Abstract
With an estimated 40% of the world population at risk, dengue poses a significant threat to human health, especially in tropical and subtropical regions. Preventative and curative efforts, such as vaccine development and drug discovery, face additional challenges due to the occurrence of four antigenically distinct serotypes of the causative dengue virus (DEN1 to -4). Complex immune responses resulting from repeat assaults by the different serotypes necessitate simultaneous targeting of all forms of the virus. One of the promising targets for drug development is the highly conserved two-component viral protease NS2B-NS3, which plays an essential role in viral replication by processing the viral precursor polyprotein into functional proteins. In this paper, we report the 2.1-Å crystal structure of the DEN1 NS2B hydrophilic core (residues 49 to 95) in complex with the NS3 protease domain (residues 1 to 186) carrying an internal deletion in the N terminus (residues 11 to 20). While the overall folds within the protease core are similar to those of DEN2 and DEN4 proteases, the conformation of the cofactor NS2B is dramatically different from those of other flaviviral apoprotease structures. The differences are especially apparent within its C-terminal region, implicated in substrate binding. The structure reveals for the first time serotype-specific structural elements in the dengue virus family, with the reported alternate conformation resulting from a unique metal-binding site within the DEN1 sequence. We also report the identification of a 10-residue stretch within NS3pro that separates the substrate-binding function from the catalytic turnover rate of the enzyme. Implications for broad-spectrum drug discovery are discussed.
Dengue fever (DF) affects tens of millions of people each year, with an average mortality rate of 5%, comprising mostly young children. The increasing spread and frequency of global epidemics of this disease have heightened the urgency for developing effective strategies for prevention, diagnosis, and treatment. Though several groups are involved in vaccine development (17, 23, 25), dengue presents a unique, complex challenge. Four antigenically distinct serotypes of the causative dengue virus (DENV) (DEN1 to -4) occur in nature, with differing pathogenicities in partially overlapping geographic locations. In individuals previously exposed to a certain serotype, repeat assault by a different serotype can lead to potentially fatal complications of the disease, dengue hemorrhagic fever and dengue shock syndrome. The presence of subneutralizing levels of antibodies against the first serotype, resulting in antibody-dependent enhancement (ADE), is believed to be a causative mechanism underlying this complication. Forty percent of the world population lives in dengue risk areas, mainly in tropical and subtropical regions, which necessitates the development of vaccines and therapeutics that can simultaneously protect against all four serotypes (24).
DEN1 to -4 belong to the family Flaviviridae (genus Flavivirus) of positive-stranded RNA viruses transmitted by Aedes aegypti mosquitoes. Upon infection, the genomic RNA is translated by the host cell machinery into a 370-kDa polyprotein, which is subsequently cleaved and processed into 10 distinct structural (C, E, and prM) and nonstructural (NS1, 2A, 2B, 3, 4A, 4B, and 5) proteins. A majority of this processing (junctions of NS2A/2B, 2B/3, 3/4A, and 4B/5, as well as internal sites within C, 2A, 3, and 4A) is carried out by a virus-encoded two-component protease, NS2B-NS3. Additional processing, especially of sites toward the N terminus (junctions of C/prM, prM/E, E/NS1, NS1/NS2A, and NS4A/4B), employs cellular proteases, such as signalase. The NS2B-NS3-mediated cleavage forms an essential step in the viral replicative cycle, as evidenced by the lack of production of infectious virions in mutants carrying inactivating substitutions in the protease. Of the duo, the N terminus of NS3 encodes the enzymatic core, while a hydrophilic core within NS2B provides an essential cofactor function (9).
Owing to its essential nature in viral replication and the promise and success of drugs targeting the proteases in the treatment of human immunodeficiency virus (HIV) and hepatitis C virus (HCV) infections (4), several recent studies have concentrated on identifying inhibitors of the flaviviral protease (5, 10, 21, 26-28). An added advantage of targeting the protease is that it is highly conserved between the serotypes (63 to 74%). For such structure-based drug design efforts, the availability of three-dimensional (3D) structures is an essential prerequisite, and given the differing pathogenicities and immune complexity generated by the multiple serotypes, it is a prudent first step to determine the structures of all four DENV proteases. The crystal structures of the DEN2 and DEN4 proteases in complex with various lengths of NS2B are available (7, 15). In this paper, we present the structures of an optimal construct of DEN1pro in complex with the essential hydrophilic core of NS2B (NS2Bc), as well as its active-site mutant. These structures reveal a novel and unexpected serotype-specific structural element embedded in the DENV genome, with implications for the discovery of broad-spectrum drugs. We also report the identification of a 10-residue stretch within the NS3 sequence that allows the separation of the substrate-binding function of the enzyme from its catalytic efficiency.
