATTT | 5′-C998AGTCATCAGAGGCCACAAA | 863 |
−, number of nucleotides upstream of the start codon; +, number of nucleotides downstream of the stop codon.
Visualization of PBPs with Bocillin FL.
First, membrane-enriched proteins were extracted from one liter of bacterial culture in the late exponential phase of growth (optical density at 620 nm [OD620] of ca. 0.7) as described previously (12). The concentration of membrane proteins were determined using the Bio-Rad protein assay (Bio-Rad Laboratories, Richmond, CA), with bovine serum albumin as a standard. Then, 10 μg of membrane proteins were mixed with 15 μM Bocillin FL (Invitrogen) and incubated at 37°C for 45 min. The reaction was stopped by adding 4 μl of a rinsing solution (unlabeled penicillin [60 mg/ml] and the detergent Sarkosyl [10%]), followed by 15 min of incubation at room temperature before the samples were resuspended in loading buffer (1:1 ratio; 2% sodium dodecyl sulfate [SDS], 10% glycerol, 0.1% bromophenol blue in 50 mM Tris-HCl [pH 6.8], and 100 mM dithiothreitol) and heated at 95°C for 3 min. The labeled PBPs were separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) with 6% gels and were visualized directly by fluorography using a Typhoon Trio+ variable-mode imager (excitation at 488 nm and emission at 520 nm; GE Healthcare). Images were analyzed using the ImageQuant TL software program (GE Healthcare). Gels were then stained with Coomassie brilliant blue to visualize the standard and all membrane proteins.
RESULTS
S. uberis PBPs.
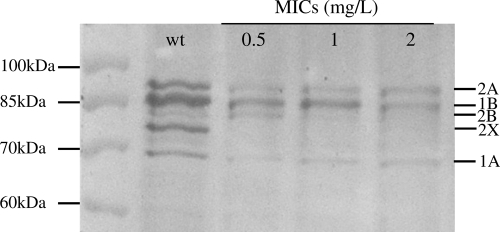
Five PBPs were detected in silico, which were named according to their pneumococcal homologues. The presence of these five high-molecular-weight PBP genes was confirmed in S. uberis by PCR, and the expression of their protein products was assessed by gel separation of Bocillin-FL labeled membranes (Fig. 1).
FIG. 1.
PBP profiles of S. uberis ATCC 19436 wild type and its derivatives, presenting increasing MICs. Proteins were separated by SDS-PAGE, and Bocillin-PBP complexes were visualized by fluorography. Molecular mass markers are indicated at the left of the fluorogram, and PBP numbers, named after their homologues in S. pneumoniae, are at the right. The wild type (wt) and cycled mutants with increased penicillin MICs are indicated at the top.
Selection for penicillin resistance.
The MIC of the S. uberis ATCC 19436 reference strain was 0.032 mg/liter. When the strain was cycled with increasing concentrations of penicillin G, the MIC increased from the wild-type level up to 2 mg/liter over 30 cycles (Table 2), representing a 60-fold increase. The 2-mg/liter level constituted a plateau, since despite continuous cycling (up to 50 cycles) the MIC did not increase any more. According to the recommendations of either the Antibiogram Committee of the French Society for Microbiology (CA-SFM; sensitive, ≤0.25 mg/liter; resistant, >16 mg/liter) or of the CLSI (sensitive, ≤0.12 mg/liter; resistant, >2 mg/liter), these mutants are considered intermediately resistant.
TABLE 2.
