Abstract
The M2 blockers amantadine and rimantadine and the neuraminidase (NA) inhibitors (NAIs) oseltamivir and zanamivir are approved by the FDA for use for the control of influenza A virus infections. The 2009 pandemic influenza A (H1N1) viruses (H1N1pdm) are reassortants that acquired M and NA gene segments from a Eurasian adamantane-resistant swine influenza virus. NAI resistance in the H1N1pdm viruses has been rare, and its occurrence is mainly limited to oseltamivir-exposed patients. The pyrosequencing assay has been proven to be a useful tool in surveillance for drug resistance in seasonal influenza A viruses. We provide a protocol which allows the detection of adamantane resistance markers as well as the I43T change, which is unique to the H1N1pdm M2 protein. The protocol also allows the detection of changes at residues V116, I117, E119, Q136, K150, D151, D199, I223, H275, and N295 in the NA, known to alter NAI drug susceptibility. We report on the detection of the first cases of the oseltamivir resistance-conferring mutation H275Y and the I223V change in viruses from the United States using the approach described in this study. Moreover, the assay permits the quick identification of the major NA group (V106/N248, I106/D248, or I106/N248) to which a pandemic virus belongs. Pyrosequencing is well suited for the detection of drug resistance markers and signature mutations in the M and NA gene segments of the pandemic H1N1 influenza viruses.
In the spring of 2009, an antigenically novel influenza A virus (H1N1) was detected in North America (7). The rapid widespread transmission of the virus resulted in the declaration of an influenza pandemic by the World Health Organization (WHO) (42). The 2009 pandemic influenza A (H1N1) virus (H1N1pdm) was determined to be a reassortant with a combination of gene segments that had not been previously described (12, 21). Phylogenetic analysis of the full genome sequences revealed that in the late 1990s, reassortment between seasonal influenza A virus (H3N2), classical swine influenza virus, and North American avian influenza viruses led to the appearance of triple-reassortant H3N2 and H1N2 swine influenza viruses that have since circulated in pigs in North America (40). The pandemic virus was a result of further reassortment between a triple-reassortant swine influenza virus and a Eurasian avian influenza virus-like swine influenza virus, resulting in the acquisition of two gene segments, coding for the M protein and neuraminidase (NA), from the Eurasian avian influenza virus-like swine influenza virus lineage. Recent genome sequence analysis performed with pandemic viruses collected in different regions found variants with characteristic amino acid changes, including 2 amino acid changes in the NA (21, 29). The reports identified three NA variants among the H1N1pdm viruses: one variant group has V106 and N248 (referred to as the A/California/04/2009 group); the second variant, named the A/Osaka/164/2009 group, is characterized by I106 and N248; and the third NA variant group contains I106 and D248, such as the A/New York/18/2009 strain.
Currently circulating triple-reassortant swine influenza viruses in the United States do not contain any known markers of adamantane resistance (L26F, V27A, A30V, A30T, S31N, and G34E) (10, 25), whereas the Eurasian avian-like influenza viruses as well as the pandemic virus contain the adamantane resistance-conferring change S31N in the M2 protein. Currently, two classes of antiviral drugs are approved for use by the FDA for the control of influenza virus infections: adamantanes (M2 blockers) and neuraminidase inhibitors (NAIs). Resistance to adamantanes makes the NAIs oseltamivir and zanamivir the only pharmaceutical options available for use for the control of infections caused by the pandemic virus. Monitoring of resistance to NAIs is mainly based on the NA inhibition assay (23, 39, 41), which allows the detection of resistance conferred by known and novel mutations. However, the NA inhibition assay requires virus isolation and propagation, and the detection of resistance by the NA inhibition assay requires confirmation by sequencing of the NA gene segment to identify the markers of resistance and their presence in the original clinical material.
Prior to the 2007-2008 influenza season, the frequency of resistance to NAIs had been very low (<0.5%) among field isolates (28, 35, 36). During the 2007-2008 influenza season, seasonal H1N1 viruses resistant to oseltamivir emerged and spread globally (3, 17, 31, 39), and by April of 2009, the majority of the H1N1 viruses were resistant to oseltamivir but sensitive to zanamivir. Of note, nearly all of the 2009 pandemic H1N1 viruses were sensitive to NAIs (8); only sporadic cases of oseltamivir-resistant viruses with the H275Y mutation in the NA gene segment were reported to the WHO, and they were mainly detected following antiviral drug treatment (5, 6, 42). The H275Y mutation is equivalent to the H274Y mutation in the N2 subtype amino acid numbering. Throughout the text, amino acids are described with the N1 numbering, and the corresponding N2 amino acid numbering is shown in parentheses, when it differs from the N1 numbering. Recent reports on the emergence of oseltamivir resistance highlight the need for close monitoring of the susceptibility of the pandemic H1N1 virus to the available drugs (5, 6, 42). Such information is needed to make informed decisions on measures aimed at managing pandemic virus infections.
The molecular markers of NAI resistance are type and subtype specific and are also drug specific (1, 23). The H275Y (H274Y) change is the most commonly reported mutation conferring resistance to oseltamivir in the N1 subtype of NA. This change has been reported not only in seasonal H1N1 viruses but also in highly pathogenic H5N1 viruses (13, 22, 23, 31, 33). The H275Y (H274Y) mutation is also known to reduce susceptibility to the investigational NAI peramivir (23). The amino acid replacement N295S (N294S) in N1 has also been shown to reduce susceptibility to oseltamivir and zanamivir (33, 43). In addition, recent studies have demonstrated that mutations in other residues located in and around the NA active site can alter the susceptibilities of viruses to NAIs. For instance, changes at residues V116, I117, E119, Q136, D199 (D198), and I223 (I222) were associated with reduced susceptibility to NAIs in both seasonal and H5N1 viruses (26-28, 30, 32, 39). Moreover, crystal structure studies with the NAs of H1N1 and H5N1 viruses (9, 37) suggested that mutations at amino acids Q136, K150, and D151 (37) may affect susceptibility to oseltamivir and zanamivir, presumably by interfering with the binding of the drug to the NA. Changes at these residues were reported to reduce the susceptibilities to NAIs of viruses with the N1 enzyme (34; CDC, Atlanta, GA, unpublished data).