MATERIALS AND METHODS
Construct design.
DNA constructs were sequence optimized and synthesized to remove codon bias and increase heterologous expression in bacterial hosts (DNA 2.0 Inc.). For protease constructs covalently linked to the cofactor, the hydrophilic core of DEN1 NS2B (residues 49 to 95; NS2Bc) was fused to the protease domain of NS3 (residues 1 to 186; NS3pro) through a previously reported 9-residue flexible linker, Gly4-Ser-Gly4 (14). Constructs were cloned into a modified pET28a vector (Novagen) with an N-terminal 6-His tag, followed by a TEV protease cleavage site (20). Deletions and mutations were carried out through site-directed mutagenesis PCR using Pfu Ultra II polymerase (Stratagene).
Expression and purification.
Fresh transformants of E. coli BL21(DE3) cells (Invitrogen Inc.) were grown at 37°C in LB medium containing 30 μg/liter kanamycin to an optical density at 600 nm (OD600) of 0.5 to 0.8. The cultures were then cooled and induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) overnight at 18°C. The cells were pelleted and extracted using a Microfluidizer Processor (Microfluidics International Corp.) in buffer containing 50 mM Tris, pH 8.0, 250 mM NaCl, 2.5 mM MgCl2, 10% glycerol, 0.5 g/liter lysozyme, and 100 μl/liter OmniCleave endonuclease (Epicentre Biotechnologies). The extracts were clarified by ultracentrifugation at 45,000 × g and passed over Ni-nitrilotriacetic acid (NTA) columns (Qiagen Inc.). The columns were washed with buffer containing 50 mM Tris, pH 8.0, and 250 mM NaCl, followed by buffer with an additional 25 mM imidazole to remove nonspecifically and loosely bound contaminants. Bound His-tagged protein was eluted in 50 mM Tris, pH 8.0, 250 mM NaCl, and 200 mM imidazole.
The N-terminal 6-His tag was cleaved by incubating the protein with a noncleavable His-tagged TEV protease overnight at 4°C (1:50 [mass/mass] ratio); desalted in Tris, pH 8.0; and passed over Ni-NTA to remove uncleaved protein and His-TEV protease. It was then subjected to anion exchange using 0.1 M sodium cacodylate at pH 6.5 with a gradient of 0 to 0.5 M sodium sulfate. Size exclusion chromatography (0.1 M sodium cacodylate, 0.25 M sodium sulfate) and matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) were used to verify the presence of a single monomeric species of the purified protein. A final concentration of 40 mg/ml was used for crystallization trials.
Protein stability (Tm) assay.
