Abstract
Hepatitis B virus (HBV) antiviral drug resistance mutations prevent successful outcome of treatment and lead to worsening of liver disease. Detection of its emergence permits opportune treatment with alternative drugs. Unfortunately, the use of newly approved antivirals, including adefovir dipivoxil, emtricitabine, and telbivudine, is also associated with the development of drug resistance, albeit to a lesser extent than the use of lamivudine. The objectives of this work were to assess the performance characteristics (sensitivity and accuracy) of an updated drug resistance test, the INNO-LiPA HBV DR v2, which includes detection of mutations associated with lamivudine, adefovir, emtricitabine, and telbivudine resistance, and to compare the results with consensus sequencing of serum samples from patients treated with HBV antivirals. Diagnostic sensitivity, defined as detection of a positive amplification line on the line probe assay (LiPA) strip, was 94.8% (95% confidence interval [CI], 89.7 to 97.9) after initial testing, increasing to 96.3% (95% CI, 91.6 to 98.8) after repeat test 1 and to 100% (95% CI, 97.3 to 100.0) after repeat test 2. In diagnostic accuracy determinations, full concordance was observed between sequencing and LiPA for 77.0% of the codons tested (620/805 codons [95% CI, 74.0 to 79.9]), whereas LiPA and sequencing were partially concordant 22% of the time (177/805 codons). In 167 out of 177 cases, LiPA detected a wild-type/mutant mixture whereas sequencing detected only one of the two results. Performance testing of the new LiPA test, the INNO-LiPA HBV DR v2, showed convincing diagnostic sensitivity and accuracy. The ability of the test to detect mixed infections and minority viral populations associated with resistance to the current generation of antivirals, including adefovir, emtricitabine, and telbivudine, makes it a useful tool for HBV therapy monitoring.
For the 350 million persons chronically infected with hepatitis B virus (HBV), the two therapeutic approaches presently available to control infection and its sequelae are the use of immunomodulatory agents and/or antiviral chemotherapy. These treatments aim at interrupting the progression and clinical outcomes of the disease (cirrhosis, hepatocellular carcinoma) by stimulating the anti-HBV-specific host immune response or by markedly decreasing viral replication. Of these two approaches, antiviral therapy has been the more frequently chosen option for long-term treatment, especially because of its relative lack of side effects or when immunomodulation therapy has been unsuccessful.
Unfortunately, the sustained efficacy of antiviral agents with respect to halting disease progression can readily be compromised by the frequent occurrence of viral mutations. In the course of treatment using approved antivirals such as lamivudine, adefovir, and emtricitabine, HBV mutants are often selected from the preexisting pool of circulating quasispecies. Over time, one or several of these mutants will become the dominant species as a result of a variety of factors (21). In particular, resistance to lamivudine, the most frequently prescribed antiviral, is practically unavoidable, and the inexorable development of lamivudine drug resistance has been a major clinical impediment to its extended use. In pooled results from four multicenter controlled trials involving patients receiving lamivudine monotherapy, HBV variants associated with drug resistance were detected in 24% of patients after 1 year, a proportion that rose to 42% after 2 years (4, 9, 10).
In contrast, the emergence of mutants with resistance to adefovir has been somewhat slower in naïve compared to lamivudine-resistant patients. Adefovir has a low resistance profile, with 3%, 9%, 18%, and 28% of patients showing resistance after 2, 3, 4, and 5 years, respectively (6). For its part, emtricitabine is associated with a high rate of drug resistance, even in treatment-naïve patients, which tends to preclude its use as monotherapy (15). Finally, the use of telbivudine has also been associated with drug resistance for about 5% of patients after only 12 months of treatment (11). This is not surprising, given that it shares the same resistance profile as lamivudine, especially at rtM204I within the YMDD motif of the viral polymerase (24).
Due to the increasing use and expanding repertoire of nucleotide or nucleoside analogues to treat HBV, drug resistance mutations are becoming more problematic clinically. Unfortunately, the emergence of drug resistance often leads to a worsening of liver disease (13). Furthermore, cases of severe hepatitis reactivation due to infections by drug-resistant virus, resulting in hepatic decompensation and even mortality, have been previously reported (5, 14, 17). Cases of severe reactivation after withdrawal of antiviral therapy have also been previously described (7). Such unsatisfactory outcomes of drug resistance could be prevented by early detection of its emergence, thereby permitting an opportune alteration of treatment with appropriate alternatives.
The CE-marked INNO-LiPA HBV DR v2 (Innogenetics, Gent, Belgium) (see Fig. 1) is an in vitro, reverse hybridization line probe assay (LiPA) used to detect the presence of different genetic variants of HBV in human serum or plasma samples. The test—an update of the INNO-LiPA HBV DR, which detected only lamivudine-related mutations—includes new and clinically relevant wild-type and mutant motifs for codons L80V/I, V/G173L, L180M, A181T/V, M204V/I/S, and N236T, located in the HBV polymerase protein, that confer resistance to lamivudine, adefovir dipivoxil, emtricitabine, and telbivudine.
