Abstract
Foamy viruses are a member of the spumavirus subfamily of retroviruses with unique mechanisms of virus replication. Foamy virus replication is cell cycle dependent; however, the genome is found in the nuclei of cells arrested in the G1/S phase. Despite the presence of genome in the nuclei of growth-arrested cells, there is no viral gene expression, thus explaining its dependency on cell cycle. This report shows that the foamy virus genome remains unintegrated in G1/S phase-arrested cells. The foamy virus genome is detected by confocal microscopy in the nuclei of both dividing and growth-arrested cells. Alu PCR revealed foamy virus-specific DNA amplification from genomic DNA isolated in cycling cells at 24 h postinfection. In arrested cells no foamy virus DNA band was detected in cells harvested at 1 or 7 days after infection, and a very faint band that is significantly less than DNA amplified from cycling cells was observed at day 15. After these cells were arrested at the G1/S phase for 1, 7, or 15 days they were allowed to cycle, at which time foamy virus-specific DNA amplification was readily observed. Taken together, these results suggest that the foamy virus genome persists in nondividing cells without integrating. We have also established evidence for the first time that the foamy virus genome and Gag translocation into the nucleus are dependent on integrase in cycling cells, implicating the role of integrase in transport of the preintegration complex into the nucleus. Furthermore, despite the presence of a nuclear localization signal sequence in Gag, we observed no foamy virus Gag importation into the nucleus in the absence of integrase.
Foamy viruses are found in many mammalian species and appear to be nonpathogenic in their natural hosts, even though they have a wide tissue range and induce extensive cytopathic effects in cell culture. The lack of disease by foamy virus infection has prompted their potential utility as a safe vector system for gene therapy applications. Principles for the design of foamy virus vectors are established, and several foamy virus vectors that efficiently transduce a variety of cell types from different species are available (39). Integration analysis of foamy virus vectors into the host chromosome reveal that they have a reduced risk of mutating or activating cellular genes compared to that of human immunodeficiency virus (HIV) or murine leukemia virus (MuLV) vectors (22, 60). The murine retrovirus-based gene transfer systems require dividing cells for efficient transduction, thus to target hematopoietic stem cells one must activate them to cycle ex vivo by use of cytokines, which may limit their engrafting ability (14, 15). Lentivirus-based gene transfer systems do not require cell division; therefore, efficient gene transfer to human hematopoietic stem cells in the absence of any ex vivo cytokine stimulation is possible (14, 15). Foamy virus vectors transduce hematopoietic stem cells with high efficiency (24, 26, 70). The level of foamy virus vector transduction in unstimulated hematopoietic stem cells is equivalent to that of lentivirus-based gene transfer systems (24, 32, 70). Transplantation of leukocyte adhesion-deficient dogs with hematopoietic stem cells transduced with a foamy virus vector containing the canine CD18 gene was able to reverse the defect (3).
Foamy virus vectors are capable of transducing stationary-phase human fibroblasts, albeit less efficiently than dividing fibroblasts (51, 59). Furthermore, the efficiency of gene transduction by foamy virus was shown to be higher than that of the MuLV. Mitosis is required for MuLV genome to translocate to the nucleus and subsequently depends on cell division for gene expression (35, 41, 50). Similarly, foamy virus gene expression requires mitosis (47, 59). Although there is no viral gene expression, the foamy virus DNA genome can enter the nuclei of cells arrested in the G1/S phase (47, 52, 59). Lentiviruses, on the other hand, are able to replicate in postmitotic nondividing cells and G1/S phase-arrested cells (6, 20, 34, 35, 42, 43, 65). However, it is proposed that due to the inefficiency of reverse transcription, import of the preintegration complex to the nucleus, or limited resource availability of transcription factors lentivirus vectors are not effective in quiescent/G0 cells (6, 7, 28, 29, 48). Recently, it was demonstrated that in quiescent cells incoming HIV subviral complexes are restricted to the centrosome and reside there until cell activation (68). The foamy virus preintegration complex (PIC) is also associated with the centrosome in quiescent cells similar to that of HIV (31). The localization and restriction of the PIC to the centrosome explains the blockage at the postentry level and the lack of effective replication by both HIV and foamy viruses in quiescent cells.
Since foamy virus genome was identified in the nuclei of G1/S phase-arrested cells, the reason(s) for the lack of viral gene expression or transgene expression with the foamy vector system is not known. The accumulation of large amounts of Gag in the nucleus is a hallmark of foamy viruses. A nuclear localization signal (NLS) sequence has been identified in the Gag protein and was shown to be responsible for its translocation into the nucleus (53, 67). The integrase domain of the Pol protein also has an NLS, which explains its import into the nucleus (23). It is not known whether these NLSs, the finding of Gag proteins in the nucleus, and/or the importation of Pol into the nucleus have a role in genome translocation and integration in G1/S phase-arrested cells. Furthermore, there is no information available as to whether foamy virus genome in the nucleus is integrated into the host chromosome or remains unintegrated in G1/S phase-arrested cells. In this report we provide evidence that the foamy virus genome remains unintegrated in G1/S phase-arrested cells and that integrase is required for nuclear import of foamy virus genome and Gag in cycling cells.
MATERIALS AND METHODS
Cell culture and growth arrest.
The 293T (human kidney fibroblasts) and HSF (diploid human skin fibroblasts) cell lines were obtained from American Type Culture Collection (Rockville, MD). The HSF cells were maintained in minimal essential medium with Earles salt containing 2 mM l-glutamine, 1 mM sodium pyruvate, 0.1 mM nonessential amino acids, 1.5 mg of sodium bicarbonate/ml, and 10% fetal calf serum (FCS). The 293T cells were grown in Dulbecco modified Eagle medium containing 10% FCS. In order to arrest the HSF cells at the G1/S phase of the cell cycle, cells were grown to confluence and then treated with 20 μg of aphidicolin/ml, as described previously (35). Cell arrest at the G1/S phase was verified by flow cytometry for DNA content by propidium iodide staining as described previously (64).
Plasmids.
Foamy virus vectors were constructed by using the simian foamy virus vector, isolated from rhesus macaque (SFVmac). Construction of the recombinant SFVmac packaging plasmids (pCIgag, pCIpol, and pCIenv) and vector plasmids (pEGFPD) were described previously (45, 46). The integrase defective Pol packaging construct was created by introducing a deletion in the integrase domain from pCIpol. The MuLV vector system plasmids, pCLampho (packaging plasmid) and pCLMFG-lacZ (MuLV vector containing the lacZ gene), were gifts from Inder M. Verma of the Salk Institute, San Diego, CA. The FIV vector system plasmids (pFLX-5CG, pCPRΔEnv, and pCI-VSV) were kindly provided by Garry Nolan of Stanford University.
Vector production and infection.
