Abstract
The adenovirus (Adv) oncoprotein E1A stimulates cell proliferation and inhibits differentiation. These activities are primarily linked to the N-terminal region (exon 1) of E1A, which interacts with multiple cellular protein complexes. The C terminus (exon 2) of E1A antagonizes these processes, mediated in part through interaction with C-terminal binding proteins 1 and 2 (CtBP1/2). To identify additional cellular E1A targets that are involved in the modulation of E1A C-terminus-mediated activities, we undertook tandem affinity purification of E1A-associated proteins. Through mass spectrometric analysis, we identified several known E1A-interacting proteins as well as novel E1A targets, such as the forkhead transcription factors, FOXK1/K2. We identified a Ser/Thr-containing sequence motif in E1A that mediated interaction with FOXK1/K2. We demonstrated that the E6 proteins of two beta-human papillomaviruses (HPV14 and HPV21) associated with epidermodysplasia verruciformis also interacted with FOXK1/K2 through a motif similar to that of E1A. The E1A mutants deficient in interaction with FOXK1/K2 induced enhanced cell proliferation and oncogenic transformation. The hypertransforming activity of the mutant E1A was suppressed by HPV21 E6. An E1A-E6 chimeric protein containing the Ser/Thr domain of the E6 protein in E1A interacted efficiently with FOXK1/K2 and inhibited cell transformation. Our results suggest that targeting FOXK1/K2 may be a common mechanism for certain beta-HPVs and Adv5. E1A exon 2 mutants deficient in interaction with the dual-specificity kinases DYRK1A/1B and their cofactor HAN11 also induced increased cell proliferation and transformation. Our results suggest that the E1A C-terminal region may suppress cell proliferation and oncogenic transformation through interaction with three different cellular protein complexes: FOXK1/K2, DYRK(1A/1B)/HAN11, and CtBP1/2.
Adenovirus (Adv) is a model DNA tumor virus that has been widely used to decipher critical pathways of oncogenesis. The adenovirus early gene E1A is an intensely investigated viral oncogene and has been instrumental in uncovering common cell cycle-regulatory pathways shared by other DNA tumor virus oncogenes such as human papillomavirus (HPV) E7 (reviewed in reference 43) and simian virus 40 (SV40) T antigen (Ag) (reviewed in reference 9). E1A promotes cellular entry into S phase by deregulating the cell cycle and activates other early viral genes to facilitate viral replication. As a consequence of nonproductive infection, E1A immortalizes rodent cells and also oncogenically transforms these cells in cooperation with other viral or cellular oncogenes (27, 32, 52, 62).
The E1A gene encodes two major protein isoforms that are expressed from two mRNAs (13S and 12S) generated by alternative RNA splicing. The 13S mRNA encodes a 289-amino-acid ([aa] 289R) protein while the 12S mRNA encodes a 243-amino-acid (243R) protein. The 289R and 243R proteins differ by a 46-amino-acid region that is unique to 289R. The 243R protein is not required for viral replication; however, it is sufficient to mitigate cell cycle deregulation and to induce cell transformation. These activities of E1A have been linked to interaction with different cellular protein complexes, which are mediated through distinct sequence motifs in E1A (reviewed in reference 50). Many of these interactions occur with the N-terminal half of E1A encoded by exon 1. These cellular proteins include histone acetyltransferases, p300/CBP (2, 16), and the TRRAP/p400/GCN5 multiprotein chromatin remodeling complex (11, 25, 36) and the retinoblastoma (Rb) family proteins (18, 31, 37, 42, 60). The retinoblastoma tumor suppressor, pRb, is one of the best-characterized cellular targets of E1A. E1A binds pRb through an LXCXE motif, which results in the displacement of pRb from E2F family transcription factors (7). Relief of pRb-mediated transcriptional repression allows E2F transcription factors to activate S-phase genes, resulting in cell cycle progression. This is a common mechanism shared by other DNA tumor virus transforming proteins such as HPV E7 and SV40 T Ag that also bind pRb through the LXCXE motif (10, 15).
Although the role of E1A target proteins in cell transformation has been intensely investigated (reviewed in reference 24), the complex mechanisms through which E1A exerts its effects on cell transformation cannot be explained through interactions with known cellular proteins alone. We have only limited understanding of the potential role of cellular proteins and the E1A C-terminal region (encoded by exon 2) in modulating cell transformation and oncogenesis. Deletion of the E1A C-terminal region results in “hypertransformed” cells in Ras-cooperative transformation assays (6, 13, 20, 54, 56). In addition, the transformed cells expressing E1A with deletions in the C-terminal region are more tumorigenic, invasive, and metastatic (38, 56). These results are consistent with the view that the C-terminal region of E1A suppresses oncogenic transformation, tumorigenesis, and metastasis (8). However, the mechanisms underlying the tumor-suppressive effect of the E1A C-terminal region are not fully understood. Our laboratory identified and cloned a cellular protein, C-terminal binding protein (CtBP), that interacts with a conserved sequence motif, PLDLS, in the C-terminal region of E1A (6, 54). At least part of the tumor-suppressive effect of the E1A C-terminal region appears to be linked to the interaction with CtBP via relief of CtBP-mediated transcriptional repression of genes involved in epithelial to mesenchymal transformation and apoptosis regulation (29). In addition to CtBP, the C-terminal region has also been reported to interact with the yeast dual-specificity Ser/Thr kinase Yak1p (homolog of mammalian DYRK1A/1B) in yeast two-hybrid screening (64). The functional consequence of a Yak1p interaction with E1A is not known.
In an attempt to identify novel E1A targets that could enhance our knowledge of the role of the C-terminal region, we isolated E1A protein complexes by tandem affinity purification (TAP) and identified E1A-associated proteins by mass spectrometry (MS). Here, we report the association of the forkhead family transcription factors, FOXK1 and FOXK2, in addition to CtBP1/2 and DYRK1A/1B, with the C-terminal region of E1A. We also present evidence that suggests that E1A interaction with FOXK1/K2 inhibits E1A-induced proliferation and transformation. We have discovered that the E6 proteins of certain low-oncogenic-potential cutaneous β-HPVs such as HPV14 and HPV21 (HPV14/21) also target FOXK1/K2 through a distinct sequence motif shared with Adv5 E1A. Our results suggest that two different viral proteins, E1A and HPV14/21 E6 may suppress cell transformation by a common mechanism. Additionally, our results also suggest that interaction of E1A with the dual-specificity kinases DYRK1A/1B and their cofactor HAN11 may result in suppression of cell proliferation and transformation.
MATERIALS AND METHODS
Cells.
Suspension cultures of KB cells were maintained in Joklik modified minimal essential medium (MEM) containing 5% horse serum (Sigma). A549, 293, and HeLa cells were maintained in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum. BRK (baby rat kidney) cells were prepared as previously described (56) and maintained in DMEM containing 10% fetal bovine serum.
Viruses.
For construction of Adv5FH-E1A (Adv5 expressing an E1A fusion protein tagged with Flag and hemagglutinin [HA]), a double-stranded oligonucleotide (5′-CGCGTGACTACAAGGATGACGATGACAAGGCTTACCCTTATGACGTCCCCGATTACGCCAGCCTCGAGA and 5′-CTAGTCTCGAGGCTGGCGTAATCGGGGACGTCATAAGGGTAAGCCTTGTCATCGTCATCCTTGTAGTCA) coding for the Flag (DYKDDDDK) and HA(YPYDVPDYA) epitopes at the N terminus of E1A was cloned into two unique restriction sites (MluI and SpeI) located immediately after the ATG codon of EIA in a plasmid containing the left end of Adv5 DNA (nucleotides [nt]1 to 2804). The plasmid pFH-E1A was further extended to include the E1B sequences to generate a single transfer vector (nt 1 to 5788). Adv5FH-E1A recombinant virus was generated by cotransfection of the left-end transfer vector and the Adv5 genomic plasmid pacAd5 9.5-100 (nt 3320 to 33935) (1). The recombinant virus was plaque purified, amplified, and banded in CsCl. The Adv5 exon 1 and exon 2 deletion mutants (based on dl520 that expresses only the 243R protein) have been previously described (17, 34, 46).
Plasmids.