PBP mutations in laboratory-evolved S. uberis strains
| Isolate | No. of cycles | MIC (μg/ml) | Mutations |
||
|---|---|---|---|---|---|
| PBP1A | PBP2B | PBP2X | |||
| ATCC 19436 | 0 | 0.032 | —a | — | — |
| ATCC 19436 | 15 | 0.5 | A227T | N366I/T402I | E381K/Q554E |
| ATCC 19436 | 18 | 1 | A227T | N366I/T402I | E381K/D524A/Q554E |
| ATCC 19436 | 30 | 2 | A227T/G386R | N366I/T402I | E381K/D524A/Q554E |
| 8749 | 0 | 0.016 | — | N366I/T402I/V570A/P575S | A492E |
| 8749 | 16 | 0.5 | R273H | N366I/T402I | E381K/Q554E |
| 8749 | 28 | 1 | A227T/G386R | N366I/T402I | E381K/D524A/Q554E |
| 8749 | 37 | 2 | A227T/G386R | N366I/T402I | E381K/D524A/Q554E |
| 9529 | 0 | 0.128 | — | N366I/T402I | E381K/Q554E |
| 9529 | 11 | 0.5 | R273P | N366I/T402I | E381K/Q554E |
| 9529 | 28 | 1 | R273P | N366I/T402I | E381K/Q554E |
| 9529 | 39 | 2 | A227T/G386R | N366I/T402I | E381K/D524A/Q554E |
—, no detected mutation.
The whole cycling experiment was repeated with two additional isolates tested independently. One of them had a basal MIC of 0.016 to 0.032 mg/liter (strain 8749), and one had an increased basal MIC of 0.128 mg/liter (strain 9529), which would correspond to intermediate resistance according to CLSI criteria. Interestingly, this strain already carried four consensus resistance mutations in PBP2X and PBP2B (see details in the next section).
In all three strains (ATCC 19436, 8749, and 9529), the MIC increased step by step at a steady rate. The rapidity with which the different steps were reached differed slightly between the isolates, but the general pattern remained unchanged and reproducible.
Resistance mutations in penicillin-resistant derivatives from a single culture.
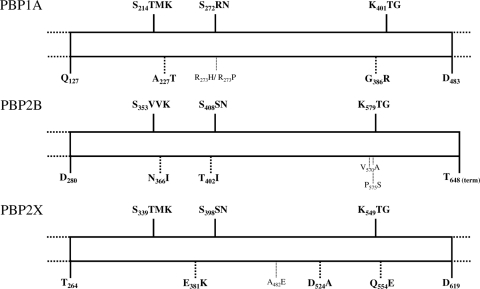
Mutations were first sought in the transpeptidase domains of PBP2B, -2X, and -1A of the three S. uberis cycled isolates (ATCC 19436, 8749, and 9529). The relevance of the detected mutations was validated by the absence of alterations in the two control genes, fruA, which is not implicated in the penicillin-dependent pathway, and pepC, which is located close to PBP1A but does not belong to the same operon, both in the wild-type strains and their resistant derivatives. Table 2 presents the mutations found in the parent strains and at MIC levels of 0.5, 1, and 2 mg/liter. The specific locations of all PBP mutations are schematically represented in Fig. 2.
FIG. 2.
Schematic representation of the transpeptidase domains of PBP1A, PBP2B, and PBP2X. The three conserved motifs of the active site (SXXK, SXN, and KXG) are indicated at the tops of the schemes. The mutations detailed in Table 2 are indicated at the bottoms of the schemes. Bold characters correspond to mutations detected in all laboratory-evolved isolates.
At baseline, no sequence differences were observed between the analyzed PBP domains of the ATCC 19436 wild-type strain and the nonannotated genome available on the Sanger website (www.sanger.ac.uk/Projects/S_uberis/). In this strain, the first PBP mutations were detected at an MIC of 0.5 mg/liter, and three PBPs were affected. First, class B PBP2X, which is a main determinant of penicillin resistance in S. pneumoniae, carried the E381K and Q554E mutations, which were both located close to a conserved motif (Fig. 2). Second, class B PBP2B also carried two substitutions (N366I and T402I) that were located in the vicinity of conserved domains. Third, class A PBP1A already carried an A227T mutation at this early resistance step, although it is reputed to be mutated only in pneumococci with high-level resistance.