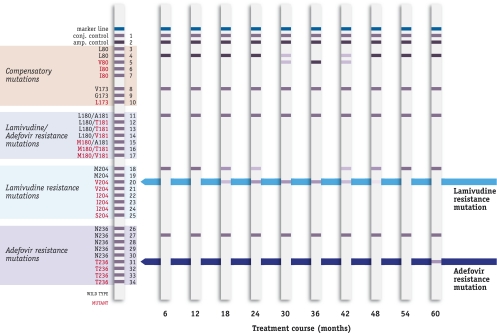
FIG. 1.
Example of monitoring resistance with the INNO-LiPA HBV DR v2 strip. The patient was receiving lamivudine treatment and developed an M204V resistance mutation and later a compensatory L80V mutation. The patient was switched to adefovir therapy in month 36 and in week 60 developed a new adevofir resistance mutation (N236T).
The need for new-generation assays to detect resistance mutations and tailor therapy with respect to virological status is clinically relevant, given the emergence of complex patterns of resistance mutations which may impact treatment decisions (2, 23, 22, 25). While the previous version of the test covered only mutations in the HBV polymerase B and C domains, some of the new mutations associated with recently approved antivirals are present in other HBV polymerase domains. Therefore, a new assay was developed, extending coverage to HBV polymerase domains A through F. Because of this change and the fact that nested PCR technology is less well accepted in the market, the new amplicon was developed for use in a single-round PCR. This assay has been extensively challenged during the development phase. A design verification procedure was performed with 100 HBV DNA positive clinical samples. All these samples were tested with LiPA and sequenced. Mixtures which were not confirmed by sequencing were investigated through a method called “reduced clonal analysis.” These samples were cloned, and two to four clones were picked up and plated several times to produce monoclonal clones. Each clone was sequenced, amplified two to four times, and tested on LiPA strips. The identities of the components of the mixture were considered to have been confirmed when all LiPA results had been confirmed by sequencing of the clones. Data of the design verification procedure are not shown, but the results of this study are described in the INNO-LiPA HBV DR v2 kit insert. We report here the performance characteristics of the INNO-LiPA HBV DR v2 and compare the results with those obtained using direct sequencing.
MATERIALS AND METHODS
Aim.
The objectives of this study were twofold: an evaluation of test sensitivity and calculation of the diagnostic accuracy of the new assay. The assessment of diagnostic sensitivity was carried out using HBV-positive human blood samples. The number of positive test results was determined by detection of a positive amplification control (AC) line on the LiPA strip. The sensitivity was calculated as the number of positive test results versus the total number of samples tested.
For its part, the accuracy of the INNO-LiPA HBV DR v2 was determined at the codon level (positions 80, 173, 180/181, 204, and 236) by determining the percentages of codons with fully concordant, partially concordant, or completely discordant results obtained by LiPA versus sequencing results. Determination of accuracy at the sample level was performed by calculating the percentages of samples with discordant results between LiPA and sequencing.
INNO-LiPA HBV DR v2.
For both internal testing at Innogenetics and external testing at other sites, HBV DNA was isolated from the serum samples by using a commercially available QIAamp DNA blood mini kit (Qiagen GmbH, Hilden, Germany). Samples from sites 1 and 3 (internal testing) were extracted at Innogenetics and eluted in 200 μl of buffer; samples from site 2 (external testing) were eluted in 60 μl of buffer. HBV DNA was amplified in a single-round PCR with INNO-LiPA HBV DR v2 primers (Innogenetics) according to the manufacturer's instructions (Innogenetics). The biotinylated amplified product of 867 bp was denaturated and hybridized to specific oligonucleotide probes coated onto the INNO-LiPA HBV DR v2 strip. After hybridization, unhybridized DNA was washed from the strip. Streptavidin labeled with alkaline phosphatase was added and bound to any biotinylated hybrid previously formed. Incubation with BCIP/NBT chromogen resulted in a purple/brown precipitate. The INNO-LiPA HBV DR v2 strip contains 1 blue marker line, 2 control lines, and 32 parallel probe lines.
Sequencing.
In order for the analysis for accuracy to be performed, a sequencing reference result was required for each sample. The sequence analysis was performed with using an AB377 sequencer and a BigDye Terminator V3.1 kit (Applied Biosystems, Foster City, CA). This was done on the basis of dideoxynucleotide DNA sequencing (25 cycles of 10 s at 95°C, 5 s at 50°C, and 4 min at 60°C), with the sense and antisense primers (HBV nucleotides 255 to 278 and 1121 to 1099 [based on the sequence with GenBank accession number X70185]) amplifying HBV polymerase domains A to F. The procedure was conducted in a completely blinded manner with respect to data available to the technician for the expected test outcome in terms of comparative assays when performing and interpreting the test. The results of the alternative typing method at Innogenetics (LiPA) were completed by a person who was not involved in the practical testing during this study.
Samples and test sites.
To achieve the above-mentioned objectives, stored samples from HBV patients were tested at Innogenetics (internal site) and at external sites. Samples tested at Innogenetics were made available by the liver clinic of Hospital Vall d′Hebron (M. Buti, Barcelona, Spain [site 1]) and by the laboratory of F. Zoulim (Lyon, France [site 3]). Patient samples for sites 1 and 3 were provided under conditions of informed consent. Samples for testing at the external sites were selected by and tested at the site of H. G. Niesters (Rotterdam, the Netherlands [site 2]). Informed consent was not necessary for site 2 (routine diagnostics). Statistical evaluation was separately carried out for testing at Innogenetics and the external sites and also for the pooled data of all sites. Subject confidentiality was maintained by removal of all identifiers from the patient samples.