It is important to develop the tools necessary for the rapid detection of NA markers known or suspected of affecting susceptibility to NAIs. Pyrosequencing has previously been shown to provide a rapid and high-throughput method for the detection of molecular markers of drug resistance in seasonal as well as highly pathogenic avian influenza viruses (4, 8, 15, 16, 19, 30, 31, 38).
Here we report on the design and validation of pyrosequencing assays for the detection of signature markers in the M2 and NA gene segments of the pandemic H1N1 viruses.
MATERIALS AND METHODS
Viruses.
Viruses were collected and submitted to the WHO Collaborating Center for Surveillance, Epidemiology, and Control of Influenza at the CDC. Pandemic H1N1 influenza viruses A/Massachusetts/12/2009, A/New York/72/2009, A/North Carolina/15/2009, A/North Carolina/16/2009, A/North Dakota/04/2009, A/Tennessee/08/2009, A/Texas/41/2009, A/Utah/34/2009, A/Wisconsin/53/2009, and A/Washington/29/2009 as well as triple-reassortant swine A/Ohio/01/2007 and A/Illinois/09/2007 influenza viruses were analyzed for molecular markers of drug resistance and signature amino acid changes. The triple-reassortant swine influenza viruses were used mainly for comparison. Unless they are identified as a triple-reassortant swine influenza virus strain, all viruses referred to in the study are pandemic H1N1 strains.
Pyrosequencing assay primer design.
The NA sequences from the pandemic H1N1 viruses available from databases in the public domain were aligned by using BioEdit software (version 5.0.6; North Carolina State University). Similarly, M genes from these viruses were also aligned by using BioEdit software. Consensus sequences were generated from both data sets and were used to design pyrosequencing primers, as described elsewhere (4, 15). Table 1 summarizes the pyrosequencing-reverse transcription-PCR (pyro-RT-PCR) and sequencing primers used in the present study.
TABLE 1.
Pyro-RT-PCR and sequencing primers designed in the present study
| Primer purpose and primer | Sequence | Target gene or residue(s) |
|---|---|---|
| Pyro-RT-PCR | ||
| H1N1pdm-M2-F670 | 5′-AGCTCCAGTGCTGGTCTGAAAG-3′ | H1N1pdm-M2 |
| H1N1pdm-M2-R900-biot | 5′-GACTCAGGCACTCCTTCCGTAGAA-3′ | |
| H1N1pdm-N1-F256 | 5′-GCSGGCAATTCCTCTCTYTG-3′ | H1N1pdm-N1 fragment A |
| H1N1pdm-N1-R730-biot | 5′-CGGTCATTACAGTAAARCAHGAACC-3′ | |
| H1N1pdm-N1-F524 | 5′-ARTCAGTYGCTTGGTCAGCAAG-3′ | H1N1pdm-N1 fragment B |
| H1N1pdm-N1-R908-biot | 5′-CAYGGTCGATTCGARSCATG-3′ | |
| H1N1pdm-N1-F780 | 5′-GGGGAAGATTGTYAAATCAGTYGA-3′ | H1N1pdm-N1 fragment Ca |
| H1N1pdm-N1-R1273-biot | 5′-CWACCCAGAARCAAGGYCTTATG-3′ | |
| Pyrosequencing | ||
| H1N1pdm-N1-F292 | 5′-GCTATATACAGTAAAGAC-3′ | V106b,c |
| H1N1pdm-N1-F325 | 5′-GGTTCYAARGGGGA-3′ | V116, I117, E119c |
| H1N1pdm-N1-F386 | 5′-GCAGAACCTTYTTCYTG-3′ | Q136c |
| H1N1pdm-N1-F427 | 5′-AARCATTCCAATGGAAC-3′ | K150, D151c |
| H1N1pdm-N1-F568 | 5′-TGG CTA ACA AT-3′ | D198c |
| H1N1pdm-N1-F647 | 5′-TCAAGAGTTGGAGAAACA-3′ | I222c |
| H1N1pdm-N1-F723 | 5′-AATGACYGATGGMCC-3′ | N247b,c |
| H1N1pdm-N1-F804 | 5′-GYTGAATGCMCCTAATT-3′ | H274c |
| H1N1pdm-N1-F864 | 5′-CACRTGTGTKTGCAG-3′ | N294c |
| H1N1pdm-M2-F747 | 5′-GCGATTCAAGTGATCC-3′ | L26, V27, A30, S31, G34d |
| H1N1pdm-M2-F801 | 5′-TGA TAT TGT GGA-3′ | L43d |
Set of primers published in 2009 by the CDC (8).
Residues are used for NA variant signatures and not antiviral resistance markers.
NA target residue(s).
M2 target residue(s).
RT-PCR and pyrosequencing.
Viral RNAs were extracted either directly from clinical specimens or from the supernatants of isolates propagated in MDCK cells or in the allantoic cavities of 9- to 11-day-old embryonated chicken eggs. Amplifications were performed with a SuperScript III one-step RT-PCR system with Platinum Taq high-fidelity enzyme (Invitrogen, Carlsbad, CA) for 45 cycles, with the primers being used at 20 μM.
Three sets of RT-PCR primers were used to generate three amplicons of the NA gene segment covering the sequences encoding the target residues. One pair of primers was used to amplify a 230-bp fragment containing the sequences coding for the 5 amino acids responsible for resistance to adamantanes (Table 1). The pyrosequencing reactions were performed as described previously (4, 15). Briefly, biotinylated PCR products were washed through a series of buffers, and single-stranded DNA was generated and used as a template for hybridization to residue-specific sequencing primers, which were used at a concentration of 100 μM.
RESULTS
Detection of signature markers in M2 protein of pandemic H1N1 viruses.
The testing of the first pandemic viruses in April 2009 showed that the M-gene pyrosequencing primers previously developed and used at the CDC for the detection of adamantane resistance in seasonal influenza A and H5N1 viruses (4, 14, 15) failed to amplify most of the M genes (8). Therefore, new RT-PCR and pyrosequencing primers specific to swine-origin viruses were designed by using pyrosequencing assay design software (Biotage, Uppsala, Sweden) (Table 1). The newly designed primers were tested and successfully used to amplify all viruses (n = 518) tested at the CDC.