The E1A plasmids (dl520-based) containing exon 2 deletions have been described previously (46). The exon 2 mutants were also constructed in retroviral plasmid vectors pBABE-Puro (45) or pLPC (53) expressing Adv2 12S cDNA. E1A mutants S219A, T229A, and S231A were constructed by oligonucleotide-directed mutagenesis using a Quick-Change kit from Stratagene. For the construction of HPV E6 expression plasmids, the E6-encoding regions (with Flag tags at the C terminus) were amplified by PCR from plasmids pZNeo21E6PH, pZneo20E6PH, pZneo14E6PH (35), and pDONR5E6 (59) and cloned into pTet-GFP (where GFP is green fluorescent protein) vector. For induced expression of pTet-E6 plasmids, they were cotransfected with pTet-TAK as described previously (63). Mutations in the E6 open reading frame (ORF) were introduced by a QuickChange Site-Directed Mutagenesis Kit (Stratagene). pE1A-HPV21-E6 was constructed by replacing the E1A sequences coding for residues 224 to 237 (14 aa) with HPV21 E6 sequences coding for the N-terminal 14-aa region (1 to 14). For transformation assays, the HPV21 E6 ORF was cloned in the vector pLPC. The FOXK1 expression plasmid pAS2256 (FOXK1) was a gift from A. D. Sharrocks (21). Mutants in the FOXK1 ORF were generated by PCR and recloned into p3XFlagCMV7.1 (Sigma). Plasmid pcDNA-S-HAN11 was a gift from A. V. Skurat (55).
TAP and mass spectrometry.
To isolate cellular protein complexes associated with E1A, a suspension culture of 1.0 × 109 KB cells was either mock infected or infected with Adv5FH-E1A at 5 PFU/cell. After 17 to 18 h of infection, whole-cell lysates were prepared in a lysis buffer (20 mM Tris-HCl, pH 8.0, 0.3 M KCl, 5 mM MgCl2, 10% glycerol, 0.1% Tween 20, 10 mM 2-mercaptoethanol, 0.2 mM phenylmethylsulfonyl fluoride [PMSF] supplemented with a protease inhibitor tablet). The cells were lysed by three freeze-thaw cycles and incubation on ice for 30 min. Lysates were cleared by centrifugation, and the supernatant was subsequently diluted with 2 volumes of lysis/dilution buffer (20 mM Tris-HCl, pH 8.0, 10% glycerol, 0.1% Tween 20, 7 mM 2-mercaptoethanol, and 0.25 mM PMSF supplemented with a protease inhibitor tablet). The lysates were recleared by centrifugation, and the supernatant was then incubated with protein A-agarose. E1A-associated protein complexes were isolated by TAP using anti-Flag-conjugated agarose beads (Sigma) and anti-HA-conjugated agarose beads (Roche). First, lysates were added to the anti-Flag beads and left rotating at 4°C overnight. The protein complexes were washed three times with wash buffer (20 mM Tris HCl, pH 8.0, 0.1 M KCl, 5 mM MgCl2, 0.2 mM EDTA, 10% glycerol, 0.1% Tween 20, 7 mM 2-mercaptoethanol, and 0.25 mM PMSF supplemented with a protease inhibitor tablet [Roche]). Protein complexes were eluted with 0.2 mg/ml of the Flag peptide (Sigma) for 1 h and subsequently purified with anti-HA beads. Protein complexes were extensively washed with wash buffer and eluted with 2× SDS sample buffer. The isolated protein complexes were boiled and resolved by a 4 to 12% SDS-NuPAGE gradient gel (Invitrogen). Protein bands were either visualized by staining with silver stain (Bio-Rad) or with SYPRO Ruby protein gel stain (Invitrogen) and subsequently excised and trypsinized for identification by mass spectrometric analysis. Liquid chromatography-tandem MS (LC-MS/MS) was performed at the Donald Danforth Plant Science Center Proteomics and Mass Spectrometry facility.
Immunoprecipitation, Western blot analysis, and antibodies.
Cells were typically collected at 20 to 24 h post-viral infection or at 24 to 48 h posttransfection (using Lipofectamine 2000 reagent; Invitrogen). Cells were washed, lysed, and cleared by centrifugation. Cell lysates were then cleared with protein A-agarose beads and bound to antibody-conjugated beads. After precipitation, protein complexes were extensively washed and eluted with 2× SDS sample buffer, resolved by SDS-PAGE, and subjected to Western blotting. The following antibodies were used for immunoprecipitation: anti-Flag (A2220; Sigma), anti-HA (11815016001; Roche), anti-E1A (M73; sc-25AC; Santa Cruz), anti-E1A (M58; catalog number 554155; BD Biosciences). The following antibodies were used for Western blotting: anti-TRRAP (T17; Santa Cruz), anti-p300 (sc-554; Santa Cruz), anti-p130 (C-20; Santa Cruz), anti-p107 (C-18; Santa Cruz), anti-Rb (IF-8; Santa Cruz), anti-DYRK1A (ab54944; abcam), anti-DYRK1B (2703; Cell Signaling), anti-FOXK1 (ab18196; abcam), anti-FOXK2 (P100774-100; Aviva Systems Biology), anti-E1A (M73 sc-25; Santa Cruz), anti-E1A (M73; 05-599; Upstate), anti-E1A (M58; 554155; BD), anti-HAN11 (provided by A. V. Skurat), anti-B23 (ab24412; abcam), anti-cyclin E (CC05; Calbiochem), anti-Flag (A9469; Sigma), anti-S tag (18588; abcam), anti-pSer219E1A (antibody to phosphorylated Ser of Adv2 E1A; sc-16713; Santa Cruz). The CtBP1 monoclonal antibody (MAb) was raised in our laboratory, and the CtBP2 MAb (612044) was from BD Biosciences.
Phosphatase treatment.
E1A was immunoprecipitated from either 293 or HeLa cells using anti-E1A (M73)-conjugated agarose beads (M73; sc-25AC; Santa Cruz). Immunoprecipitates were washed twice with EDTA-free wash buffer (20 mM Tris-HCl, pH 8.0, 0.1 M KCl, 5 mM MgCl2, 10% glycerol, and 0.1% Tween 20) and once with 1× NEBuffer, provided with a phosphatase kit (P0753S; New England Biolabs). Precipitates were then treated with either the phosphatase buffer (1× NEBuffer and 1 mM MnCl2) alone or with the phosphatase buffer containing 400 units of lambda protein phosphatase at 30°C for 30 min. The pellet and supernatant were separated by centrifugation. The pellet was washed twice with wash buffer (20 mM Tris-HCl, pH 8.0, 0.1 M KCl, 5 mM MgCl2, 0.2 mM EDTA, 10% glycerol, and 0.1% Tween 20). The pellet was then mixed with 200 μl of lysate from HeLa cells transfected with Flag-FOXK1 and reprecipitated. Immunoprecipitates were washed extensively with the wash buffer and eluted with 2× SDS sample buffer. Eluted protein was analyzed by Western blotting with the anti-Flag antibody (A2220; Sigma).
Cell proliferation assay.
Approximately 5.0 × 10 5 primary BRK cells were plated into 25-cm2 flasks and infected with different Adv mutants at 50 PFU/cell. Infected cells were maintained for 20 h in DMEM containing 2% fetal bovine serum. The cells were labeled with bromodeoxyuridine (BrdU) for 4 h in medium containing 2% serum. A commercial kit (BD Pharmingen FITC BrdU Flow kit; item 559619) was used for BrdU labeling and flow cytometric analysis.
Transformation and tumorigenesis assays.
Primary BRK cells were plated in six-well plates 1 day prior to transfection. Plasmids expressing Adv5 E1A, HPV E6, and the E1A-E6 chimera (0.5 μg/well; in vectors containing Neo or Puro markers) were transfected along with a plasmid expressing Ras (1.0 μg/well) into cells using jetPEI according to the manufacturer's specifications. At 24 h posttransfection, the medium was replaced with fresh medium containing G418 (50 μg/ml) or puromycin (0.5 μg/ml). Medium was refreshed every 4 days for 10 to 12 days. Transformed colonies were stained with crystal violet or trypsinized to establish pooled cell lines for tumor studies. For tumor induction, 1 × 106 transformed BRK cells were injected subcutaneously into athymic (nude) mice. The injection site was monitored for tumor formation for up to 2 weeks. Tumor sizes were determined by measurement of tumor length, width, and depth with a Vernier caliper. At least four mice were used per group.
RESULTS
Identification of E1A-interacting proteins.
