Abstract
The RNA-dependent RNA polymerase (NS5B) of hepatitis C virus (HCV) is an unusually attractive target for drug discovery since it contains five distinct drugable sites. The success of novel antiviral therapies will require nonnucleoside inhibitors to be active in at least patients infected with HCV of subtypes 1a and 1b. Therefore, the genotypic assessment of these agents against clinical isolates derived from genotype 1-infected patients is an important prerequisite for the selection of suitable candidates for clinical development. Here we report the 1a/1b subtype profiling of polymerase inhibitors that bind at each of the four known nonnucleoside binding sites. We show that inhibition of all of the clinical isolates tested is maintained, except for inhibitors that bind at the palm-1 binding site. Subtype coverage varies across chemotypes within this class of inhibitors, and inhibition of genotype 1a improves when hydrophobic contact with the polymerase is increased. We investigated if the polymorphism of the palm-1 binding site is the sole cause of the reduced susceptibility of subtype 1a to inhibition by 1,5-benzodiazepines by using reverse genetics, X-ray crystallography, and surface plasmon resonance studies. We showed Y415F to be a key determinant in conferring resistance on subtype 1a, with this effect being mediated through an inhibitor- and enzyme-bound water molecule. Binding studies revealed that the mechanism of subtype 1a resistance is faster dissociation of the inhibitor from the enzyme.
One of the major challenges to overcome in the development of hepatitis C virus (HCV)-directed antivirals is the high propensity of the virus to mutate. This is due to the lack of proofreading capacity of the HCV NS5B RNA-dependent RNA polymerase (RdRp), which replicates the HCV RNA strand with an error rate of 10−2 to 10−3 nucleotide substitutions per site per year (17). The diversity of HCV has been recognized as six phylogenetically distinct groups, referred to as genotypes and, within each genotype, as subtypes (a, b, c, d, etc.) (44). HCV subtype 1b is the most prevalent genotype in the world, and subtype 1a is widely distributed in northern Europe and in the United States; subtypes 2a and 2b are common in North America, Europe, and Japan, and subtype 2c is found predominantly in Northern Italy; HCV genotype 3a is more prevalent in the Far East and has recently increased in Europe and in the United States, possibly due to the spread of the virus through intravenous drug use (11, 17, 18, 44, 46). Of the other genotypes, genotype 4 is common in Africa and to a lesser extent in Europe (11, 39), whereas genotypes 5 and 6 are found predominantly in southern Africa and Southeast Asia, respectively. Despite the availability of a standard of care for the treatment of hepatitis C, a combination of pegylated alpha interferon and ribavirin, many HCV-infected patients cannot be cured because of the frequent failure of the treatment, particularly in patients with genotype 1 and 4 infections, and perhaps also in those with genotype 6 infections (12, 35). In addition, tolerability issues associated with the standard of care lead to discontinuation of therapy in many patients (31). Therefore, major efforts have been made toward developing novel oral therapeutics that target a specific step of the HCV life cycle (45), with particular attention to HCV subtypes 1a and 1b, as they are the most common genotypes underserved by the current standard of care. Subtypes 1a and 1b are estimated to account for 57% and 17% of the HCV-infected patients in the United States (1) versus approximately 11% and 45% in Europe (11), respectively.
The development of HCV polymerase nonnucleoside inhibitors (NNIs) has been successfully validated in phase II clinical trials (21, 24, 41). From the extensive screening of NS5B inhibitors that has been performed to date, several chemotypes have emerged as promising scaffolds, namely, the indole, thiophene, benzothiadiazine, and benzofuran analogs. Each of these NNIs targets four different binding pockets of the HCV polymerase, thumb-1 NNI-1 (10), thumb-2 NNI-2 (29, 48), palm-1 NNI-3 (9), and palm-2 NNI-4 (19), respectively. Historically, the screen for novel NS5B inhibitors was limited to representatives of genotype 1b only (3, 28) because the tools to target other genotypes were not yet available (16, 38, 40). Further assessment of these analogs, using enzyme isolates and intergenotypic chimeric replicons derived from clinical isolates, revealed that the genotypic coverage of the NNI-1 and -4 analogs extends beyond genotype 1, unlike the NNI-2 and -3 derivatives that typically inhibit genotype 1 only (16, 38). An additional drawback stems from the lower genetic barrier of the NNI-2 and -3 analogs in genotype 1 (25) and the reduced susceptibility in subtype 1a of the NNI-3 series (7, 16, 34, 38, 43). This effect was mostly attributed to the Y415F polymorphism observed in the NNI-3 binding site in subtype 1a (38).
Here we report the 1a/1b subtype profiling of 1,5-benzodiazepine (1,5-BZD) HCV polymerase inhibitors that bind to the NNI-3 site (32, 36) using a panel of enzyme and chimeric replicons derived from clinical isolates, X-ray crystallography, and surface plasmon resonance (SPR) studies and compare these inhibitors to the four classes of NNIs by thoroughly assessing a representative of each nonnucleoside binding site in subtypes 1a and 1b.
MATERIALS AND METHODS
Design of NS5B shuttle vector.
The HCV replicon vector cassette designed to shuttle NS5B was generated from plasmid pFKi341Luc_NS3-3′-ET (23). The replicon derived from this plasmid consists of the NS3 to 3′ untranslated region sequence of the HCV genotype 1b Con1 strain and carries three cell culture adaptive mutations (23). Two AflII restriction sites were generated with the QuikChange site-directed mutagenesis kit from Stratagene (La Jolla, CA) using the pFKi341Luc_NS3-3′-ET plasmid, the first in the 3′ noncoding region directly after the stop codon of NS5B and the second eight amino acids upstream of the NS5A/NS5B cleavage site. Because the ScaI site, necessary for linearization of the plasmid prior to in vitro transcription, was found to be present in the polymerase sequence of some clinical isolates, it was mutated to XbaI. The endogenous XbaI site of the firefly luciferase gene was removed by the introduction of a silent mutation.
Construction of NS5B chimeric replicons.
The cDNA encoding the C terminus of NS5A (residues 440 to 447) and full-length NS5B (residues 1 to 591) was amplified from clinical isolates using subtype-specific primers with a 5′ part (16 nucleotides) complementary to the shuttle vector and a 3′ part (13 to 19 nucleotides) designed to be complementary to clinical isolate sequences. The amplicons were cloned into the NS5B shuttle vector by the In-Fusion cloning method (In-Fusion Dry Down PCR cloning kit; Clontech). Individual clones or pools of all clones obtained after transformation were used for in vitro transcription and tested in the transient replicon assay.
Transient replicon assay.
Ten micrograms of in vitro-transcribed linear replicon RNA was transfected into Huh7-cure cells (27), and replicon replication was quantitated after 48 h of incubation (37°C, 5% CO2) by measurement of luminescence after the addition of a luciferase substrate (Steady Lite Plus; Perkin-Elmer). The concentration of a compound necessary to inhibit the expression of the replicon reporter gene by 50% compared to the untreated control (50% effective concentration [EC50]) was measured in a nine-point dilution series, and the n-fold change in EC50 compared to the reference HCV ET replicon was determined. To search for determinants responsible for the reduced susceptibility of genotype 1a to compound 3 and compound 4, the clinical isolates were ranked according to the increase in the fold change in the EC50. Then, for each sequence position, the isolates were categorized as identical or not identical to H77 at that position, and the Mann-Whitney U test was used to determine whether the distribution of ranks between the two categories differed significantly (P < 0.1).
Cloning, expression, and purification of NS5B.
The coding sequence for NS5B (genotype 1b consensus strain Con1, genotype 1a consensus strain H77, or genotype 1a representative 1a_18) lacking 21 C-terminal residues was amplified from plasmid pFKI389/ns3-3′ (GenBank accession no. AJ242654), from plasmid pCV-H77C (GenBank accession no. AF011751), or from the cDNA template of the 1a_18 clinical isolate and subcloned into the pET21b plasmid as described previously (38). The NS5BΔC21 expression constructs were transformed into Escherichia coli Rosetta 2(DE3) (Novagen, Madison, WI). One hundred milliliters of LB medium supplemented with carbenicillin (50 μg/ml) and chloramphenicol (34 μg/ml) was inoculated with one colony, grown overnight, and transferred to fresh LB medium supplemented with 3% ethanol, carbenicillin, and chloramphenicol at a ratio of 1:200. The remainder of the procedure was as described previously (38), except that the column used for ion-exchange chromatography was a 6-ml Resource S column (GE Healthcare) and that protein concentrations were determined with the Nanodrop (Nanodrop Technologies, Wilmington, DE).
RdRp assay.
Fifty percent inhibitory concentrations (IC50s) were essentially determined as described previously (38), using a primer-dependent transcription assay. Following a 10-min preincubation with the inhibitor, the purified NS5B enzyme (20 nM Con1b, 20 nM 1a_H77, 50 nM 1b J4, or 200 nM 1a_18) was incubated for 10 min with 150 nM 5′-biotinylated oligo(rG13) primer preannealed with 15 nM poly(rC) template, 19 mM Tris-HCl, 5 mM MgCl2, 17 mM NaCl, 21 mM KCl, and 2.5 mM dithiothreitol (DTT). Next, 600 nM GTP and 0.13 μCi of [3H]GTP were added to initiate the 40-μl reaction mixture, which was then incubated at room temperature for 2 h before the reaction was stopped by addition of 40 μl streptavidin-coated SPA beads as described previously (38). It was necessary to use a higher concentration of the 1a_18 enzyme isolate because of its lower catalytic competency.