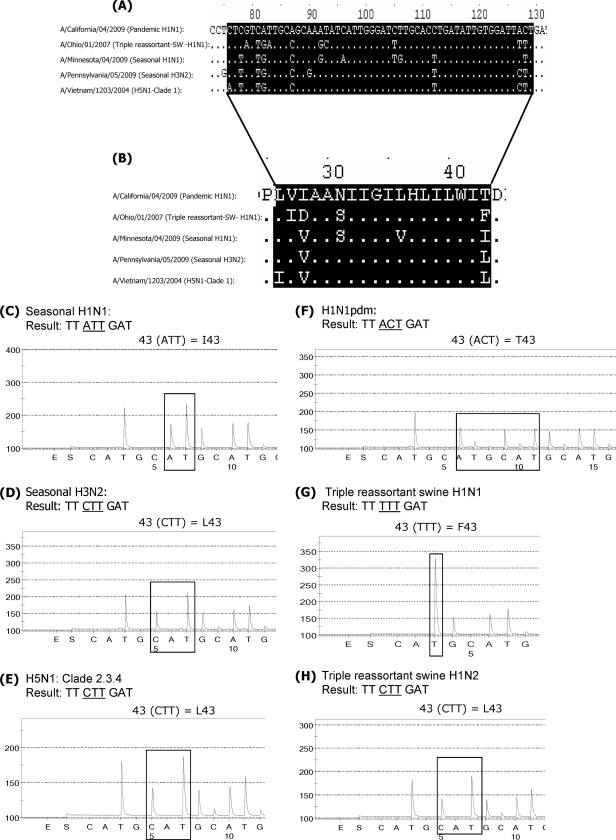
Despite the highly conserved nature of the M-gene sequence among influenza A viruses (18), the region encompassing the 5 amino acid markers of adamantane resistance in M2 is variable among the different genetic lineages and subtypes (14). While the assay was primarily designed for the detection of markers of adamantane resistance, the variability in the target sequences also provides information on the M-gene lineage [i.e., if the M gene is of seasonal human A(H1N1), A(H3N2), swine, or avian A(H5N1) origin (14)]. It is important to note, however, that the M-gene sequence alone cannot be used to infer the virus hemagglutinin (HA) subtype due to possible intersubtype reassortment. Figure 1 summarizes the alignment of the partial sequence of the M2 protein-coding region (Fig. 1A) as well as the alignment of the corresponding amino acids from residues 25 to 44 of the M2 protein sequence (Fig. 1B). All pandemic viruses contain the change I43T in M2 which is not present in seasonal, triple-reassortant swine or H5N1 influenza viruses. The amino acid at residue 43 therefore allows distinction of the pandemic H1N1 viruses from viruses of other subtypes and lineages. Figure 1C to H represents pyrograms of the different influenza virus subtypes used to analyze sequences at this position.
FIG. 1.
The variability in M2 protein-coding sequences among influenza A viruses can be used to differentiate between the various subtypes. (A) Partial nucleotide sequence alignment of the M2 protein-coding region from viruses of different subtypes: a pandemic virus, A/California/04/2009; a triple-reassortant swine influenza virus, A/Ohio/01/2007; seasonal H1N1 and H3N2 viruses, A/Minnesota/04/2009 and A/Pennsylvania/05/2009, respectively; and an H5N1 virus, A/Vietnam/1203/2004. (B) Partial amino acid sequence alignment of the M2 protein. Highlighted are amino acids 26 through 43. (C to H) Analysis of the sequences at residue 43. Seasonal H1N1 virus shows the presence of ATT (isoleucine) at residue 43 (I43) (C). Seasonal H3N2 virus, avian H5N1 virus, and a triple-reassortant swine H1N2 virus contain CTT (leucine) at this position, which corresponds to residue L43 (D, E, and H). Another triple-reassortant swine H1N1 virus has TTT (phenylalanine) at this position (G). The pandemic viruses have the unique sequence ACT (threonine) at residue 43 (F).
Figure S1 in the supplemental material presents examples of the detection of adamantane resistance markers. Figure S1A in the supplemental material illustrates the results for the adamantane-sensitive virus A/Ohio/01/2007, which has wild-type sequences at all five positions. Figure S1B in the supplemental material shows the pyrograms of the adamantane-resistant pandemic virus with sequences CTC (leucine) at residue 26, GTC (valine) at residue 27, GCA (alanine) at residue 30, AAT (asparagine) at residue 31, and GGG (glycine) at residue 34. All pandemic viruses tested so far contain the adamantane resistance-conferring mutation S31N. The only exception was found in a single virus, A/Wisconsin/53/2009, which was sensitive to adamantanes and which had AGT at position 31; it appears to have lost the S31N resistance marker due to a reversion (see Fig. S1C in the supplemental material). This virus was detected by pyrosequencing during routine surveillance for adamantane resistance. To our knowledge, this is the first and only adamantane-sensitive pandemic H1N1 virus reported.
Detection of signature markers in the neuraminidase of pandemic H1N1 viruses.
The target region for NA analysis is a gene region spanning over 600 nucleotides. For optimal assay design on the basis of the manufacturer's recommendations, three pyro-RT-PCR amplification primer sets were used to generate three fragments (fragments A, B, and C). Position-specific sequencing primers were then used to determine the sequences encompassing the residues of interest (Table 1). The fragment A amplicon was used to analyze the sequences at residues V106, V116, I117, E119, Q136, K150, D151, and D199 (D198). Fragment B was used to detect changes at residues I223 (I222) and N248 (N247), and fragment C was used to analyze changes at H275 (H274) and N295 (N294). It is worth noting that assays are not limited to a specific strain, and a set of 12 viruses was tested with each primer. We present here only the results for a representative strain for each site tested.
(i) Detection of oseltamivir resistance marker H275Y (H274Y).