Transfections of 293T cells were carried out in T75 cell culture flasks by the calcium phosphate method. Seeded cells were transfected with 20 μg of vector and packaging plasmids pCIgag (20 μg), pCIpol (4 μg), and pCIenv (2 μg). Viral supernatants were harvested 4 days posttransfection and were clarified by centrifugation at 5,000 rpm for 20 min and then by filtering through a 0.45-μm-pore-size filter. The vector particles were further concentrated 100-fold by using a 150-kDa Apollo centrifugal spin concentrator (Orbital Biosciences, Topsfield, MA). The titers of SFVmac vector produced were determined on fresh 293T cells plated at a density of 2.5 × 104 per well in 24-well plates. At 72 h postinfection the cells were monitored and scored for green fluorescent protein (GFP) fluorescence or for LacZ-positive cells. Transduction of HSF cells was performed by spin inoculation as previously described (40). Briefly, HSF cells were seeded in six-well plates with 300 μl of medium containing SFVmac vector particles (multiplicity of infection [MOI] = 10). The cells were then spun at 1,500 rpm for 1 h as described previously (2, 8).
Fluorescence in situ hybridization and immunohistochemistry in combination with BrdU labeling.
For confocal microscopy studies, bromodeoxyuridine (BrdU)-labeled (10 μM) HSF cells or unlabeled cells were harvested, washed with phosphate-buffered saline, and fixed in 4% paraformaldehyde for 10 min at 4°C as described previously (12). Fixed cells were placed onto poly-lysine-coated glass slides by spinning at 1,000 rpm for 5 min, followed by air drying for 20 min. The cells were permeabilized by incubating slides in a mixture of 0.5% Triton-x 100 and 0.5% saponin for 20 min. Permeabilized cells were probed with SFVmac DNA fragment to identify foamy virus genome in the nucleus. A 1,550-base gag DNA fragment was labeled with digoxigenin by standard nick translation method using a nick translation kit as described by the manufacturer (Roche Diagnostics, Indianapolis, IN). The slides were hybridized in cocktail containing 50% formamide, 10% dextran sulfate, 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 0.2 μg of sonicated herring sperm DNA/μl, 0.2 μg of yeast tRNA/μl, and 2 ng of labeled probe DNA/μl. Labeled hybridized probe was detected by using primary mouse monoclonal anti-digoxigenin antibody (1 μg/ml; Roche), followed by a secondary antibody (1 μg/ml; Roche) specific for mouse antibody, which itself was conjugated with digoxigenin, and the digoxigenin label was detected by rhodamine-tagged anti-digoxigenin (1 μg/ml; Roche). Immunohistochemistry for foamy virus Gag was performed by labeling with primary antibody specific to Gag (1:2,000), followed by the fluorescence-tagged secondary antibody (2 μg/ml) Alexa 594-conjugated donkey anti-rabbit IgG (Molecular Probes, Eugene, OR). Anti-Gag SFVmac antibody was kindly provided by Axel Rethwilm of the Institut fuer Virologie im MTZ, Dresden, Germany. BrdU incorporation was detected by using Alexa Fluor 647-conjugated mouse anti-BrdU monoclonal IgG at 100 μg/ml (Molecular Probes) for 30 min at 37°C. Nuclei were counterstained with DAPI (4′,6′-diamidino-2-phenylindole) at 1 μg/ml in a phenylenediamine antifade solution or by labeling the nucleoporin with 4 μg of anti-Nup153 antibody (Santa Cruz, Santa Cruz, CA)/ml and then detected with a fluorescence-labeled secondary antibody (40 μg/ml), Alexa 350-conjugated donkey anti-goat antibody (Molecular Probes).
Nested Alu PCR.
To amplify integrated foamy virus DNA, we used a nested Alu PCR method as described previously (13). Briefly, 30 ng of genomic DNA was amplified by using Taq polymerase and 20 pmol of each primer. The primers for PCR were 5′-TCCCAGCTACTGGGGAGGCTGAGG-3′ (forward, AluI-specific primer) and 5′-ATGCCACGTAAGCGAAACTATCACTGGTATAACTCACTC3′ (5′ U3 long terminal repeat [LTR] with a lambda sequence tagged at the 5′ end). The PCR was performed by first denaturing at 94°C for 3 min, followed by 22 cycles of denaturing at 94°C for 30 s, annealing at 55°C for 30 s, polymerization at 72°C for 3 min and 1 cycle of final extension at 72°C for 10 min. A second round of amplification was performed using DNA from the first-round amplification with forward primer specific to 5′ U3 of SFVmac LTR (5′-TGTATAGGACCAGAGGAGG-3′) within the amplified DNA and a reverse primer with a sequence specific to the lambda sequence only (5′-ATGCCACGTAAGCGAAACT-3′). The second PCR was performed by first denaturing at 94°C for 3 min, followed by 29 cycles of denaturing at 94°C for 30 s, annealing at 55°C for 30 s, polymerization at 72°C for 30 s and 1 final cycle extension at 72°C for 7 min. Each of the amplified products was electrophoresed on a 1.2% agarose gel and probed with radioactive [γ-32P]ATP-labeled LTR-specific sequence (5′-ATGAGCTTCTACCCCCTGGTTTC-3′) using a prehybridization buffer (5× SSC, 5× Denhardt solution, 0.5% sodium dodecyl sulfate, and 100 μg of denatured salmon sperm DNA/ml) to determine specificity.
RESULTS
Infection of G1/S-arrested cells with foamy virus vector.
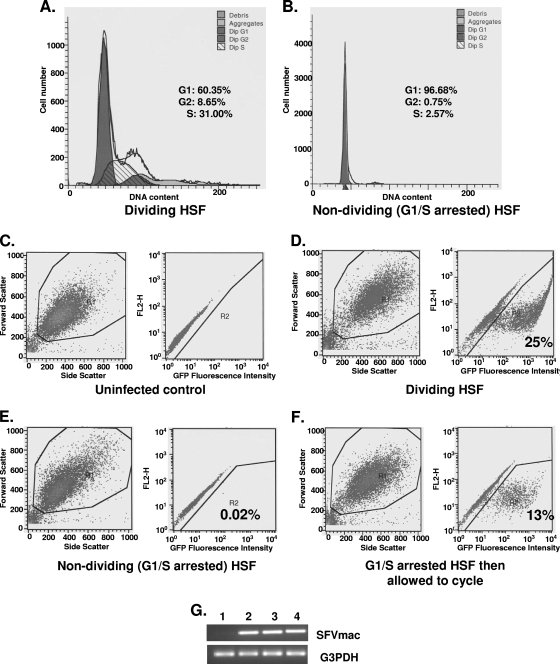
To demonstrate the fate of foamy virus genome in the nuclei of G1/S phase-arrested cells, diploid HSF cells were first arrested by growing the cells to confluence, followed by aphidicolin treatment. As shown in Fig. 1B, 97% of the cells were arrested in the G1/S phase. Cells were transduced with MuLV vector expressing LacZ or SFVmac vector expressing GFP after 24 h with or without aphidicolin exposure. The medium was replenished daily with or without aphidicolin for an additional 48 h prior to analysis for GFP expression. Efficiency of transduction by the SFVmac vector was calculated as a percentage of GFP-positive cells by fluorescence-activated cell sorting (FACS) analysis. Consistent with previous reports (35), the MuLV vector was inefficient at transducing growth-arrested cells, with <1% of the arrested cell population staining positive for β-galactosidase compared to the transduction of dividing cells (data not shown). The extent of infection of G1/S phase-arrested cells by SFVmac at an MOI of 10, as calculated by transgene expression, was 0.02% (Fig. 1E). The transduction efficiency in cycling cells was 25% (Fig. 1D). Similar results were also obtained previously using the prototype foamy virus (47, 52, 59). The levels of foamy virus DNA genome determined by semiquantitative PCR were comparable in both cycling and G1/S-arrested cells (Fig. 1G). When arrested cells were induced to cycle (Fig. 1F) by removing the aphidicolin medium and culturing them in fresh growth medium, the transgene expression was evident (13%), thus indicating that the steps blocking subsequent transgene or SFVmac gene expression was relieved when the cells were allowed to cycle.