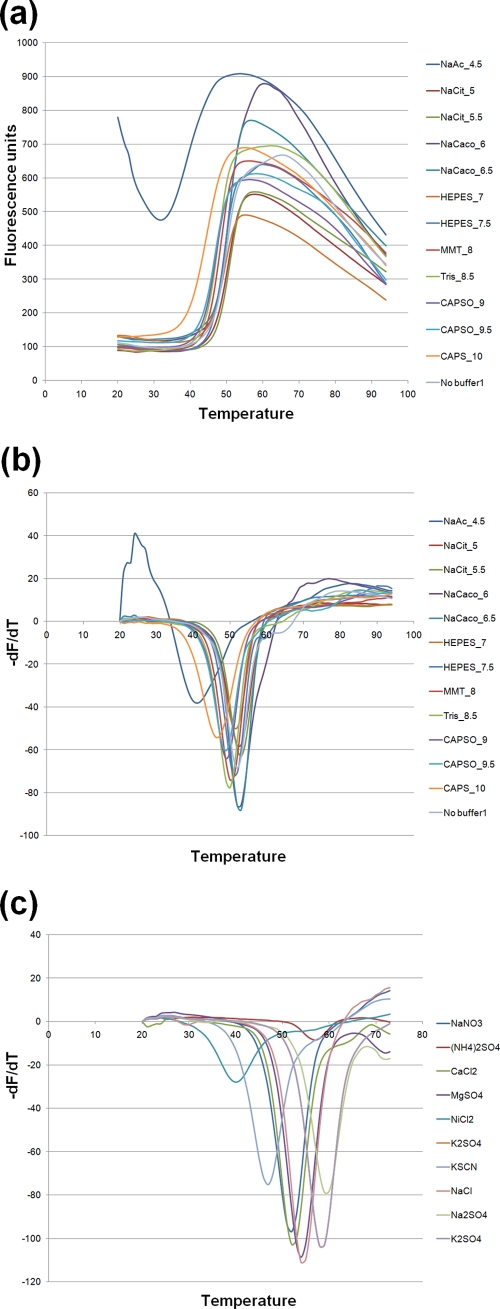
Thermal-stability experiments were performed based on the principles outlined by Ericsson and colleagues (8) to identify stabilizing buffers and additives prior to crystallization trials. Briefly, 20 μg His-tagged protein was mixed with 0.1 M buffers ranging from pH 4 to pH 10, along with a 1:2,500 dilution of Sypro-Orange dye (Invitrogen) in 50-μl reaction mixtures. The mixture was subjected to thermal ramping from 20°C to 94°C at 1°C/min in a thermal cycler (myIQ; Bio-Rad), and the binding of Sypro-Orange was measured as fluorescence at 575 nm (excitation at 490 nm). The negative derivatives (−dF/dT) were plotted, and the melting temperature (Tm) was determined as the temperature at the point of inflection of the curve (the midpoint for protein unfolding). To determine the effects of various salts, the assay was repeated in a 96-well format with the Salt Rx and Additives suites (Hampton Research) in the presence of the identified optimal buffer. The buffer (sodium cacodylate, pH 6.5) and salt (sodium sulfate) giving the maximum increase in Tm were then used in the purification process (Fig. 1).
FIG. 1.
Thermostability assays to identify the optimal buffer and salt for DEN1pro. (a) Increase in fluorescence upon binding of Sypro-Orange dye to the DEN1pro protein when subjected to thermal ramping from 20°C to 95°C in the presence of various buffers. A pH range of 4 to 10.5 was tested, with a representative set of 12 buffers out of a total 32 tested shown here. (b) Negative-derivative plot (−dF/dT) of panel a, with the temperature at the point of inflection giving the Tm of the protein. (c) Effects of various salts on the Tm of DEN1pro in the presence of the optimal buffer identified in panel b, sodium cacodylate, pH 6.5. Shown is a negative-derivative plot of a fluorescence assay similar to that in panel a, with 10 out of 24 salts tested shown for clarity.
Crystallization.
Purified Den1proΔ11-20 protein (see below) at 40 mg/ml was screened in sitting drop vapor diffusion crystallization trials. Unusual pebble-shaped crystals appeared after 2 days in a well containing 0.2 μl of protein and 0.2 μl of the reservoir solution (0.1 M HEPES, pH 7.5, 12% polyethylene glycol [PEG] 3350, and 0.005 M [each] NiCl2, CdCl2, MgCl2, and CoCl2). We noticed that many of the crystals formed in protein precipitate at the junction of the protein and well solutions. Crystals of the Den1proΔ11-20 S135A mutant were grown under the same crystallization conditions with an additional 0.1 M potassium sodium tartrate tetrahydrate.
Data collection and refinement.
Crystals were flash frozen in liquid nitrogen with reservoir solution containing 20% glycerol as a cryoprotectant. DEN1proΔ11-20 crystals were screened remotely at the Stanford Synchrotron Radiation Lightsource (SSRL) 11-1 Beamline using the Blu-Ice interface, and a 2.2-Å data set was collected. DEN1proΔ11-20 S135A crystals were similarly frozen, and a 2.1-Å data set was collected remotely using Blu-Ice at Advanced Photon Source (APS) Beamline GM/CA-CAT. The reflections were indexed (Spacegroup I222), integrated, and scaled using HKL2000 software (22). Initial phases were obtained through molecular replacement using Phaser within CCP4 (3, 18), using the NS3 protease core region of DEN2pro as a search model. The NS2B region, which differs substantially from that of the published structure, was then built manually using the Windows version of Coot (6). The model was then refined and improved using Refmac5 (19), simulated annealing in Phenix (1), and Win Coot. The details are summarized in Table 1. To minimize model bias, the built model was also verified against selenomethionine-derived data for DEN1proΔ11-20 and was found to be essentially identical (root mean square deviation [RMSD], 0.25). All figures were generated using PyMol (DeLano Scientific LLC).