NS5B-inhibitor interaction studies.
Measurements of interactions between the NS5BΔC21 enzyme isolates and the inhibitors were performed using a Biacore T100 instrument (GE Healthcare). Recombinant NS5BΔC21 protein (Con1, 1b J4, 1a_18, or 1a_H77) was immobilized noncovalently on a Ni2+-loaded nitrilotriacetic acid chip (GE Healthcare) in immobilization buffer (20 mM morpholinepropanesulfonic acid [MOPS, pH 7.4], 500 mM NaCl, 0.005% Tween P20, 1 mM DTT, 50 μM EDTA) to allow for the removal of the tightly bound NS5B/inhibitor complex after each cycle. The proteins were dissolved at 50 to 500 nM in immobilization buffer and immobilized at a flow rate of 10 μl/min and a contact time that varied between 1 and 5 min. Immobilization levels of 2,000 to 10,000 resonance units (RU) were reached routinely. Interaction studies were all performed at 25°C with a flow rate of 10 μl/min. Inhibitors were serially diluted in running buffer (20 mM Tris-HCl [pH 7.4], 150 mM NaCl, 50 μM EDTA, 1 mM DTT, 0.005% Tween P20) containing 5% dimethyl sulfoxide (DMSO). Next, the inhibitor was injected onto the chip by single-cycle kinetics, a method that enables the sequential injection of an analyte concentration series without any regeneration steps (22). Five increasing concentrations of compound were injected for a period of 5 min each in one single cycle, and dissociation was monitored for a period of 20 min. The sensor surface was regenerated between cycles by injecting a regeneration buffer containing 350 mM EDTA (pH 8), and the flow system was washed with 50% DMSO. The use of single-cycle kinetics not only increases the throughput of the assay but also permits the accurate determination of koff values for tight binders within a reasonable time span.
Biacore data analysis.
Data were analyzed using simultaneous nonlinear regression analysis (global fitting) adapted for single-cycle kinetics with Biacore T100 BiaEval evaluation software 2.0 (GE Healthcare). The individual association and dissociation rate constants (kon and koff, respectively) and a derived affinity constant, Kd (koff/kon), were determined by a kinetic evaluation of the sensorgrams. A 1:1 Langmuir binding model that assumes binding homogeneity was used preferentially. For indole 55 and thiophene 2, the 1:1 model mismatched the kinetic data due to additional binding components, particularly at high compound concentrations. In that case, the heterogeneous ligand binding model was used and the parameters of the single component that most closely resembled the overall binding kinetics were given. When the binding kinetics were too fast to determine (as was the case for, e.g., the binding of thiophene 2 and GSK625433 to 1a_18), a steady-state affinity Kd value was calculated based on equilibrium levels at the end of each injection step. In that case, no individual rate constants could be calculated. The kinetic models accounted for bulk and limited mass transport effects. Every analysis was performed at least in two independent experiments, and it was verified that the fit values for kon, koff, or Kd were reproduced within 1 order of magnitude. The significance of the kinetic parameters from the 1:1 binding model was confirmed using a Biacore software tool (BiaEval evaluation software 2.0). The residuals (i.e., the difference between the experimental and fitted values for each data point in the sensorgram) were confirmed to lie within reasonable limits and to scatter around zero. The dissociation rate of a kinetic interaction can be translated into a compound residence time or dissociation half-life [t1/2 = ln(2)/koff], which is representative of the interaction time between the polymerase and its inhibitors.
NS5B-RNA interaction studies.
RNA interaction studies were performed as described previously (14), using a Biacore T100 instrument equipped with a streptavidin-coated sensor chip (GE Healthcare) in a running buffer containing 20 mM HEPES (pH 7.4), 150 mM NaCl, 50 μM EDTA, 0.005% Tween P20, and 0.1% DMSO. A biotinylated 36-mer RNA (5′-AUAUUAUCCUAUAUACCGGCUUGCAUAGCAAGCCGG-3′) was immobilized to a surface density of 80 RU by injecting the RNA solution at 8 nM for 60 s at a flow rate of 10 μl/min. Purified Con1 NS5BΔC21 polymerase was diluted to 5 nM in running buffer and supplemented with a 200-fold excess (1 μM) of compound. Mixtures were preincubated at 25°C for at least 2 h prior to injection to allow full association of the inhibitor with the polymerase. Samples containing running buffer alone, 1 μM compound, or 5 nM NS5B were used as controls. All samples were injected for 200 s at a flow rate of 90 μl/min, and dissociation was monitored for 150 s. The RNA surface was regenerated between sample injections by using a 30-s injection of running buffer supplemented with 2 M NaCl. The flow system was washed between sample injections with 50% DMSO. Signals (expressed in RU) were calibrated using a reference flow cell.
Protein crystallography.
Crystallography was performed by Proteros Biostructures GmbH (Martinsried, Germany). Protein was crystallized by vapor diffusion with 17.5% polyethylene glycol and 300 mM ammonium sulfate buffered at pH 5.0. Hexahedral (genotype 1a) and stick-shaped (genotype 1b) crystals grew within 5 days. Inhibitor solutions were in DMSO, and NS5B-inhibitor complexes were formed by cocrystallization or soaking.
Data collection and processing.
Crystals were flash frozen with glycerol as a cryoprotectant, and diffraction data were collected at 100K at the Swiss Light Source (Villigen, Switzerland) under cryogenic conditions. The crystals belong to space groups P21 (genotype 1a) and P212121 (genotype 1b), with three and two molecules per asymmetric unit, respectively. Data were processed using the programs XDS and XSCALE.
Structure modeling and refinement.
The phase information necessary to determine and analyze the structure was obtained by molecular replacement using the previously determined structure of HCV NS5B as a search model (37). Model building and refinement were performed as described previously (36).
Nucleotide sequence and protein structure accession numbers.
The 1a_18 NS5B sequence used for X-ray crystallography has been deposited in EMBL under accession number FN598877. The atomic coordinates and structure factors have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/), under accession numbers 3HKW and 3HKY for the complex with HCV polymerase subtypes 1a and 1b, respectively.
RESULTS
Differences in 1,5-BZD inhibition against subtypes 1a and 1b.
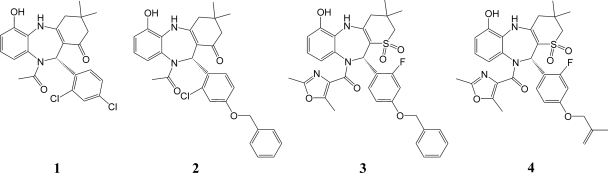
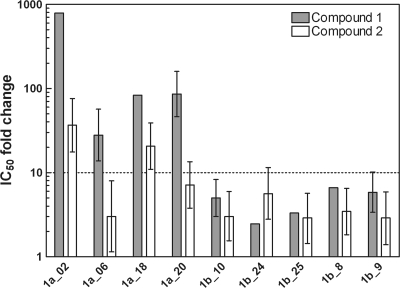
Recently, we disclosed the discovery of 1,5-BZDs 1 and 2 (Fig. 1), novel nonnucleoside polymerase inhibitors of HCV (32, 36). These two compounds differ mainly by the presence of an O-benzyl moiety at the para position of the 1,5-BZD phenyl group in compound 2, which is attached to the chiral center of the BZD. We have shown that 1,5-BZD inhibition of the HCV polymerase is highly stereodependent and limited to genotype 1 (36). To gain better knowledge of the profile of compounds 1 and 2 in genotypes 1a and 1b, we extended the panel of enzyme isolates reported previously (38) to four 1a and five 1b enzyme isolates. The inhibitory effects measured for each enzyme isolate are reported as n-fold changes in IC50s relative to the reference enzyme NS5BΔC21 1b J4 (Fig. 2). We found that both analogs maintained inhibition in subtype 1b. However, in subtype 1a, the inhibition of 1,5-BZDs was notably reduced for compound 2 and severely impaired for compound 1. These data indicate that the O-benzyl 1,5-BZD analog compound 2 is a better inhibitor of genotype 1a than the dichloro-phenyl analog compound 1.
FIG. 1.
Structural representation of the 1,5-BZDs used in this study. According to the Cahn-Ingold-Prelog nomenclature rules, the configuration at the chiral center is R for compounds 1 and 2 and S for compounds 3 and 4.
FIG. 2.
Inhibitory activities of compounds 1 and 2 against the NS5B enzyme isolate panel. Gray bars represent compound 1, and white bars represent compound 2. Inhibitory activities determined using a primer-dependent RdRp assay are reported as n-fold changes in the IC50 relative to the IC50 for NS5BΔC21 1b J4. The 1a_2, 1a_6, 1b_9, and 1b_10 enzyme isolates were previously described, and the other enzymes were cloned and overexpressed as described previously (38). Compounds with a change in the IC50 of >10-fold were considered to be less active (dotted line). The experiment was performed between 1 and 10 times in quadruplicate. Fold changes are reported as median values ± interquartile ranges.