At the second step of resistance (MIC of 1 mg/liter), these five mutations were conserved, and an additional one was detected in PBP2X (D524A). At the last step (MIC of 2 mg/liter), a second substitution in PBP1A appeared (G386R), which was also located in the vicinity of conserved motifs (Fig. 2). Both PBP1A modifications led to the conversion of simple hydrophobic amino acids (alanine or glycine) into a more complex hydrophilic threonine or basic arginine.
This progressive accumulation of mutations paralleling the step-by-step increase in the MIC was a feature of each strain, and most interestingly, the same seven consensus mutations were systematically detected in all of them at the highest level of MIC (at 2 mg/liter). Nevertheless, slight differences occurred in between, as shown by the appearance of transient mutations that disappeared at higher resistance levels. For example, isolate 8749 already carried two PBP2B mutations at the basal MIC level, in addition to three other transitory alterations. This strain was cycled in triplicate to test whether these transient mutations (V570A and P575S in PBP2B and A492E in PBP2X) might be deleterious for penicillin resistance development. Yet in all triplicates, they were always detected both at the wild-type level and at an MIC of 0.5 mg/liter, suggesting that whereas they are not necessary for penicillin resistance development, they are not impeaching it either. Similarly, isolate 9529, which had an increased MIC (0.128 mg/liter) and already carried four mutations in PBP2B and PBP2X at the basal level (Table 2), also displayed a mutation in PBP1A (R273P) that disappeared after it had reached an MIC of 2 mg/liter and developed its whole set of consensus mutations.
Mutations were also sought in the murMN operon, at the basal level and at an MIC of 2 mg/liter. In all three strains, two mutations were detected in the murN gene in comparison to the Sanger sequence, both at the basal level and in laboratory-evolved mutants. Two additional substitutions in murM were also transiently present in the noncycled ATCC 19436 strain. However, these modifications were not linked to penicillin resistance development, since they were already present at the basal level.
Impact of mutations on the affinity for penicillin.
The PBPs of wild-type ATCC 19436 and the two associated mutants presenting increasing MICs to penicillin were analyzed on Bocillin-FL-labeled gels (Fig. 1). A progressive fading of all PBPs could be observed, but the ones corresponding to nonmutated PBP2A and -1B seemed less impacted. On the contrary, the bands corresponding to PBP2B and -2X had already completely disappeared in the mutant presenting an MIC of 1 mg/liter.
Resistance mutations in environmental S. uberis.
The transpeptidase domains of PBP1A, -2B, and -2X of seven noncycled strains isolated from bovine mastitis cases and presenting a decreased susceptibility to penicillin (MICs of 0.25 to 05 mg/liter) were sequenced. No mosaic PBPs were identified, as assessed by the correct PCR amplification of their transpeptidase domains and the absence of internal fragments presenting highly diverging sequences compared to both the nonannotated Sanger sequence and the sequences of the other isolates tested in this study. On the other hand, all mastitis isolates carried ≥2 of the consensus resistance mutations found in the laboratory-cycled strains (Table 3). The two alterations (E381K and Q554E) in PBP2X were systematically detected. Mutations in PBP1A and -2B were not found in all isolates, but whenever present, they were in conformity with the consensus (G386R in PBP1A, N366I and T402I in PBP2B). Consequently, very similar PBP mutations could be observed between laboratory-evolved mutants and the natural strains with decreased penicillin susceptibility.
TABLE 3.
PBP mutations in S. uberis isolates naturally presenting decreased susceptibility to penicillin
| Isolate | Origin | MIC (μg/ml) | Mutation(s) |
||
|---|---|---|---|---|---|
| PBP1A | PBP2B | PBP2X | |||
| ALP8092 | Switzerland | 0.25 | G386R | —a | E381K/Q554E |
| ALP 8144 | Switzerland | 0.25 | — | — | E381K/Q554E |
| BL246 | Switzerland | 0.25 | — | N366I/T402I | E381K/Q554E |
| 1.11 | Holland | 0.25 | G386R | T593A | E381K/Q554E |
| 20568 | France | 0.5 | G386R | N366I/T402I/V570A/P575S | E381K/Q554E |
| 20592 | France | 0.5 | — | — | E381K/Q554E |
| 20618 | France | 0.25 | G386R | — | E381K/Q554E |
—, no detected mutation.