Proficiency testing.
Prior to test validation, a suitability check using a proficiency panel consisting of four serum samples positive for wild-type HBV (confirmed by sequencing and INNO-LiPA HBV DR), a blank sample for extraction (HBV-negative sample), and a negative control for amplification (water) was performed prior to sample testing to ensure that laboratory personnel would be able to perform the assay appropriately. The proficiency panel was the same for the internal and the external testing sites and was assessed with validation lot 1 of the INNO-LiPA HBV DR v2 (code 80348, batch 143 832). As expected, the results of proficiency testing were found to be good.
Design of validation study.
Stored samples from different HBV-positive individuals who were currently undergoing or had received nucleoside analogue treatment were tested. All samples were tested using the INNO-LiPA HBV DR v2 (code 80348, batch 143 832) and an Auto-LiPA instrument (Innogenetics; same kits and instrumentation as used for the proficiency panel). The LiPA results were visually interpreted and compared to sequencing results that either were generated at the R&D facilities of Innogenetics or were available from the external investigator's site.
Discrepant specimen analysis/repeat testing.
Taking into consideration that sequencing has heretofore been considered the gold standard, the results were divided into four classes with respect to sequencing: fully concordant, partially concordant, completely discordant, or indeterminate. Results were considered fully concordant when LiPA and direct sequencing showed the same results at the amino acid level, i.e., a wild-type, a mutant, or a mixed sequence with respect to codons 80, 173, 180, 181, 204, and 236. Results were considered partially concordant when (i) LiPA provided additional information compared to sequencing (i.e., when the LiPA results showed a mixture of wild-type and mutant sequences whereas sequencing showed either a wild-type or a mutant sequence only) or (ii) sequencing showed a mixture of wild-type and mutant sequence whereas LiPA showed only one of the two results or (iii) both methods showed mixes but the combination of amino acids detected by LiPA and sequencing shared at least one amino acid but not all of the amino acids. Results were considered completely discordant if the amino acids detected by LiPA were completely different from those determined by sequencing; i.e., one test showed a mutant and the other test showed a wild type. Finally, an indeterminate result was considered to have occurred when either LiPA or sequencing gave no results for a codon.
In cases of test failure or a discrepancy between assay results and sequencing, the test was repeated according to a predetermined study algorithm based on the following guidelines. When negative results occurred on the strip (i.e., when no positive amplification line was seen), the amplification was repeated. When results were repeatedly negative, the sample was extracted a second time. If the results were indeterminate for one codon (i.e., if the lines on the LiPA remained blank for both the wild type and the mutant of this codon, with the HBV control line on the strip giving a positive result), verification of the electropherogram was performed (at Innogenetics). If results were indeterminate for more than one codon, a repeat test procedure starting from the amplification step was performed. When the results be repeatedly indeterminate, a restart of the procedure from the sample extraction step was carried out. If all lines were reactive, the test was repeated from the extraction step. Finally, if the results from LiPA and sequencing were discordant, Innogenetics checked the electropherogram. If the results were still discordant, sequencing on the LiPA amplicon was performed by Innogenetics.
With respect to diagnostic sensitivity, the term “initial result” refers to the LiPA typing result obtained from hybridization of the amplicon of a first amplification of extracted DNA. “Repeat result” refers to the final LiPA typing result obtained upon repeating this LiPA one (R1) or two (R2) times.
RESULTS
Disposition of blood samples.
A total of 139 archived serum samples from 129 patients were selected for the study. Three samples were subsequently rejected: one sample contained no volume, and two samples did not meet the inclusion criterion of the presence of a viral load exceeding 1,000 copies/ml. These rejections resulted in an actual number of tested samples of 136.
Samples from seven patients were included twice. This was allowed because the collection times were spread in time and/or a different therapy was given at the time of blood sampling.
Of the total sample pool, 100 samples were taken from patients who received treatment at the time of blood sampling. Thirty of the patients were being treated with lamivudine, 11 with lamivudine-interferon, 3 with lamivudine-telbivudine, 7 with adefovir-lamivudine, 43 with adefovir only, 1 with interferon, and 1 with famciclovir. Treatment details were not documented for four patients. For 36 patients, it was not known whether treatment was still being given at the time of blood sampling.
Sequencing data.
DNA (originating from 200 μl of serum) used for sequencing and validation was eluted in 200 μl of buffer for all 39 and 47 samples at sites 1 and site 3, respectively. Elution was done in 60 μl of buffer for all 50 samples at external site 2.
A reference sequencing result could be determined for all 136 samples sequenced at the Research and Development facilities of Innogenetics (39 samples from site 1, 47 from site 3, and 50 from site 2). For two samples from site 1, the reference result was obtained by sequencing the LiPA amplicon, while for the other samples, sequencing was performed on the same DNA as was used for LiPA. For five samples from site 2, sequencing and LiPA were performed on different templates, implying that a second batch of extracted DNA was used.