Based on the results in the 1,5-BZD O-benzyl series, structure-based design was used to guide medicinal chemistry and improve upon the potency of the initial leads (47). These efforts resulted in the identification of the 1,1-dioxo-1-thia-5,10-diazadibenzocycloheptene analogs compounds 3 and 4 (Fig. 1), potent inhibitors of genotype 1b replication (Table 1). The luciferase reporter data from the replicon assay were generally confirmed by direct quantitative reverse transcription-PCR quantification of replicon RNA with EC50s of <100 nM (Table 1). To assess if the 1,5-BZD subtype coverage improves in genotype 1 with these sulfone analogs, we used replicons derived from genotype 1a (H77) and genotype 1b (Con1). Compounds 3 and 4 were found to be potent inhibitors of genotype 1b but were notably less active against genotype 1a (Table 1). The genotype 1a EC50 fold change values of compounds 3 and 4 relative to the subgenomic Con1b replicon (Table 1, 5.5- and 7.5-fold changes, respectively) or relative to the Huh7-Luc replicon (10.2- and 13.1-fold changes, respectively) are in the same range as the 1a IC50 fold changes of compound 2 relative to 1b J4 in the enzyme isolate panel (Fig. 2). Notably, no major cytotoxicity in various cell lines, including MT4, Huh7, HepG2, Vero, and HEK293, was observed with compounds 3 and 4, with 50% cytotoxic concentration [CC50] values of >32 μM (Table 2). Thus, despite increased potency, the 1,5-BZD sulfone analogs still remained less active against subtype 1a.
TABLE 1.
Antiviral activities of compounds 3 and 4
| Assay | Genotype | Compound 3 |
Compound 4 |
||
|---|---|---|---|---|---|
| EC50, EC90 (μM)a | IQRsb (μM) | EC50, EC90 (μM) | IQRs (μM) | ||
| Huh7-Lucc | 1b | 0.019, 0.118 | 0.013-0.024, 0.094-0.186 | 0.014, 0.082 | 0.010-0.019, 0.071-0.120 |
| Huh7-Lucd | 1b | 0.036, 0.263 | 0.022-0.067, 0.170-0.427 | 0.024, 0.203 | 0.010-0.065, 0.116-0.353 |
| Huh7-SGcon1bd | 1b | 0.067, 0.929 | 0.027-0.109, 0.350-1.217 | 0.042, 0.211 | 0.006-0.105, 0.144-7.375 |
| Huh7-SG1a H77d | 1a | 0.368, 3.772 | 0.157-0.926, 1.866-8.257 | 0.314, 3.378 | 0.098-0.626, 1.480-8.263 |
EC50 and EC90 values for cell-based replicons are expressed as median values from at least six independent experiments.
IQR, interquartile range (Q1 to Q3).
Luciferase readout.
Quantitative reverse transcription-PCR readout. No significant effect of compounds 3 and 4 on the cellular ribosomal protein RPL13A transcript level was observed in any of the cell lines tested, supporting specificity for HCV rather than an impact on replicon cell viability or division (data not shown).
TABLE 2.
Cytotoxicity of compounds 3 and 4
| Cell line | Compound 3 |
Compound 4 |
||
|---|---|---|---|---|
| CC50a (μM) | IQRb (μM) | CC50 (μM) | IQR (μM) | |
| MT4 | 54 | 52-56 | 88 | 83-93 |
| Huh7 | 60 | 50-73 | 56 | 42-74 |
| HepG2 | 53 | 48-65 | >100 | NAc |
| HEK293T | 40 | 39-46 | 99 | NA |
| Vero | 74 | 71-77 | >98 | NA |
CC50s are expressed as median values from two to nine independent experiments performed in quadruplicate. Cytotoxicity was measured by determining resazurin staining.
IQR, interquartile range (Q1 to Q3).
NA, not applicable.
Structural comparison between NS5B polymerases of genotypes 1a and 1b.
Given the global prevalence of subtype 1a, it is important to understand why the 1,5-BZD inhibitors are less potent against this genotype. We determined the crystal structures of the complexes formed between compound 4 and the 1a_18 and 1b J4 polymerases C-terminally truncated by 21 residues at 1.90 Å and 1.55 Å resolutions, respectively. To our knowledge, this represents the first publicly disclosed three-dimensional structure of a genotype 1a NS5B polymerase in complex with an inhibitor (F. Gray and colleagues at GlaxoSmithKline have reported a genotype 1a NS5B-inhibitor complex structure [13], but this structure is not publicly available). The primary sequences of our 1a and 1b enzymes are 88% identical (data not shown). The characteristic right-hand shape with finger, palm, and thumb domains encircling the active site is strictly conserved in the new structure (Fig. 3A), and various structural superimpositions establish that the 1a and 1b structures reported here are similar to previous 1b structures (6, 26, 32, 36, 37, 47). The three monomers of the asymmetric unit of the new 1a structure are fully consistent, with pairwise Cα root mean square deviations (RMSDs) of ≤0.23 Å. Similarly, the two monomers of the asymmetric unit of the new 1b structure superimpose with a Cα RMSD of 0.27 Å. The six possible 1a-1b monomer-monomer pairs from the new structures all superimpose with Cα RMSDs in the range of 0.45 to 0.55 Å (Fig. 3A). Another useful frame of reference is comparison to our previous 1b structures with inhibitors of the same 1,5-BZD chemotype (compounds 1 and 2) bound at the same site (32, 47). The various monomer-monomer superimpositions of these two previous structures with the new 1b structure are within the RMSD range of 0.31 to 0.65 Å. Thus, the 1a-4 complex structure is as similar to the 1b-4 complex structure as the 1b-4 complex structure is to the 1b-1 and 1b-2 complexes.
FIG. 3.
X-ray structure of the 1.90 Å and 1.55 Å resolution crystal structure of BZD 4 bound to HCV NS5B of genotypes 1a and 1b. (A) Coil representation of an overlay of HCV NS5B of genotype 1a and genotype 1b. Genotype 1b is shown in purple by atom, and genotype 1a is shown in white by atom. An arrow indicates the position of each NNI binding site. The areas color coded cyan, yellow, blue, and green represent the NNI-1, -2, -3, and -4 binding sites. (B) Overlay of the 1,5-BZD binding sites of genotypes 1a and 1b. Genotype 1b is shown in purple by atom, and genotype 1a is shown in white by atom. The highlighted hydrogen bonds are of genotype 1b. This hydrogen bonding network of the inhibitor hydroxyl has also been observed in previously reported BZD-NS5B bound structures (Protein Data Bank codes 3GOL, 3GNV, and 3GNW). (C and D) 1,5-BZD binding site of genotype 1b and genotype 1a, respectively. The side chain of Arg386 and its hydrogen bond with the inhibitor hydroxyl have been removed for a better view of residue 415. The two polymorphic amino acid side chains Y415F and Q446E are highlighted in green.
Similar to previous 1b-1,5-BZD complexes, we have described (32, 36, 47), the NS5B-4 complex is observed in the closed active conformation for both genotypes 1a and 1b. On the basis of our earlier studies, we previously concluded that binding of 1,5-BZDs did not lead to a significant conformational change in NS5B (32, 36, 47). Our present results are consistent with those of our earlier study and allow us to tentatively extend this conclusion to include the 1a enzyme. This is distinct from what has been observed for the NNI-1 and -2 inhibitors, where inhibitor binding involves significant conformational adjustment of NS5B (4, 5, 10).
Comparison of NS5B 1a and NS5B 1b BZD interactions.
Bound compound 4 occupies the NNI-3 site of genotype 1b NS5B, near the enzyme active site, with a binding mode very similar to that described for compound 4c in a previous report, Protein Data Bank identification code 3GNW (47). The alkoxyphenyl group is the most deeply buried part of the bound inhibitor, making extensive hydrophobic contact with the NNI-3 pocket (Fig. 3B). The inhibitor sulfone group hydrogen bonds with Tyr448:N and Gly449:N, and the inhibitor hydroxyl, involved in an extensive and partially water-mediated hydrogen bonding network that also includes Arg386 and Tyr415, hydrogen bonds directly with Ser367:OG. The oxazole O of bound compound 4 is positioned to form a weak hydrogen bond with Ser556:OG.
The major binding interactions noted above for the 1b-4 complex are largely conserved in the 1a-4 complex. Y415F and Q446E represent the two residue differences in the NNI-3 sites of genotypes 1a and 1b occupied by bound 1,5-BZDs (Fig. 3C and D). These changes do not affect the side chain conformations at these two positions. Interestingly, the Y415F change does not disturb the inhibitor binding mode at all. This is perhaps surprising, given the close and extensive contact between this side chain and the bound inhibitor. Furthermore, and perhaps even more surprising, the Y415F change allows conservation of a bound water molecule (Fig. 3D) that hydrogen bonds with the inhibitor and Tyr415 hydroxyls in the 1b structure (Fig. 3C). Loss of the later contact with Y415F explains the decrease in BZD potency observed in genotype 1a, as the binding site is less complementary in the absence of the Tyr hydroxyl moiety. How Q446E influences inhibitor potency remains unclear.
Analysis of NNI-1, -2, and -4.
Comparison of the 1a and 1b complexes with compound 4 indicates that the other NNI binding sites, NNI-1, -2, and -4, are highly structurally conserved between these two NS5B complexes (Fig. 3A). The NNI-1 site is almost identical for the two enzymes. In the absence of an NNI-1 ligand, helix A of the mobile Λ1 loop occupies the known indole-binding site (10, 20), as has been observed in other structures (6, 26, 32, 47). The change from Thr (1b) to Ala (1a) at position 499 may impact the conformation of the side chain of Arg503. The guanidine of Arg503 makes an electrostatic binding contact in the available structures of NNI-1 indole-NS5B complexes (10, 20), so modulating conformational preference at this position could impact inhibitor affinity; this is, however, highly speculative and requires further structural studies for clarification. Interestingly, the Ala499 variant has been suggested as a possible cause of benzimidazole resistance (42). The NNI-2 sites of the 1a and 1b complexes with compound 4 are also identical, with the exception of the side chain conformations of a couple of solvent-exposed polar side chains that do not make direct binding contacts with inhibitors at this site (4). Two key NNI-2 residues, Met423 and Leu497, are in identical conformations in the 1a-4, 1b-4, and 1b-NNI-2 inhibitor complexes (4). Finally, the NNI-4 sites of the 1a-4 and 1b-4 structures are also highly structurally conserved, with no significant differences observed between the two. The key structural elements of the NNI-4 sites of the two new structures, including the active-site loop, the serine-rich loop, and helix M, also superimpose well with those features of the HCV-796-NS5B-bound complex (19). In the absence of an NNI-4 ligand, in both the 1a-4 and 1b-4 complexes, Arg200 adopts a more extended conformation that overlaps with the HCV-796 binding site (19).
