Abstract
Scavenger receptor CD163 is a key entry mediator for porcine reproductive and respiratory syndrome virus (PRRSV). To identify the CD163 protein domains involved in PRRSV infection, deletion mutants and chimeric mutants were created. Infection experiments revealed that scavenger receptor cysteine-rich (SRCR) domain 5 (SRCR 5) is essential for PRRSV infection, while the four N-terminal SRCR domains and the cytoplasmic tail are not required. The remaining CD163 protein domains need to be present but can be replaced by corresponding SRCR domains from CD163-L1, resulting in reduced (SRCR 6 and interdomain regions) or unchanged (SRCR 7 to SRCR 9) infection efficiency. In addition, CD163-specific antibodies recognizing SRCR 5 are able to reduce PRRSV infection.
Porcine reproductive and respiratory syndrome (PRRS) is one of the most devastating viral pig diseases worldwide (17, 26). The causative agent, PRRS virus (PRRSV), has a restricted host and cell tropism, with porcine alveolar macrophages as important target cells (7, 13, 25). PRRSV entry into these macrophages has been studied extensively (6, 15, 16, 28, 31), and to date, two macrophage-specific molecules are known as PRRSV entry mediators: the siglec sialoadhesin and scavenger receptor CD163 (2, 29, 30). The interaction between PRRSV and its internalization receptor, sialoadhesin, has been the subject of intensive investigation, with recently identification of the M/GP5 complex as a viral ligand interacting with the N-terminal immunoglobulin-like domain of sialoadhesin (1, 4, 5, 27). In contrast, our understanding of the specific contribution of CD163 during PRRSV infection is still in its infancy. So far, it has been demonstrated that CD163 is not involved in virus binding and internalization in macrophages but most likely acts during PRRSV uncoating (30). Most recently, viral minor glycoproteins GP2 and GP4 were shown to interact with CD163 (3). Further, the two N-terminal scavenger receptor cysteine-rich (SRCR) domains are not involved, but the transmembrane domain is essential for CD163 to sustain PRRSV infection (2). To get more insight into the role of CD163 during PRRSV infection, this study aimed to identify the CD163 protein domains involved in PRRSV infection.
CD163 deletion mutants.
CD163 is a type I membrane protein composed of a signal peptide followed by nine SRCR domains, with a 35-amino-acid proline-serine-threonine (PST)-rich region separating SRCR domain 6 (SRCR 6) and SRCR 7. A second PST-rich region connects SRCR 9 with the transmembrane domain and a short cytoplasmic tail, which contains a functional internalization motif (Fig. 1a) (12, 18, 19, 21, 22). To define the protein domains of CD163 that are essential during PRRSV infection, a fusion PCR was used to construct CD163 deletion mutants lacking all nine extracellular SRCR domains or the intracellular cytoplasmic tail, as depicted in Fig. 1b (see also Fig. S1, Table S2, and Protocol S3 in the supplemental material) (23, 33). All mutants retained the N-terminal signal peptide and were fused to a C-terminal V5-His tag to enable detection of all constructs. Figure 1c and d show a Western blot analysis of the constructs expressed and indicate whether the recombinant proteins are present at the cell surface (see also Fig. S4 in the supplemental material), respectively. To evaluate the potential of the different mutants to sustain PRRSV infection, nonpermissive HEK293T cells were transfected with sialoadhesin in combination with different CD163 constructs, since expression of both entry mediators offers a model for virus entry and infection similar to the primary target cells, macrophages (30). Relative percentages of infected cells were calculated, with the original CD163 construct as a reference (Fig. 1e), which showed that full-length CD163 constructs with and without the V5-His tag behave similarly (mutants A and B). The essential domains seem to be present in the extracellular part of CD163, since deletion of the cytoplasmic tail had no influence on infection, in contrast to deletion of all extracellular SRCR domains, which resulted in a complete loss of infection (mutant D and C, respectively). Refinement of the deletions of the extracellular part shows that the three N-terminal SRCR domains are not needed in PRRSV infection (mutant E). Deletion of the small PST I interdomain region resulted in reduced PRRSV infection (mutant G). In contrast, no infection was observed for mutants lacking SRCR domains 4, 5, and 6, domains 7, 8, and 9, or the PST II domain (mutants F, H, and I, respectively).