FIG. 1.
Transduction and transgene expression efficiency by SFVmac in cycling and G1/S phase-arrested HSF cells. Cell cycle analysis results in cycling and arrested cells are shown in panels A and B, respectively. The populations of cells at different phases of the cell cycle are given as a percentage of the total population. Cells in G1/S phase were at a 97% level. FACS analyses were performed at the same time for GFP expression. (C) FACS results for GFP expression in uninfected dividing cells. (D and E) FACS results of dividing and growth-arrested GFP-expressing cells, respectively, infected with SFVmac vector. (F) GFP expression in cells where medium containing aphidicolin was removed from the culture and replaced with fresh medium without aphidicolin, allowing cells to undergo cell division. (G) DNA amplified by PCR from SFVmac vector-transduced HSF cells. Lane 1 is mock transduced cells, lane 2 is cycling HSF cells transduced with SFVmac vector, and lane 3 represents G1/S phase-arrested cells transduced with foamy vector. PCR results from growth-arrested cells infected with foamy virus vector and released into cycle 24 h later are shown in lane 4. DNA was amplified using pol-specific forward 5′-TGTAATACCACTCCAAGCCTGGAT-3′ and reverse 5′-GACTTTCAGAAAAGTAGCGTCTCG-3′ primers.
Fate of foamy virus genome in the nuclei of G1/S phase-arrested cells.
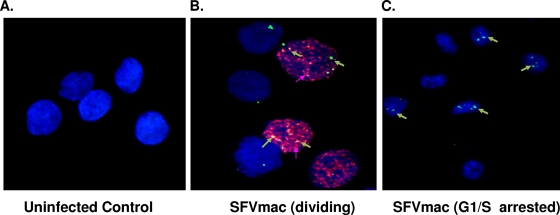
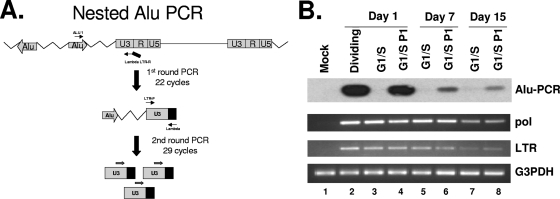
Like lentiviruses the foamy virus genome can enter the nuclei of G1/S-arrested cells with no gene expression. To address the reason for the lack of gene expression in growth-arrested cells while the genome is present in the nucleus, we needed to determine whether the foamy genome can integrate into the host genome or remain as an episome in the nucleus. In order to identify resting cells containing the SFVmac genome in the nucleus, we performed in situ hybridization in conjunction with BrdU incorporation and examined the results by using confocal microscopy. A DNA probe of the gag region of SFVmac was labeled with digoxigenin-dUTP and then fluorescence tagged and used for in situ hybridization of resting cells. The foamy virus genome is found in the nuclei of both dividing and arrested cells (Fig. 2). To determine whether the foamy virus genome remains unintegrated in the nuclei or integrated into the chromosomes of G1/S phase-arrested cells, we isolated DNA from SFVmac vector transduced cycling and growth-arrested cells and then performed nested Alu PCR amplification, as shown in Fig. 3. Genomic DNA obtained from cycling or arrested cells was copied by using an AluI-specific primer and amplified with a foamy virus-specific primer tagged with a lambda sequence (Table 1). The amplified DNA was subjected to a second-round PCR with SFVmac LTR forward and reverse lambda-specific primers and then hybridized to radioactive labeled foamy virus-specific probe to determine the specificity. Amplification of the genome using primers specific to AluI repeats and foamy virus showed an expected size DNA band from samples of cells that are in cycle. We were able to amplify the foamy genome at 1, 7, or 15 days posttransduction. In contrast, there was no foamy virus DNA amplification from genomic DNA obtained from cells arrested in G1/S phase at days 1 and 7. There is a very faint band present in the arrested cells at day 15 (Fig. 3B, lane 7) that is significantly less than in cells that were allowed to cycle (lane 8). The presence of a faint band at day 15 in the arrested cells could be due to genome integration in a few cells where the block in cell arrest is incomplete. When the aphidicolin was removed from the arrested cells and then placed in regular medium, allowing the cells to undergo a normal cell cycle, we were able to detect the corresponding DNA size band by Alu PCR. Although less than at day 1, the presence of foamy virus-specific band was evident in cells that were kept for 7 or 15 days in G1/S phase and then released into the cycle. This therefore shows an ability of the unintegrated genome to remain within the nucleus for at least 15 days posttransduction. These results confirm that the foamy virus genome can persist as an episome and is capable of integrating into the host genome when the cells are released from G1/S phase into a normal cell cycle.
FIG. 2.
SFVmac genome localization in dividing and growth-arrested cells. For confocal microscopy cells were labeled with BrdU (red) to distinguish between dividing and growth-arrested cells. SFV-1 genome was identified with in situ hybridization using gag-specific probe (green). The nucleus was counterstained with DAPI (blue). (A) In situ staining of uninfected control cells. (B and C) Dividing and growth-arrested cells, respectively, transduced with foamy virus vector.
FIG. 3.
Alu PCR of genomic DNA isolated from SFVmac vector-infected dividing or growth-arrested cells. (A) Schematic representation of an Alu PCR methodology. DNA was copied using an Alu sequence-specific primer and amplified with an SFVmac LTR-specific primer tagged with a lambda sequence. (B) Alu PCR with samples isolated from dividing and growth-arrested cells (top panel). The Alu PCR-amplified products were hybridized to radioactive [γ-32P]ATP-labeled LTR-specific probe. Lane 1 (mock) contains PCR results from uninfected control cells. Lane 2 (dividing) is the PCR product with DNA isolated from SFVmac-transduced dividing cells 24 h after transduction. Lanes 3, 5, and 7 represent PCR results from G1/S-arrested cells harvested at days 1, 7, and 15 posttransduction, respectively. Lanes 4, 6, and 8 are PCR products for cells that were arrested in G1/S for 1, 7, and 15 days, respectively, and then allowed to resume a normal cycle for 72 h. The second and third panels represent corresponding semiquantitative regular PCR detecting the pol and LTR region of SFVmac. For level of sample recovery, the DNAs were amplified with primers specific to G3PDH (glyceraldehyde-3-phosphate dehydrogenase) (bottom panel).