Diagnostic sensitivity. (i) Internal testing.
Eighty-six samples were tested with INNO-LiPA HBV DR v2 (data not shown). Two out of the 86 samples gave no band on 2% agarose gel and also showed an initial hybridization failure, as seen by a nonreactive AC line on the LiPA strip. Amplification was repeated (R1) for these two samples and resulted in positive results for gel and LiPA for one of the two samples. For the other sample, no band was visible on the gel and the AC line remained negative after this repeat amplification. A repeat extraction (R2) was then performed for this one sample, and initial amplification of this new DNA sample resulted in a positive gel result and an interpretable hybridization result.
(ii) External testing.
Fifty samples were tested with INNO-LiPA HBV DR v2 (data not shown). Six out of the 50 samples gave no band on a 2% agarose gel, and 5 of those 6 also showed an initial hybridization failure (nonreactive LiPA strip AC line). For these five samples, new DNA had to be extracted, as initial elution had been carried out in 60 μl of buffer only and insufficient DNA remained for a repetition of the amplification using the initial DNA batch. After repeat extraction, only one out of five samples had a detectable band on a 2% agarose gel, and only this sample had an interpretable typing result on the LiPA strip (R1). Repeat amplification on DNA of R1 resulted in all four remaining samples giving positive results for both the gel and the LiPA strips (R2).
The overall diagnostic sensitivity of the INNO-LiPA HBV DR v2, defined as the presence of a positive amplification line on the strip, is summarized in Table 1. Overall sensitivity was 94.8% (95% confidence interval [CI], 89.7 to 97.9) after initial testing. Sensitivity increased to 96.3% (95% CI, 91.6 to 98.8) after repeat test 1 (implying a second extraction and/or reamplification) and to 100% (95% CI, 97.3 to 100.0) after repeat test 2.
TABLE 1.
Overall diagnostic sensitivity of the INNO-LiPA HBV DR v2
| Testing site | No. of samples | No. and % of successes for indicated test/total no. of tests |
|||||
|---|---|---|---|---|---|---|---|
| Initiala |
Repeat 1b |
Repeat 2c |
|||||
| LiPA | % | LiPA | % | LiPA | % | ||
| Internal | 86 | 84/86 | 97.7 | 1/2 | 98.8 | 1/1 | 100 |
| External | 50 | 45/50 | 90.0 | 1/5 | 92.0 | 4/4 | 100 |
| Overall | 136 | 129/136 | 94.8 | 2/7 | 96.3 | 5/5 | 100 |
95% CI, 89.7 to 97.9.
95% CI, 91.6 to 98.8.
95% CI, 97.3 to 100.0.
Indeterminate results.
A total of five indeterminate codon test results were observed out a total of 816 codons tested for the 136 samples. This means only 3.7% indeterminate results at the sample level (95% CI, 1.2 to 8.4) and a rate of 0.6% indeterminate results (95% CI, 0.2 to 1.4), taking all 816 codons into account.
Three of the indeterminate results were produced by codon 173 tests (2.2% indeterminate results for codon 173 [95% CI, 0.5 to 6.3]), while the other two indeterminates were located at codon 204 (1.5% indeterminates for codon 204; 95% CI, 0.2 to 5.2). One of the indeterminate results produced by codon 173 tests was due to a mutation in this sample, resulting in a methionine at position 173. The other four indeterminate results could be explained by a polymorphism in an adjacent codon (Tables 2, 3, and 4). There was no correlation of the indeterminate results with either genotype or viral load.
TABLE 2.
Diagnostic accuracy at the codon level: results of initial and repeat hybridization compared to sequencing (Innogenetics internal validation)
| Codon | No. (%) of tests with indicated resulta |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Full concordanceb |
Partial concordance |
IND on LiPA | Complete discordance | Total | ||||||
| Total | WT | MUT | MIX | MIX LiPA no MIX Seq | MIX Seq no MIX LiPA | Different MIX | ||||
| L80V/I | 44 | 39 | 3 | 2 | 31 | 0 | 0 | 0 | 2 | 77 |
| V/G173L | 70 | 70 | 0 | 0 | 12 | 0 | 0 | 2 | 0 | 84 |
| L180M | 57 | 31 | 21 | 5 | 28 | 1 | 0 | 0 | 0 | 86 |
| A181T/V | 72 | 68 | 1 | 3 | 12 | 1 | 1 | 0 | 0 | 86 |
| M204V/I/S | 50 | 16 | 34 | 0 | 30 | 1 | 3 | 1 | 1 | 86 |
| N236T | 83 | 82 | 0 | 1 | 3 | 0 | 0 | 0 | 0 | 86 |
| Total | 376 (74.5) | 306 (60.6) | 59 (11.7) | 11 (2.2) | 116 (23.0) | 3 (0.6) | 4 (0.8) | 3 (0.6) | 3 (0.6) | 505 (100) |
WT, wild type; MUT, mutant; MIX, wild type and mutant both present with respect to codons 80, 173, 180, 181, 204, and 236; Mix LiPA no MIX Seq, detection of both wild type and mutant by LiPA versus either wild type or mutant detected by sequencing; MIX Seq no MIX LiPA, detection of both wild type and mutant by sequencing versus either wild type or mutant detected by LiPA; Different MIX, different mixture between LiPA and sequencing of both wild type and mutant present with at least one shared amino acid by both methods; IND on LiPA, indeterminate result with LiPA.