DISCUSSION
In this study, the capacity of S. uberis to develop penicillin resistance was questioned, first, because S. uberis is one of the principal causes of streptococcal mastitis; second, because penicillin is widely used to treat intramammary infections in animals; and third, because field cases of S. uberis strains presenting decreased susceptibility to this drug were reported to our laboratory. In addition, S. uberis is phylogenetically linked to S. pyogenes, which is reputedly incapable of developing penicillin resistance. The results obtained in vitro demonstrated that a step-by-step increase in the MIC can occur for S. uberis. This was exemplified by the 60-fold increase in the MIC that progressively developed over 30 cycles of penicillin exposure in the reference strain ATCC 19436 and in the two other strains studied.
PBP alterations have been extensively described as the major mechanism leading to penicillin resistance in S. pneumoniae (5). Additionally, PBP alterations were shown to be implicated in decreased susceptibility in Streptococcus gordonii and more recently in S. agalactiae (13, 17). Here the development of laboratory-evolved mutants, which presented MICs that were increasing concomitantly to the accumulation of mutations in both class A and class B PBPs, demonstrates that S. uberis shared the same resistance mechanism common to all streptococci. In PBP2B, two recurrent mutations were detected in the vicinity of the SVVK and SSN domains, both generating an isoleucine residue that might play a central role in protein stability because of its critical importance for binding of ligands to proteins. In PBP2X, all mutants systematically harbored three mutations at an MIC of 2 mg/liter, among which was the Q554E alteration. This substitution is characteristic of all penicillin-resistant streptococci described so far (4, 13, 19, 21) and was shown to be one of the major genetic determinants of pneumococcal resistance (23). Interestingly, two PBP2X mutations and two PBP2B mutations were already found in the wild-type 9529 isolate, which displayed an MIC of 0.128 mg/liter. Conserved mutations were also observed in PBP1A, close—but not inside—the STMK and KTG motifs. In coherence with the presence of point mutations in class B and class A PBPs, a likely decrease in the affinity of the proteins for penicillin was evidenced with Bocillin FL-labeled gels. Thus, all of these mutations were probably responsible for the resistance of the isolates, though to different extents.
All mutations were highly conserved between laboratory-evolved mutants, apart from a few marginal alterations that were transiently detected, as already observed in S. gordonii (data not published) and probably reflecting the high mutability of these PBP proteins upon selection by penicillin. Additionally, the same mutations were present in isolates from bovine mastitis that presented decreased susceptibilities to penicillin. More precisely, two PBP2X mutations were systematically found (E381K and Q554E), alone or in association with PBP2B or PBP1A substitutions. On the contrary, no mosaic proteins were detected in these natural isolates, since their transpeptidase domain could be amplified by PCR and presented sequences quasi-identical to that of the wild-type ATCC reference strain. This high conservation between mutations from unrelated isolates (laboratory and environmental strains from diverse origins) seems to be specific to S. uberis. Indeed, in pneumococci, as in other Streptococcus spp., high-level MICs of penicillin are generated by a diversity of different patterns of mutations (6, 13, 16, 21). This specificity of S. uberis might argue for a strong pressure of selection on very precise regions of the protein and probably represents a quasiobligatory pathway for the development of decreased susceptibility to penicillin in this particular species. Moreover, constraint seems particularly strong considering the fact that all the resistant environmental isolates from France, Holland, and Switzerland carried the same types of mutations. Such a stringent selection mechanism, combined with the ecological niche on a usually sterile mammary gland, which decreases the chances of horizontal gene transfer with other species, might explain why no S. uberis penicillin-resistant strains have been reported yet.