FIG. 1.
CD163 deletion constructs used to identify the essential domains involved in PRRSV infection. (a) Structural domain organization of CD163 (nine extracellular SRCR domains, two PST-rich domains, a transmembrane region, and an intracellular cytoplasmic tail). (b) Domain organization of CD163 deletion mutants. (c) Detection of recombinant CD163 deletion variants in HEK293T cell lysates. Cell lysates were subjected to reducing SDS-PAGE prior to electroblotting and immunodetection with a V5-specific MAb (GenScript). (d) Expression profile of CD163 mutants by using MAb 2A10 (AbD Serotec), PAb AF1607 (R&D Systems), and a V5-specific MAb (GenScript). Immunofluorescence staining was performed first on nonpermeabilized cells to visualize CD163 variants at the cell surface, followed by permeabilization and visualization of surface as well as intracellular recombinant CD163 variants. +, surface and intracellular expression; −, no surface expression, only intracellular expression; NA, not applicable. (e) Twenty-four hours prior to inoculation, HEK293T cells were transfected with sialoadhesin combined with one of the CD163 deletion variants. Transfected cells were inoculated with PRRSV, and 24 h later, cells were fixed with ice-cold methanol and stained. Transfected as well as infected cells were visualized, and the relative percentage of infected cells was calculated, with the original full-length porcine CD163 as a reference. The average numbers of infected cells counted for each CD163 variant were 280 (A), 286 (B), 0 (C), 271 (D), 296 (E), 0 (F), 185 (G), 0 (H), and 0 (I), respectively. (See the text for descriptions of the variants.) Each value represents the mean ± standard deviation for three experiments. Different lowercase letters indicate statistically significant differences between different CD163 mutants (one-way analysis of variance; Tukey B test, α < 0.05).
Chimeric mutants with CD163 backbone.
Since the deletion of protein domains might influence the conformation of the remaining molecule, replacing the domains with similar ones is a good complementary strategy. Intriguingly, besides swine CD163, human, monkey, mouse, and canine homologues have also been described as enabling PRRSV infection in vitro (2), making them inappropriate for domain swapping. A possible candidate, however, was found in CD163-L1, which is also known as CD163b or M160 and is structurally very similar to CD163 (Fig. 2a). Both CD163 and CD163-L1 were cloned from the human cell line U937, and their potential to sustain PRRSV infection was evaluated as described above (Fig. 2b to e). Although less efficient than porcine CD163, human CD163 renders nonpermissive cells permissive for PRRSV infection (mutant J), as was reported before (2). CD163-L1 is not able to sustain PRRSV infection, making it a good source of SRCR domains in the construction of chimeric mutants (mutant K). On the basis of the results obtained with the deletion mutants, we created chimeric constructs with a CD163 backbone in which specific protein domains were replaced with corresponding domains from CD163-L1 (Fig. 2b; see also Fig. S1, Table S2, and Protocol S3 in the supplemental material). As for the deletion mutants, expression was analyzed via Western blotting and immunofluorescence staining (Fig. 2c and d; see also Fig. S4), and the potential to sustain PRRSV infection in HEK293T cells expressing sialoadhesin combined with different CD163 constructs was analyzed (Fig. 2e). Simultaneous replacement of CD163 SRCR 4, 5, and 6 with the corresponding domains of CD163-L1 resulted in a loss of infectivity (mutant L). To evaluate the contribution of each of the three domains, all three were replaced separately. Substitution of CD163 SRCR 4 had no influence on infection (mutant P), while swapping CD163 SRCR 5 completely inhibited infection (mutant Q). Replacing CD163 SRCR 6 resulted in reduced infection efficiency (mutant R). Like SRCR 6, both PST I and II seemed not to be essential, but the presence of the corresponding domains of CD163-L1 reduced infection (mutants M and O). Interestingly, CD163 SRCR domains 7, 8, and 9, closest to the plasma membrane, could be replaced by the corresponding domains from CD163-L1 without significantly influencing PRRSV infection (mutant N).
FIG. 2.