The data for full concordance between LiPA and direct sequencing are divided into three groups: the WT group, MUT group, and MIX group.
TABLE 3.
Diagnostic accuracy at the codon level: results of initial and repeat hybridizations compared to sequencing (validation at external sites)
| Codon | No. (%) of tests with indicated resulta |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Full concordanceb |
Partial concordance |
IND on LiPA | Complete discordance | Total | ||||||
| Total | WT | MUT | MIX | MIX LiPA no MIX Seq | MIX Seq no MIX LiPA | Different MIX | ||||
| L80V/I | 39 | 34 | 3 | 2 | 11 | 0 | 0 | 0 | 0 | 50 |
| V/G173L | 44 | 41 | 3 | 0 | 5 | 0 | 0 | 1 | 0 | 50 |
| L180M | 32 | 14 | 18 | 0 | 18 | 0 | 0 | 0 | 0 | 50 |
| A181T/V | 49 | 48 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 50 |
| M204V/I/S | 32 | 10 | 21 | 1 | 14 | 0 | 3 | 1 | 0 | 50 |
| N236T | 48 | 48 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 50 |
| Total | 244 (81.3) | 195 (65.0) | 46 (15.3) | 3 (1.0) | 51 (17) | 0 (0) | 3 (1.0) | 2 (0.7) | 0 (0) | 300 (100) |
WT, wild type; MUT, mutant; MIX, wild type and mutant both present with respect to codons 80, 173, 180, 181, 204, and 236; Mix LiPA no MIX Seq, detection of both wild type and mutant by LiPA versus either wild type or mutant detected by sequencing; MIX Seq no MIX LiPA, detection of both wild type and mutant by sequencing versus either wild type or mutant detected by LiPA; Different MIX, different mixture between LiPA and sequencing of both wild type and mutant present with at least one shared amino acid by both methods; IND on LiPA, indeterminate result with LiPA.
The data for full concordance between LiPA and direct sequencing are divided into three groups: the WT group, MUT group, and MIX group.
TABLE 4.
Diagnostic accuracy at the codon level: results of initial and repeat hybridizations compared to sequencing (overall internal and external site validation)
| Codon | No. (%) of tests with indicated resulta |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Full concordanceb |
Partial concordancec |
IND on LiPA | Complete discordanced | Totale | ||||||
| Total | WT | MUT | MIX | MIX LiPA no MIX Seq | MIX Seq no MIX LiPA | Different MIX | ||||
| L80V/I | 83 | 73 | 6 | 4 | 42 | 0 | 0 | 0 | 2 | 127 |
| V/G173L | 114 | 111 | 3 | 0 | 17 | 0 | 0 | 3 | 0 | 134 |
| L180M | 89 | 45 | 39 | 5 | 46 | 1 | 0 | 0 | 0 | 136 |
| A181T/V | 121 | 116 | 2 | 3 | 13 | 1 | 1 | 0 | 0 | 136 |
| M204V/I/S | 82 | 26 | 55 | 1 | 44 | 1 | 6 | 2 | 1 | 136 |
| N236T | 131 | 130 | 0 | 1 | 5 | 0 | 0 | 0 | 0 | 136 |
| Total | 620 (77.0) | 501 (62.2) | 105 (13.0) | 14 (1.7) | 167 (20.7) | 3 (0.4) | 7 (0.9) | 5 (0.7) | 3 (0.4) | 805 (100) |
WT, wild type; MUT, mutant; MIX, wild type and mutant both present with respect to codons 80, 173, 180, 181, 204, and 236; Mix LiPA no MIX Seq, detection of both wild type and mutant by LiPA versus either wild type or mutant detected by sequencing; MIX Seq no MIX LiPA, detection of both wild type and mutant by sequencing versus either wild type or mutant detected by LiPA; Different MIX, different mixture between LiPA and sequencing of both wild type and mutant present with at least one shared amino acid by both methods; IND on LiPA, indeterminate result with LiPA.
The data for full concordance between LiPA and direct sequencing are divided into three groups: the WT group, MUT group, and MIX group. 95% CI, 74.0 to 79.9 (Total), 58.8 to 65.6 (WT), 10.8 to 15.6 (MUT), 1.0 to 2.9 (MIX).
95% CI, 18.0 to 23.7 (MIX LiPA no MIX Seq), 0.1 to 1.1 (MIX Seq no MIX LiPA), 0.4 to 1.8 (Different MIX). The partial concordance results are expanded in Table 5 below.
95% CI, 0.1 to 1.1.
95% CI, 99.6 to 100.0.
Diagnostic accuracy. (i) Internal testing.
As seen in Table 2, a total of 505 codons were tested internally. Full or partial concordance between LiPA and sequencing results was found for 74.5% and 24.4% of the codons tested, respectively. Importantly, 116 cases (23% of total codons) were observed where LiPA results showed a mixture of wild-type and mutant sequences whereas sequencing showed either a wild-type or mutant sequence result only. The reverse situation occurred for only three codons (0.6% of positive samples). An indeterminate result for LiPA or complete discordance between the tests was found for three codons (0.6%) each.