In addition to PBP-related mutations, a few other genes have been implicated in penicillin resistance (8, 9, 14). Here the murMN operon was sequenced, but the only detected alterations were either already present in the wild-type strain or transient, which indicates that they were most probably not related to resistance development. However, the fact that the same set of mutations can be found in two successive laboratory-evolved mutants with different MICs (strains 8749 and 9529, MICs of 1 mg/liter and 2 mg/liter; Table 2), as well as the existence of environmental isolates with the same pattern but different MICs (ALP 8144 and 20592; Table 3) or with different patterns but the same MIC (20568 and 20592), indicates that other still-unknown genetic determinants have an important function.
Potential genetic exchanges are undoubtedly less frequent for bacteria living in the open environment or on the normally sterile mammary gland than for an organism living in the oral cavity, like S. pneumoniae, in close contact with thousands of other species. Yet each step toward decreased susceptibility might be a risk for the emergence of new resistant phenotypes. Consequently, attention should be paid to any shift of population toward progressively increased MICs.
An ultimate practical question relates to the phenotypic and genotypic definition of resistance. In S. uberis, a simultaneous E381K/Q554E substitution in PBP2X reliably correlated with a penicillin MIC of ≥0.125 mg/liter, while partner mutations could differ at this stage. Furthermore, a set of 7 consensus mutations in PBP1A, -2B, and -2X was reproducibly observed at an MIC of 2 mg/liter. Although this correlation needs to be confirmed with larger numbers of isolates, it could provide a genetically based marker for newly defined intermediate resistance (MIC ≥ 0.125 mg/liter) and high-level resistance (MIC ≥ 2 mg/liter), which would conform with CLSI criteria, compared to the CA-SFM criteria (MIC ≥ 0.25 mg/liter and MIC ≥ 16 mg/liter for intermediate and full resistance, respectively). Moreover, it would also help in rationalizing the conceptual differences between epidemiological cutoff values, which are defined to differentiate a wild-type population from a diverging one, and clinical breakpoints, which are related primarily to the therapeutic chances of success.
In conclusion, the present work demonstrates that S. uberis can develop decreased susceptibilities to penicillin. This development involves mechanisms that are in part common to all streptococci (accumulation of PBP mutations, leading to a decreased affinity for the drug) and in part specific to S. uberis (accumulation of highly conserved alterations). Numerous and precise mutations related to penicillin resistance acquisition were described, which might contribute to the pool of resistance determinants exchangeable between streptococci—for instance, between S. uberis and S. agalactiae, which are both causative agents of intramammary infections in cattle. Yet the use of penicillin as a first-line therapeutic choice can still be recommended in veterinary medicine. But the use of epidemiological cutoff values that would be more stringent, combined with molecular tools, seems mandatory for the survey of the evolution of resistance in S. uberis strains causing bovine mastitis and for decreasing the contribution of animals to the pool of penicillin-resistant streptococci shared by animals and humans.
Acknowledgments
We thank Vincent Perreten and Dik Mevius for providing the strains from Switzerland and Holland, respectively.
Footnotes
Published ahead of print on 11 January 2010.
REFERENCES
- 1.Amsterdam, D. 1996. Susceptibility testing of antimicrobials in liquid medium. In V. Lorian (ed.), Antibiotics in laboratory medicine, 4th ed. Williams & Wilkins, Baltimore, MD.