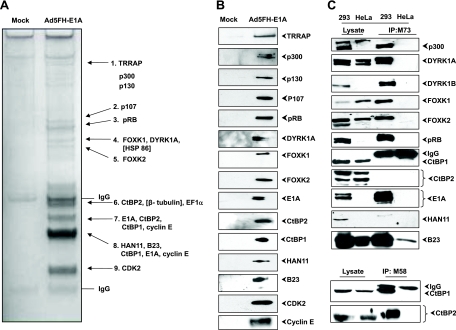
To identify novel cellular proteins associated with E1A during viral infection, we used a TAP (tandem affinity purification) approach. We generated a recombinant Adv5 that expresses an E1A fusion protein tagged with Flag and HA epitopes. The recombinant virus (Adv5FH-E1A) replicated and formed plaques similar to the wild-type (wt) Adv5 in human A549 cells, indicating that the TAP tag did not significantly disrupt E1A functions (data not shown). In order to isolate E1A-protein complexes, human KB cells growing in suspension culture were either mock infected or infected with Adv5FH-E1A. E1A-associated protein complexes were isolated from whole-cell lysates, purified by TAP, and resolved by SDS-PAGE. Proteins present specifically in the protein complex purified from cells infected with Adv5FH-E1A were selected and identified by LC-MS/MS. This analysis identified several previously described E1A-interacting proteins: TRRAP, pRb, p107, CtBP1/2, CDK2, and cyclin E. We also identified novel E1A-interacting proteins including FOXK1/K2, HAN11, and B23. Additionally, the human homolog of Yak1p, DYRK1A, was identified (Fig. 1A). The TAP process followed by mass spectrometric analysis was repeated three times with a consistent profile of E1A-interacting proteins. Proteins identified by mass spectrometry were confirmed by Western blot analysis of the TAP eluate (Fig. 1B). Two previously known E1A-binding proteins, p300 and p130, were not detected by mass spectrometry but were detected by Western blot analysis of TAP protein complexes.
FIG. 1.
Identification of E1A-interacting proteins. (A) Silver-stained image of E1A protein complexes. Protein bands corresponding to previously known and newly identified E1A-associated proteins are shown. (B) Western blot analysis of the TAP eluate. (C) Immunoprecipitation analysis of HeLa or 293 cells. The cell lysates were immunoprecipitated with anti-E1A M73 antibody (upper panel) or with anti-E1A M58 antibody (lower panel). Proteins were eluted and resolved on a 10% SDS-PAGE gel and analyzed by Western blotting.
In order to further verify the interaction of E1A with the newly identified E1A-interacting proteins, we purified the E1A-cellular protein complex from 293 cells (which constitutively express Adv5 E1A and E1B proteins) using anti-E1A (M73) antibody-conjugated beads followed by Western blot analysis (Fig. 1C). Since the epitope for the E1A M73 antibody maps in the CtBP1/2 binding site, the anti-E1A (M58) antibody was used for immunoprecipitation in the bottom panel to demonstrate the E1A interaction with CtBP1/2. All the newly identified proteins (FOXK1/K2, HAN11, and B23) were detected in the E1A protein complex immunoprecipitated with E1A antibodies from 293 cells (which express E1A and E1B) and not from HeLa cells. Between the two related dual-specificity kinases DYRK1A and DYRK1B, only DYRK1A was detected in mass spectrometric analysis of the protein complex isolated from KB cells (Fig. 1A and B). However, immunoprecipitation analysis of proteins isolated from 293 cells revealed association of both kinases with E1A (Fig. 1C). Among the E1A-interacting proteins identified here, we chose to focus on FOXK1/K2 transcription factors since the yeast homologs, Fkh1 and Fkh2 (Fkh1/2) (reviewed in reference 5) and mammalian FOXK1, have been shown to regulate cell cycle and differentiation (reviewed in reference 41). Due to our specific interest in the proteins interacting with the C-terminal region of E1A, we also chose to map the E1A sequences involved in DYRK1A/1B and HAN11 binding.
Mapping of FOXK1/K2 and DYRK1A/HAN11 interaction domains in E1A.
To map the E1A sequences involved in interaction with the newly identified E1A-interacting proteins, we infected HeLa cells with a large panel of previously characterized Adv5 mutants (Fig. 2A). The E1A protein was immunoprecipitated with M73-conjugated beads and analyzed by Western blotting. All exon 1 mutants interacted with FOXK1/K2 and DYRK1A (Fig. 2B). (The low level of pRb interaction with mutant dl1108 [conserved region 2 (CR2)] may reflect the presence of the second Rb-binding site in CR1). Similarly, these E1A-associated proteins also interacted with both the 243R (12S) and the 289R amino acid (13S) proteins (data not shown), ruling out interaction with the CR3 region. These results suggested that FOXK1/K2, DYRK1A, and HAN11 interacted with the E1A exon 2-encoded region. Immunoprecipitation analysis revealed that the deletion mutant dl1132, which lacks amino acids 224 to 238 in the C-terminal region, exhibited a greatly reduced interaction with FOXK1/K2. The mutant dl1132 retained the ability to interact with the other E1A C-terminal binding proteins, CtBP2 and DYRK1A, as well as with pRb, which is an E1A N-terminal binding protein (Fig. 2C). (In Fig. 2C, E1A protein was not immunoprecipitated from cells infected with dl1135 as the epitope for M73 antibody maps within the sequences deleted in the mutant [6, 46]). Our results also showed that DYRK1A and HAN11 were deficient in interaction with the E1A C-terminal deletion mutants dl1133 (deletion of aa 241 to 254) and dl1134 (deletion of aa 255 to 270) (Fig. 2D). Both mutants appeared to be specifically deficient in interaction with DYRK1A as they retained the ability to interact with FOXK1/K2 (Fig. 2C) and CtBP2 (Fig. 2D). To map the region of E1A required for interaction with HAN11, we transfected S-tagged HAN11 in HeLa cells and infected them with Adv5 E1A exon 2 deletion mutants. Protein complexes were immunoprecipitated with anti-E1A (M58) antibody, and the Western blots were probed with S-tag antibody (Fig. 2D). HAN11 exhibited an interaction pattern similar to that of DYRK1A (i.e., deficient in interaction with dl1133 and dl1134). These results agree with previous results which identified HAN11 as a cofactor of DYRK1A (55). Based on our current results and previously known results, the interaction of major cellular proteins with E1A is summarized in Fig. 7.
FIG. 2.
Mapping of FOXK1/K2 and DYRK1A/HAN11 interaction domains in E1A. (A) Schematic of adenovirus exon 1 and exon 2 deletion mutants. All mutants shown in panel A express only the 243R protein. However, exon 2 deletion coordinates are based on the 289R protein. The E1A-null mutant dl312 is not depicted. (B and C) Mapping of FOXK1/K2 and DYRK1A interaction domains. HeLa cells were infected with the indicated Adv5 E1A mutants for 24 h, and the cell lysates were immunoprecipitated (IP) with anti-E1A M73 antibody, resolved on 10% SDS-PAGE, and analyzed by Western blotting with the various antibodies. (D) Mapping of the HAN11 interaction domain. HeLa cells were transfected with S-tagged HAN11. After 24 h, the cells were infected with the indicated Adv5 E1A mutants. After 24 h, the cell lysates were immunoprecipitated with anti-E1A M58 antibody and probed by Western blotting using anti-DYRK1A, anti-CtBP2, anti-S tag, anti-E1A (M58) antibodies.
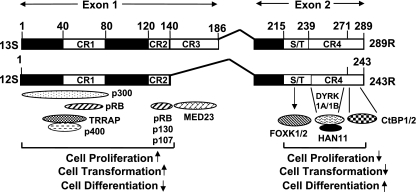
FIG. 7.
Interaction of cellular proteins with E1A. The interaction of the N-terminal region with previously known protein complexes and the functional consequences are indicated. The interaction of FOXK1/K2 and DYRK(1A/1B)/HAN11 and CtBP1/2 and the potential functional consequences are indicated.
Defining the FOXK1/K2 interaction motif on E1A.
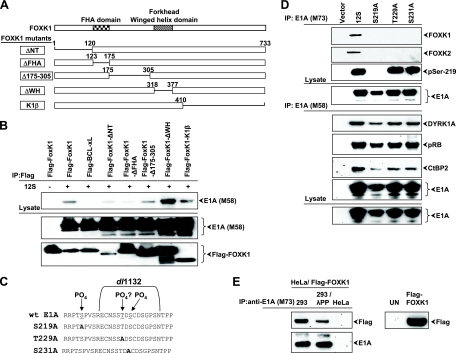
In addition to the winged-helix DNA binding domain that is characteristic of all forkhead family members, FOXK1 and FOXK2 contain an FHA (forkhead-associated) domain. FOXK1/K2 are the only members of the forkhead transcription factors that have an FHA domain (reviewed in reference 14). FHA domains are phosphoprotein interaction modules that can specifically recognize phosphorylated residues within diverse proteins (reviewed in reference 39). We hypothesized that FOXK1 may interact with E1A through this unique domain. To investigate this possibility, we made a series of Flag-tagged FOXK1 deletion mutants (Fig. 3A). When coexpressed with E1A-12S in HeLa cells, the FOXK1 deletion mutant lacking the FHA domain (ΔFHA) showed a greatly reduced association with E1A, as detected by coimmunoprecipitation (Fig. 3B). Additionally, deletion of the N-terminal region of FOXK1 reduced the level of interaction with E1A, but the interaction was detectable. Considering the lower-level expression of this mutant protein (Fig. 3B, lower panel), it appears that the relative level of interaction of this mutant with E1A may be more significant than with the ΔFHA mutant.