(ii) External testing.
Results of 300 codon tests indicated the same general pattern seen with internal testing (Table 3). Complete concordance occurred for 81.3% of the results. LiPA detected 51 mixtures of wild types and mutants (17% of total codons), whereas the reverse situation did not occur with sequencing. A different mixture was seen for only three codons. Two indeterminate results were observed with LiPA, and there were no instances of complete discordance between LiPA and sequencing.
Table 4 summarizes the overall results of diagnostic accuracy at the codon level. Taken together, those results showed that full concordance was observed for 77.0% of codons tested (95% CI, 74.0 to 79.9) whereas LiPA and sequencing were partially concordant 22% of the time (177 out of 805 results). An overview of these results is presented in Table 5.
TABLE 5.
Details of overall results after initial and repeat hybridizations for partially concordant results
| Codon | No. of results with indicated mutation(s)a |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| WT Seq |
MUT Seq |
MIX Seqb |
Total | ||||||
| WT + MUT LiPA |
WT + MUT LiPA |
MUT LiPA | WT + MUT LiPA | MUT LiPA | WT LiPA | ||||
| Total | Total | ||||||||
| L80V/I | 17 (V), 5 (V+I), 2 (I) | 24 | 4 (V), 6 (V+I), 1 (I) | 11 | 7 (V+I) | 0 | 0 | 0 | 42 |
| V/G173L | 14 | 14 | 3 | 3 | 0 | 0 | 0 | 0 | 17 |
| L180M | 9 | 9 | 37 | 37 | 0 | 0 | 11 | 0 | 47 |
| A181T/V | 6 (T), 4 (V) | 10 | 2 (T), 1 (V) | 3 | 0 | 1 (T+V)3 | 0 | 12 | 15 |
| M204V/I/S | 5 (V), 3 (I), 2 (V+I) | 10 | 10 (V), 2 (V+I), 3 (I) | 15 | 19 (V+I) | 1 (V+I)5 | 1 (V)4,5, (V+I)6,7-10 | 0 | 51 |
| N236T | 4 | 4 | 1 | 1 | 0 | 0 | 0 | 0 | 5 |
| Total | 71 | 70 | 26 | 2 | 7 | 1 | 177 | ||
WT Seq, wild-type codon detected by sequencing; MUT Seq, mutant codon detected by sequencing; MIX Seq, wild-type and mutant codons both present with respect to codons 80, 173, 180, 181, 204, 236; WT + MUT LiPA, detection of both wild type and mutant by LiPA versus either wild type or mutant detected by sequencing; MUT LiPA, two or more mutant codons detected by LiPA; WT LiPA, wild type detected by LiPA.
Superscripted numbers refer to results that showed a mix of wild types and mutants as detailed in Table 6.
Three main groups of partially concordant results could be distinguished according to the sequencing results. In the first group, sequencing revealed a wild-type codon, but the LiPA result indicated the presence of both wild-type and mutant codons (71 codons). When more than one mutant was detected, the mix could contain one or more mutant amino acids. The second group of results comprised situations in which sequencing revealed a mutant amino acid whereas the LiPA result included this mutant either in a mix of two mutants (26 codons) or in combination with a wild-type result (70 cases). Finally, as detailed in Table 6, the third group (partial concordance) was made up of sequencing results that showed a mix of wild types and mutants (10 codons) as opposed to LiPA results showing the presence of a mutant (7 codons), a wild type (1 codon), or both (2 codons).
TABLE 6.
Partially concordant codons with mixed results by sequencing
| Codon and superscripted no. in Table 5 | Sequencing result | LiPA result |
|---|---|---|
| L180 M | ||
| 1 | L/M | M |
| A181T/V | ||
| 2 | A/Ta | A |
| 3 | A/T | A/T/V |
| M204V/I/S | ||
| 4 | M/V | V |
| 5 | M/V | M/V/I |
| 6 | M/I | V/I |
| 7-10 | M/V/I | V/I |
This mutation was found in a sample with a discordant test result. Upon sequencing of the LiPA amplicon, only the WT A amino acid was observed at this position.
In the course of the study, two results that showed discordance between LiPA and sequencing were observed at codon 80 and one result showed discordance at codon 204. These three discordant results were present in two different specimens. After the LiPA amplicon was sequenced, the LiPA result was confirmed for all three discordant codons.
Finally, upon follow-up analysis, no correlation was found between diagnostic accuracy and either viral load or viral genotype.
DISCUSSION
Performance testing of the new INNO-LiPA HBV DR v2 reverse hybridization LiPA, designed for the detection of wild-type and mutant motifs for codons L80V/I, V/G173L, L180M, A181T/V, M204V/I/S, and N236T, demonstrated the test's ability to detect wild type and mutants at the above-mentioned codon positions with a very high rate of success. Upon initial testing, a diagnostic sensitivity of 94.8% was observed. Reamplification and/or reextraction of seven specimens eventually led to a diagnostic sensitivity of 100% after the repeat testing.