- 2.Botrel, M.-A., M. Haenni, E. Morignat, P. Sulpice, J.-Y. Madec, and D. Calavas. Distribution and antimicrobial resistance of clinical and subclinical mastitis pathogens in dairy cows in Rhône-Alpes, France. Foodborne Pathog. Dis., in press. [DOI] [PubMed]
- 3.Bradley, A. J., K. A. Leach, J. E. Breen, L. E. Green, and M. J. Green. 2007. Survey of the incidence and aetiology of mastitis on dairy farms in England and Wales. Vet. Rec. 160:253-258. [DOI] [PubMed] [Google Scholar]
- 4.Dahesh, S., M. E. Hensler, N. M. Van Sorge, R. E. Gertz, Jr., S. Schrag, V. Nizet, and B. W. Beall. 2008. Point mutation in the group B streptococcal pbp2x gene conferring decreased susceptibility to beta-lactam antibiotics. Antimicrob. Agents Chemother. 52:2915-2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Denapaite, D., F. Chi, P. Maurer, O. Nolte, and R. Hakenbeck. 2007. Mechanism of penicillin resistance in Streptococcus pneumoniae: targets, gene transfer, and mutations, p. 290-303. In R. Hakenbeck and G. S. Chhatwal (ed.), Molecular biology of streptococci. Horizon Biosciences, Wymondham, Norfolk, United Kingdom.
- 6.Dias, R., D. Felix, and M. Canica. 2008. Diversity of penicillin binding proteins among clinical Streptococcus pneumoniae strains from Portugal. Antimicrob. Agents Chemother. 52:2693-2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Entenza, J. M., J. Vouillamoz, M. P. Glauser, and P. Moreillon. 1997. Levofloxacin versus ciprofloxacin, flucloxacillin, or vancomycin for treatment of experimental endocarditis due to methicillin-susceptible or -resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 41:1662-1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Filipe, S. R., E. Severina, and A. Tomasz. 2002. The murMN operon: a functional link between antibiotic resistance and antibiotic tolerance in Streptococcus pneumoniae. Proc. Natl. Acad. Sci. U. S. A. 99:1550-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guenzi, E., A. M. Gasc, M. A. Sicard, and R. Hakenbeck. 1994. A two-component signal-transducing system is involved in competence and penicillin susceptibility in laboratory mutants of Streptococcus pneumoniae. Mol. Microbiol. 12:505-515. [DOI] [PubMed] [Google Scholar]
- 10.Guerin-Faublee, V., F. Tardy, C. Bouveron, and G. Carret. 2002. Antimicrobial susceptibility of Streptococcus species isolated from clinical mastitis in dairy cows. Int. J. Antimicrob. Agents 19:219-226. [DOI] [PubMed] [Google Scholar]
- 11.Gutmann, L., and A. Tomasz. 1982. Penicillin-resistant and penicillin-tolerant mutants of group A streptococci. Antimicrob. Agents Chemother. 22:128-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haenni, M., P. A. Majcherczyk, J. L. Barblan, and P. Moreillon. 2006. Mutational analysis of class A and class B penicillin-binding proteins in Streptococcus gordonii. Antimicrob. Agents Chemother. 50:4062-4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haenni, M., and P. Moreillon. 2006. Mutations in penicillin-binding protein (PBP) genes and in non-PBP genes during selection of penicillin-resistant Streptococcus gordonii. Antimicrob. Agents Chemother. 50:4053-4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hakenbeck, R., T. Grebe, D. Zahner, and J. B. Stock. 1999. Beta-lactam resistance in Streptococcus pneumoniae: penicillin-binding proteins and non-penicillin-binding proteins. Mol. Microbiol. 33:673-678. [DOI] [PubMed] [Google Scholar]
- 15.Hoban, D., F. Baquero, V. Reed, and D. Felmingham. 2005. Demographic analysis of antimicrobial resistance among Streptococcus pneumoniae: worldwide results from PROTEKT 1999-2000. Int. J. Infect. Dis. 9:262-273. [DOI] [PubMed] [Google Scholar]