FIG. 3.
Role of Ser/Thr residues of E1A on interaction with FOXK1/K2. (A) Domains of FOXK1. The FHA and DNA binding (forkhead) domains are illustrated. Numbers indicate amino acid residues that are deleted in each FOXK1 deletion mutant. (B) Interaction of FOXK1 mutants with E1A. HeLa cells were cotransfected with either vector alone or E1A (12S) and Flag-BCL-XL (a nonspecific protein control), Flag-FOXK1 wt, or mutants. The proteins were immunoprecipitated (IP) with anti-Flag antibody, and Western blots were probed with anti-E1A (M58) or anti-Flag antibodies. (C) Schematic of the E1A-FOXK1 interaction domain. Amino acids that are deleted in the E1A mutant that is deficient in FOXK1/K2 interaction (dl1132) are highlighted with brackets. E1A mutants in Ser and Thr residues are also shown. The amino acid coordinates are based on the 289R protein; however, all viruses express the 243R protein only. (D) Interaction of FOXK1/K2 with Ser/Thr mutants. HeLa cells were transfected with vectors that expressed wt E1A (12S) or indicated mutants. Cell lysates were immunoprecipitated with anti-E1A (M73 or M58) antibody and analyzed by Western blotting with the indicated antibodies. The anti-serine 219 (residue 173 in 12S) E1A antibody was used to confirm that this residue is indeed phosphorylated. (E) 293 or HeLa cells were incubated with anti-E1A antibody-coated agarose beads. Immunoprecipitates were washed and resuspended in phosphatase buffer with or without λ protein phosphatase. Samples were incubated at 30°C for 30 min. The pellets were separated by centrifugation, washed, and incubated with HeLa cell lysates from cells transfected with Flag-FOXK1 and reprecipitated. The bound protein was washed extensively, eluted, and analyzed by Western blotting with the indicated antibodies. The panel at right is a Western blot of HeLa cells either transfected with Flag-FOXK1 or untransfected (UN).
There are multiple serine and threonine residues within the C-terminal FOXK1/K2-interacting domain on E1A which could mediate interaction with the FHA domain of FOXK1/K2. Serine 219 (residue 173 in 12S) and S231 (residue 185 in 12S) in the exon 2 region have been shown to be phosphorylated (57, 58). In order to investigate the requirement of E1A C-terminal serine and threonine residues for FOXK1 interaction, we made Ser-219 → Ala, Thr-229 → Ala, and Ser-231 → Ala substitution mutants within the FOXK1/K2 interaction site (Thr-229 and Ser-231) and the adjoining upstream sequence (Ser-219) (Fig. 3C). The E1A mutant constructs were then transfected into HeLa cells, E1A was immunoprecipitated with M73-conjugated antibody beads, and E1A-associated proteins were probed by Western blot analysis with antibodies specific to FOXK1 or FOXK2. Our results indicated that mutating Ser-219, Thr-229, or Ser-231 abolished FOXK1/K2 interaction with E1A (Fig. 3D). These results indicated that all three residues were critical for the interaction of FOXK1/K2 with E1A. It should be noted that Ser-219 is located upstream of the E1A region deleted in dl1132. Therefore, it appears that sequences deleted in dl1132 as well as some additional upstream sequences that include Ser-219 may be required for FOXK1/K2 interaction with E1A. In order to confirm that the serine and threonine point mutations did not cause gross structural changes in E1A, we determined the interaction of the mutant proteins with cellular proteins that interact with the N-terminal region (pRb) or the C-terminal region (DYRK1A and CtBP2) of E1A. E1A was immunoprecipitated with M58 antibody and analyzed by Western blotting. This analysis showed efficient interaction of the mutant proteins with pRb, DYRK1A, and CtBP2 (Fig. 3D). Lastly, we investigated the effect of E1A phosphorylation on FOXK1 binding by treating immunoprecipitated E1A with λ protein phosphatase before combining E1A with the cell extract containing Flag-tagged FOXK1. The level of interaction of FOXK1 with phosphatase-treated E1A was reduced (Fig. 3E), suggesting that phosphorylation of E1A enhances interaction with FOXK1.
Suppression of cell proliferation, transformation, and tumor formation by E1A C-terminal protein interaction modules.
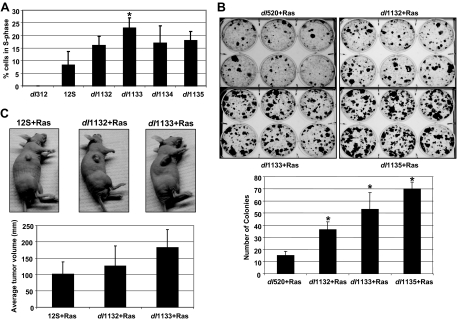
Reports from our laboratory (6, 54, 56) and from the Quinlan laboratory (13, 20) have indicated that the C terminus of E1A negatively regulates E1A-induced cell transformation in the presence of activated Ras. Since the vertebrate and the yeast homologs of FOXK1/K2 have been reported to regulate the cell cycle, we investigated the role of the FOXK1-interaction domain on E1A-induced cell proliferation of primary BRK cells. BRK cells were infected with Adv5 dl312 (E1A minus), Adv5-12S, or different E1A C-terminal deletion mutants (dl1132, dl1133, dl1134, and dl1135). At 24 h postinfection, cells were labeled with BrdU and 7-aminoactinomycin D (7-AAD), and the number of cells in S phase was determined by fluorescence-activated cell sorter (FACS) analysis (Fig. 4A). Our results indicated that all of the E1A C-terminal deletion mutants that we tested, including dl1132, induced increased cell proliferation. Since dl1132 is deficient in FOXK1 interaction [yet retains the ability to interact with CtBP1/2 and DYRK(1A/1B)HAN11], these results suggest that the E1A-FOXK1/K2 interaction inhibits E1A-induced proliferation. Similarly, mutants dl1133 and dl1134, deficient in interaction with DYRK(1A/1B)/HAN11, also induced enhanced cell proliferation.
FIG. 4.
Effect of E1A C-terminal mutants on cell proliferation and Ras-cooperative transformation. (A) Effect of Adv mutants on BRK cell proliferation. The experiment was repeated in triplicates. Each data set was normalized to the percentage of cells in S phase induced by dl312. Data are the average percentage of cells in S-phase relative to the total number of cells. (B) Effect on transformation. Colonies of BRK cells transformed with plasmids expressing either E1A dl520 (which expresses 12S only) and Ras or the indicated E1A C-terminal deletion mutants and Ras. Transformed cells were stained with crystal violet and counted. The number of colonies per well was determined. The experiments were repeated at least twice, and the results of one experiment are shown in the figure. All data are represented as means ± standard deviations. *, P < 0.05. Statistical significance was examined using a Student's t test for panels A and B. (C) Effect on tumorigenesis. Athymic mice were injected subcutaneously with pooled transformed BRK cells. Transformed cells were established from cells transfected with vectors expressing Adv2 12S cDNA or mutants along with a Ras expression vector. The average tumor volume for at least four mice per group is indicated, and representative mice are pictured.
Next, we determined the effect of E1A-FOXK1/K2 interaction on E1A-Ras cooperative transformation. BRK cells were transfected with plasmids expressing either wt E1A 12S (dl520) and Ras or the FOXK1/K2 interaction-deficient mutant, dl1132, and Ras. The numbers and sizes of colonies produced in cells transfected with dl1132 and Ras were greater than those generated by wt 12S and Ras-transfected cells (Fig. 4B). These results suggest that the E1A-FOXK1/K2 interaction domain inhibits E1A-induced transformation. As expected, E1A mutants dl1133 and dl1134 as well as dl1135 resulted in enhanced Ras-cooperative transformation. Therefore, E1A interactions with FOXK1/K2 as well as DYRK(1A/1B)/HAN11 and CtBP likely play significant roles in the negative regulation of E1A-induced transformation.
Lastly, we determined the effect of the FOXK1/K2 interaction domain of E1A on tumor formation. Transformed cells induced by either E1A 12S and Ras, dl1132 and Ras, or dl1133 and Ras were tested for tumorigenicity in athymic mice. As expected, mice injected with cells transformed with dl1132 plus Ras and dl1133 plus Ras produced larger tumors than those injected with 12S plus Ras (Fig. 4C). Collectively, these results suggest that the E1A-FOXK1/K2 interaction may negatively regulate cell proliferation, transformation, and tumorigenicity. Therefore, FOXK1/K2 likely play a significant role in the negative regulation of E1A-induced tumorigenicity. Interestingly, dl1133-Ras induced tumors were larger than dl1132-Ras induced tumors. These results are consistent with the effect of dl1133 [defective interaction with DYRK(1A/1B)/HAN11] on cell proliferation and transformation. Thus, our results also suggest that the E1A region involved in DYRK(1A/1B)/HAN11 interaction may be a more potent negative regulator of E1A-induced oncogenesis than the FOXK1/K2 interaction domain.