Reverse genetics of the NNI-3 binding site.
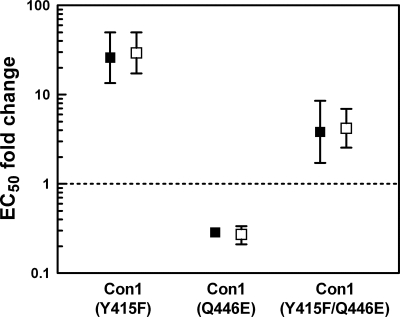
To investigate if the polymorphism of the 1,5-BZD binding site is the sole cause of the observed loss of inhibition of genotype 1a, we mutated Tyr415 and Gln446 in the wild-type Con1 NS5B sequence of the genotype 1b ET replicon (27) to Phe415 and Glu446. The resulting Y415F and Q446E single mutations and the Y415F/Q446E double mutation were introduced into a genotype 1b replicon that allows transient replicon replication when transfected into permissive Huh7 Lunet cells (23). Consistent with our previous findings with an acyl pyrrolidine NNI-3 inhibitor (38), compounds 3 and 4 lost significant activity against the Y415F single mutant (Fig. 4). However, the Q446E mutation resulted in a slight increase in sensitivity to inhibition. Against the Y415F/Q446E double mutant, compounds 3 and 4 showed an intermediate decrease in activity of about fourfold. Consistent with the double-mutant data, the fold change value with the H77 1a replicon relative to the subgenomic Con1b replicon is in the same range (Table 1, 5.5- and 7.5-fold changes for compounds 3 and 4, respectively). Altogether, these data indicate that the Y415F variant is a key determinant in conferring resistance in subtype 1a.
FIG. 4.
Inhibitory activities of compounds 3 and 4 against replicon NS5B mutants. Filled symbols represent compound 3, and empty symbols represent compound 4. Inhibitory activities are reported as n-fold changes in EC50 relative to the EC50 obtained against the ET replicon. Compounds with n-fold EC50 changes above the dotted line were considered to be less active than against the ET reference strain.
HCV NS5B chimeric replicons from clinical isolates of subtypes 1a and 1b show different NNI susceptibilities.
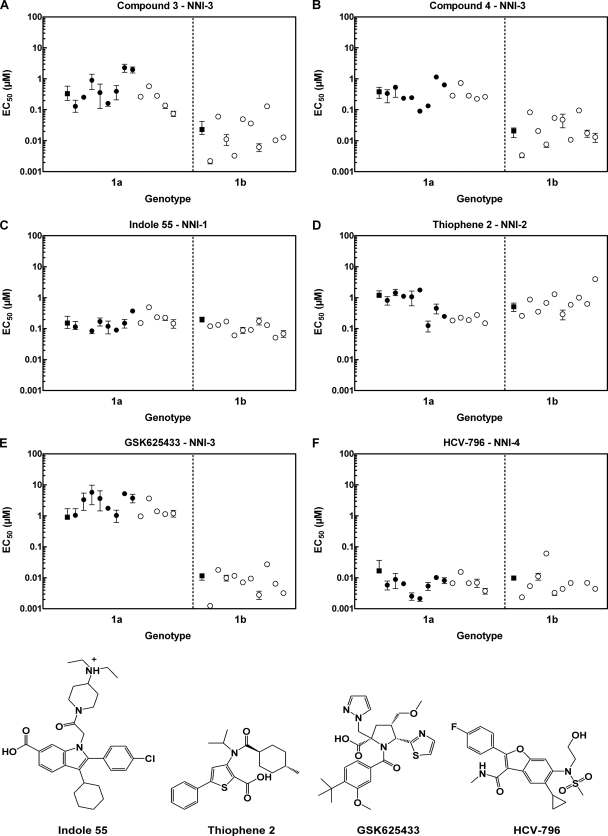
The NS5B enzymatic genotypic panel described in Fig. 2 is not appropriate to profile potent inhibitors such as compounds 3 and 4 because of the high enzyme concentration required to perform this assay. Therefore, we decided to generate chimeric NS5B replicons from clinical isolates representing a broad set of circulating 1a and 1b genotypes. We assembled 13 isolates of genotype 1a and 10 of genotype 1b from a collection of clinical sera as single clones or pools of clones (25, 34). When the inhibitory activities of the 1,5-BZDs against this panel were assessed, we found that compounds 3 and 4 maintained their potency across the genotype 1b isolates, albeit with some fluctuation for four 1b isolates (Fig. 5A and B). Sequencing revealed that three out of these four isolates bear the G556S variant, a mutation which results in the loss of an H bond with the oxazole moiety of the 1,5-BZDs (Fig. 3B).
FIG. 5.
Genotypic profiling of compounds 3 and 4 and reference compounds against HCV NS5B chimeric replicons. The black filled symbols refer to the chimeras representing single clones, and the empty white symbols refer to pools of clones. Circles, replicon NS5B chimeric isolate from clinical sera; squares, ET replicon for subtype 1b (23) and H77 chimera for subtype 1a. Inhibitory activities are reported as median EC50s (μM). The median EC50s of each inhibitor for genotypes 1a and 1b are listed in Table 3.
In the genotype 1a panel, the pattern of 1,5-BZD inhibition was quite variable and reminiscent of that of A-782759, a potent benzothiadiazine inhibitor (34). The EC50s ranged between 0.074 and 2.19 μM for compound 3 and between 0.09 and 1.15 μM for compound 4. The median EC50 across all genotype 1a NS5B chimeric replicons was 0.28 μM for both compounds (Table 3).
TABLE 3.
Median EC50s of HCV NS5B NNIs for genotype 1a and 1b chimeric replicons
| NNI | Genotype 1a |
Genotype 1b |
||
|---|---|---|---|---|
| EC50a (μM) | IQRb (μM) | EC50 (μM) | IQR (μM) | |
| Compound 3 | 0.28 | 0.16-0.58 | 0.012 | 0.007-0.046 |
| Compound 4 | 0.28 | 0.24-0.53 | 0.019 | 0.011-0.052 |
| Indole 55 | 0.15 | 0.12-0.23 | 0.11 | 0.07-0.13 |
| Thiophene 2 | 0.28 | 0.19-1.07 | 0.65 | 0.41-0.98 |
| GSK625433 | 1.75 | 1.13-3.66 | 0.008 | 0.004-0.011 |
| HCV-796 | 0.007 | 0.005-0.008 | 0.005 | 0.004-0.007 |
The median EC50 values for genotypes 1a and 1b were calculated from the median EC50 values of each individual NS5B chimeric replicon obtained from 13 clinical isolates and 10 clinical isolates, respectively. Each EC50 results from at least two independent experiments performed in quadruplicate.
IQR, interquartile range (Q1 to Q3).
To compare the BZD inhibition pattern to those of other NNIs, we synthesized the reference compounds indole 55 (NNI-1), thiophene 2 (NNI-2), GSK625433 (NNI-3), and HCV-796 (NNI-4) (36) and assessed their inhibitory activities in our HCV NS5B replicon chimeric panel. Except for NNI-3, the inhibition was well maintained across genotype 1 NS5B isolates. The inhibitory activity of these compounds was affected the least by the heterogeneity of the NS5B isolates in the following order: indole 55, HCV-796, thiophene 2, and GSK625433 (Fig. 5C to F). The inhibition by the NNI-1 and -4 analogs was consistent across the isolates, except for one 1b sample, where HCV-796 displayed an 8.6-fold change relative to ET. NS5B sequencing of this particular isolate revealed the presence of the C316N variant, a mutation known to confer resistance to HCV-796 (19). In NNI-2, the response was slightly more variable, with EC50s ranging from 0.18 to 1.43 μM (Fig. 5D). Finally, the NNI-3 acyl pyrrolidine analog GSK625433 lost considerable inhibitory activity against genotype 1a (Fig. 5E). The EC50s ranged between 0.96 and 5.69 μM in genotype 1a and between 0.0012 and 0.027 μM in genotype 1b. These results indicate that inhibition of genotype 1a polymerases can differ across and within nonnucleoside binding sites.
Mechanism of subtype 1a resistance in NNI-3.