TABLE 1.
Data collection and refinement statistics
| Parameter | Valuea |
|
|---|---|---|
| DEN1proΔ11-20 | DEN1proΔ11-20 S135A | |
| Data collection statistics | ||
| Wavelength (Å) | 0.9497 | 0.9493 |
| Spacegroup | I222 | I222 |
| Unit cell dimensions (Å) | 56.721, 60.848, 161.644 | 56.868, 60.894, 160.616 |
| α = 90°, β = 90°, γ = 90° | α = 90°, β = 90°, γ = 90° | |
| Mosaicity | 0.284-0.481 | 0.171-0.348 |
| Total no. of observations | 56,543 | 118,008 |
| No. of unique reflections | 14,424 (1,337) | 16,495 (1,062) |
| Resolution range (Å) | 40.0-2.2 (2.28-2.20) | 50.0-2.1 (2.15-2.1) |
| Completeness (%) | 99.1 (94.0) | 99.8 (98.4) |
| Redundancy | 3.9 (3.2) | 7.2 (5.9) |
| Rsymb | 0.045 (0.437) | 0.068 (0.388) |
| Mean I/σ | 22.4 (2.56) | 28.95 (4.55) |
| Refinement statistics | ||
| No. of refined residues | 181 | 194 |
| Rworkc | 21.50 | 19.94 |
| Rfreed | 25.54 | 24.37 |
| RMSD bond lengths (Å) | 0.009 | 0.012 |
| RMSD bond angles (°) | 1.187 | 1.323 |
| Ramachandran statistics | ||
| Most favored [no. (%)] | 173 (95.58) | 180 (92.78) |
| Additionally allowed [no. (%)] | 8 (4.42) | 13 (8.33) |
| Outliers [no. (%)] | 0 (0) | 0 (0) |
Values in parantheses correspond to data for outermost shell.
Rsym = Σhkl[(ΣjIj − <I>)/ΣjIj].
Rwork = ΣhklFo − Fc/ΣhklFo, where Fo and Fc are the observed and calculated structure factors, respectively.
Five percent of randomly chosen reflections were used in the calculation of Rfree.
Enzyme activity assay.
In vitro enzyme assays were carried out essentially as described previously (14) using a commercial (Anaspec Inc.) or custom-synthesized (Biomatik Corp.) hexapeptide substrate, FAAGRK (corresponding to the NS3/4A junction), fused to para-nitroanilide (pNA). Desalted protein was used, as salt has been reported to be inhibitory to the reaction (11). Reactions were carried out at 37°C in buffer containing 0.1 M Tris, pH 9.0, 20% glycerol, 1 mM CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}, and 200 nM protease, with substrate concentrations ranging from 50 μM to 4 mM. Triplicates of 100-μl reactions for each condition were carried out in 96-well plates, and the release of pNA was monitored by measuring the absorbance at 405 nm using a multimode detector (Genios Pro; Tecan). Absorbance units were converted to the concentration of pNA released using a standard curve with free pNA (BioVision Inc.). Values were analyzed using the Michaelis-Menten kinetics equation, v = Vmax·[S]/(Km+[S]), in GraphPad Prism software.
Protein structure accession numbers.
The coordinates of the structures have been deposited in the Protein Data Bank under accession codes 3L6P and 3LKW.
RESULTS AND DISCUSSION
Purification and crystallization.
The N-terminal region of the hydrophilic core of NS2B is known to play a chaperone-like role in stabilizing the expression of the NS3 protease, while its C terminus is required for catalytic activity (7, 14). To produce stable, enzymatically active DEN1pro, we adopted a construct design that fused the hydrophilic core of NS2B (NS2Bc) to the protease domain of NS3 (NS3pro) through a flexible, noncleavable Gly4-Ser-Gly4 linker, as previously described for other DENV proteases (7, 14, 15). Sequence optimization to remove codon bias and improve expression in Escherichia coli resulted in very high yields of soluble expression (∼70 mg/liter).