With respect to diagnostic accuracy, LiPA and sequencing identified the same amino acids 77% of the time. In contrast, for 22% of the codons (177/805), LiPA and sequencing gave only partially concordant results. In these cases, the difference between the reverse hybridization technique and sequencing was especially striking: LiPA detected the presence of a mixture of wild-type and mutant codons in no less than 20.7% of cases compared to sequencing. This difference has important ramifications, since monitoring the evolution of primary and compensatory mutations during long-term antiviral treatment remains a clinically relevant issue. Indeed, previous studies (1, 3, 8, 12, 16, 18) have indicated the ability of reverse hybridization tests such as LiPA to detect minority virus populations that are not (or not yet) picked up by sequencing. In some cases, mutations amounting to 30% of the entire HBV population may fail to be detected by sequencing (1). This implies that HBV antiviral treatment efficacy can be maximized by changing or adding drugs at the onset of genotypic resistance as monitored by reverse hybridization tests such as LiPA (12). Anticipation of the need for therapy modification through regular testing can likely prevent progression of liver fibrosis and attenuate the risk of liver decompensation.
Aside from the fact that hybridization tests are generally more sensitive than sequencing for early detection of minority viral populations, the interpretation of emerging resistance using sequencing is affected by such factors as the experience of the operators and the software used to interpret the results. Sequencing also tends to be time-consuming, labor-intensive, and not readily adaptable to high-throughput screening (19). For their part, the limitation of hybridization tests lies in their single-base discrimination. Specificity can be influenced by the sequences adjacent to a polymorphic site or possibly by interference from secondary structures (20). Three indeterminate results in the present study (Table 2) could be explained by the presence of a polymorphism in an adjacent codon.
In conclusion, performance testing of the new version of the LiPA test, the INNO-LiPA HBV DR v2, showed convincing diagnostic sensitivity and accuracy. The test's ability to detect mixed infections and minority viral populations associated with resistance to the current generation of antivirals, including adefovir, emtricitabine, and telbivudine, makes it a practical and useful tool for HBV therapy monitoring.
Acknowledgments
This work was part of the activity of the ViRgil network of excellence suppported by the European Community (ViRgil grant LSHM-CT-2004-503359 to F.Z. and M.B.).
Footnotes
Published ahead of print on 11 January 2010.
REFERENCES
- 1.Aberle, S. W., J. Kletzmayr, B. Watschinger, B. Schmied, N. Vetter, and E. Puchhammer-Stockl. 2001. Comparison of sequence analysis and the INNO-LiPA HBV DR line probe assay for detection of lamivudine-resistant hepatitis B virus strains in patients under various clinical conditions. J. Clin. Microbiol. 39:1972-1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brunelle, M. N., A. C. Jacquard, C. Pichoud, D. Durantel, S. Carrouee-Durantel, J. P. Villeneuve, C. Trepo, and F. Zoulim. 2005. Susceptibility to antivirals of a human HBV strain with mutations conferring resistance to both lamivudine and adefovir. Hepatology 41:1391-1398. [DOI] [PubMed] [Google Scholar]
- 3.Degertekin, B., M. Hussain, J. Tan, K. Oberhelman, and A. S. Lok. 2009. Sensitivity and accuracy of an updated line probe assay (HBV DR v. 3) in detecting mutations associated with hepatitis B antiviral resistance. J. Hepatol. 50:42-48. [DOI] [PubMed] [Google Scholar]
- 4.Dienstag, J. L., E. R. Schiff, T. L. Wright, R. P. Perrillo, H. W. Hann, Z. Goodman, et al. 1999. Lamivudine as initial treatment for chronic hepatitis B in the United States. N. Engl. J. Med. 341:1256-1263. [DOI] [PubMed] [Google Scholar]
- 5.Hadziyannis, S. J., G. V. Papatheodoridis, E. Dimou, A. Laras, and C. Papaioannou. 2000. Efficacy of long-term lamivudine monotherapy in patients with hepatitis B e antigen-negative chronic hepatitis B. Hepatology 32(Pt. 1):847-851. [DOI] [PubMed] [Google Scholar]
- 6.Hadziyannis, S. J., N. C. Tassopoulos, E. J. Heathcote, T. T. Chang, G. Kitis, M. Rizzetto, P. Marcellin, S. G. Lim, Z. Goodman, J. Ma, C. L. Brosgart, K. Borroto-Esoda, S. Arterburn, S. L. Chuck, and the Adefovir Dipivoxil 438 Study Group. 2006. Long-term therapy with adefovir dipivoxil for HBeAg-negative chronic hepatitis B for up to 5 years. Gastroenterology 131:1743-1751. [DOI] [PubMed] [Google Scholar]
- 7.Honkoop, P., R. A. de Man, H. G. Niesters, P. E. Zondervan, and S. W. Schalm. 2000. Acute exacerbation of chronic hepatitis B virus infection after withdrawal of lamivudine therapy. Hepatology 32:635-639. [DOI] [PubMed] [Google Scholar]