To gain further insights into the difference in the inhibition of NNI-3 analogs in subtypes 1a and 1b, we studied the binding kinetics of compounds 3 and 4 and GSK625433 to NS5B by SPR to determine the dissociation rate constants kon, koff, and Kd and t1/2. Figure 6A exemplifies the trace of compound 4 binding to the wild-type 1b J4 protein, and its binding specificity for the NS5B NNI-3 site was confirmed by SPR using the 1b J4 (M414T) mutant polymerase (Fig. 6B). Binding kinetics against recombinant NS5BΔC21 (Con1, 1b J4, 1a_18, and 1a_H77) are reported in Table 4. We found that 1,5-BZDs 3 and 4 bind to the genotype 1b enzymes with higher affinity than to the genotype 1a enzymes (compare Kd values). Binding kinetic analysis revealed that the kon values of compounds 3 and 4 were similar for all NS5B enzymes, unlike the koff values, which were found to be higher for the 1a enzymes. The koff value is an indicator of the stability of the ligand-analyte complex and can be converted into t1/2, which defines the ligand residence time (representative of the interaction time between the inhibitor and NS5B). The residence times of compounds 3 and 4 indicate that the decrease in 1,5-BZD inhibition in subtype 1a is due to a lower stability of the 1,5-BZD/NS5B complex and not to a difference in recognition of the NNI-3 site, which would be reflected by lower kon values. The binding kinetics of GSK625433, less potent than compound 4 against the genotype 1a NS5B chimeric replicons (median EC50, 1.75 μM), could not be measured against the 1a enzymes because they were too fast. Despite this limitation, a binding affinity could still be calculated by using a steady-state model (see Materials and Methods) and was found to be in the same range as that of compound 4. These data suggest that GSK625433 is still able to bind to the enzyme but with such rapid kinetics that genotype 1a inhibition is even more impaired than in the case of the 1,5-BZD analogs.
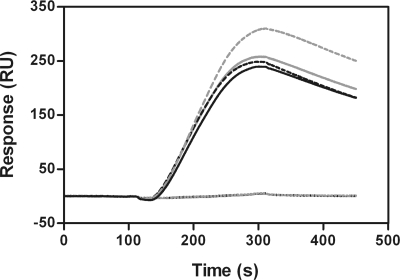
FIG. 6.
Representative sensorgrams for the binding of compound 4 to NS5BΔC21 (A) wild-type 1b J4 and (B) NNI-3 mutant 1b J4 M414T. Increasing concentrations of compound 4 (15.6 nM to 250 nM for 1b J4, 156 nM to 2.5 μM for 1b J4 M414T) were sequentially injected onto the immobilized proteins. The double-reference subtracted data (gray traces) were fitted by using a 1:1 model (black traces). The M414T mutant does not affect the recognition of the compound (kon value) for the NNI-3 binding site (so no major conformational change of the binding pocket is expected) but instead strongly reduces the stability of the complex and the compound residence time (t1/2 = 12 min versus 2 h 57 min for 1b J4).
TABLE 4.
Values of dissociation constants kon, koff, and Kd and t1/2 of HCV NS5B NNIs determined by SPR
| Compound and NS5BΔ21 enzyme | Parameter |
|||
|---|---|---|---|---|
| kon (M−1s−1)a | koff (s−1)a | Kd (nM)a | t1/2 (s) | |
| Compound 3 | ||||
| Con1 | (4.6 ± 0.4)E+04 | (7.6 ± 0.0)E−05 | 1.6 ± 0.1 | 9,150 |
| 1a_18 | (4.7 ± 1.6)E+04 | (5.7 ± 1.4)E−04 | 14 ± 8 | 1,211 |
| 1b J4 | (3.5 ± 0.1)E+04 | (7.1 ± 2.6)E−05 | 2.1 ± 0.8 | 9,831 |
| 1a_H77 | (3.6 ± 1.7)E+04 | (3.0 ± 8.3)E−04 | 8.6 ± 1.7 | 2,321 |
| Compound 4 | ||||
| Con1 | (3.1 ± 0.4)E+04 | (1.2 ± 0.1)E−04 | 4.0 ± 0.9 | 5,764 |
| 1a_18 | (3.5 ± 1.3)E+04 | (2.7 ± 0.5)E−03 | 80 ± 15 | 256 |
| 1b J4 | (2.8 ± 0.1)E+04 | (6.5 ± 4.6)E−05 | 2.3 ± 1.7 | 10,602 |
| 1a_H77 | (2.3 ± 0.1)E+04 | (8.8 ± 0.3)E−04 | 38 ± 0.5 | 786 |
| GSK625433 | ||||
| Con1 | (6.5 ± 1.5)E+04 | (6.6 ± 3.6)E−05 | 0.98 ± 0.3 | 10,501 |
| 1a_18b | NAd | NA | 110 ± 90 | NA |
| 1b J4 | (5.6 ± 0.9)E+04 | (5.8 ± 5.6)E−05 | 0.98 ± 0.9 | 11,925 |
| 1a_H77c | (1.1 ± 0.1)E+05 | (1.2 ± 0.0)E−02 | 110 ± 4.2 | 60 |
| HCV-796 | ||||
| Con1 | (7.2 ± 0.6)E+03 | (1.2 ± 0.3)E−04 | 16 ± 3 | 6,011 |
| 1a_18 | (1.4 ± 0.1)E+05 | (1.4 ± 0.1)E−03 | 10 ± 2 | 485 |
| 1b J4 | (2.2 ± 0.3)E+03 | (3.3 ± 0.4)E−04 | 150 ± 0.4 | 2,117 |
| 1a_H77 | (2.3 ± 0.2)E+05 | (3.4 ± 0.2)E−03 | 15 ± 1 | 204 |
Values are means ± standard deviations from at least two independent experiments.
Data were analyzed using the steady-state model (yielding only a global Kd) because the kinetics were too fast to fit properly using a kinetic model (see Materials and Methods).
Data were analyzed using a heterogeneous ligand binding model because the standard 1:1 binding model did not properly fit the data (see Materials and Methods).
NA, not applicable.
As expected, HCV-796 bound with similar affinities to the genotype 1a and 1b enzymes (Table 4), except to 1b J4, which contains the C316N variant (19). The Kd fold change of HCV-796 against 1b J4 relative to Con1 (9.4-fold) was in good agreement with the data of the NS5B chimeric replicon isolate bearing the C316N variant (Fig. 5F, 8.6-fold). Consistent with a previous report (15), HCV-796 displayed slow kinetics of binding to the genotype 1b enzymes, with slow association and dissociation rates. However, we found that the rate of association of HCV-796 with NS5B was faster in genotype 1a but that the overall binding affinity remained similar because of the concomitant increase in the dissociation rate. Thus, despite similar binding affinities, the differences in the binding kinetics of HCV-796 against these subtypes result in a shorter residence time in genotype 1a, which potentially could have consequences for the duration of the pharmacological effect of the drug in clinical settings (8). Taken together, these data indicate that the mechanism of subtype 1a resistance to NNI-3 analogs is faster dissociation of the inhibitor from the enzyme.
Mechanism of inhibition of 1,5-BZDs.
We have shown previously that the mode of 1,5-BZD RdRp inhibition is noncompetitive with regard to GTP (36). Using a gel-based assay, we have also shown that these compounds prevent the formation of the first phosphodiester bond during the polymerization cycle. Here we used an SPR method reported previously (14) to assess if the 1,5-BZDs can form a ternary complex with HCV NS5B and RNA. The response difference shown in Fig. 7 corresponds to a direct measurement of the change in the molecular weight of the RNA tethered to the streptavidin chip surface. The differential for 1,5-BZDs 3 and 4, GSK625433, and the benzothiadiazine analog reported previously (14) was 0 RU, indicating that these compounds do not bind nonspecifically to the template. As reported (14), the response differential of the benzothiadiazine analog was higher than HCV NS5B alone, thereby indicating that NS5B is able to bind to the chip-tethered RNA and that the addition of this compound further increased the affinity of NS5B for the RNA. However, the response differentials of 1,5-BZDs 3 and 4 were similar to that of NS5B alone. These data indicate that the addition of 1,5-BZDs does not interfere with the interaction of the polymerase to the RNA template. Hence, the 1,5-BZDs most likely prevent translocation of the quaternary initiation complex once it is assembled at the catalytic site.
FIG. 7.
Effects of NNIs on the binding of HCV NS5B to RNA by SPR. The baseline curves consist of the RNA template with or without NNIs. The black curve represents NS5B only, the dotted black curve represents compound 4, the gray curve represents compound 3, and the gray dotted curve represents the benzothiadiazine analog previously described (14).
Binding affinities of HCV polymerase NNIs correlate with inhibitory activities obtained in the replicon, but not in the biochemical RdRp assay.
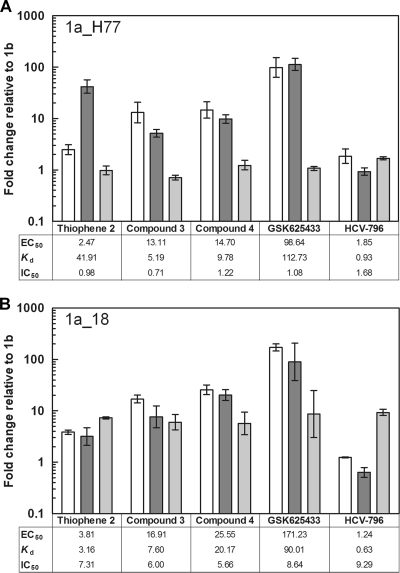
A recent study has shown that the IC50s of NNI-3 and -4 binding derivatives correlate well with their respective Kd values when tested against the NS5BΔC21 Con1 enzyme (15). Therefore, we used the Con 1 strain as a 1b reference to compare to the 1a/1b subgenotypic profile of our NNI analogs. The EC50s, IC50s, and Kd values of 1,5-BZDs 3 and 4 and three reference compounds against NS5B 1a_H77 and 1a_18 were obtained with the NS5B chimeric replicon system, the biochemical RdRp assay, and the SPR binding assay, respectively, and plotted as n-fold changes relative to the Con1 reference. Unexpectedly, no change in IC50 relative to that of Con1 was observed against the 1a_H77 enzyme for any of the compounds tested (Fig. 8A). In contrast, with the 1a_18 enzyme (Fig. 8B), a reduction of inhibitory activity (5- to 10-fold) was observed for all of the compounds tested. However, it should be noted that the data for this enzyme were probably biased because of the higher enzyme concentration (200 nM) required for the RdRp assay, due to the lower catalytic competency of 1a_18. Therefore, 1a_H77 is more appropriate to measure the changes in IC50 because it can be used at the same concentration as the Con1 enzyme (20 nM). We found that the EC50 and Kd fold changes correlated well for all of the inhibitors with both the 1a_H77 and 1a_18 enzymes (Fig. 8), except for thiophene 2: this compound showed a Kd fold change which was unexpectedly too high against the 1a_H77 enzyme. This result may be due to the low level of binding (maximum binding level, <10 RU), which prevented accurate Kd determination. Altogether, these data suggest that the biochemical RdRp assay should be used with caution for 1a/1b subtype profiling and that the measurement of binding affinities for recombinant enzymes offers a valid surrogate.