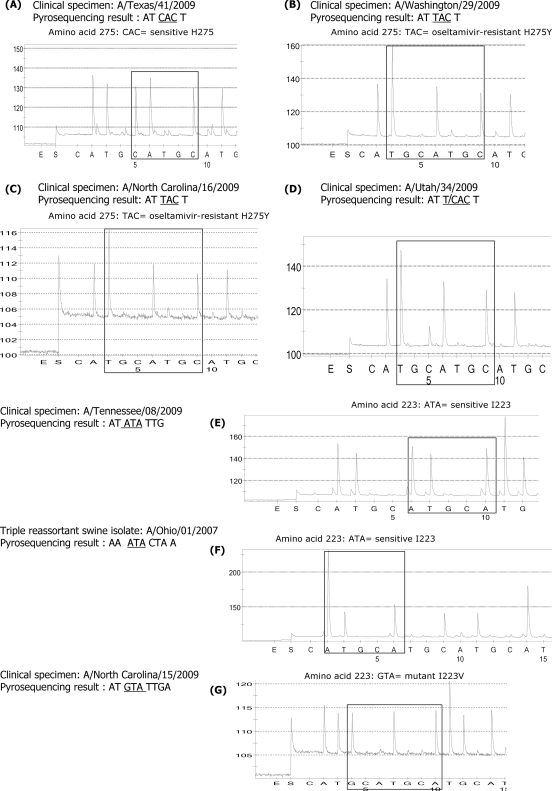
The primers designed generated sequences that allowed a clear distinction between the wild-type sequence, CAC (histidine) (A/Texas/41/2009), and oseltamivir resistance-conferring mutation TAC (tyrosine) in A/Washington/29/2009 and A/North Carolina/16/2009 viruses (Fig. 2A to C). For this test, viral RNA was extracted from clinical specimens, which expedited the detection of oseltamivir resistance (5, 6). These were the first reported cases of oseltamivir resistance in pandemic viruses. Moreover, the assay allowed the detection of a mixture of the wild-type H275 sequence and oseltamivir resistance-conferring mutation H275Y in A/Utah/34/2009 virus. Figure 2D shows the presence of T/CAC at codon 275.
FIG. 2.
Detection of molecular markers for oseltamivir resistance among the H1N1pdm viruses. (A) The wild-type CAC (histidine) sequence was detected in A/Texas/41/2009 virus. The oseltamivir resistance-conferring mutation CAC (histidine) to TAC (tyrosine) was detected in A/Washington/29/2009 (B) and A/North Carolina/16/2009 (C). A mixture of wild-type sequence CAC (histidine) and oseltamivir resistance-conferring mutation TAC (tyrosine) was detected in A/Utah/34/2009 (D). (E to G) Detection of changes at I223 (I222). Wild-type sequences ATA (isoleucine) at codon 223 (I222) were present, as expected, in both A/Tennessee/08/2009 and the triple-reassortant swine influenza A/Ohio/01/2007 viruses. However, the change from ATA (isoleucine) to GTA (valine) at residue 223 (residue 222) was detected in A/North Carolina/15/2009 virus (G).
The amplicon generated from primer set H1N1pdm-N1-F780 and H1N1pdm-N1-R1273-biot (where biot represents biotin; fragment C) was also used as a DNA template to analyze changes at residue N295 (N294), which is known to confer resistance to oseltamivir. Figure S2A in the supplemental material represents pyrograms for oseltamivir-sensitive virus A/Tennessee/08/2009. The wild-type sequence, AAT (asparagine), is also shown for a triple-reassortant swine influenza virus A/Illinois/09/2007 (H1N1) (see Fig. S2B in the supplemental material).
It is worth noting that the NA sequences of the pandemic H1N1 viruses and those of North American triple-reassortant H1N1 viruses differ in the regions encoding residues H275 (H274) and N295 (N294). Moreover, primers were designed so that they allowed the analysis of changes in other residues previously reported to affect susceptibility to NAIs, although the clinical relevance of some of these changes has yet to be determined.
(ii) Detection of I223V (I222V) change conferring reduced susceptibility to NAIs.
A primer was designed to generate a sequence around the highly conserved residue 223 (residue 222) in the enzyme active site. The DNA product from fragment B was used as the template for sequencing. The wild-type sequence, ATA (isoleucine), was detected in the A/Tennessee/08/2009 virus as well as in a triple-reassortant swine influenza virus, A/Ohio/01/2007 (Fig. 2E and F). The pyrogram generated allows the differentiation between pandemic 2009 viruses and North American triple-reassortant swine influenza viruses. Significantly, in addition to the oseltamivir resistance-conferring mutation H275Y (H274Y), the change from ATA (isoleucine) to GTA (valine) was detected in the clinical specimen with A/North Carolina/15/2009 (Fig. 2G). This is the first time that a substitution at highly conserved residue I223 (I222) has been detected in pandemic 2009 viruses. Because the virus isolate was not available, it remains unknown at this time whether the I223V replacement had an effect on susceptibility to NAIs. The individual from whom the clinical specimen was obtained was receiving oseltamivir prophylaxis (6), which raises the possibility that this mutation was selected under drug pressure (6). It is essential to note that no evidence of a mixed viral population at residue I223 (I222) was noticed on the pyrograms from clinical specimens collected from the patient.
(iii) Residue D199 (D198).
The detection of changes at residue D199 (D198) was assessed by using A/Washington/29/2009 virus, and the results indicate that the virus contains wild-type sequence GAC (glutamic acid) at this position (see Fig. S2C in the supplemental material). Of note, a variation at this position was recently found in a pandemic virus recovered from an oseltamivir-treated patient (Kirsten St. George, unpublished data). The D199E (D198E) change has also been detected in seasonal H1N1 viruses with reduced susceptibility to oseltamivir (CDC, unpublished data).
(iv) Residues V116, I117, and E119.
The pyrograms presented in Fig. S2D in the supplemental material reveal the presence of GTC (valine), ATA (isoleucine), and GAA (glutamic acid) at positions 116, 117, and 119, respectively, in A/North Dakota/04/2009 virus.
(v) Residues Q136, K150, and D151.