Determining optimal protein purification conditions is often a crucial rate-limiting step in crystallization studies. To address this, we employed a systematic approach using the melting temperature of the protein as an indicator of its stability in the presence of various buffers and additives, as previously described (8). We then devised a purification protocol incorporating the optimal buffer and salt identified through these screens (described above). The DEN1pro construct subsequently crystallized under several PEG-containing conditions within 1 to 2 days. However, these crystals diffracted poorly (∼10 Å), and various methods, including dehydration, altering the cryoprotection conditions, methylation of surface lysines using dimethylamino-borane complex, and screens for non-PEG conditions, proved unsuccessful.
With the aim of improving diffraction by reducing molecular disorder within the crystals, a series of deletions and truncations were made in the DEN1pro sequence based on the disordered loops in the published DEN2pro structure (Protein Data Bank [PDB] code, 2FOM). Three main regions were identified: between residues 77 and 84 in NS2Bc and between residues 1 and 18 and residues 167 and 185 of NS3pro. Of these, deletions within amino acids 77 to 84 of NS2Bc were not pursued, as this region is critically implicated in substrate binding.
A deletion mutant lacking residues 11 to 20 of DEN1 NS3pro (designated DEN1proΔ11-20) crystallized into unusual pebble-shaped crystals under PEG-containing conditions with four different metal salts, NiCl2, CdCl2, MgCl2, and CoCl2. It was observed that all four salts were necessary for obtaining well-formed crystals. Further fine screens with additives and salts yielded crystals that diffracted to 2.2 Å. The data collection and refinement statistics are summarized in Table 1. An active-site mutant (S135A) of DEN1proΔ11-20 was also generated and crystallized under similar conditions with the aim of using it in future cocrystallization studies of enzyme-substrate complexes. Of the catalytic triad, Ser135 was chosen for mutagenesis over His51 and Asp75 to preserve the substrate-binding pocket while abolishing substrate hydrolysis and subsequent dissociation of the complex.
Similarities and differences between the NS3pros of the DENVs.
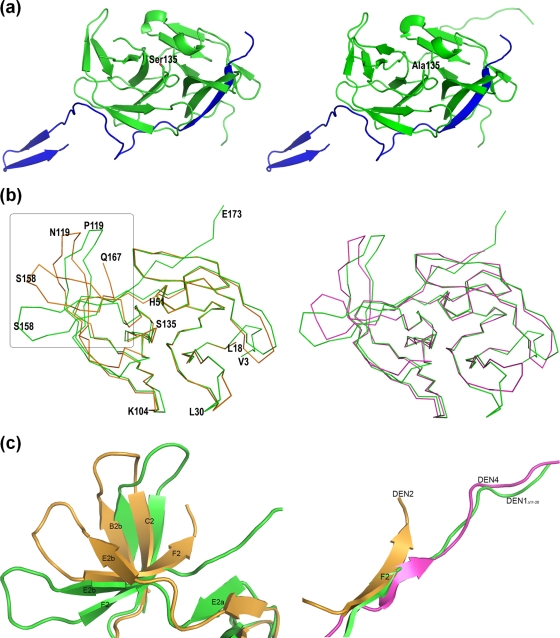
The overall structures of DEN1proΔ11-20 and its S135A mutant are shown in Fig. 2A. The structure was solved through molecular replacement using just the NS3 protease core of DEN2 as a search model. As with the other flaviviral proteases, DEN1proΔ11-20 has an overall structure of two β barrels made of six β sheets each, with the active site located in the cleft between the barrels. The NS2Bc cofactor contributes one of the N-terminal β sheets. The structure of the S135A mutant was essentially identical, with an RMSD of 0.431 Å for 212 Cα positions. However, in general, it was more ordered, possibly reflected in the more regular cuboidal shape of the crystals. The NS3pro region superimposes closely on the corresponding domain of similar constructs from DEN2 (7) and DEN4 (15), with RMSDs of 0.77 Å and 0.75 Å for 129 out of 144 Cα positions, respectively (Fig. 2B).
FIG. 2.