FIG. 8.
Correlation of EC50s, Kds, and IC50s of HCV NS5B NNIs between subtypes 1a and 1b using (A) the 1a_H77 strain and (B) the 1a_18 clinical isolate. 1a_18 and 1a_H77 are identical in sequence to NS5B of the enzyme isolates used in Fig. 2 and of the Huh7-SG1a H77 replicon (Table 1), respectively. The n-fold changes in EC50, IC50, and Kd were calculated relative to the HCV NS5B Con1 isolate (genotype 1b). White bars, EC50; dark gray bars, Kd; light gray bars, IC50.
DISCUSSION
Here we report the 1a/1b subtype profiling of HCV NS5B NNIs obtained with a panel of NS5B chimeric replicons derived from clinical isolates and compare the differences in NNI susceptibilities across the various NNI binding sites. We show that NNIs generally maintain inhibition across these subtypes, except for NNI-3 analogs, whose potency in genotype 1a decreases to a degree that is dependent on both the chemotype and the nature of the clinical isolate. More specifically, we have focused our study on 1,5-BZD analogs, using reverse genetics, SPR studies, and X-ray crystallography, with the aim to understand the reasons for these fluctuations.
Subtype specificity in NNI-3 is not confined within a chemotype, as exemplified by 1,5-BZD 2, an analog that shows a dramatic difference in potency against genotype 1a compared to that of its counterpart 1,5-BZD 1. These data indicate that the O-benzyl moiety is an important determinant to improve the potency of the BZDs in genotype 1a and suggest that specificity in NNI-3 within a chemotype can be modulated by changing some part of the molecule despite the polymorphism of the binding site. Indeed, structural analysis revealed that the binding interactions of compound 2 to the HCV polymerase NS5BΔC21 1b J4 are identical to these of compound 1, except for the O-benzyl moiety, which is deeply buried in an NNI-3 subpocket defined by Pro197, Arg200, Leu384, Met414, Tyr415, and Tyr448 (47). We conclude that the increased hydrophobic and aromatic interaction of the O-benzyl moiety in this lipophilic pocket differentially improves inhibitor potency against genotypes 1a and 1b. On the other hand, the higher loss of potency observed with GSK625433 in genotype 1a indicates that subtype coverage in NNI-3 can also differ across chemotypes or behave similarly, as reported for the benzothiadiazine analog A-782759 (34).
To better understand these differences, we mutated the polymorphic residues of the Con1 subtype 1b replicon to subtype 1a and assessed if these mutations could account for the decrease in 1,5-BZD inhibition we observed with the H77 subgenomic and NS5B chimeric replicons. This led us to the finding that Y415F has a severe effect on 1,5-BZD inhibition. On the other hand, Q446E had the opposite effect and resulted in a slight increase in inhibition. This result was not expected, since E446Q was found to contribute to F415Y in restoring acyl pyrrolidine inhibition when the NNI-3 polymorphic residues of the 1a_6 enzyme isolate were reverted back to genotype 1b (38). Likewise, when the 1,5-BZDs were tested against a panel of NS5B chimeric replicons derived from genotype 1a clinical isolates, no variations other than the expected polymorphisms could be found in the panel of clinical isolates to explain the variability in 1,5-BZD inhibition. To investigate this further (see transient replicon assay in Materials and Methods), we searched the complete aligned NS5B primary sequences of the 1a panel for potential determinants beyond the NNI-3 site. This analysis implicated L184Q and K212R as mutations with a potentially biased distribution of fold change in EC50 for both compounds 3 and 4. However, the three-dimensional structure of subtype 1a reveals that the side chains of these residues are solvent exposed and remote from the binding pocket, arguing against a direct impact on 1,5-BZD binding.
The X-ray analysis of compound 4 in complex with the 1a_18 and 1b J4 polymerases explains, at least partially, the reverse genetic data. Interestingly, the direct inhibitor binding contacts of the 1b-4 complex are strictly conserved in the 1a-4 complex, and the position of a bound water molecule that appears to hydrogen bond with the hydroxyls of the inhibitor and Tyr415 in the subtype 1b complex is conserved in the subtype 1a complex despite the loss of the Y415F side chain hydroxyl in the 1a enzyme. The reduced complementarity of the subtype 1a inhibitor (plus water) binding site, due to the Y415F change, is consistent with the observed decrease in inhibitor potency against the 1a enzyme. Furthermore, the biophysical investigation of the binding kinetics of 1,5-BZDs to NS5B by SPR studies revealed that the binding interactions of compounds 3 and 4 with the 1a polymerase are weaker than those with the 1b enzyme. Altogether, these data suggest that the strength of the inhibitor hydroxyl hydrogen bonding network is weaker in genotype 1a. Even if the decrease in 1,5-BZD potency observed with 1a_18 in the N5SB chimeric replicon correlates well with the SPR studies, and a rationale to explain these differences could be found in the biophysical and X-ray studies, we cannot exclude the possibility that the variation in 1,5-BZD inhibition across 1a shuttle isolates results from an altered conformation of the polymerase not seen in the 1a polymerase structure described in this study. In summary, our data indicate that the factors that impact 1,5-BZD subgenotypic coverage include (i) a bound water molecule that mediates inhibitor contact with Tyr415, (ii) hydrophobic and aromatic interactions in the deep NNI-3 lipophilic pocket, (iii) lower stability of the NS5B-BZD complex in genotype 1a, and (iv) factors more subtle than direct binding interactions with the polymerase that we were not able to identify with the tools described here.
Last, we found that the RdRp biochemical assay must be used with caution when assessing HCV NS5B NNI analogs in these subtypes. As described previously with other NNI analogs (38), HCV NS5B recombinant enzymes were used successfully to demonstrate differences in genotype 1a susceptibilities to 1,5-BZDs 1 and 2. However, the results obtained when testing compounds 3 and 4 against the enzyme isolate panel were not consistent with the replicon and SPR data. This could be due to the enzyme concentration required to perform the RdRp assay, which may have exceeded the potency of compounds 3 and 4. Stable preannealed double-stranded RNAs are poor substrates for HCV NS5B. Hence, a solution to circumvent this issue may be to use di- or trinucleotides as primers, since they are known to catalyze primer extension more efficiently (49). In any case, the HCV NS5B chimeric replicon panel and SPR method described here are complementary tools for the genotypic and/or phenotypic assessment of NNI-3 analogs against clinical isolates. Alternatively, the tryptophan fluorescence quenching assay can perhaps also be used as an alternative to SPR studies (15).
The intrinsic heterogeneity of HCV polymerase in genotype 1-infected patients is a concern for the successful clinical development of NNIs. We and others (16, 25) have shown that the inhibitory activity of HCV NS5B NNIs is affected the least by the heterogeneity of the NS5B isolates in subtypes 1a and 1b in the following order: NNI-1, NNI-4, NNI-2, and NNI-3. Despite impressive genotypic versatility, HCV-796 has suffered limited clinical efficacy because of the presence of the C316N variant in a large proportion of genotype 1b-infected patients (19). Not surprisingly, the NNI-3 ANA598 was recently reported to be less efficacious in patients infected with subtype 1a in early clinical studies (2). Incidentally, F415Y has been reported as a mutation resulting from the use of ribavirin in monotherapy trials of genotype 1a HCV-infected patients, thereby reversing the key binding site polymorphism back to genotype 1b (30). It will be interesting to see if the NS5B chimeric replicon data described here for the 1,5-BZD series and GSK625433 will also translate into large variability in viral load decline when ANA598 and ABT-333 (33) are tested in a larger cohort of patients.
Acknowledgments
We are most grateful to Hugo Ceulemans for the alignment and analysis of the NS5B clinical isolates, to Inky De Baere for technical assistance in the plasmon resonance experiments, and to Sabine Höppner (Proteros Biostructures GmbH) for managing the X-ray crystallography studies. We thank Anita Howe and Luc Geeraert for critical reading of the manuscript.
Footnotes
Published ahead of print on 13 January 2010.
REFERENCES
- 1.Alter, M. J., D. Kruszon-Moran, O. V. Nainan, G. M. McQuillan, F. Gao, L. A. Moyer, R. A. Kaslow, and H. S. Margolis. 1999. The prevalence of hepatitis C virus infection in the United States, 1988 through 1994. N. Engl. J. Med. 341:556-562. [DOI] [PubMed] [Google Scholar]
- 2.Anadys Pharmaceuticals, Inc. 9 January 2009. ANA598 demonstrates potent antiviral activity in an early clinical study in HCV-infected patients. http://www.medicalnewstoday.com/articles/134837.php.