A/New York/72/2009 virus was used to analyze the sequences at residue 136, and the results show the presence of CAA (glutamine), which corresponds to the wild-type sequence at this position (Q136) (see Fig. S2E in the supplemental material).
The wild-type sequences AAA (lysine) at residue 150 and GAC (aspartic acid) at 151 were detected in A/Tennessee/08/2009 virus (see Fig. S2F in the supplemental material).
Detection of NA molecular markers that distinguish between the three NA variants of the currently circulating H1N1pdm viruses.
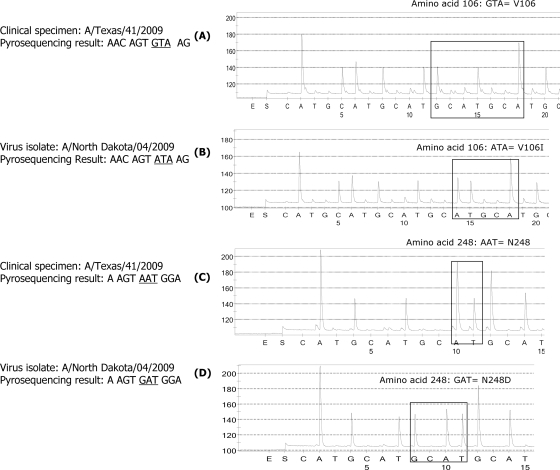
A set of viruses was used to demonstrate the utility of the assay for the detection of amino acid changes at residues V106 and N248 (N247). Figure 3A and C represents pyrograms showing the presence of GTA (valine) at residue 106 and AAT (asparagine) at residue 248 (residue 247) in A/Texas/41/2009 virus; while Fig. 3B and D reveals the presence of ATA (isoleucine) at residue 106 and GAT (aspartic acid) at residue 248 (residue 247) in A/North Dakota/04/2009 virus.
FIG. 3.
Analysis of sequences at residues 106 and 247 for distinction between the NAs from the three major groups of the pandemic H1N1 viruses. (A) Valine (GTA) at position 106 in A/Texas/41/2009 virus; (B) mutation from GTA to ATA (isoleucine) at the same residue in viral isolate A/North Dakota/04/2009; (C) A/Texas/41/2009 with AAT (asparagine) at position 248 (N247); (D) change from AAT to GAT (aspartic acid) at the same residue, N248D (N247D), in A/North Dakota/04/2009 virus.
DISCUSSION
Monitoring of the susceptibilities of the pandemic H1N1 viruses to anti-influenza drugs, especially to NAIs, is critical, since these viruses are resistant to adamantanes (8). NAIs, most commonly, oseltamivir, have been stockpiled by many countries for use in a pandemic. The widespread use of NAIs can lead to the emergence of resistant viruses and is a serious public health concern. For influenza surveillance laboratories, pyrosequencing offers a valuable tool for the rapid high-throughput detection of signature mutations linked to drug resistance (15, 16, 19, 38). We previously designed primers for the detection of the most common resistance markers in seasonal H1N1 viruses (16), but due to differences in the NA sequences between the seasonal H1N1 viruses and 2009 pandemic viruses (21), a new set of primers needed to be designed and validated.
Immediately after the first cases of the H1N1 pandemic outbreak, we designed a method for detection of the most common mutation, H275Y, and posted a description of the procedure on the WHO website (8). In the present study, we provide evidence that the assay continues to perform well (over 1,100 virus isolates and clinical specimens have been tested to date) (http://www.cdc.gov/flu/weekly/). It is worth noting that the primers designed to generate fragment C in N1 of pandemic viruses cross amplified N1 from seasonal H1N1 strains (n = 12) and generated partial NA sequences; however, these sequences are distinguishable from those of the H1N1pdm viruses.
We demonstrated in the present study the detection of the H275Y (H274Y) mutation in pandemic viruses A/Washington/29/2009, A/North Carolina/15/2009, and A/North Carolina/16/2009. In addition, we describe the detection of the I223V (I222V) change in the A/North Carolina/15/2009 virus. These are among the first reported cases of oseltamivir-resistant pandemic viruses carrying the H275Y change in the United States. To our knowledge, this is also the first report of the I223V (I222V) mutation in pandemic viruses. Amino acid replacements at this residue have previously been shown to affect susceptibility to NAIs among seasonal H1N1 (27, 36), H3N2 (2), H5N1 (27), as well as type B influenza (36, 39) viruses.
We also describe a modification of the pyrosequencing protocol made to include markers previously associated with NAI resistance or reduced susceptibility in viruses of the N1 subtype (26-28, 30, 32, 35, 39). However, the relevance of some of these changes remains to be established. For example, it is possible that the Q136K change that caused zanamivir resistance in cell culture-grown seasonal H1N1 viruses in the NA inhibition assay (27; CDC, unpublished data) may have the same effect in the pandemic H1N1 viruses. While the D199E (D198E) change was shown to affect drug susceptibility in seasonal H1N1 viruses (CDC, unpublished data), the effect of the recently detected D199N (D198N) mutation among the pandemic viruses needs further investigation. The prudent approach to the detection of NAI resistance is to utilize the functional NI assay complemented by confirmation by conventional sequencing or pyrosequencing for the identification of a novel or a known mutation(s). However, in many instances, an NAI assay is not possible because of the unavailability of a virus isolate. In such cases, pyrosequencing provides an alternative means for the detection of known markers of resistance in clinical specimens. Furthermore, virus passaging in cell culture has been shown to lead to an alteration in the ratios of sensitive to resistant viral populations, and pyrosequencing provides an invaluable tool for the analysis of changes in such ratios (16).
The NA is a surface antigen and is constantly evolving under host immune pressure (20). Pyrosequencing could aid with the tracking of changes associated with the evolution of the pandemic H1N1 viruses. For this purpose, pyrosequencing was successfully used to determine the NA clade of viruses tested on the basis of previously identified variant-specific amino acid signatures (21, 29). It is worth noting that previously reported M2 pyrosequencing primers (4) did not amplify all pandemic viruses tested. Consequently, new pyrosequencing primers specific to the swine influenza virus M gene were also designed for the detection of adamantane resistance markers. We note that in addition to the pandemic and triple-reassortant swine influenza viruses, the newly designed primers were able to amplify all seasonal and H5N1 viruses tested (n = 25), although more viruses need to be tested to demonstrate reliable coverage.