- 16.Izdebski, R., J. Rutschmann, J. Fiett, E. Sadowy, M. Gniadkowski, W. Hryniewicz, and R. Hakenbeck. 2008. Highly variable penicillin resistance determinants PBP 2x, PBP 2b, and PBP 1a in isolates of two Streptococcus pneumoniae clonal groups, Poland23F-16 and Poland6B-20. Antimicrob. Agents Chemother. 52:1021-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kimura, K., S. Suzuki, J.-I. Wachino, H. Kurokawa, K. Yamane, N. Shibata, N. Nagano, H. Kato, K. Shibayama, and Y. Arakawa. 2008. First molecular characterization of group B streptococci with reduced penicillin susceptibility. Antimicrob. Agents Chemother. 52:2890-2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levy, C. S., P. Kogulan, V. J. Gill, M. B. Croxton, J. G. Kane, and D. R. Lucey. 2001. Endocarditis caused by penicillin-resistant viridans streptococci: 2 cases and controversies in therapy. Clin. Infect. Dis. 33:577-579. [DOI] [PubMed] [Google Scholar]
- 19.Mouz, N., A. M. Di Guilmi, E. Gordon, R. Hakenbeck, O. Dideberg, and T. Vernet. 1999. Mutations in the active site of penicillin-binding protein PBP2x from Streptococcus pneumoniae. Role in the specificity for beta-lactam antibiotics. J. Biol. Chem. 274:19175-19180. [DOI] [PubMed] [Google Scholar]
- 20.Nagai, K., T. A. Davies, M. R. Jacobs, and P. C. Appelbaum. 2002. Effects of amino acid alterations in penicillin-binding proteins (PBPs) 1a, 2b, and 2x on PBP affinities of penicillin, ampicillin, amoxicillin, cefditoren, cefuroxime, cefprozil, and cefaclor in 18 clinical isolates of penicillin-susceptible, -intermediate, and -resistant pneumococci. Antimicrob. Agents Chemother. 46:1273-1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nagano, N., Y. Nagano, K. Kimura, K. Tamai, H. Yanagisawa, and Y. Arakawa. 2008. Genetic heterogeneity in pbp genes among clinically isolated group B streptococci with reduced penicillin susceptibility. Antimicrob. Agents Chemother. 52:4258-4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perez-Trallero, E., C. Fernandez-Mazarrasa, C. Garcia-Rey, E. Bouza, L. Aguilar, J. Garcia-de-Lomas, and F. Baquero. 2001. Antimicrobial susceptibilities of 1,684 Streptococcus pneumoniae and 2,039 Streptococcus pyogenes isolates and their ecological relationships: results of a 1-year (1998-1999) multicenter surveillance study in Spain. Antimicrob. Agents Chemother. 45:3334-3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pernot, L., L. Chesnel, A. Le Gouellec, J. Croize, T. Vernet, O. Dideberg, and A. Dessen. 2004. A PBP2x from a clinical isolate of Streptococcus pneumoniae exhibits an alternative mechanism for reduction of susceptibility to beta-lactam antibiotics. J. Biol. Chem. 279:16463-16470. [DOI] [PubMed] [Google Scholar]
- 24.Rossitto, P. V., L. Ruiz, Y. Kikuchi, K. Glenn, K. Luiz, J. L. Watts, and J. S. Cullor. 2002. Antibiotic susceptibility patterns for environmental streptococci isolated from bovine mastitis in central California dairies. J. Dairy Sci. 85:132-138. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook, J., E. J. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 26.Tuohy, M., and J. A. Washington. 1997. Antimicrobial susceptibility of viridans group streptococci. Diagn. Microbiol. Infect. Dis. 29:277-280. [DOI] [PubMed] [Google Scholar]
- 27.Ward, P., M. Holden, J. Leigh, N. Lennard, A. Bignell, A. Barron, L. Clark, M. Quail, J. Woodward, B. Barrell, S. Egan, T. Field, D. Maskell, M. Kehoe, C. Dowson, N. Chanter, A. Whatmore, S. Bentley, and J. Parkhill. 2009. Evidence for niche adaptation in the genome of the bovine pathogen Streptococcus uberis. BMC Genomics 10:54. [DOI] [PMC free article] [PubMed] [Google Scholar]