Construction of chimeric constructs via domain swapping between CD163 and CD163-L1 to identify the essential domains involved in PRRSV infection. (a) Structural domain organization of CD163 (white) and CD163-L1 (black). CD163 has been suggested to emerge from CD163-L1 by gene duplication, a process during which three of the first six SRCR domains of CD163-L1 were lost (10, 24). They share six consecutive SRCR domains in the structure k-[b-c-d-e-d]. (b and f) Domain organization of chimeric constructs. Porcine CD163, human CD163, and human CD163-L1 are represented in white, gray, and black, respectively. (c and g) Detection of recombinant chimeric variants in HEK293T cell lysates. Cell lysates were subjected to reducing SDS-PAGE prior to electroblotting and immunodetection with a V5-specific MAb (GenScript). (d and h) Expression profile of CD163 mutants using antibodies MAb 2A10 (AbD Serotec), PAb AF1607 (R&D Systems) and a V5-specific MAb (GenScript). Immunofluorescence staining was performed first on nonpermeabilized cells to visualize CD163 variants at the cell surface, followed by permeabilization and visualization of surface as well as intracellular recombinant CD163 variants. (e and i) Twenty-four hours prior to inoculation, HEK293T cells were transfected with sialoadhesin combined with one of the CD163 chimeric constructs. Transfected cells were inoculated with PRRSV, and 24 h later, cells were fixed with ice-cold methanol and stained. Transfected as well as infected cells were visualized, and the relative percentage of infected cells was calculated, with the full-length porcine CD163 with a V5-His tag as a reference. The average numbers of infected cells counted for each CD163 variant in panel e were 283 (B), 165 (J), 0 (K), 0 (L), 175 (M), 268 (N), 189 (O), 287 (P), 0 (Q), and 107 (R), while the average numbers in panel i were 279 (B), 0 (S), 168 (T), and 243 (U), respectively. (See the text for descriptions of the variants.) Each value represents the mean ± standard deviation for three experiments. Different lowercase letters indicate statistically significant differences between different CD163 mutants (one-way analysis of variance; Tukey B test, α < 0.05).
Chimeric mutants with CD163-L1 backbone.
Since CD163 domain SRCR 5 seems to be vital for PRRSV infection, this domain was used to replace its corresponding domain in a CD163-L1 background (Fig. 2f to i; see also Fig. S1, Table S2, Protocol S3, and Fig. S4 in the supplemental material). However, this was not sufficient to enable PRRSV infection (mutant S). Therefore, additional domains were replaced, starting with the domains that were not essential but for which replacement resulted in reduced infection efficiency, like SRCR 6 and both PST I and PST II domains. Replacement of SRCR 5 combined with replacement of SRCR 6 or the PST I or PST II domain in a CD163-L1 background did not restore infectivity (data not shown). Only the CD163-L1 construct in which SRCR 5, SRCR 6, and the PST I and II domains were replaced with CD163 sequences was able to sustain PRRSV infection, albeit less efficiently than the full-length CD163 (mutant T). Infection was almost fully restored when the three SRCR domains closest to the plasma membrane were also replaced (mutant U).
CD163-specific antibodies blocking PRRSV infection.
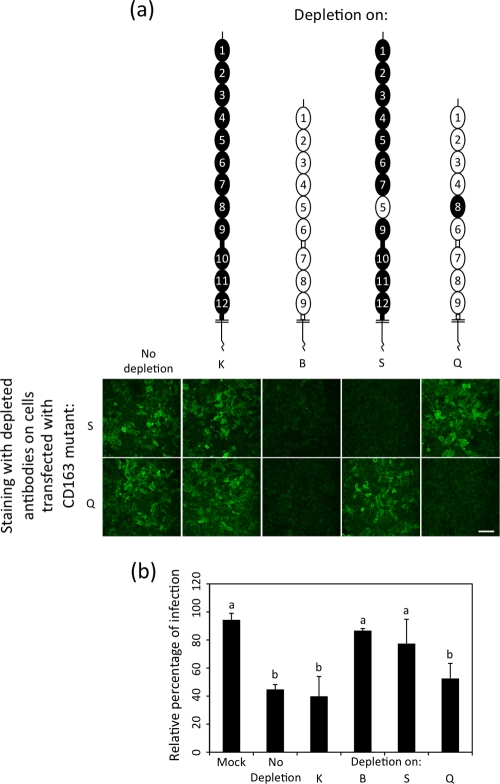
Previously, it was shown that polyclonal antibody (PAb) AF1607 (R&D Systems) is able to block PRRSV infection in primary porcine alveolar macrophages, in contrast to the monoclonal antibody (MAb) 2A10 (AbD Serotec) (30). Both CD163-specific antibodies were tested for binding to the panel of CD163 constructs to map their recognition sites. The epitope recognized by MAb 2A10 is present in one of the three N-terminal SRCR domains. The PAb AF1607 recognizes several epitopes, with at least one of them being present in SRCR domain 5. Both the PAb and the MAb recognize porcine CD163, while human CD163 is recognized only by the PAb and not by the MAb. Neither of the two antibodies cross-reacted with human CD163-L1 (data not shown). Based on the results of the CD163 mutants indicating that SRCR 5 is essential and since this domain is recognized by the PAb able to block PRRSV infection, it was hypothesized that the blocking effect of the PAb was due to the antibodies recognizing an