TABLE 1.
Probe and primers
| Primer or probe | Sequence (5′-3′)a |
|---|---|
| ALU1 | TCCCAGCTACTGGGGAGGCTGAGG |
| LTR-F | TGTATAGGACCAGAGGAGG |
| Lambda LTR-R | ATGCCACGTAAGCGAAACTATCACTGGTATAACTCACTC |
| Lambda | ATGCCACGTAAGCGAAACT |
| pol-F | TGTAATACCACTCCAAGCCTGGAT |
| pol-R | GACTTTCAGAAAAGTAGCGTCTCG |
| G3PDH | ATCCCATCACCATCTTCCAGGAG |
| G3PDHrH | AATGAGCCCCAGCCTTCTCC |
| SFV U3 (probe) | ATGAGCTTCTACCCCCTGGTTTC |
The lambda sequence is underlined.
Foamy virus genome nuclear localization is dependent on integrase in both arrested and cycling cells.
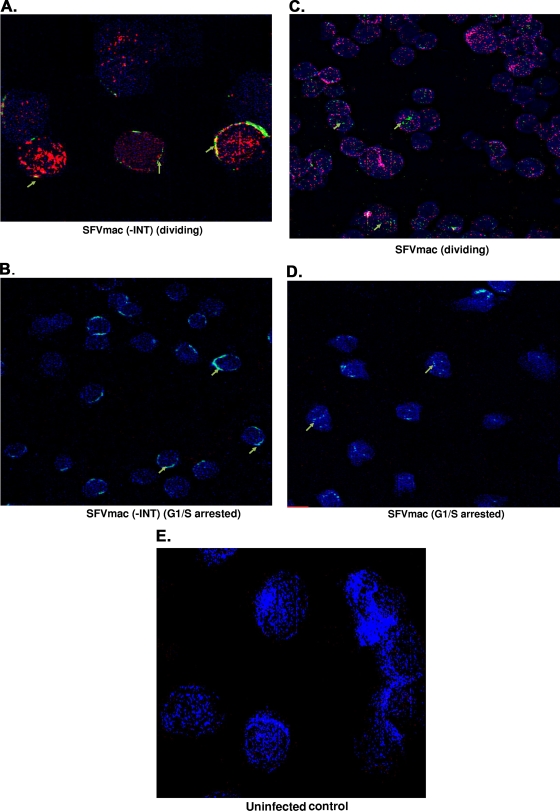
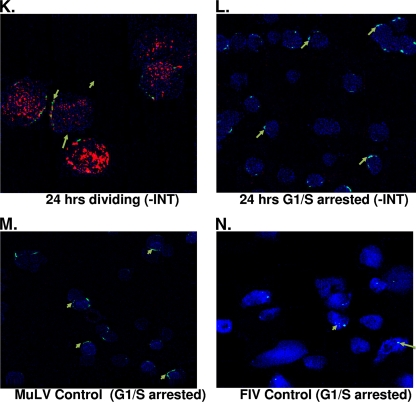
NLS sequences have been identified in Gag and integrase of the foamy virus. However, whether both Gag and integrase are involved in nuclear localization of the foamy virus genome has not been previously determined. In order to identify the viral protein involved in nuclear localization of the foamy virus genome in both cycling and G1/S phase-arrested cells, we introduced a deletion in the integrase domain of the pol gene, which was then used as a packaging construct, along with gag and env expression cassettes. Foamy virus vector particles were generated and used to infect cells that were dividing or arrested at G1/S phase. Immunofluorescence staining for nucleoporin, along with BrdU labeling for cell division and in situ hybridization for foamy virus genome, was performed 24 h posttransduction. No nuclear localization of foamy virus genome was observed with the defective integrase vector transduction (Fig. 4A and B). In situ analysis revealed that the foamy virus genome was around the rim of the nucleus and yet outside of the nuclear membrane for both dividing and arrested cells. In contrast, the foamy virus genome is present in the nuclei of both cycling and growth-arrested cells transduced with vector containing intact integrase (Fig. 4C and D).
FIG. 4.
SFVmac genome localization in the absence of integrase. Cycling cells or growth-arrested cells were transduced with SFVmac vector prepared in the absence of integrase. Permeabilized cells were labeled with anti-Nup153 for nucleoporin 153 of the nuclear membrane (blue). Cells were also labeled with BrdU to identify dividing cells (red), and in situ hybridization was performed to label SFVmac genome (green). (A) Dividing cells transduced with SFVmac vector with an integrase deficient SFVmac vector. (B) Nondividing cells transduced with the same SFVmac vector. (C and D) Images representative of dividing and growth-arrested cells, respectively, transduced with an SFVmac vector with intact integrase. An uninfected control is shown in panel E. “-INT” indicates integrase-defective SFVmac vector.
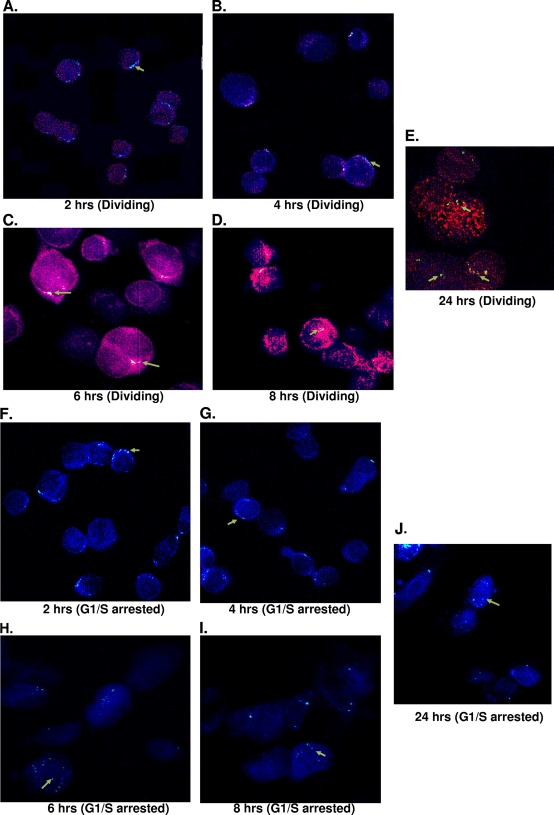
To further confirm the importance of integrase, we examined the movement of foamy virus genome at different time points and compared our findings to those of the defective integrase at 24 h posttransduction. The genome appears at the nuclear membrane at 2 and 4 h postinfection in both arrested and dividing cells (Fig. 5A, B, F, and G). By 6 h the foamy virus genome was localized inside the nucleus, indicating it was within the nuclear membrane of both dividing and arrested cells (Fig. 5C and H). Subsequently, at 8 and 24 h postinfection the genome is found inside the nucleus (Fig. 5D, E, I, and J), demonstrating the translocation of foamy virus genome into the nuclei of growth-arrested cells. Since FIV translocates to the nucleus in arrested cells, we obtained similar results when the FIV vector was used to infect G1/S-arrested cells (Fig. 5N). In the samples where the integrase was defective, the genome of foamy virus was found outside of the nucleus and around the rim of the nuclear membrane in both cycling and arrested cells (Fig. 5K and L). As expected with the control MuLV, which the genome requires cell division to enter the nucleus, we observed no viral genome inside the nucleus in growth-arrested cells in the presence of intact integrase (Fig. 5M). These results establish evidence that foamy virus integrase is important for nuclear localization of the viral genome.