A variety of assays have been developed to monitor resistance by sequence analysis of certain positions. Some of them are based on single nucleotide polymorphism (SNP) analysis, such as restriction fragment length polymorphism analysis and real-time PCR (11, 24). The pyrosequencing approach described here, which uses the PyroMark IdentiFire apparatus (Biotage), both allows the detection of SNPs and provides partial sequences. We believe that its ability to generate sequences, albeit partial sequences, is valuable especially for virus surveillance laboratories because it provides highly informative data in regard to virus origin and genetic lineages. The pyrosequencing assays were performed directly with clinical specimens prior to cell culture; this not only reduces the time required for drug resistance and susceptibility testing but also provides a tool for investigating the mechanisms of resistance and allows the differentiation between changes that occur after virus isolation.
In conclusion, the timely determination of drug susceptibility is critical and helps with making informed decisions about antiviral usage in the control of influenza virus infections. Pyrosequencing provides a high-throughput and rapid platform and can be an invaluable tool in achieving such goals.
Supplementary Material
Acknowledgments
We are grateful to the Influenza Division staff for their contributions to this project, especially Michael Shaw for fruitful discussion. We thank the state laboratories, the national influenza centers in many countries, and WHO collaborating centers for submitting their specimens for testing.
This work was funded by the Centers for Disease Control and Prevention. T.G.S. received financial support for this work from the Oak Ridge Institute for Science and Education, Oak Ridge, TN.
The findings and conclusions presented in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention.
We declare that we have no conflicts of interest.
Footnotes
Published ahead of print on 12 December 2009.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Abed, Y., M. Baz, and G. Boivin. 2006. Impact of neuraminidase mutations conferring influenza resistance to neuraminidase inhibitors in the N1 and N2 genetic backgrounds. Antivir. Ther. 11:971-976. [PubMed] [Google Scholar]
- 2.Baz, M., Y. Abed, J. McDonald, and G. Boivin. 2006. Characterization of multidrug-resistant influenza A/H3N2 viruses shed during 1 year by an immunocompromised child. Clin. Infect. Dis. 43:1555-1561. [DOI] [PubMed] [Google Scholar]
- 3.Besselaar, T. G., D. Naidoo, A. Buys, V. Gregory, J. McAnerney, J. M. Manamela, L. Blumberg, and B. D. Schoub. 2008. Widespread oseltamivir resistance in influenza A viruses (H1N1), South Africa. Emerg. Infect. Dis. 14:1809-1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bright, R. A., M. J. Medina, X. Xu, G. Perez-Oronoz, T. R. Wallis, X. M. Davis, L. Povinelli, N. J. Cox, and A. I. Klimov. 2005. Incidence of adamantane resistance among influenza A (H3N2) viruses isolated worldwide from 1994 to 2005: a cause for concern. Lancet 366:1175-1181. [DOI] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. 2009. Oseltamivir-resistant novel influenza A (H1N1) virus infection in immunosuppressed patients receiving oseltamivir therapy. MMWR Morb. Mortal. Wkly. Rep. 58:893-896. [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. 2009. Oseltamivir-resistant pandemic influenza A (H1N1) virus infection among summer camp attendees in the setting of liberal oseltamivir prophylaxis for influenza-like illness—North Carolina, 2009. MMWR Morb. Mortal. Wkly. Rep. 58:969-972. [PubMed] [Google Scholar]
- 7.Centers for Disease Control and Prevention. 2009. Swine influenza A (H1N1) infection in two children—Southern California, March-April 2009. MMWR Morb. Mortal. Wkly. Rep. 58:400-402. [PubMed] [Google Scholar]
- 8.Centers for Disease Control and Prevention. 2009. Update: drug susceptibility of swine-origin influenza A (H1N1) viruses, April 2009. MMWR Morb. Mortal. Wkly. Rep. 58:433-435. [PubMed] [Google Scholar]
- 9.Cheng, L. S., R. E. Amaro, D. Xu, W. W. Li, P. W. Arzberger, and J. A. McCammon. 2008. Ensemble-based virtual screening reveals potential novel antiviral compounds for avian influenza neuraminidase. J. Med. Chem. 51:3878-3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheung, C. L., J. M. Rayner, G. J. Smith, P. Wang, T. S. Naipospos, J. Zhang, K. Y. Yuen, R. G. Webster, J. S. Peiris, Y. Guan, and H. Chen. 2006. Distribution of amantadine-resistant H5N1 avian influenza variants in Asia. J. Infect. Dis. 193:1626-1629. [DOI] [PubMed] [Google Scholar]
- 11.Chutinimitkul, S., K. Suwannakarn, T. Chieochansin, l. Q. Mai, S. Damrongwatanapokin, A. Chaisingh, A. Amonsin, O. Landt, T. Songserm, A. Theamboonlers, and Y. Poovorawan. 2007. H5N1 oseltamivir-resistance detection by real-time PCR using two high sensitivity labeled TaqMan probes. J. Virol. Methods 139:44-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dawood, F. S., S. Jain, L. Finelli, M. W. Shaw, S. Lindstrom, R. J. Garten, L. V. Gubareva, X. Xu, C. B. Bridges, and T. M. Uyeki. 2009. Emergence of a novel swine-origin influenza A (H1N1) virus in humans. N. Engl. J. Med. 360:2605-2615. [DOI] [PubMed] [Google Scholar]