epitope present in SRCR 5. To evaluate this hypothesis, the PAb was passaged on transfected cells expressing different mutants to deplete the PAb of antibodies recognizing specific epitopes. The antibodies were first examined to confirm depletion (Fig. 3a) and were then used to block PRRSV infection on macrophages (Fig. 3b). When passaged on CD163-L1, no antibodies were removed and PRRSV infection on macrophages could be reduced to a level similar to that effected by the control antibody that was not passaged on transfected cells. In contrast, when passaged on CD163, all antibodies recognizing different epitopes were depleted, resulting in the loss of the ability to influence PRRSV infection. The PAb incubated on cells expressing mutant S was depleted of antibodies recognizing CD163 SRCR 5, which resulted in the loss of the ability to influence PRRSV infection. However, in the case of the PAb incubated on cells expressing mutant Q, all antibodies except the ones recognizing CD163 SRCR 5 were depleted, and the remaining antibodies were still able to reduce PRRSV infection to the same level as the original PAb, confirming that the antibodies recognizing SRCR 5 are responsible for the observed blocking effect.
FIG. 3.
Usage of CD163 variants to deplete a goat polyclonal antibody able to block PRRSV infection in macrophages. (a) The goat PAb AF1607 (R&D Systems) was passaged 16 times on transfected cells expressing different CD163 variants. Depletion of specific antibodies was confirmed via an immunofluorescence staining. Images represent an overlay of several z-sections throughout whole cells. Bar, 50 μm. (b) Depleted antibodies were used to treat macrophages for 1 h at 37°C prior to and during inoculation with PRRSV. Ten hours postinoculation, cells were fixed and infected cells were visualized using MAb P3/27 (32). The relative percentage of infected cells was calculated, with untreated cells as a reference. The average numbers of infected cells counted for each condition were 63 (mock), 30 (no depletion), 27 (depletion on K), 58 (depletion on B), 52 (depletion on S), and 35 (depletion on Q), respectively. Each value represents the mean ± standard deviation for three experiments. Different lowercase letters indicate statistically significant differences between treatments (one-way analysis of variance; Tukey B test, α < 0.05).
In conclusion, the CD163 cytoplasmic tail is dispensable for PRRSV infection, as are the four N-terminal extracellular SRCR domains. The essential domains are more centrally located, with SRCR 5 as a key component. So far, only SRCR 2 and SRCR 3 have been found involved in biological processes. Hemoglobin-haptoglobin (HbHp) complexes are internalized upon binding to SRCR 3 of CD163 (11, 14). Previously, it was shown that HbHp complexes are not able to reduce PRRSV infection (30), which is in agreement with the results obtained with the mutants described here, showing that the four N-terminal SRCR domains are not involved. In addition, a 13-amino-acid motif within SRCR 2 has been identified as a putative interaction site that mediates erythroblast binding (8) and interaction with both Gram-positive and -negative bacteria (9). In addition to PRRSV, one other virus, African swine fever virus (ASFV), is known to use CD163 to enter its target cells (20). For ASFV, MAb 2A10 has been shown to inhibit both ASFV infection and viral particle binding to macrophages. Here we showed that MAb 2A10 recognizes an epitope present in SRCR 1, 2, or 3, indicating that ASFV interacts with one of the three N-terminal SRCR domains. Interestingly, MAb 2A10 is not able to influence PRRSV infection (30), indicating that PRRSV does not interact with one of the three N-terminal SRCR domains. This is consistent with our observation that the four N-terminal SRCR domains of CD163 are not involved during PRRSV infection. Another difference between ASFV and PRRSV is that for ASFV, CD163 is suggested to be involved in virus-cell attachment, since CD163-specific antibodies can block ASFV attachment to macrophages (20). In contrast, for PRRSV, CD163 is suggested to be involved during a later step in virus entry, i.e., virus uncoating (30). In conjunction with the data obtained with MAb 2A10, these observations suggest that the two viruses interact with CD163 differently during infection.