FIG. 5.
SFVmac genome localization in the nuclei of nucleoporin labeled dividing and growth-arrested cells at different time points after infection with SFVmac vector. BrdU (red)-labeled cells were permeabilized, and in situ hybridization was performed by probing for SFVmac genome (green). The nucleus was defined by immunostaining with antibody to nucleoporin, Nup153 (blue). (A to E) In situ hybridization for foamy virus genome with immunolabeling for Nup153 at the indicated time points in dividing cells. (F to J) Genome localization at different time points in cells arrested at the G1/S phase. Foamy virus genome localization in dividing and growth-arrested cells infected with integrase defective vector at 24 h posttransduction is shown in panels K and L, respectively. In situ hybridization for MuLV and FIV controls at 24 h posttransduction are represented in panels M and N, respectively.
Gag translocates into the nuclei of G1/S phase-arrested cells, and the Gag translocation is dependent on integrase.
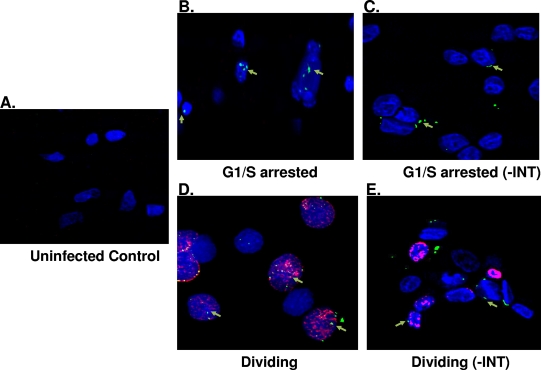
A hallmark of foamy viruses is the large accumulation of Gag in the nucleus, and yet the role of this accumulation is not known. Both foamy genome and Gag were shown to be absent in the nuclei of cells at the G0 phase (31). We tested whether Gag translocates into the nuclei of G1/S phase-arrested cells. Using immunohistochemistry with antibody specific to Gag and BrdU labeling for cell proliferation, we found that the Gag protein is present in the nuclei of both cycling and G1/S phase-arrested cells (Fig. 6B and D). An NLS sequence has been identified in Gag and is responsible for its translocation into the nucleus (53, 67). To demonstrate whether Gag can localize to the nucleus independent of integrase, we used an integrase deficient SFVmac vector to infect both cycling and growth-arrested cells. In both cases we could only detect Gag outside of the nucleus or around the rim of the nucleus, suggesting that integrase is important for PIC translocation to the nucleus (Fig. 6C and E). Taken together, these results suggest that integrase is critical for importation of the foamy virus PIC into the nucleus.
FIG. 6.
SFV Gag localization in the absence of integrase. Dividing and nondividing cells were transduced with SFVmac vector defective for integrase, and immunohistochemistry was performed with antibody against Gag (green) and nucleoporin with anti-Nup153 (blue). BrdU labeling is represented by a red stain showing dividing cells. (A) Uninfected control. (B and C) Growth-arrested cells transduced with wild-type SFVmac vector and integrase defective vector, respectively. Gag localization in dividing cells are represented in panels D and E. “-INT” indicates integrase-defective SFVmac vector.
DISCUSSION
Foamy viruses are capable of efficient transduction of their genome into cells arrested in G1/S phase, but transgene or viral gene expression is restricted. In this report we demonstrate that the foamy virus genome remains unintegrated in the nuclei of G1/S-arrested cells. When foamy virus-infected cells are released from G1/S phase arrest to undergo cell cycle, the genome integrates into the chromosome and gene expression is evident. These results suggest that cell cycle is necessary for genome integration and that efficient gene expression requires viral genome integration into the host chromosome. The viral genome and Gag translocation to the nucleus requires integrase in cycling cells. This suggests an integrase dependent mechanism for the translocation of the PIC into the nucleus.
Retroviruses such as MuLV require at least one round of cell division for proviral integration into target cells (41, 50, 56). More specifically, they require cell division, in which the nuclear envelope breakdown allows genome translocation into the nucleus (20, 35, 41, 50). Consequently, vectors based on these viruses are inadequate to stably transduce nondividing and/or terminally differentiated cells. This severely restricts their potential utility for clinical gene transfer. In contrast, terminally differentiated macrophages, mucosal dendritic cells, and quiescent T lymphocytes are nonproliferating targets of critical importance for the transmission, propagation and spread of HIV in an infected host (18, 33). Lentiviruses can therefore productively infect postmitotic cells or cells arrested in the G1/S phase of the cycle, suggesting that the PIC traverses an intact nuclear membrane (17, 25, 54, 57). Our results show that foamy virus genome and Gag can enter the nuclei of growth-arrested cells, which indicates an ability for the foamy virus PIC to cross an intact nuclear membrane similar to that of lentiviruses. Observing no Gag or viral genome within the nuclei of cells transduced with an integrase-deficient vector suggests that Gag alone is not critical for PIC transport to the nucleus, whereas integrase is an absolute requirement. An NLS is present in both the Gag and integrase proteins of foamy viruses (1, 23, 30, 53, 67). The NLS in Gag can serve as a nuclear transport signal. However, foamy virus replication is not dependent on the Gag NLS (67), thus supporting our findings. In infected cells, Gag may enter the nucleus during RNA assembly. Our results show, whether Gag enters the nucleus during RNA assembly or after the infection of new cells, that Gag and genome translocation into the nucleus is dependent on integrase. The matrix (MA) domain of Gag and Vpr of lentiviruses have NLS sequences and are thought to be important for PIC importation into the nuclei of growth-arrested cells (5, 11, 21). This notion, however, remains controversial since viruses with mutated MA and Vpr NLS sequences can infect nondividing cells, thus refuting a major role for MA and Vpr in PIC translocation to the nucleus (19, 27). Similar to our findings for foamy viruses, the integrase in lentiviruses is also central for nucleus importation of the viral genome (9, 57).
Genetic studies in lentiviruses with the introduction of mutations in integrase reveal that defects in integrase can affect viral packaging, processing of viral proteins, and reverse transcription (4, 37, 49, 61, 66, 69). Similarly, an active integrase is required for foamy virus replication (16). Efficient transient gene expression, however, has been accomplished with nonintegrating HIV vectors using an integrase-deficient system, which involves the introduction of mutations to disable the integrase and to alter the integrase recognition sequences (att) in the viral LTR (44). The presence of foamy virus Gag and proviral genome that we observed outside of the nucleus, when an integrase defective foamy virus vector was used, establishes vector packaging in the absence of integrase from the transient expression of the foamy viral vector system.