Interaction of FOXK1/K2 with the E6 protein of cutaneous HPVs.
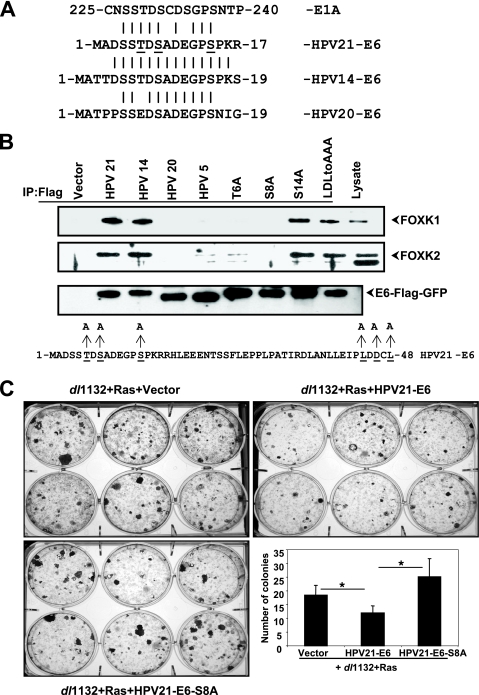
The results shown in Fig. 2C suggested that FOXK1/K2 interacted with E1A sequences encompassing residues 224 to 238 (i.e., sequences deleted in mutant dl1132). Prompted by previously known findings that other DNA tumor virus oncoproteins, such as HPV oncoproteins E7 and E6, target proteins that are targeted by Adv oncoproteins E1A (reviewed in reference 43) and E4(HAdv9)-Orf1 (26), we undertook a data bank search to identify sequence motifs similar to the FOXK1/K2-interacting sequences of E1A in other viral oncoproteins. This search identified a striking homology with the E6 proteins of cutaneous β-HPVs, HPV14 and HPV21 (Fig. 5A). A related motif was also detected in the E6 protein of HPV20. HPVs 21, 14, and 20 are highly related and belong to a subgroup of β-HPVs (12, 59). The E6 proteins of these HPVs are substantially similar to the prototypical member of the β-HPVs, HPV5. Additionally, they contain a unique N-terminal domain that is similar to the E1A C-terminal domain involved in interaction with FOXK1/K2. Among the E6 proteins of this subgroup, the E6 protein of HPV20 contains an amino acid substitution for the conserved Thr residue (Thr → Glu). To determine whether the E6 proteins of these HPVs interact with FOXK1/K2, we transfected plasmids that expressed Flag-tagged versions of different E6 proteins in 293 cells. The proteins were immunoprecipitated with the Flag antibody and probed with antibodies specific to FOXK1/K2 (Fig. 5B). HPV21 E6 and HPV14 E6 readily interacted with FOXK1/K2 proteins while the related HPV20 E6 as well as HPV5 E6 did not interact. Similar to E1A, mutations in the conserved Thr and Ser residues in HPV21 E6 abolished interaction with FOXK1/K2. Mutations elsewhere in HPV21 E6, S14 → A or L44/D46/L48 → AAA (Fig. 5B), did not affect the interaction. These results suggest that Adv5 E1A and HPV21/14 E6 proteins interact with FOXK1/K2 transcription factors through a conserved Thr-Ser-containing motif.
FIG. 5.
Interaction of FOXK1/K2 with HPV E6 proteins and effect of HPV21-E6 on E1A transformation. (A) FOXK1/K2 interaction domains in E1A and E6 proteins. Sequence alignments of the E1A-FOXK1/K2 interaction domain and the N-terminal domain of HPVs 21, 14, and 20 are shown. The conserved Ser/Thr residues that are critical for FOXK1/K2 interaction are underlined. (B) Interaction of FOXK1 with β-HPV E6 protein. Human 293 cells were transfected with plasmids expressing either vector or Flag-tagged E6 proteins of HPV serotypes 5, 21, 14, and 20 or HPV21 E6 mutants. Cells lysates were immunoprecipitated with anti-Flag antibody and analyzed by Western blotting. Lysates from HPV21 E6-transfected 293 cells (without immunoprecipitation) were used as markers for FOXK1/K2. (C) Inhibition of transformation by HPV21 E6. BRK cells were transfected with plasmids expressing E1A dl1132 and Ras along with HPV21 E6 or the HPV E6 S8A mutant. The number of colonies per well was determined. All data are represented as means ± standard deviations. *, P < 0.05. Statistical significance was examined using a Student's t test.
Suppression of cell transformation by HPV21 E6.
To determine whether HPV21 E6 can suppress cell transformation, we carried out a cooperative transformation assay in primary BRK cells by cotransfection of E1A dl1132 and Ras along with HPV21 E6 or a mutant (S8 → A) defective in interaction with FOXK1/K2. Cotransfection of HPV21 E6 with E1A dl1132 suppressed transformation while the S8 → A mutant did not (Fig. 5C). The mutant E6 induced an increase in transformation (compared to dl1132 and Ras), possibly due to an inherent transforming activity of E6 unmasked as a result of the mutation (S8 → A) to the N-terminal domain. These results suggest that HPV21 E6 possesses a transformation suppression activity encoded by the unique N-terminal domain that is absent in the E6 protein of other β-HPVs and high-risk HPVs.
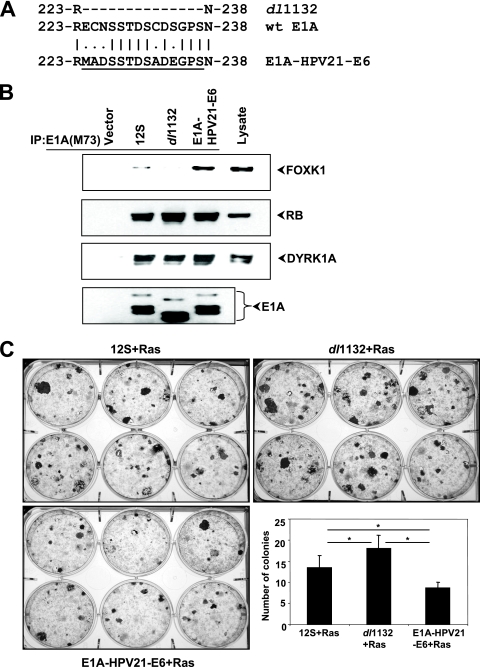
We also determined the transformation suppression activity of the N-terminal domain of HPV21 E6 by domain substitution in E1A. We constructed an E1A-E6 chimeric gene by substituting the E6 domain for the corresponding E1A sequence (Fig. 6A). The E1A-E6 chimeric protein interacted with known E1A-interacting proteins such as pRb and DYRK1A as well as FOXK1, showing that the chimeric protein is functional (Fig. 6B). Interestingly, the chimeric protein interacted with FOXK1 more strongly than wt E1A. The E1A-E6 chimeric construct inhibited transformation compared to wt E1A or dl1132 (Fig. 6C). These results strongly suggest that the E1A sequences encompassing the deletion region in dl1132 and the HPV21 E6 sequences in the unique N-terminal domain are functionally similar with regard to interaction with FOXK1 and suppression of cell transformation.
FIG. 6.
Functional domain substitution in E1A. (A) Schematic of E1A-E6. The FOXK1/K2 interaction motif of HPV21 E6 was cloned into the region that is deleted in the E1A dl1132 mutant. (B) HeLa cells were transfected with plasmids expressing vector, E1A 12S, E1A dl1132, and the E1A-E6 chimera. Cell lysates were immunoprecipitated with anti-E1A M73 antibody and analyzed by Western blotting. Lysates from HPV21 E6-transfected HeLa cells (without immunoprecipitation) were used as markers for FOXK1. (C) Transforming activity of the E1A-E6 chimeric construct. BRK cells were transformed with plasmids expressing E1A 12S and Ras, E1A dl1132 and Ras, or the E1A-E6 chimera and Ras. The number of colonies per well was determined. All data are represented as means ± standard deviations. *, P < 0.05. Statistical significance was examined using a Student's t test.