Nucleotide sequence accession number.
The cDNA sequence for the human CD163-L1 used in this study is available under GenBank accession number GQ397482.
Supplementary Material
Acknowledgments
We gratefully thank Carine Boone, Chantal Vanmaercke, Lieve Sys, and Dries Helderweirt for excellent technical assistance.
This work was supported by the Industrial Research Fund (IOF) of Ghent University.
We declare that we have no conflict of interest.
Footnotes
Published ahead of print on 23 December 2009.
REFERENCES
- 1.An, T.-Q., Z.-J. Tian, Y.-X. He, Y. Xiao, Y.-F. Jiang, J.-M. Peng, Y.-J. Zhou, D. Liu, and G.-Z. Tong. 5 December 2009, posting date. Porcine reproductive and respiratory syndrome virus attachment is mediated by the N-terminal domain of the sialoadhesin receptor. Vet. Microbiol. doi: 10.1016/j.vetmic.2009.11.006. [DOI] [PubMed]
- 2.Calvert, J. G., D. E. Slade, S. L. Shields, R. Jolie, R. M. Mannan, R. G. Ankenbauer, and S. K. Welch. 2007. CD163 expression confers susceptibility to porcine reproductive and respiratory syndrome viruses. J. Virol. 81:7371-7379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Das, P. B., P. X. Dinh, I. H. Ansari, M. de Lima, F. A. Osorio, and A. K. Pattnaik. 2010. The minor envelope glycoproteins GP2a and GP4 of porcine reproductive and respiratory syndrome virus interact with the receptor CD163. J. Virol. 84:1731-1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Delputte, P. L., and H. J. Nauwynck. 2004. Porcine arterivirus infection of alveolar macrophages is mediated by sialic acid on the virus. J. Virol. 78:8094-8101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Delputte, P. L., W. Van Breedam, I. Delrue, C. Oetke, P. R. Crocker, and H. J. Nauwynck. 2007. Porcine arterivirus attachment to the macrophage-specific receptor sialoadhesin is dependent on the sialic acid-binding activity of the N-terminal immunoglobulin domain of sialoadhesin. J. Virol. 81:9546-9550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delputte, P. L., N. Vanderheijden, H. J. Nauwynck, and M. B. Pensaert. 2002. Involvement of the matrix protein in attachment of porcine reproductive and respiratory syndrome virus to a heparinlike receptor on porcine alveolar macrophages. J. Virol. 76:4312-4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duan, X., H. J. Nauwynck, and M. B. Pensaert. 1997. Virus quantification and identification of cellular targets in the lungs and lymphoid tissues of pigs at different time intervals after inoculation with porcine reproductive and respiratory syndrome virus (PRRSV). Vet. Microbiol. 56:9-19. [DOI] [PubMed] [Google Scholar]
- 8.Fabriek, B. O., M. M. Polfliet, R. P. Vloet, R. C. van der Schors, A. J. Ligtenberg, L. K. Weaver, C. Geest, K. Matsuno, S. K. Moestrup, C. D. Dijkstra, and T. K. van den Berg. 2007. The macrophage CD163 surface glycoprotein is an erythroblast adhesion receptor. Blood 109:5223-5229. [DOI] [PubMed] [Google Scholar]
- 9.Fabriek, B. O., R. van Bruggen, D. M. Deng, A. J. Ligtenberg, K. Nazmi, K. Schornagel, R. P. Vloet, C. D. Dijkstra, and T. K. van den Berg. 2009. The macrophage scavenger receptor CD163 functions as an innate immune sensor for bacteria. Blood 113:887-892. [DOI] [PubMed] [Google Scholar]
- 10.Gronlund, J., L. Vitved, M. Lausen, K. Skjodt, and U. Holmskov. 2000. Cloning of a novel scavenger receptor cysteine-rich type I transmembrane molecule (M160) expressed by human macrophages. J. Immunol. 165:6406-6415. [DOI] [PubMed] [Google Scholar]