The lack of foamy genome integration in G1/S phase-arrested cells suggests a role for a cellular protein(s), which may have been downregulated, in viral DNA integration into the host genome. Along with viral proteins several other cellular factors are found to be associated with other retroviral PICs (63). These include barrier-to-autointegration factors (BAF), high mobility groups (HMGs), Ku, lamina-associated polypeptides 2α (LAP2α), and lens-epithelium-derived growth factor (LEDGF/p75). LEDGF/p75, which forms a specific nuclear complex with HIV integrase, has been found to be critical to HIV genome integration into the host chromosome (36, 55, 62), as well as target selection into transcriptional units (10, 38, 55). Cellular proteins involved in foamy virus genome integration are yet to be uncovered. One study reports that a domain in the C terminus of the foamy virus Gag protein interacts with H2A/H2B core histones for efficient binding to host chromosome (58). Introduction of a mutation in the Gag domain that binds to H2A/H2B does not affect the late stages of virus replication. However, such a mutation in the foamy Gag influences genome integration into the host due to the lack of tethering for the PICs onto the host chromosome. Our finding shows impairment of proviral integration into growth-arrested cells while Gag is present in the nucleus. Therefore, the role H2A/H2B in PIC tethering in arrested cells, along with the identification of other potential host proteins required for integration, will give insight into the mechanisms of the block of foamy virus genome integration in growth-arrested cells.
Understanding the limiting factors of retrovirus replication in growth-arrested cells is important for the development of an efficient viral vector system. Restriction of both foamy and lentiviruses to the centrosome in G0-arrested cells is a major stumbling block to developing a vector that can access quiescent cells in vivo efficiently. Unraveling the mechanisms that restrict these viruses to the centrosome will help in the development of effective vectors that would relieve such blocks for clinical applications. Similarly, the finding that the foamy virus remains unintegrated in G1/S phase-arrested cells will establish a basis for solutions to address the additional constraints of developing an ideal foamy virus vector system.
Acknowledgments
This research was supported by a grant from the National Institute of Health (AI39126) to A.M.
Footnotes
Published ahead of print on 23 December 2009.
REFERENCES
- 1.An, D. G., U. Hyun, and C. G. Shin. 2008. Characterization of nuclear localization signals of the prototype foamy virus integrase. J. Gen. Virol. 89:1680-1684. [DOI] [PubMed] [Google Scholar]
- 2.Bahner, I., K. Kearns, Q. L. Hao, E. M. Smogorzewska, and D. B. Kohn. 1996. Transduction of human CD34+ hematopoietic progenitor cells by a retroviral vector expressing an RRE decoy inhibits human immunodeficiency virus type 1 replication in myelomonocytic cells produced in long-term culture. J. Virol. 70:4352-4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bauer, T. R. J., J. M. Allen, M. Hai, L. M. Tuschong, I. F. Khan, E. M. Olson, R. L. Adler, T. H. Burkholder, Y. C. Gu, D. W. Russell, and D. D. Hickstein. 2008. Successful treatment of canine leukocyte adhesion deficiency by foamy virus vectors. Nat. Med. 14:93-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bukovsky, A., and H. Gottlinger. 1996. Lack of integrase can markedly affect human immunodeficiency virus type 1 particle production in the presence of an active viral protease. J. Virol. 70:6820-6825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bukrinsky, M. I., S. Haggerty, M. P. Dempsey, N. Sharova, A. Adzhubel, L. Spitz, P. Lewis, D. Goldfarb, M. Emerman, and M. Stevenson. 1993. A nuclear localization signal within HIV-1 matrix protein that governs infection of non-dividing cells. Nature 365:666-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bukrinsky, M. I., N. Sharova, M. P. Dempsey, T. L. Stanwick, A. G. Bukrinskaya, S. Haggerty, and M. Stevenson. 1992. Active nuclear import of human immunodeficiency virus type 1 preintegration complexes. Proc. Natl. Acad. Sci. U. S. A. 89:6580-6584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bukrinsky, M. I., T. L. Stanwick, M. P. Dempsey, and M. Stevenson. 1991. Quiescent T lymphocytes as an inducible virus reservoir in HIV-1 infection. Science 254:423-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bunnell, B. A., L. M. Muul, R. E. Donahue, R. M. Blaese, and R. A. Morgan. 1995. High-efficiency retroviral-mediated gene transfer into human and nonhuman primate peripheral blood lymphocytes. Proc. Natl. Acad. Sci. U. S. A. 92:7739-7743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ciuffi, A., and F. D. Bushman. 2006. Retroviral DNA integration: HIV and the role of LEDGF/p75. Trends Genet. 22:388-395. [DOI] [PubMed] [Google Scholar]
- 10.Ciuffi, A., M. Llano, E. Poeschla, C. Hoffmann, J. Leipzig, P. Shinn, J. R. Ecker, and F. Bushman. 2005. A role for LEDGF/p75 in targeting HIV DNA integration. Nat. Med. 11:1287-1289. [DOI] [PubMed] [Google Scholar]
- 11.Connor, R. I., B. K. Chen, S. Choe, and N. R. Landau. 1995. Vpr is required for efficient replication of human immunodeficiency virus type 1 in mononuclear phagocytes. Virology 206:935-944. [DOI] [PubMed] [Google Scholar]
- 12.Cremer, M., F. Grasser, C. Lanctôt, S. Müller, M. Neusser, R. Zinner, I. Solovei, and T. Cremer. 2008. Multicolor 3D fluorescence in situ hybridization for imaging interphase chromosomes. Methods Mol. Biol. 463:205-239. [DOI] [PubMed] [Google Scholar]
- 13.Delelis, O., A. Brussel, and P. Sonigo. 2005. Quantification of HFV-integrated DNA in human cells by Alu-LTR real-time PCR. Methods Mol. Biol. 304:155-170. [DOI] [PubMed] [Google Scholar]
- 14.Douglas, J., P. Kelly, J. T. Evans, and J. V. Garcia. 1999. Efficient transduction of human lymphocytes and CD34+ cells via human immunodeficiency virus-based gene transfer vectors. Hum. Gene Ther. 10:935-945. [DOI] [PubMed] [Google Scholar]
- 15.Douglas, J. L., W. Y. Lin, M. L. Panis, and J. V. Garcia. 2001. Efficient human immunodeficiency virus-based vector transduction of unstimulated human mobilized peripheral blood CD34+ cells in the SCID-hu Thy/Liv model of human T-cell lymphopoiesis. Hum. Gene Ther. 12:401-413. [DOI] [PubMed] [Google Scholar]