- 13.de Jong, M. D., T. T. Tran, H. K. Truong, M. H. Vo, G. J. Smith, V. C. Nguyen, V. C. Bach, T. Q. Phan, Q. H. Do, Y. Guan, J. S. Peiris, T. H. Tran, and J. Farrar. 2005. Oseltamivir resistance during treatment of influenza A (H5N1) infection. N. Engl. J. Med. 353:2667-2672. [DOI] [PubMed] [Google Scholar]
- 14.Deyde, V. M., and L. V. Gubareva. 2009. Influenza genome analysis using pyrosequencing method: current applications for a moving target. Expert Rev. Mol. Diagn. 9:493-509. [DOI] [PubMed] [Google Scholar]
- 15.Deyde, V. M., T. Nguyen, R. A. Bright, A. Balish, B. Shu, S. Lindstrom, A. I. Klimov, and L. V. Gubareva. 2009. Detection of molecular markers of antiviral resistance in influenza A (H5N1) viruses using pyrosequencing method. Antimicrob. Agents Chemother. 53:1039-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deyde, V. M., M. Okomo-Adhiambo, T. G. Sheu, T. R. Wallis, A. Fry, N. Dharan, A. I. Klimov, and L. V. Gubareva. 2009. Pyrosequencing as a tool to detect molecular markers of resistance to neuraminidase inhibitors in seasonal influenza A viruses. Antivir. Res. 81:16-24. [DOI] [PubMed] [Google Scholar]
- 17.Dharan, N. J., L. V. Gubareva, J. J. Meyer, M. Okomo-Adhiambo, R. C. McClinton, S. A. Marshall, K. St. George, S. Epperson, L. Brammer, A. I. Klimov, J. S. Bresee, and A. M. Fry. 2009. Infections with oseltamivir-resistant influenza A(H1N1) virus in the United States. JAMA 301:1034-1041. [DOI] [PubMed] [Google Scholar]
- 18.Donofrio, J. C., J. D. Coonrod, J. N. Davidson, and R. F. Betts. 1992. Detection of influenza A and B in respiratory secretions with the polymerase chain reaction. PCR Methods Appl. 1:263-268. [DOI] [PubMed] [Google Scholar]
- 19.Duwe, S., and B. Schweiger. 2008. A new and rapid genotypic assay for the detection of neuraminidase inhibitor resistant influenza A viruses of subtype H1N1, H3N2, and H5N1. J. Virol. Methods 153:134-141. [DOI] [PubMed] [Google Scholar]
- 20.Fitch, W. M., J. M. Leiter, X. Q. Li, and P. Palese. 1991. Positive Darwinian evolution in human influenza A viruses. Proc. Natl. Acad. Sci. U. S. A. 88:4270-4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garten, R. J., C. T. Davis, C. A. Russell, B. Shu, S. Lindstrom, A. Balish, W. M. Sessions, X. Xu, E. Skepner, V. Deyde, M. Okomo-Adhiambo, L. Gubareva, J. Barnes, C. B. Smith, S. L. Emery, M. J. Hillman, P. Rivailler, J. Smagala, M. de Graaf, D. F. Burke, R. A. Fouchier, C. Pappas, C. M. Alpuche-Aranda, H. Lopez-Gatell, H. Olivera, I. Lopez, C. A. Myers, D. Faix, P. J. Blair, C. Yu, K. M. Keene, P. D. Dotson, Jr., D. Boxrud, A. R. Sambol, S. H. Abid, K. St. George, T. Bannerman, A. L. Moore, D. J. Stringer, P. Blevins, G. J. Demmler-Harrison, M. Ginsberg, P. Kriner, S. Waterman, S. Smole, H. F. Guevara, E. A. Belongia, P. A. Clark, S. T. Beatrice, R. Donis, J. Katz, L. Finelli, C. B. Bridges, M. Shaw, D. B. Jernigan, T. M. Uyeki, D. J. Smith, A. I. Klimov, and N. J. Cox. 2009. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science 325:197-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gubareva, L. V., L. Kaiser, M. N. Matrosovich, Y. Soo-Hoo, and F. G. Hayden. 2001. Selection of influenza virus mutants in experimentally infected volunteers treated with oseltamivir. J. Infect. Dis. 183:523-531. [DOI] [PubMed] [Google Scholar]
- 23.Gubareva, L. V., R. G. Webster, and F. G. Hayden. 2002. Detection of influenza virus resistance to neuraminidase inhibitors by an enzyme inhibition assay. Antivir. Res. 53:47-61. [DOI] [PubMed] [Google Scholar]
- 24.Guo, L., R. J. Garten, A. S. Foust, W. M. Sessions, M. Okomo-Adhiambo, L. V. Gubareva, A. I. Klimov, and X. Xu. 2009. Rapid identification of oseltamivir-resistant influenza A(H1N1) viruses with H274Y mutation by RT-PCR/restriction fragment length polymorphism assay. Antivir. Res. 82:29-33. [DOI] [PubMed] [Google Scholar]
- 25.Hay, A. J., M. C. Zambon, A. J. Wolstenholme, J. J. Skehel, and M. H. Smith. 1986. Molecular basis of resistance of influenza A viruses to amantadine. J. Antimicrob. Chemother. 18(Suppl. B):19-29. [DOI] [PubMed] [Google Scholar]
- 26.Hurt, A. C., J. K. Holien, and I. G. Barr. 2009. In vitro generation of neuraminidase inhibitor resistance in A(H5N1) influenza viruses. Antimicrob. Agents Chemother. 53:4433-4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hurt, A. C., J. K. Holien, M. Parker, A. Kelso, and I. G. Barr. 2009. Zanamivir-resistant influenza viruses with a novel neuraminidase mutation. J. Virol. 83:10366-10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hurt, A. C., P. Selleck, N. Komadina, R. Shaw, L. Brown, and I. G. Barr. 2007. Susceptibility of highly pathogenic A(H5N1) avian influenza viruses to the neuraminidase inhibitors and adamantanes. Antivir. Res. 73:228-231. [DOI] [PubMed] [Google Scholar]