DISCUSSION
In an attempt to identify new cellular targets of Adv5 E1A, we utilized the proteomic approach of TAP. Our investigation identified several well-established cellular targets of E1A in addition to novel E1A-interacting proteins, the forkhead transcription factors FOXK1 and FOXK2. E1A interaction with FOXK1/K2 occurs through the C-terminal region of E1A (exon 2). The E1A sequences involved in the interaction of FOXK1/K2 are conserved only among the species C (nononcogenic) Advs (4). Unlike the FOXK1/K2-interacting module, the DYRK(1A/1B)/HAN11 binding sequences are located within the CR4 region conserved among different species of human Advs. Interaction with DYRK(1A/1B)/HAN11, like interaction with CtBP1/2, may be a common feature of E1A proteins of all primate Advs. Interestingly, the E6 proteins of low-risk β-HPVs (HPV21 and HPV14) also interacted with FOXK1/K2 through a sequence similar to that of Adv5 E1A.
Our results suggest that interaction of FOXK1/K2 with E1A may be mediated, at least in part, through the FHA (forkhead-associated) domain. The FHA domain is a phosphoprotein binding domain that predominantly recognizes phospho-Thr ([p-Thr] in addition to p-Ser and p-Tyr) residues within diverse proteins (39). We have identified a Thr-Ser-containing sequence motif within Adv5 E1A and HPV21/14 E6 that is essential for interaction with FOXK1/K2 transcription factors. Our results show that both Thr and Ser residues are essential. Between these critical residues in E1A, Ser-219 and Ser-231 were reported to be phosphorylated (57, 58). We note that the phosphorylation site at Ser-219 is the consensus phosphorylation site for DYRK1A/1B, which suggests that DYRK1A/1B could possibly alter E1A activities by phosphorylation. Different phosphorylation prediction algorithms suggest that Thr-229 is not an optimal phosphorylation site, consistent with our inability to detect phosphorylation at this site (data not shown). Nonetheless, the critical importance of the Thr residue is evident from our mutational analyses of E1A and of HPV21 E6 and from the inability of HPV20 E6 (which contains a Glu substitution instead of Thr) to interact with FOXK1/K2 (Fig. 3 and 5).
Previous results from our laboratory reported enhanced transforming, tumor-inducing, and metastasis activities associated with E1A exon 2 mutants (56). These results led us to forge the concept that E1A exon 2 possesses a transformation- and oncogenesis-restraining activity (8). Part of this activity was linked to the extreme C-terminal region of E1A which interacts with CtBP (6, 54). Our present studies have linked two other cellular protein complexes, FOXK1/K2 and DYRK(1A/1B)/HAN11 (in addition to CtBP1/2), with transformation- and tumorigenesis-restraining activities of the E1A C-terminal (exon 2) region. Our results suggest that E1A interaction with FOXK1/K2 results in retardation of E1A-induced cell proliferation and oncogenic transformation. The E1A mutant (dl1132) deficient in interaction with FOXK1/K2 consistently induced increased proliferation, transformation, and tumor formation compared to wt E1A, albeit not as robustly as mutants defective in interaction with DYRK(1A/1B)/HAN11 and CtBP1/2 (dl1133, dl1134, and dl1135). The present results on the transforming activity of mutant dl1132 (defective in interaction with FOXK1/K2) are in good agreement with a different mutant that contained a deletion encompassing the region deleted in dl1132 (6). In the present study, we demonstrate that deletion mutants dl1133 and dl1134, defective in interaction with DYRK(1A/1B)/HAN11 are hypertransforming, suggesting that E1A interaction with the DYRK(1A/1B)/HAN11 complex may inhibit cell proliferation and transformation. Certain hypertransforming E1A mutants previously identified by the Quinlan laboratory (13, 51) map in the C-terminal region deleted in mutants dl1133 and dl1134 (aa 241 to 270). It would be interesting to determine whether the E1A mutants reported by the Quinlan group are deficient in interaction with DYRK(1A/1B)/HAN11. Thus, E1A interaction of all three cellular protein complexes [i.e., FOXK1/K2, DYRK(1A/1B)/HAN11, and CtBP1/2] may additively contribute to the suppression of transformation and tumorigenesis since deletion encompassing all three protein interaction regions of the E1A C terminus resulted in the most potent transforming activity (56).
The Adv E1A C-terminal region may exert its inhibitory effect on cell transformation by deregulation of cell differentiation-related activities via recruitment of the three different cellular protein complexes (Fig. 7). Steve Frisch reported that E1A was able to suppress the tumorigenic activity of several human cancer cell lines by engendering an epithelial phenotype (23). Subsequently, the mesenchymal-to-epithelial transformation activity of E1A was linked to interaction with CtBP (29) and relief of transcriptional repression of cellular genes involved in the regulation of cell proliferation, differentiation, and apoptosis (28, 29).
Like the CtBP1/2 complex, the FOXK1/K2 complex may modulate cellular pathways involved in cell proliferation, differentiation, and apoptosis. In mammalian cells, FOXK1 appears to be a stem cell maintenance factor (reviewed in reference 41). Disruption of FOXK1 was reported to result in upregulation of differentiation-related genes in a muscle stem cell population (44). In contrast to FOXK1, there are no significant studies of the activities of FOXK2. The yeast forkhead proteins Fkh1 and Fkh2 appear to be homologs of mammalian FOXK1/K2 because they both contain FHA domains, which are not typically found in other forkhead family members. Several reports indicate that Fkh1/2 have a definitive role in transcriptional regulation of the genes involved in the G2/M transition (65). In addition, the activities of Fkh1/2 have been linked to pseudohyphal growth, a form of cell transformation in yeast (65). Although transcriptional repression and activation functions of mammalian FOXK1/K2 are possible, only the transcriptional repression activity of FOXK1 has been studied in some detail (21, 61). Thus, Adv5 E1A and HPV21/14 E6 may deregulate transcriptional activities of FOXK1/K2 either by sequestration or by redirection to other target genes to facilitate cell cycle withdrawal and promotion of cell differentiation.
The DYRK(1A/1B)/HAN11 complex may also function at the crossroads of cellular proliferation and differentiation. Mammalian DYRK1A has also been shown to interfere with the Notch signaling pathway and the NFAT and Gli transcriptional programs, which are linked to cell fate and differentiation activities (3, 19, 30, 40). DYRK1B has been shown to regulate differentiation and apoptosis (reviewed in reference 22). Lastly, the activity of yeast Yak1p has been linked to pseudohyphal growth (64). Genetic studies of zebra fish have linked the DYRK1A cofactor HAN11 with certain differentiation programs (47). Therefore, the E1A C-terminal region may exert its effects on cell proliferation and transformation by disruption of the cellular activities of these protein kinases and HAN11. As evidenced from genetic studies of yeast which revealed significant cross talk between Yak1p and Fkh1/2 pathways (33), there may be cross talk between all three pathways deregulated by the E1A C-terminal region to exert a concerted effect on cell proliferation and transformation.
Of considerable significance is our discovery of a commonality between Adv5 E1A and HPV21/14 E6 proteins which target FOXK1/K2 through a conserved S/T sequence motif to suppress cell transformation. We have shown that HPV21 E6 can dampen Ras-cooperative transformation induced by the hypertransforming E1A mutant that is deficient in interaction with FOXK1/K2 (dl1132) while an HPV21 E6 mutant (S8 → A) that is unable to interact with FOXK1/K2 is not able to do so. These results suggest that the E6 proteins of HPV that complex with FOXK1/K2 may also suppress transformation by retarding cell proliferation. What might be the functional consequence of such an activity in the natural history of these HPVs? HPV21 and HPV14, in addition to various other HPVs, are associated with benign cutaneous epidermodysplasia verruciformis (EV) lesions (9, 48). The E6 proteins of HPV21 and HPV14 may promote differentiation by inhibiting cell proliferation to facilitate replication of these epithelium-specific viruses. Since about half of the EV patients develop squamous cell carcinoma (49), the transformation-restraining activities of the N-terminal region of HPV21/14 E6 may also suppress the oncogenic activities of other HPVs present in EV lesions to promote productive viral infection rather than oncogenic transformation. The E6 proteins of HPV21/14 and Adv5 E1A might target FOXK1/K2 to manipulate the delicate balance between proliferation and differentiation to create an optimal cellular state for viral replication in epithelial cells. The shared mechanism may be important in the natural history of these different epithelium-specific viruses.
Acknowledgments
This study was supported by research grants CA-84941 and CA-33616.
We thank A. D. Sharrocks, A. V. Skurat, Denise Galloway, and T. Kiyono for their kind gifts of reagents.
Footnotes
Published ahead of print on 6 January 2010.