- 16.Enssle, J., A. Moebes, M. Heinkelein, M. Panhuysen, B. Mauer, M. Schweizer, D. Neumann-Haefelin, and A. Rethwilm. 1999. An active foamy virus integrase is required for virus replication. J. Gen. Virol. 80:1445-1452. [DOI] [PubMed] [Google Scholar]
- 17.Fassati, A. 2006. HIV infection of non-dividing cells: a divisive problem. Retrovirology 3:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fauci, A. S. 1996. Host factors and the pathogenesis of HIV-induced disease. Nature 384:529-534. [DOI] [PubMed] [Google Scholar]
- 19.Gallay, P., T. Hope, D. Chin, and D. Trono. 1997. HIV-1 infection of non-dividing cells through the recognition of integrase by the importin/karyopherin pathway. Proc. Natl. Acad. Sci. U. S. A. 94:9825-9830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hatziioannou, T., and S. P. Goff. 2001. Infection of nondividing cells by Rous sarcoma virus. J. Virol. 75:9526-9531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heinzinger, N. K., M. I. Bukinsky, S. A. Haggerty, A. M. Ragland, V. Kewalramani, M. A. Lee, H. E. Gendelman, L. Ratner, M. Stevenson, and M. Emerman. 1994. The Vpr protein of human immunodeficiency virus type 1 influences nuclear localization of viral nucleic acids in nondividing host cells. Proc. Natl. Acad. Sci. U. S. A. 91:7311-7315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hendrie, P. C., Y. Huo, R. B. Stolitenko, and D. W. Russell. 2008. A rapid and quantitative assay for measuring neighboring gene activation by vector proviruses. Mol. Ther. 16:534-540. [DOI] [PubMed] [Google Scholar]
- 23.Imrich, H., M. Heinkelein, O. Herchenroder, and A. Rethwilm. 2000. Primate foamy virus Pol proteins are imported into the nucleus. J. Gen. Virol. 81:2941-2947. [DOI] [PubMed] [Google Scholar]
- 24.Josephson, N. C., G. Vassilopoulos, G. D. Trobridge, G. V. Priestley, B. L. Wood, T. Papayannopoulou, and D. W. Russell. 2002. Transduction of human NOD/SCID-repopulating cells with both lymphoid and myeloid potential by foamy virus vectors. Proc. Natl. Acad. Sci. U. S. A. 99:8295-8300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Katz, R. A., J. G. Greger, and A. M. Skalka. 2005. Effects of cell cycle status on early events in retroviral replication. J. Cell. Biochem. 94:880-889. [DOI] [PubMed] [Google Scholar]
- 26.Kiem, H. P., J. Allen, G. Trobridge, E. Olson, K. Keyser, L. Peterson, and D. W. Russell. 2007. Foamy-virus-mediated gene transfer to canine repopulating cells. Blood 109:65-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kootstra, N. A., and H. Schuitemaker. 1999. Phenotype of HIV-1 lacking a functional nuclear localization signal in matrix protein of Gag and Vpr is comparable to wild-type HIV-1 in primary macrophages. Virology 253:170-180. [DOI] [PubMed] [Google Scholar]
- 28.Korin, Y. D., and J. A. Zack. 1999. Nonproductive human immunodeficiency virus type 1 infection in nucleoside-treated G0 lymphocytes. J. Virol. 73:6526-6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Korin, Y. D., and J. A. Zack. 1998. Progression to the G1b phase of the cell cycle is required for completion of human immunodeficiency virus type 1 reverse transcription in T cells. J. Virol. 72:3161-3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee, H. S., S. Y. Kang, and C. G. Shin. 2005. Characterization of the functional domains of human foamy virus integrase using chimeric integrases. Mol. Cells 19:246-255. [PubMed] [Google Scholar]
- 31.Lehmann-Che, J., N. Renault, M. L. Giron, P. Roingeard, E. Clave, J. Tobaly-Tapiero, P. Bittoun, A. Toubert, H. de Thé, and A. Saib. 2007. Centrosomal latency of incoming foamy viruses in resting cells. PLoS Pathog. 3:e74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leurs, C., M. Jansen, K. E. Pollok, M. Heinkelein, M. Schmidt, M. Wissler, D. Lindemann, C. V. Kalle, A. Rethwilm, D. A. Williams, and H. Hanenberg. 2003. Comparison of three retroviral vector systems for transduction of nonobese diabetic/severe combined immunodeficiency mice repopulating human CD34+ cord blood cells. Hum. Gene Ther. 10:509-519. [DOI] [PubMed] [Google Scholar]
- 33.Levy, J. 1993. Pathogenesis of human immunodeficiency virus infection. Microbiol. Rev. 57:183-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lewis, P., M. Hensel, and M. Emerman. 1992. Human immunodeficiency virus infection of cells arrested in the cell cycle. EMBO J. 11:3053-3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lewis, P. F., and M. Emerman. 1994. Passage through mitosis is required for oncoretroviruses but not for the human immunodeficiency virus. J. Virol. 68:510-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Llano, M., D. T. Saenz, A. Meehan, P. Wongthida, M. Peretz, W. H. Walker, W. Teo, and E. M. Poeschla. 2006. An essential role for LEDGF/p75 in HIV integration. Science 314:461-464. [DOI] [PubMed] [Google Scholar]
- 37.Lu, R., A. Limon, E. Devroe, P. A. Silver, P. Cherepanov, and A. Engelman. 2004. Class II integrase mutants with changes in putative nuclear localization signals are primarily blocked at a postnuclear entry step of human immunodeficiency virus type 1 replication. J. Virol. 78:12735-12746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marshall, H. M., K. Ronen, C. Berry, M. Llano, H. Sutherland, D. Saenz, W. Bickmore, E. Poeschla, and F. D. Bushman. 2007. Role of PSIP1/LEDGF/p75 in lentiviral infectivity and integration targeting. PLoS One 2:e1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mergia, A., and M. Heinkelein. 2003. Foamy virus vectors. Curr. Top. Microbiol. Immunol. 277:131-159. [DOI] [PubMed] [Google Scholar]
- 40.Mergia, A., C. Soumya, D. L. Kolson, M. M. Goodenow, and T. Ciccarone. 2001. The efficiency of simian foamy virus vector type-1 (SFV-1) in non-dividing cells and in human PBLs. Virology 280:243-252. [DOI] [PubMed] [Google Scholar]
- 41.Miller, D. G., M. A. Adam, and A. D. Miller. 1990. Gene transfer by retroviral vectors occurs only in cells that are actively replicating at the time of infection. Mol. Cell. Biol. 10:4239-4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miyake, K., N. Suzuki, H. Matsuoka, T. Tohyama, and T. Shimada. 1998. Stable integration of human immunodeficiency virus-based retroviral vectors into the chromosomes of nondividing cells. Hum. Gene Ther. 9:467-475. [DOI] [PubMed] [Google Scholar]
- 43.Naldini, L., U. Blomer, P. Gallay, D. Ory, R. Mulligan, F. H. Gage, I. M. Verma, and D. Trono. 1996. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 272:263-267. [DOI] [PubMed] [Google Scholar]