- 8.Hussain, M., S. Fung, E. Libbrecht, E. Sablon, C. Cursaro, P. Andreone, et al. 2006. Sensitive line probe assay that simultaneously detects mutations conveying resistance to lamivudine and adefovir. J. Clin. Microbiol. 44:1094-1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lai, C. L., R. N. Chien, N. W. Leung, T. T. Chang, R. Guan, D. I. Tai, et al., for the Asia Hepatitis Lamivudine Study Group. 1998. A one-year trial of lamivudine for chronic hepatitis B. N. Engl. J. Med. 339:61-68. [DOI] [PubMed] [Google Scholar]
- 10.Lai, C. L., J. Dienstag, E. Schiff, N. W. Leung, M. Atkins, C. Hunt, et al. 2003. Prevalence and clinical correlates of YMDD variants during lamivudine therapy for patients with chronic hepatitis B. Clin. Infect. Dis. 36:687-696. [DOI] [PubMed] [Google Scholar]
- 11.Lai, C. L., N. Leung, E. K. Teo, M. Tong, F. Wong, H. W. Hann, et al. 2005. A 1-year trial of telbivudine, lamivudine, and the combination in patients with hepatitis B e antigen-positive chronic hepatitis B. Gastroenterology 129:528-536. [DOI] [PubMed] [Google Scholar]
- 12.Lampertico, P., M. Vigano, E. Manenti, M. Iavarone, G. Lunghi, and M. Colombo. 2005. Adefovir rapidly suppresses hepatitis B in HBeAg-negative patients developing genotypic resistance to lamivudine. Hepatology 42:1414-1419. [DOI] [PubMed] [Google Scholar]
- 13.Leung, N. W., C. L. Lai, T. T. Chang, R. Guan, C. M. Lee, K. Y. Ng, et al., on behalf of the Asia Hepatitis Lamivudine Study Group. 2001. Extended lamivudine treatment in patients with chronic hepatitis B enhances hepatitis B e antigen seroconversion rates: results after 3 years of therapy. Hepatology 33:1527-1532. [DOI] [PubMed] [Google Scholar]
- 14.Liaw, Y. F., R. N. Chien, C. T. Yeh, S. L. Tsai, and C. M. Chu. 1999. Acute exacerbation and hepatitis B virus clearance after emergence of YMDD motif mutation during lamivudine therapy. Hepatology 30:567-572. [DOI] [PubMed] [Google Scholar]
- 15.Lim, S. G., T. M. Ng, N. Kung, Z. Krastev, M. Volfova, P. Husa, et al. 2006. A double-blind placebo-controlled study of emtricitabine in chronic hepatitis B. Arch. Intern. Med. 166:49-56. [DOI] [PubMed] [Google Scholar]
- 16.Lok, A. S., F. Zoulim, S. Locarnini, A. Mangia, G. Niro, H. Decraemer, G. Maertens, F. Hulstaert, K. De Vreese, and E. Sablon. 2002. Monitoring drug resistance in chronic hepatitis B virus (HBV)-infected patients during lamivudine therapy: evaluation of performance of INNO-LiPA HBV DR assay. J. Clin. Microbiol. 40:3729-3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nafa, S., S. Ahmed, D. Tavan, C. Pichoud, F. Berby, L. Stuyver, et al. 2000. Early detection of viral resistance by determination of hepatitis B virus polymerase mutations in patients treated by lamivudine for chronic hepatitis B. Hepatology 32:1078-1088. [DOI] [PubMed] [Google Scholar]
- 18.Osiowy, C., J. P. Villeneuve, E. J. Heathcote, E. Giles, and J. Borlang. 2006. Detection of rtN236T and rtA181V/T mutations associated with resistance to adefovir dipivoxil in samples from patients with chronic hepatitis B virus infection by the INNO-LiPA HBV DR line probe assay (version 2). J. Clin. Microbiol. 44:1994-1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sablon, E., and F. Shapiro. 2005. Advances in molecular diagnosis of HBV infection and drug resistance. Int. J. Med. Sci. 2:8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Southern, E., K. Mir, and M. Shchepinov. 1999. Molecular interaction on microarrays. Nat. Genet. 21(Suppl.):5-9. [DOI] [PubMed] [Google Scholar]
- 21.Tillmann, H. L. 2007. Antiviral therapy and resistance with hepatitis B virus infection. World J. Gastroenterol. 13:125-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Villet, S., A. Ollivet, C. Pichoud, L. Barraud, J. P. Villeneuve, C. Trepo, and F. Zoulim. 2007. Stepwise process for the development of entecavir resistance in a chronic hepatitis B virus infected patient. J. Hepatol. 46:531-538. [DOI] [PubMed] [Google Scholar]
- 23.Villet, S., C. Pichoud, J. P. Villeneuve, C. Trepo, and F. Zoulim. 2006. Selection of a multiple drug-resistant hepatitis B virus strain in a liver-transplanted patient. Gastroenterology 131:1253-1261. [DOI] [PubMed] [Google Scholar]
- 24.Yang, H., X. Qi, A. Sabogal, M. Miller, S. Xiong, and W. E. Delaney IV. 2005. Cross-resistance testing of next-generation nucleoside and nucleotide analogues against lamivudine-resistant HBV. Antivir. Ther. 10:625-633. [PubMed] [Google Scholar]
- 25.Yim, H. J., M. Hussain, Y. Liu, S. N. Wong, S. K. Fung, and A. S. Lok. 2006. Evolution of multi-drug resistant hepatitis B virus during sequential therapy. Hepatology 44:703-712. [DOI] [PubMed] [Google Scholar]