REFERENCES
- 1.Anderson, R. D., R. E. Haskell, H. Xia, B. J. Roessler, and B. L. Davidson. 2000. A simple method for the rapid generation of recombinant adenovirus vectors. Gene Ther. 7:1034-1038. [DOI] [PubMed] [Google Scholar]
- 2.Arany, Z., D. Newsome, E. Oldread, D. M. Livingston, and R. Eckner. 1995. A family of transcriptional adaptor proteins targeted by the E1A oncoprotein. Nature 374:81-84. [DOI] [PubMed] [Google Scholar]
- 3.Arron, J. R., M. M. Winslow, A. Polleri, C. P. Chang, H. Wu, X. Gao, J. R. Neilson, L. Chen, J. J. Heit, S. K. Kim, N. Yamasaki, T. Miyakawa, U. Francke, I. A. Graef, and G. R. Crabtree. 2006. NFAT dysregulation by increased dosage of DSCR1 and DYRK1A on chromosome 21. Nature 441:595-600. [DOI] [PubMed] [Google Scholar]
- 4.Avvakumov, N., A. E. Kajon, R. C. Hoeben, and J. S. Mymryk. 2004. Comprehensive sequence analysis of the E1A proteins of human and simian adenoviruses. Virology 329:477-492. [DOI] [PubMed] [Google Scholar]
- 5.Bahler, J. 2005. Cell-cycle control of gene expression in budding and fission yeast. Annu. Rev. Genet 39:69-94. [DOI] [PubMed] [Google Scholar]
- 6.Boyd, J. M., T. Subramanian, U. Schaeper, M. La Regina, S. Bayley, and G. Chinnadurai. 1993. A region in the C-terminus of adenovirus 2/5 E1a protein is required for association with a cellular phosphoprotein and important for the negative modulation of T24-ras mediated transformation, tumorigenesis and metastasis. EMBO J. 12:469-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chellappan, S., V. B. Kraus, B. Kroger, K. Munger, P. M. Howley, W. C. Phelps, and J. R. Nevins. 1992. Adenovirus E1A, simian virus 40 tumor antigen, and human papillomavirus E7 protein share the capacity to disrupt the interaction between transcription factor E2F and the retinoblastoma gene product. Proc. Natl. Acad. Sci. U. S. A. 89:4549-4553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chinnadurai, G. 1992. Adenovirus E1a as a tumor-suppressor gene. Oncogene 7:1255-1258. [PubMed] [Google Scholar]
- 9.DeCaprio, J. A. 2009. How the Rb tumor suppressor structure and function was revealed by the study of adenovirus and SV40. Virology 384:274-284. [DOI] [PubMed] [Google Scholar]
- 10.DeCaprio, J. A., J. W. Ludlow, J. Figge, J. Y. Shew, C. M. Huang, W. H. Lee, E. Marsilio, E. Paucha, and D. M. Livingston. 1988. SV40 large tumor antigen forms a specific complex with the product of the retinoblastoma susceptibility gene. Cell 54:275-283. [DOI] [PubMed] [Google Scholar]
- 11.Deleu, L., S. Shellard, K. Alevizopoulos, B. Amati, and H. Land. 2001. Recruitment of TRRAP required for oncogenic transformation by E1A. Oncogene 20:8270-8275. [DOI] [PubMed] [Google Scholar]
- 12.de Villiers, E. M. 1989. Heterogeneity of the human papillomavirus group. J. Virol. 63:4898-4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Douglas, J. L., S. Gopalakrishnan, and M. P. Quinlan. 1991. Modulation of transformation of primary epithelial cells by the second exon of the Ad5 E1A12S gene. Oncogene 6:2093-2103. [PubMed] [Google Scholar]
- 14.Durocher, D., and S. P. Jackson. 2002. The FHA domain. FEBS Lett. 513:58-66. [DOI] [PubMed] [Google Scholar]
- 15.Dyson, N., P. M. Howley, K. Munger, and E. Harlow. 1989. The human papilloma virus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science 243:934-937. [DOI] [PubMed] [Google Scholar]
- 16.Eckner, R., M. E. Ewen, D. Newsome, M. Gerdes, J. A. DeCaprio, J. B. Lawrence, and D. M. Livingston. 1994. Molecular cloning and functional analysis of the adenovirus E1A-associated 300-kD protein (p300) reveals a protein with properties of a transcriptional adaptor. Genes Dev. 8:869-884. [DOI] [PubMed] [Google Scholar]
- 17.Egan, C., T. N. Jelsma, J. A. Howe, S. T. Bayley, B. Ferguson, and P. E. Branton. 1988. Mapping of cellular protein-binding sites on the products of early-region 1A of human adenovirus type 5. Mol. Cell. Biol. 8:3955-3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ewen, M. E., Y. G. Xing, J. B. Lawrence, and D. M. Livingston. 1991. Molecular cloning, chromosomal mapping, and expression of the cDNA for p107, a retinoblastoma gene product-related protein. Cell 66:1155-1164. [DOI] [PubMed] [Google Scholar]
- 19.Fernandez-Martinez, J., E. M. Vela, M. Tora-Ponsioen, O. H. Ocana, M. A. Nieto, and J. Galceran. 2009. Attenuation of Notch signalling by the Down-syndrome-associated kinase DYRK1A. J. Cell Sci. 122:1574-1583. [DOI] [PubMed] [Google Scholar]
- 20.Fischer, R. S., and M. P. Quinlan. 1998. The C terminus of E1A regulates tumor progression and epithelial cell differentiation. Virology 249:427-439. [DOI] [PubMed] [Google Scholar]
- 21.Freddie, C. T., Z. Ji, A. Marais, and A. D. Sharrocks. 2007. Functional interactions between the Forkhead transcription factor FOXK1 and the MADS-box protein SRF. Nucleic Acids Res. 35:5203-5212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Friedman, E. 2007. Mirk/Dyrk1B in cancer. J. Cell Biochem. 102:274-279. [DOI] [PubMed] [Google Scholar]
- 23.Frisch, S. M. 1991. Antioncogenic effect of adenovirus E1A in human tumor cells. Proc. Natl. Acad. Sci. U. S. A. 88:9077-9081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frisch, S. M., and J. S. Mymryk. 2002. Adenovirus-5 E1A: paradox and paradigm. Nat. Rev. Mol. Cell Biol. 3:441-452. [DOI] [PubMed] [Google Scholar]
- 25.Fuchs, M., J. Gerber, R. Drapkin, S. Sif, T. Ikura, V. Ogryzko, W. S. Lane, Y. Nakatani, and D. M. Livingston. 2001. The p400 complex is an essential E1A transformation target. Cell 106:297-307. [DOI] [PubMed] [Google Scholar]
- 26.Glaunsinger, B. A., S. S. Lee, M. Thomas, L. Banks, and R. Javier. 2000. Interactions of the PDZ-protein MAGI-1 with adenovirus E4-ORF1 and high-risk papillomavirus E6 oncoproteins. Oncogene 19:5270-5280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Graham, F. L., A. J. van der Eb, and H. L. Heijneker. 1974. Size and location of the transforming region in human adenovirus type 5 DNA. Nature 251:687-691. [DOI] [PubMed] [Google Scholar]
- 28.Grooteclaes, M., Q. Deveraux, J. Hildebrand, Q. Zhang, R. H. Goodman, and S. M. Frisch. 2003. C-terminal-binding protein corepresses epithelial and proapoptotic gene expression programs. Proc. Natl. Acad. Sci. U. S. A. 100:4568-4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grooteclaes, M. L., and S. M. Frisch. 2000. Evidence for a function of CtBP in epithelial gene regulation and anoikis. Oncogene 19:3823-3828. [DOI] [PubMed] [Google Scholar]
- 30.Gwack, Y., S. Sharma, J. Nardone, B. Tanasa, A. Iuga, S. Srikanth, H. Okamura, D. Bolton, S. Feske, P. G. Hogan, and A. Rao. 2006. A genome-wide Drosophila RNAi screen identifies DYRK-family kinases as regulators of NFAT. Nature 441:646-650. [DOI] [PubMed] [Google Scholar]
- 31.Hannon, G. J., D. Demetrick, and D. Beach. 1993. Isolation of the Rb-related p130 through its interaction with CDK2 and cyclins. Genes Dev. 7:2378-2391. [DOI] [PubMed] [Google Scholar]
- 32.Houweling, A., P. J. van den Elsen, and A. J. van der Eb. 1980. Partial transformation of primary rat cells by the leftmost 4.5% fragment of adenovirus 5 DNA. Virology 105:537-550. [DOI] [PubMed] [Google Scholar]
- 33.Jaspersen, S. L., J. F. Charles, R. L. Tinker-Kulberg, and D. O. Morgan. 1998. A late mitotic regulatory network controlling cyclin destruction in Saccharomyces cerevisiae. Mol. Biol. Cell 9:2803-2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jelsma, T. N., J. A. Howe, J. S. Mymryk, C. M. Evelegh, N. F. Cunniff, and S. T. Bayley. 1989. Sequences in E1A proteins of human adenovirus 5 required for cell transformation, repression of a transcriptional enhancer, and induction of proliferating cell nuclear antigen. Virology 171:120-130. [DOI] [PubMed] [Google Scholar]