- 44.Nightingale, S. J., R. P. Hollis, K. A. Pepper, D. Petersen, X. J. Yu, C. Yang, I. Bahner, and D. B. Kohn. 2006. Transient gene expression by nonintegrating lentiviral vectors. Mol. Ther. 13:1121-1132. [DOI] [PubMed] [Google Scholar]
- 45.Park, J., P. E. Nadeau, and A. Mergia. 2009. Activity of TAR in inducible inhibition of HIV replication by foamy virus vector expressing siRNAs under the control of HIV LTR. Virus Res. 140:112-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Park, J., P. E. Nadeau, and A. Mergia. 2002. A minimal genome simian foamy virus type 1 (SFV-1) vector system with efficient gene transfer. Virology 302:336-344. [DOI] [PubMed] [Google Scholar]
- 47.Patton, G. S., O. Erlwein, and M. O. McClure. 2004. Cell cycle dependence of foamy virus vectors. J. Gen. Virol. 85:2925-2930. [DOI] [PubMed] [Google Scholar]
- 48.Pierson, T. C., Y. Zhou, T. L. Kieffer, C. T. Ruff, C. Buck, and R. F. Siliciano. 2002. Molecular characterization of preintegration latency in human immunodeficiency virus type 1 infection. J. Virol. 76:8518-8531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Quillent, C., A. M. Borman, S. Paulous, C. Dauguet, and F. Clavel. 1996. Extensive regions of pol are required for efficient human immunodeficiency virus polyprotein processing and particle maturation. Virology 219:29-36. [DOI] [PubMed] [Google Scholar]
- 50.Roe, T., T. C. Reynolds, G. Yu, and P. O. Brown. 1993. Integration of murine leukemia virus DNA depends on mitosis. EMBO J. 12:2099-2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Russell, D. W., and A. D. Miller. 1996. Foamy virus vectors. J. Virol. 70:217-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saib, A., F. Puvion-Dutilleul, M. Schmid, J. Peries, and H. de Thé. 1997. Nuclear targeting of incoming human foamy virus Gag proteins involves a centriolar step. J. Virol. 71:1155-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schliephake, A. W., and A. Rethwilm. 1994. Nuclear localization of foamy virus Gag precursor protein. J. Virol. 68:4946-4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sherman, M. P., and W. C. Greene. 2002. Slipping through the door: HIV entry into the nucleus. Microbes Infect. 4:67-73. [DOI] [PubMed] [Google Scholar]
- 55.Shun, M. C., N. K. Raghavendra, N. Vandegraaff, J. E. Daigle, S. Hughes, P. Kellam, P. Cherepanov, and A. Engelman. 2007. LEDGF/p75 functions downstream from preintegration complex formation to effect gene-specific HIV-1 integration. Genes Dev. 21:1767-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Springett, G. M., R. C. Moen, S. Anderson, R. M. Blaese, and W. F. Anderson. 1989. Infection efficiency of T lymphocytes with amphotropic retroviral vectors is cell cycle dependent. J. Virol. 63:3865-3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Suzuki, Y., and R. Craigie. 2007. The road to chromatin: nuclear entry of retroviruses. Nat. Rev. Microbiol. 5:187-196. [DOI] [PubMed] [Google Scholar]
- 58.Tobaly-Tapiero, J., P. Bittoun, J. Lehmann-Che, O. Delelis, M. L. Giron, H. de Thé, and A. Saïb. 2008. Chromatin tethering of incoming foamy virus by the structural Gag protein. Traffic 9:1717-1727. [DOI] [PubMed] [Google Scholar]
- 59.Trobridge, G., and D. W. Russell. 2004. Cell cycle requirements for transduction by foamy virus vectors compared to those of oncovirus and lentivirus vectors. J. Virol. 78:2327-2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Trobridge, G. D., D. G. Miller, M. A. Jacobs, J. M. Allen, H. P. Kiem, R. Kaul, and D. W. Russell. 2006. Foamy virus vector integration sites in normal human cells. Proc. Natl. Acad. Sci. U. S. A. 103:1498-1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tsurutani, N., M. Kubo, Y. Maeda, T. Ohashi, N. Yamamoto, M. Kannagi, and T. Masuda. 2000. Identification of critical amino acid residues in human immunodeficiency virus type 1 IN required for efficient proviral DNA formation at steps prior to integration in dividing and nondividing cells. J. Virol. 74:4796-4806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vandekerckhove, L., F. Christ, B. V. Maele, J. D. Rijck, R. Gijsbers, C. V. de Haute, M. Witvrouw, and Z. Debyser. 2006. Transient and stable knockdown of the integrase cofactor LEDGF/p75 reveals its role in the replication cycle of human immunodeficiency virus. J. Virol. 80:1886-1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Van Maele, B., K. Busschots, L. Vandekerckhove, F. Christ, and Z. Debyser. 2006. Cellular cofactors of HIV-1 integration. Trends Biochem. Sci. 13:98-105. [DOI] [PubMed] [Google Scholar]
- 64.Vindolv, L. L., and I. J. Christensen. 1990. A review of techniques and results obtained in one laboratory by an integrated system of methods designed for routine clinical flow cytometric analysis. Cytometry 11:753-770. [DOI] [PubMed] [Google Scholar]
- 65.Weinberg, J. B., T. J. Matthews, B. R. Cullen, and M. H. Malim. 1991. Productive human immunodeficiency virus type 1 (HIV-1) infection of nonproliferating human monocytes. J. Exp. Med. 174:1477-1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu, X., H. Liu, H. Xiao, J. A. Conway, E. Hehl, G. V. Kalpana, V. Prasad, and J. C. Kappes. 1999. Human immunodeficiency virus type 1 integrase protein promotes reverse transcription through specific interactions with the nucleoprotein reverse transcription complex. J. Virol. 73:2126-2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yu, S. F., K. Edelmann, R. K. Strong, A. Moebes, A. Rethwilm, and M. L. Linial. 1996. The carboxyl terminus of the human foamy virus Gag protein contains separable nucleic acid binding and nuclear transport domains. J. Virol. 70:8255-8262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zamborlini, A., J. Lehmann-Che, E. Clave, M. L. Giron, J. Tobaly-Tapiero, P. Roingeard, S. Emiliani, A. Toubert, H. de Thé, and A. Saïb. 2007. Centrosomal pre-integration latency of HIV-1 in quiescent cells. Retrovirology 4:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhu, K., C. Dobard, and S. A. Chow. 2004. Requirement for integrase during reverse transcription of human immunodeficiency virus type 1 and the effect of cysteine mutations of integrase on its interactions with reverse transcriptase. J. Virol. 78:5045-5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zucali, J. R., T. Ciccarone, V. Kelley, J. Park, C. M. Johnson, and A. Mergia. 2002. Transduction of umbilical cord blood CD34+ NOD/SCID repopulating cells by simian foamy virus type 1 (SFV-1) vector. Virology 302:229-335. [DOI] [PubMed] [Google Scholar]