- 29.Itoh, Y., K. Shinya, M. Kiso, T. Watanabe, Y. Sakoda, M. Hatta, Y. Muramoto, D. Tamura, Y. Sakai-Tagawa, T. Noda, S. Sakabe, M. Imai, Y. Hatta, S. Watanabe, C. Li, S. Yamada, K. Fujii, S. Murakami, H. Imai, S. Kakugawa, M. Ito, R. Takano, K. Iwatsuki-Horimoto, M. Shimojima, T. Horimoto, H. Goto, K. Takahashi, A. Makino, H. Ishigaki, M. Nakayama, M. Okamatsu, K. Takahashi, D. Warshauer, P. A. Shult, R. Saito, H. Suzuki, Y. Furuta, M. Yamashita, K. Mitamura, K. Nakano, M. Nakamura, R. Brockman-Schneider, H. Mitamura, M. Yamazaki, N. Sugaya, M. Suresh, M. Ozawa, G. Neumann, J. Gern, H. Kida, K. Ogasawara, and Y. Kawaoka. 2009. In vitro and in vivo characterization of new swine-origin H1N1 influenza viruses. Nature 460:1021-1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lackenby, A., J. Democratis, M. M. Siqueira, and M. C. Zambon. 2008. Rapid quantitation of neuraminidase inhibitor drug resistance in influenza virus quasispecies. Antivir. Ther. 13:809-820. [PubMed] [Google Scholar]
- 31.Lackenby, A., O. Hungnes, S. G. Dudman, A. Meijer, W. J. Paget, A. J. Hay, and M. C. Zambon. 2008. Emergence of resistance to oseltamivir among influenza A(H1N1) viruses in Europe. Euro. Surveill. 13:pii.8026. [DOI] [PubMed] [Google Scholar]
- 32.Le, M. T., H. F. Wertheim, H. D. Nguyen, W. Taylor, P. V. Hoang, C. D. Vuong, H. L. Nguyen, H. H. Nguyen, T. Q. Nguyen, T. V. Nguyen, T. D. Van, B. T. Ngoc, T. N. Bui, B. G. Nguyen, L. T. Nguyen, S. T. Luong, P. H. Phan, H. V. Pham, T. Nguyen, A. Fox, C. V. Nguyen, H. Q. Do, M. Crusat, J. Farrar, H. T. Nguyen, M. D. de Jong, and P. Horby. 2008. Influenza A H5N1 clade 2.3.4 virus with a different antiviral susceptibility profile replaced clade 1 virus in humans in northern Vietnam. PLoS One 3:e3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Le, Q. M., M. Kiso, K. Someya, Y. T. Sakai, T. H. Nguyen, K. H. Nguyen, N. D. Pham, H. H. Ngyen, S. Yamada, Y. Muramoto, T. Horimoto, A. Takada, H. Goto, T. Suzuki, Y. Suzuki, and Y. Kawaoka. 2005. Avian flu: isolation of drug-resistant H5N1 virus. Nature 437:1108. [DOI] [PubMed] [Google Scholar]
- 34.McKimm-Breschkin, J., T. Trivedi, A. Hampson, A. Hay, A. Klimov, M. Tashiro, F. Hayden, and M. Zambon. 2003. Neuraminidase sequence analysis and susceptibilities of influenza virus clinical isolates to zanamivir and oseltamivir. Antimicrob. Agents Chemother. 47:2264-2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McKimm-Breschkin, J. L., P. W. Selleck, T. B. Usman, and M. A. Johnson. 2007. Reduced sensitivity of influenza A (H5N1) to oseltamivir. Emerg. Infect. Dis. 13:1354-1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Monto, A. S., J. L. Kimm-Breschkin, C. Macken, A. W. Hampson, A. Hay, A. Klimov, M. Tashiro, R. G. Webster, M. Aymard, F. G. Hayden, and M. Zambon. 2006. Detection of influenza viruses resistant to neuraminidase inhibitors in global surveillance during the first 3 years of their use. Antimicrob. Agents Chemother. 50:2395-2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Russell, R. J., L. F. Haire, D. J. Stevens, P. J. Collins, Y. P. Lin, G. M. Blackburn, A. J. Hay, S. J. Gamblin, and J. J. Skehel. 2006. The structure of H5N1 avian influenza neuraminidase suggests new opportunities for drug design. Nature 443:45-49. [DOI] [PubMed] [Google Scholar]
- 38.Sheu, T., V. Deyde, R. Garten, A. Klimov, and L.Gubareva. 1 November 2009. Detection of antiviral resistance and genetic lineage markers in influenza B virus neuraminidase using pyrosequencing. Antivir. Res. [Epub ahead of print.] [DOI] [PubMed]
- 39.Sheu, T., V. Deyde, M. Okomo-Adhiambo, R. Garten, X. Xu, R. A. Bright, T. R. Wallis, N. Cox, A. Klimov, and L. V. Gubareva. 2008. Surveillance for neuraminidase inhibitor resistance among human influenza A and B viruses circulating worldwide in 2004-2007. Antimicrob. Agents Chemother. 52:3284-3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shinde, V., C. B. Bridges, T. M. Uyeki, B. Shu, A. Balish, X. Xu, S. Lindstrom, L. V. Gubareva, V. Deyde, R. J. Garten, M. Harris, S. Gerber, S. Vagasky, F. Smith, N. Pascoe, K. Martin, D. Dufficy, K. Ritger, C. Conover, P. Quinlisk, A. Klimov, J. S. Bresee, and L. Finelli. 2009. Triple-reassortant swine influenza A (H1) in humans in the United States, 2005-2009. N. Engl. J. Med. 360:2616-2625. [DOI] [PubMed] [Google Scholar]
- 41.Wetherall, N. T., T. Trivedi, J. Zeller, C. Hodges-Savola, J. L. Kimm-Breschkin, M. Zambon, and F. G. Hayden. 2003. Evaluation of neuraminidase enzyme assays using different substrates to measure susceptibility of influenza virus clinical isolates to neuraminidase inhibitors: report of the neuraminidase inhibitor susceptibility network. J. Clin. Microbiol. 41:742-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.World Health Organization. 2009. Pandemic (H1N1) 2009—update 60. World Health Organization, Geneva, Switzerland. www.who.int/csr/don/2009_08_04/en/print.html. Accessed 11 August 2009.
- 43.Yen, H. L., N. A. Ilyushina, R. Salomon, E. Hoffmann, R. G. Webster, and E. A. Govorkova. 2007. Neuraminidase inhibitor-resistant recombinant A/Vietnam/1203/04 (H5N1) influenza viruses retain their replication efficiency and pathogenicity in vitro and in vivo. J. Virol. 81:12418-12426. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.