- 35.Kiyono, T., A. Hiraiwa, and M. Ishibashi. 1992. Differences in transforming activity and coded amino acid sequence among E6 genes of several papillomaviruses associated with epidermodysplasia verruciformis. Virology 186:628-639. [DOI] [PubMed] [Google Scholar]
- 36.Lang, S. E., and P. Hearing. 2003. The adenovirus E1A oncoprotein recruits the cellular TRRAP/GCN5 histone acetyltransferase complex. Oncogene 22:2836-2841. [DOI] [PubMed] [Google Scholar]
- 37.Li, Y., C. Graham, S. Lacy, A. M. Duncan, and P. Whyte. 1993. The adenovirus E1A-associated 130-kD protein is encoded by a member of the retinoblastoma gene family and physically interacts with cyclins A and E. Genes Dev. 7:2366-2377. [DOI] [PubMed] [Google Scholar]
- 38.Linder, S., P. Popowicz, C. Svensson, H. Marshall, M. Bondesson, and G. Akusjarvi. 1992. Enhanced invasive properties of rat embryo fibroblasts transformed by adenovirus E1A mutants with deletions in the carboxy-terminal exon. Oncogene 7:439-443. [PubMed] [Google Scholar]
- 39.Mahajan, A., C. Yuan, H. Lee, E. S. Chen, P. Y. Wu, and M. D. Tsai. 2008. Structure and function of the phosphothreonine-specific FHA domain. Sci. Signal. 1:re12. [DOI] [PubMed] [Google Scholar]
- 40.Mao, J., P. Maye, P. Kogerman, F. J. Tejedor, R. Toftgard, W. Xie, G. Wu, and D. Wu. 2002. Regulation of Gli1 transcriptional activity in the nucleus by Dyrk1. J. Biol. Chem. 277:35156-35161. [DOI] [PubMed] [Google Scholar]
- 41.Martin, C. M., J. L. Russell, A. Ferdous, and D. J. Garry. 2006. Molecular signatures define myogenic stem cell populations. Stem Cell Rev. 2:37-42. [DOI] [PubMed] [Google Scholar]
- 42.Mayol, X., X. Grana, A. Baldi, N. Sang, Q. Hu, and A. Giordano. 1993. Cloning of a new member of the retinoblastoma gene family (pRb2) which binds to the E1A transforming domain. Oncogene 8:2561-2566. [PubMed] [Google Scholar]
- 43.McLaughlin-Drubin, M. E., and K. Munger. 2009. The human papillomavirus E7 oncoprotein. Virology 384:335-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meeson, A. P., T. J. Hawke, S. Graham, N. Jiang, J. Elterman, K. Hutcheson, J. M. Dimaio, T. D. Gallardo, and D. J. Garry. 2004. Cellular and molecular regulation of skeletal muscle side population cells. Stem Cells 22:1305-1320. [DOI] [PubMed] [Google Scholar]
- 45.Morgenstern, J. P., and H. Land. 1990. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 18:3587-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mymryk, J. S., and S. T. Bayley. 1993. Induction of gene expression by exon 2 of the major E1A proteins of adenovirus type 5. J. Virol. 67:6922-6928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nissen, R. M., A. Amsterdam, and N. Hopkins. 2006. A zebrafish screen for craniofacial mutants identifies wdr68 as a highly conserved gene required for endothelin-1 expression. BMC Dev. Biol. 6:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Orth, G. 1986. Epidermodysplasia verruciformis: a model for understanding the oncogenicity of human papillomaviruses. Ciba Found Symp. 120:157-174. [DOI] [PubMed] [Google Scholar]
- 49.Orth, G., S. Jablonska, M. Jarzabek-Chorzelska, S. Obalek, G. Rzesa, M. Favre, and O. Croissant. 1979. Characteristics of the lesions and risk of malignant conversion associated with the type of human papillomavirus involved in epidermodysplasia verruciformis. Cancer Res. 39:1074-1082. [PubMed] [Google Scholar]
- 50.Pelka, P., J. N. Ablack, G. J. Fonseca, A. F. Yousef, and J. S. Mymryk. 2008. Intrinsic structural disorder in adenovirus E1A: a viral molecular hub linking multiple diverse processes. J. Virol. 82:7252-7263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Quinlan, M. P., and J. L. Douglas. 1992. Immortalization of primary epithelial cells requires first- and second-exon functions of adenovirus type 5 12S. J. Virol. 66:2020-2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ruley, H. E. 1983. Adenovirus early region 1A enables viral and cellular transforming genes to transform primary cells in culture. Nature 304:602-606. [DOI] [PubMed] [Google Scholar]
- 53.Samuelson, A. V., M. Narita, H. M. Chan, J. Jin, E. de Stanchina, M. E. McCurrach, M. Fuchs, D. M. Livingston, and S. W. Lowe. 2005. p400 is required for E1A to promote apoptosis. J. Biol. Chem. 280:21915-21923. [DOI] [PubMed] [Google Scholar]
- 54.Schaeper, U., J. M. Boyd, S. Verma, E. Uhlmann, T. Subramanian, and G. Chinnadurai. 1995. Molecular cloning and characterization of a cellular phosphoprotein that interacts with a conserved C-terminal domain of adenovirus E1A involved in negative modulation of oncogenic transformation. Proc. Natl. Acad. Sci. U. S. A. 92:10467-10471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Skurat, A. V., and A. D. Dietrich. 2004. Phosphorylation of Ser640 in muscle glycogen synthase by DYRK family protein kinases. J. Biol. Chem. 279:2490-2498. [DOI] [PubMed] [Google Scholar]
- 56.Subramanian, T., M. La Regina, and G. Chinnadurai. 1989. Enhanced ras oncogene mediated cell transformation and tumorigenesis by adenovirus 2 mutants lacking the C-terminal region of E1a protein. Oncogene 4:415-420. [PubMed] [Google Scholar]
- 57.Tremblay, M. L., C. J. McGlade, G. E. Gerber, and P. E. Branton. 1988. Identification of the phosphorylation sites in early region 1A proteins of adenovirus type 5 by amino acid sequencing of peptide fragments. J. Biol. Chem. 263:6375-6383. [PubMed] [Google Scholar]
- 58.Tsukamoto, A. S., A. Ponticelli, A. J. Berk, and R. B. Gaynor. 1986. Genetic mapping of a major site of phosphorylation in adenovirus type 2 E1A proteins. J. Virol. 59:14-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Underbrink, M. P., H. L. Howie, K. M. Bedard, J. I. Koop, and D. A. Galloway. 2008. E6 proteins from multiple human betapapillomavirus types degrade Bak and protect keratinocytes from apoptosis after UVB irradiation. J. Virol. 82:10408-10417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Whyte, P., K. J. Buchkovich, J. M. Horowitz, S. H. Friend, M. Raybuck, R. A. Weinberg, and E. Harlow. 1988. Association between an oncogene and an anti-oncogene: the adenovirus E1A proteins bind to the retinoblastoma gene product. Nature 334:124-129. [DOI] [PubMed] [Google Scholar]
- 61.Yang, Q., Y. Kong, B. Rothermel, D. J. Garry, R. Bassel-Duby, and R. S. Williams. 2000. The winged-helix/forkhead protein myocyte nuclear factor beta (MNF-beta) forms a co-repressor complex with mammalian sin3B. Biochem. J. 345:335-343. [PMC free article] [PubMed] [Google Scholar]
- 62.Zerler, B., B. Moran, K. Maruyama, J. Moomaw, T. Grodzicker, and H. E. Ruley. 1986. Adenovirus E1A coding sequences that enable ras and pmt oncogenes to transform cultured primary cells. Mol. Cell. Biol. 6:887-899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang, S., Y. Feng, O. Narayan, and L. J. Zhao. 2001. Cytoplasmic retention of HIV-1 regulatory protein Vpr by protein-protein interaction with a novel human cytoplasmic protein VprBP. Gene 263:131-140. [DOI] [PubMed] [Google Scholar]
- 64.Zhang, Z., M. M. Smith, and J. S. Mymryk. 2001. Interaction of the E1A oncoprotein with Yak1p, a novel regulator of yeast pseudohyphal differentiation, and related mammalian kinases. Mol. Biol. Cell 12:699-710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhu, G., P. T. Spellman, T. Volpe, P. O. Brown, D. Botstein, T. N. Davis, and B. Futcher. 2000. Two yeast forkhead genes regulate the cell cycle and pseudohyphal growth. Nature 406:90-94. [DOI] [PubMed] [Google Scholar]