- 3.Behrens, S. E., L. Tomei, and R. De Francesco. 1996. Identification and properties of the RNA-dependent RNA polymerase of hepatitis C virus. EMBO J. 15:12-22. [PMC free article] [PubMed] [Google Scholar]
- 4.Biswal, B. K., M. M. Cherney, M. Wang, L. Chan, C. G. Yannopoulos, D. Bilimoria, O. Nicolas, J. Bedard, and M. N. James. 2005. Crystal structures of the RNA-dependent RNA polymerase genotype 2a of hepatitis C virus reveal two conformations and suggest mechanisms of inhibition by non-nucleoside inhibitors. J. Biol. Chem. 280:18202-18210. [DOI] [PubMed] [Google Scholar]
- 5.Biswal, B. K., M. Wang, M. M. Cherney, L. Chan, C. G. Yannopoulos, D. Bilimoria, J. Bedard, and M. N. James. 2006. Non-nucleoside inhibitors binding to hepatitis C virus NS5B polymerase reveal a novel mechanism of inhibition. J. Mol. Biol. 361:33-45. [DOI] [PubMed] [Google Scholar]
- 6.Bressanelli, S., L. Tomei, A. Roussel, I. Incitti, R. L. Vitale, M. Mathieu, R. De Francesco, and F. A. Rey. 1999. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Proc. Natl. Acad. Sci. U. S. A. 96:13034-13039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, C. M., Y. He, L. Lu, H. B. Lim, R. L. Tripathi, T. Middleton, L. E. Hernandez, D. W. Beno, M. A. Long, W. M. Kati, T. D. Bosse, D. P. Larson, R. Wagner, R. E. Lanford, W. E. Kohlbrenner, D. J. Kempf, T. J. Pilot-Matias, and A. Molla. 2007. Activity of a potent hepatitis C virus polymerase inhibitor in the chimpanzee model. Antimicrob. Agents Chemother. 51:4290-4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Copeland, R. A., D. L. Pompliano, and T. D. Meek. 2006. Drug-target residence time and its implications for lead optimization. Nat. Rev. Drug Discov. 5:730-739. [DOI] [PubMed] [Google Scholar]
- 9.Dhanak, D., K. J. Duffy, V. K. Johnston, J. Lin-Goerke, M. Darcy, A. N. Shaw, B. Gu, C. Silverman, A. T. Gates, M. R. Nonnemacher, D. L. Earnshaw, D. J. Casper, A. Kaura, A. Baker, C. Greenwood, L. L. Gutshall, D. Maley, A. DelVecchio, R. Macarron, G. A. Hofmann, Z. Alnoah, H. Y. Cheng, G. Chan, S. Khandekar, R. M. Keenan, and R. T. Sarisky. 2002. Identification and biological characterization of heterocyclic inhibitors of the hepatitis C virus RNA-dependent RNA polymerase. J. Biol. Chem. 277:38322-38327. [DOI] [PubMed] [Google Scholar]
- 10.Di Marco, S., C. Volpari, L. Tomei, S. Altamura, S. Harper, F. Narjes, U. Koch, M. Rowley, R. De Francesco, G. Migliaccio, and A. Carfi. 2005. Interdomain communication in hepatitis C virus polymerase abolished by small molecule inhibitors bound to a novel allosteric site. J. Biol. Chem. 280:29765-29770. [DOI] [PubMed] [Google Scholar]
- 11.Esteban, J. I., S. Sauleda, and J. Quer. 2008. The changing epidemiology of hepatitis C virus infection in Europe. J. Hepatol. 48:148-162. [DOI] [PubMed] [Google Scholar]
- 12.Fried, M. W., M. L. Shiffman, K. R. Reddy, C. Smith, G. Marinos, F. L. Goncales, Jr., D. Haussinger, M. Diago, G. Carosi, D. Dhumeaux, A. Craxi, A. Lin, J. Hoffman, and J. Yu. 2002. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N. Engl. J. Med. 347:975-982. [DOI] [PubMed] [Google Scholar]
- 13.Gray, F., L. Amphlett, H. Bright, L. Chambers, A. Cheasty, R. Fenwick, D. Haigh, D. Hartley, P. Howes, R. Jarvest, F. Mirzai, F. Nerozzi, N. Parry, M. Slater, S. Smith, P. Thommes, C. Wilkinson, and E. Williams. 2007. GSK625433: a novel and highly potent inhibitor of the HCV NS5B polymerase. In 42nd Meet. Eur. Assoc. Study Liver Dis. http://www.natap.org/2007/EASL/EASL_13.htm.
- 14.Gu, B., V. K. Johnston, L. L. Gutshall, T. T. Nguyen, R. R. Gontarek, M. G. Darcy, R. Tedesco, D. Dhanak, K. J. Duffy, C. C. Kao, and R. T. Sarisky. 2003. Arresting initiation of hepatitis C virus RNA synthesis using heterocyclic derivatives. J. Biol. Chem. 278:16602-16607. [DOI] [PubMed] [Google Scholar]
- 15.Hang, J. Q., Y. Yang, S. F. Harris, V. Leveque, H. J. Whittington, S. Rajyaguru, G. Ao-Ieong, M. F. McCown, A. Wong, A. M. Giannetti, S. Le Pogam, F. Talamas, N. Cammack, I. Najera, and K. Klumpp. 2009. Slow-binding inhibition and mechanism of resistance of non-nucleoside polymerase inhibitors of hepatitis C virus. J. Biol. Chem. 284:15517-15529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herlihy, K. J., J. P. Graham, R. Kumpf, A. K. Patick, R. Duggal, and S. T. Shi. 2008. Development of intergenotypic chimeric replicons to determine the broad-spectrum antiviral activities of hepatitis C virus polymerase inhibitors. Antimicrob. Agents Chemother. 52:3523-3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hnatyszyn, H. J. 2005. Chronic hepatitis C and genotyping: the clinical significance of determining HCV genotypes. Antivir. Ther. 10:1-11. [PubMed] [Google Scholar]
- 18.Hoofnagle, J. H. 2002. Course and outcome of hepatitis C. Hepatology 36:S21-S29. [DOI] [PubMed] [Google Scholar]
- 19.Howe, A. Y., H. Cheng, S. Johann, S. Mullen, S. K. Chunduru, D. C. Young, J. Bard, R. Chopra, G. Krishnamurthy, T. Mansour, and J. O'Connell. 2008. Molecular mechanism of hepatitis C virus replicon variants with reduced susceptibility to a benzofuran inhibitor, HCV-796. Antimicrob. Agents Chemother. 52:3327-3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ikegashira, K., T. Oka, S. Hirashima, S. Noji, H. Yamanaka, Y. Hara, T. Adachi, J. Tsuruha, S. Doi, Y. Hase, T. Noguchi, I. Ando, N. Ogura, S. Ikeda, and H. Hashimoto. 2006. Discovery of conformationally constrained tetracyclic compounds as potent hepatitis C virus NS5B RNA polymerase inhibitors. J. Med. Chem. 49:6950-6953. [DOI] [PubMed] [Google Scholar]
- 21.Jacobson, I., P. Pockros, J. Lalezari, E. Lawitz, M. Rodrigues-Torres, E. DeJesus, F. Haas, C. Martorell, R. Pruitt, K. Durham, S. Srinivasan, M. Rosario, S. Jagannatha, and J. Hammond. 2009. Antiviral activity of filibuvir in combination with pegylated interferon alfa-2a and ribavirin for 28 days in treatment naive patients chronically infected with HCV genotype 1. In 44th Annu. Meet. Eur. Assoc. Study Liver. http://www.natap.org/2009/EASL/EASL_34.htm.