- 11.Kristiansen, M., J. H. Graversen, C. Jacobsen, O. Sonne, H. J. Hoffman, S. K. Law, and S. K. Moestrup. 2001. Identification of the haemoglobin scavenger receptor. Nature 409:198-201. [DOI] [PubMed] [Google Scholar]
- 12.Law, S. K., K. J. Micklem, J. M. Shaw, X. P. Zhang, Y. Dong, A. C. Willis, and D. Y. Mason. 1993. A new macrophage differentiation antigen which is a member of the scavenger receptor superfamily. Eur. J. Immunol. 23:2320-2325. [DOI] [PubMed] [Google Scholar]
- 13.Lawson, S. R., K. D. Rossow, J. E. Collins, D. A. Benfield, and R. R. Rowland. 1997. Porcine reproductive and respiratory syndrome virus infection of gnotobiotic pigs: sites of virus replication and co-localization with MAC-387 staining at 21 days post-infection. Virus Res. 51:105-113. [DOI] [PubMed] [Google Scholar]
- 14.Madsen, M., H. J. Moller, M. J. Nielsen, C. Jacobsen, J. H. Graversen, T. van den Berg, and S. K. Moestrup. 2004. Molecular characterization of the haptoglobin.hemoglobin receptor CD163. Ligand binding properties of the scavenger receptor cysteine-rich domain region. J. Biol. Chem. 279:51561-51567. [DOI] [PubMed] [Google Scholar]
- 15.Misinzo, G. M., P. L. Delputte, and H. J. Nauwynck. 2008. Involvement of proteases in porcine reproductive and respiratory syndrome virus uncoating upon internalization in primary macrophages. Vet. Res. 39:55. [DOI] [PubMed] [Google Scholar]
- 16.Nauwynck, H. J., X. Duan, H. W. Favoreel, P. Van Oostveldt, and M. B. Pensaert. 1999. Entry of porcine reproductive and respiratory syndrome virus into porcine alveolar macrophages via receptor-mediated endocytosis. J. Gen. Virol. 80:297-305. [DOI] [PubMed] [Google Scholar]
- 17.Neumann, E. J., J. B. Kliebenstein, C. D. Johnson, J. W. Mabry, E. J. Bush, A. H. Seitzinger, A. L. Green, and J. J. Zimmerman. 2005. Assessment of the economic impact of porcine reproductive and respiratory syndrome on swine production in the United States. J. Am. Vet. Med. Assoc. 227:385-392. [DOI] [PubMed] [Google Scholar]
- 18.Nielsen, M. J., M. Madsen, H. J. Moller, and S. K. Moestrup. 2006. The macrophage scavenger receptor CD163: endocytic properties of cytoplasmic tail variants. J. Leukoc. Biol. 79:837-845. [DOI] [PubMed] [Google Scholar]
- 19.Ritter, M., C. Buechler, T. Langmann, and G. Schmitz. 1999. Genomic organization and chromosomal localization of the human CD163 (M130) gene: a member of the scavenger receptor cysteine-rich superfamily. Biochem. Biophys. Res. Commun. 260:466-474. [DOI] [PubMed] [Google Scholar]
- 20.Sánchez-Torres, C., P. Gomez-Puertas, M. Gomez-del-Moral, F. Alonso, J. M. Escribano, A. Ezquerra, and J. Dominguez. 2003. Expression of porcine CD163 on monocytes/macrophages correlates with permissiveness to African swine fever infection. Arch. Virol. 148:2307-2323. [DOI] [PubMed] [Google Scholar]
- 21.Sarrias, M. R., J. Gronlund, O. Padilla, J. Madsen, U. Holmskov, and F. Lozano. 2004. The scavenger receptor cysteine-rich (SRCR) domain: an ancient and highly conserved protein module of the innate immune system. Crit. Rev. Immunol. 24:1-37. [DOI] [PubMed] [Google Scholar]
- 22.Schaer, C. A., G. Schoedon, A. Imhof, M. O. Kurrer, and D. J. Schaer. 2006. Constitutive endocytosis of CD163 mediates hemoglobin-heme uptake and determines the noninflammatory and protective transcriptional response of macrophages to hemoglobin. Circ. Res. 99:943-950. [DOI] [PubMed] [Google Scholar]
- 23.Shevchuk, N. A., A. V. Bryksin, Y. A. Nusinovich, F. C. Cabello, M. Sutherland, and S. Ladisch. 2004. Construction of long DNA molecules using long PCR-based fusion of several fragments simultaneously. Nucleic Acids Res. 32:e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stover, C. M., J. Schleypen, J. Gronlund, M. R. Speicher, W. J. Schwaeble, and U. Holmskov. 2000. Assignment of CD163B, the gene encoding M160, a novel scavenger receptor, to human chromosome 12p13.3 by in situ hybridization and somatic cell hybrid analysis. Cytogenet. Cell Genet. 90:246-247. [DOI] [PubMed] [Google Scholar]
- 25.Teifke, J. P., M. Dauber, D. Fichtner, M. Lenk, U. Polster, E. Weiland, and J. Beyer. 2001. Detection of European porcine reproductive and respiratory syndrome virus in porcine alveolar macrophages by two-colour immunofluorescence and in-situ hybridization-immunohistochemistry double labelling. J. Comp. Pathol. 124:238-245. [DOI] [PubMed] [Google Scholar]
- 26.Tian, K., X. Yu, T. Zhao, Y. Feng, Z. Cao, C. Wang, Y. Hu, X. Chen, D. Hu, X. Tian, D. Liu, S. Zhang, X. Deng, Y. Ding, L. Yang, Y. Zhang, H. Xiao, M. Qiao, B. Wang, L. Hou, X. Wang, X. Yang, L. Kang, M. Sun, P. Jin, S. Wang, Y. Kitamura, J. Yan, and G. F. Gao. 2007. Emergence of fatal PRRSV variants: unparalleled outbreaks of atypical PRRS in China and molecular dissection of the unique hallmark. PLoS One 2:e526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van Breedam, W., H. Van Gorp, J. Q. Zhang, P. Crocker, P. L. Delputte, and H. J. Nauwynck. 15 January 2010. The M/GP5 glycoprotein complex of porcine reproductive and respiratory syndrome virus binds the sialoadhesin receptor in a sialic acid-dependent manner. PLoS Pathog. 6:e1000730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vanderheijden, N., P. Delputte, H. Nauwynck, and M. Pensaert. 2001. Effects of heparin on the entry of porcine reproductive and respiratory syndrome virus into alveolar macrophages. Adv. Exp. Med. Biol. 494:683-689. [DOI] [PubMed] [Google Scholar]
- 29.Vanderheijden, N., P. L. Delputte, H. W. Favoreel, J. Vandekerckhove, J. Van Damme, P. A. van Woensel, and H. J. Nauwynck. 2003. Involvement of sialoadhesin in entry of porcine reproductive and respiratory syndrome virus into porcine alveolar macrophages. J. Virol. 77:8207-8215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Van Gorp, H., W. Van Breedam, P. L. Delputte, and H. J. Nauwynck. 2008. Sialoadhesin and CD163 join forces during entry of the porcine reproductive and respiratory syndrome virus. J. Gen. Virol. 89:2943-2953. [DOI] [PubMed] [Google Scholar]
- 31.Van Gorp, H., W. Van Breedam, P. L. Delputte, and H. J. Nauwynck. 2009. The porcine reproductive and respiratory syndrome virus requires trafficking through CD163 positive early endosomes, but not late endosomes, for productive infection. Arch. Virol. 154:1939-1943. [DOI] [PubMed] [Google Scholar]
- 32.Wieczorek-Krohmer, M., F. Weiland, K. Conzelmann, D. Kohl, N. Visser, P. van Woensel, H. J. Thiel, and E. Weiland. 1996. Porcine reproductive and respiratory syndrome virus (PRRSV): monoclonal antibodies detect common epitopes on two viral proteins of European and U.S. isolates. Vet. Microbiol. 51:257-266. [DOI] [PubMed] [Google Scholar]
- 33.Wurch, T., F. Lestienne, and P. J. Pauwels. 1998. A modified overlap extension PCR method to create chimeric genes in the absence of restriction enzymes. Biotechnol. Tech. 12:653-657. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.