- 22.Karlsson, R., P. S. Katsamba, H. Nordin, E. Pol, and D. G. Myszka. 2006. Analyzing a kinetic titration series using affinity biosensors. Anal. Biochem. 349:136-147. [DOI] [PubMed] [Google Scholar]
- 23.Krieger, N., V. Lohmann, and R. Bartenschlager. 2001. Enhancement of hepatitis C virus RNA replication by cell culture-adaptive mutations. J. Virol. 75:4614-4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lawitz, E., C. Cooper, M. Rodriguez-Torres, R. Ghalib, R. Lalonde, A. Sheikh, B. Bourgault, N. Chauret, L. Proulx, and J. McHutchison. 2009. Safety, tolerability and antiviral activity of VCH-916, a novel non-nucleoside HCV polymerase inhibitor in patients with chronic HCV genotype-1 infection. J. Hepatol. 50(Suppl.):S37. [Google Scholar]
- 25.Le Pogam, S., A. Seshaadri, A. Kosaka, S. Chiu, H. Kang, S. Hu, S. Rajyaguru, J. Symons, N. Cammack, and I. Najera. 2008. Existence of hepatitis C virus NS5B variants naturally resistant to non-nucleoside, but not to nucleoside, polymerase inhibitors among untreated patients. J. Antimicrob. Chemother. 61:1205-1216. [DOI] [PubMed] [Google Scholar]
- 26.Lesburg, C. A., M. B. Cable, E. Ferrari, Z. Hong, A. F. Mannarino, and P. C. Weber. 1999. Crystal structure of the RNA-dependent RNA polymerase from hepatitis C virus reveals a fully encircled active site. Nat. Struct. Biol. 6:937-943. [DOI] [PubMed] [Google Scholar]
- 27.Lohmann, V., S. Hoffmann, U. Herian, F. Penin, and R. Bartenschlager. 2003. Viral and cellular determinants of hepatitis C virus RNA replication in cell culture. J. Virol. 77:3007-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lohmann, V., F. Korner, J. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 29.Love, R. A., H. E. Parge, X. Yu, M. J. Hickey, W. Diehl, J. Gao, H. Wriggers, A. Ekker, L. Wang, J. A. Thomson, P. S. Dragovich, and S. A. Fuhrman. 2003. Crystallographic identification of a noncompetitive inhibitor binding site on the hepatitis C virus NS5B RNA polymerase enzyme. J. Virol. 77:7575-7581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lutchman, G., S. Danehower, B. C. Song, T. J. Liang, J. H. Hoofnagle, M. Thomson, and M. G. Ghany. 2007. Mutation rate of the hepatitis C virus NS5B in patients undergoing treatment with ribavirin monotherapy. Gastroenterology 132:1757-1766. [DOI] [PubMed] [Google Scholar]
- 31.Manns, M. P., H. Wedemeyer, and M. Cornberg. 2006. Treating viral hepatitis C: efficacy, side effects, and complications. Gut 55:1350-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McGowan, D., O. Nyanguile, M. D. Cummings, S. Vendeville, K. Vandyck, W. Van den Broeck, C. W. Boutton, H. De Bondt, L. Quirynen, K. Amssoms, J. F. Bonfanti, S. Last, K. Rombauts, A. Tahri, L. Hu, F. Delouvroy, K. Vermeiren, G. Vandercruyssen, L. Van der Helm, E. Cleiren, W. Mostmans, P. Lory, G. Pille, K. Van Emelen, G. Fanning, F. Pauwels, T. I. Lin, K. Simmen, and P. Raboisson. 2009. 1,5-Benzodiazepine inhibitors of HCV NS5B polymerase. Bioorg. Med. Chem. Lett. 19:2492-2496. [DOI] [PubMed] [Google Scholar]
- 33.Menon, R., D. Cohen, A. Nada, E. O. Dumas, Y.-L. Chiu, T. Podsadecki, W. Awni, B. Bernstein, and C. E. Klein. 2009. Pharmacokinetics, safety and tolerability of the HCV polymerase inhibitor ABT-333 following single ascending doses in healthy adult volunteers. In 44th Annu. Meet. Eur. Assoc. Study Liver. http://www.natap.org/2009/EASL/EASL_53.htm.
- 34.Middleton, T., Y. He, T. Pilot-Matias, R. Tripathi, B. H. Lim, A. Roth, C. M. Chen, G. Koev, T. I. Ng, P. Krishnan, R. Pithawalla, R. Mondal, T. Dekhtyar, L. Lu, H. Mo, W. M. Kati, and A. Molla. 2007. A replicon-based shuttle vector system for assessing the phenotype of HCV NS5B polymerase genes isolated from patient populations. J. Virol. Methods. 145:137-145. [DOI] [PubMed] [Google Scholar]
- 35.Nguyen, M. H., and E. B. Keeffe. 2005. Prevalence and treatment of hepatitis C virus genotypes 4, 5, and 6. Clin. Gastroenterol. Hepatol. 3:S97-S101. [DOI] [PubMed] [Google Scholar]
- 36.Nyanguile, O., F. Pauwels, W. Van den Broeck, C. W. Boutton, L. Quirynen, T. Ivens, L. van der Helm, G. Vandercruyssen, W. Mostmans, F. Delouvroy, P. Dehertogh, M. D. Cummings, J. F. Bonfanti, K. A. Simmen, and P. Raboisson. 2008. 1,5-Benzodiazepines, a novel class of hepatitis C virus polymerase nonnucleoside inhibitors. Antimicrob. Agents Chemother. 52:4420-4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O'Farrell, D., R. Trowbridge, D. Rowlands, and J. Jager. 2003. Substrate complexes of hepatitis C virus RNA polymerase (HC-J4): structural evidence for nucleotide import and de-novo initiation. J. Mol. Biol. 326:1025-1035. [DOI] [PubMed] [Google Scholar]
- 38.Pauwels, F., W. Mostmans, L. M. Quirynen, L. van der Helm, C. W. Boutton, A. S. Rueff, E. Cleiren, P. Raboisson, D. Surleraux, O. Nyanguile, and K. A. Simmen. 2007. Binding-site identification and genotypic profiling of hepatitis C virus polymerase inhibitors. J. Virol. 81:6909-6919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Payan, C., F. Roudot-Thoraval, P. Marcellin, N. Bled, G. Duverlie, I. Fouchard-Hubert, P. Trimoulet, P. Couzigou, D. Cointe, C. Chaput, C. Henquell, A. Abergel, J. M. Pawlotsky, C. Hezode, M. Coude, A. Blanchi, S. Alain, V. Loustaud-Ratti, P. Chevallier, C. Trepo, V. Gerolami, I. Portal, P. Halfon, M. Bourliere, M. Bogard, E. Plouvier, C. Laffont, G. Agius, C. Silvain, V. Brodard, G. Thiefin, C. Buffet-Janvresse, G. Riachi, F. Grattard, T. Bourlet, F. Stoll-Keller, M. Doffoel, J. Izopet, K. Barange, M. Martinot-Peignoux, M. Branger, A. Rosenberg, P. Sogni, M. L. Chaix, S. Pol, V. Thibault, P. Opolon, A. Charrois, L. Serfaty, B. Fouqueray, J. D. Grange, J. J. Lefrere, and F. Lunel-Fabiani. 2005. Changing of hepatitis C virus genotype patterns in France at the beginning of the third millennium: the GEMHEP GenoCII Study. J. Viral Hepat. 12:405-413. [DOI] [PubMed] [Google Scholar]
- 40.Pietschmann, T., A. Kaul, G. Koutsoudakis, A. Shavinskaya, S. Kallis, E. Steinmann, K. Abid, F. Negro, M. Dreux, F. L. Cosset, and R. Bartenschlager. 2006. Construction and characterization of infectious intragenotypic and intergenotypic hepatitis C virus chimeras. Proc. Natl. Acad. Sci. U. S. A. 103:7408-7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pockros, P., M. Rodrigues-Torres, S. Villano, E. Maller, and M. Chojkier. 2009. A phase 2, randomized study of HCV-796 in combination with pegylated-interferon (PEG) plus ribavirin versus PEG plus RBV in hepatitis C virus genotype-1 infection. J. Hepatol. 50:S7. [Google Scholar]
- 42.Rydberg, E. H., A. Cellucci, L. Bartholomew, M. Mattu, G. Barbato, S. W. Ludmerer, D. J. Graham, S. Altamura, G. Paonessa, R. De Francesco, G. Migliaccio, and A. Carfi. 2009. Structural basis for resistance of the genotype 2b hepatitis C virus NS5B polymerase to site A non-nucleoside inhibitors. J. Mol. Biol. 390:1048-1059. [DOI] [PubMed] [Google Scholar]
- 43.Shi, S. T., K. J. Herlihy, J. P. Graham, J. Nonomiya, S. V. Rahavendran, H. Skor, R. Irvine, S. Binford, J. Tatlock, H. Li, J. Gonzalez, A. Linton, A. K. Patick, and C. Lewis. 2009. Preclinical characterization of PF-00868554, a potent nonnucleoside inhibitor of the hepatitis C virus RNA-dependent RNA polymerase. Antimicrob. Agents Chemother. 53:2544-2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simmonds, P., J. Bukh, C. Combet, G. Deleage, N. Enomoto, S. Feinstone, P. Halfon, G. Inchauspe, C. Kuiken, G. Maertens, M. Mizokami, D. G. Murphy, H. Okamoto, J. M. Pawlotsky, F. Penin, E. Sablon, I. T. Shin, L. J. Stuyver, H. J. Thiel, S. Viazov, A. J. Weiner, and A. Widell. 2005. Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology 42:962-973. [DOI] [PubMed] [Google Scholar]
- 45.Sulkowski, M. S. 2007. Specific targeted antiviral therapy for hepatitis C. Curr. Gastroenterol. Rep. 9:5-13. [DOI] [PubMed] [Google Scholar]
- 46.Sy, T., and M. M. Jamal. 2006. Epidemiology of hepatitis C virus (HCV) infection. Int. J. Med. Sci. 3:41-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vandyck, K., M. D. Cummings, O. Nyanguile, C. W. Boutton, S. Vendeville, D. McGowan, B. Devogelaere, K. Amssoms, S. Last, K. Rombauts, A. Tahri, P. Lory, L. Hu, D. A. Beauchamp, K. Simmen, and P. Raboisson. 2009. Structure-based design of a benzodiazepine scaffold yields a potent allosteric inhibitor of hepatitis C NS5B RNA polymerase. J. Med. Chem. 52:4099-4102. [DOI] [PubMed] [Google Scholar]
- 48.Wang, M., K. K. Ng, M. M. Cherney, L. Chan, C. G. Yannopoulos, J. Bedard, N. Morin, N. Nguyen-Ba, M. H. Alaoui-Ismaili, R. C. Bethell, and M. N. James. 2003. Non-nucleoside analogue inhibitors bind to an allosteric site on HCV NS5B polymerase. Crystal structures and mechanism of inhibition. J. Biol. Chem. 278:9489-9495. [DOI] [PubMed] [Google Scholar]
- 49.Zhong, W., E. Ferrari, C. A. Lesburg, D. Maag, S. K. Ghosh, C. E. Cameron, J. Y. Lau, and Z. Hong. 2000. Template/primer requirements and single nucleotide incorporation by hepatitis C virus nonstructural protein 5B polymerase. J. Virol. 74:9134-9143. [DOI] [PMC free article] [PubMed] [Google Scholar]