Structure of DEN1proΔ11-20. (a) Structures of DEN1proΔ11-20 (left) and its active-site S135A mutant (right). The NS3pro core is shown in green, and the NS2B cofactor is shown in blue. (b) Superimposition of the DEN1proΔ11-20 NS3 core over DEN2pro NS3 (gold; PDB code, 2FOM) and DEN4pro (magenta; PDB code, 2VBC) showing overall similar folds. L18 and Q167 mark the N and C termini of DEN2pro. (c) Enlarged view of the boxed region in panel b showing the shift in the β hairpins between DEN1proΔ11-20 and DEN2pro (left) and the C-terminal tail of the three DENV proteases (right). DEN1proΔ11-20, green; DEN2pro, gold; DEN4pro, magenta.
The major regions of difference between the NS3 of DEN1proΔ11-20 and the other DENpro structures are in the loop regions between β sheets (Fig. 2c). There are significant shifts in the positions of β sheets B2b and C2 (residues Phe116 to Gly124) compared to both of the other DENV proteases. The loop region between these sheets is disordered in DEN1proΔ11-20 but is visible in its S135A structure. This shift in the B2b-C2 hairpin has a direct effect on the conformation of the C-terminal tail of the protein. In conjunction with the region from Gly151 (β sheets E2b to F2), it leads to an overall open conformation, leaving enough space for the NS2B C terminus, which folds into this region upon substrate binding. This is similar to the DEN4pro structure and unlike DEN2pro. The similarity to DEN4pro continues, with the C-terminal tail following the general direction of full-length DEN4 NS3, where it is directed toward the helicase domain. While residues C terminal to Glu173 are disordered in the DEN1proΔ11-20 structure, they are visible and stabilized through crystal contacts in the S135A structure.
The conformation of the catalytic site of DEN1proΔ11-20.
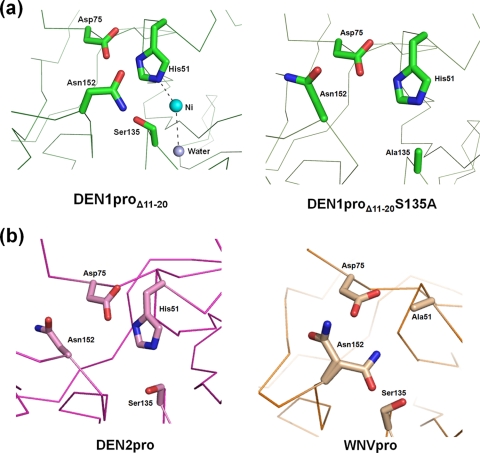
Although the members of the catalytic triad of His51, Asp75, and Ser135 occupy positions similar to those in DEN2pro, the hydrogen bond between His51 and Ser135 in DEN1proΔ11-20 is disrupted by the presence of a Ni2+ ion in the active site coordinated with the imidazole ring of the His (Fig. 3A). The resulting presence of the unbound Ser hydroxyl group leads to a complete change in the orientation of Asn152 compared not only to the other DENV proteases, but also to its S135A mutant (Fig. 3B). This alternate conformation is also noted in the related West Nile virus (WNV) protease apo structure, where the active-site His is mutated to Ala (PDB code, 2GGV) (2).
FIG. 3.
Conformation of the DEN1proΔ11-20 active site. (a) Change in conformation of Asn152 in the presence of a free hydroxyl group on Ser135 (left) compared to the S135A mutant (right). The essential hydrogen bond between the active-site His51 and Ser135 is disrupted by the presence of a Ni2+ atom (cyan) coordinated with His51 and a water molecule (gray). (b) Conformation of Asn152 in other flaviviral proteases, DEN2pro (PDB code, 2FOM) and WNVpro H51A (PDB code, 2GGV).
The essential role of the NS2Bc cofactor.
The hydrophilic core within NS2B is known to enhance the basal proteolytic activity of NS3 by 3,300- to 7,600-fold (29). Unlike the flanking transmembrane domains, which share high sequence similarity, this region is the most divergent among the members of the Flaviviridae, especially within the C-terminal region of the core. The N-terminal region of NS2Bc (residues 49 to 66) plays a chaperone-like role in stabilizing NS3. Unlike in related viruses, such as HCV, where a peptide corresponding to this region from the analogous NS4A is sufficient for cofactor activity (12), in flaviviruses, including DENV, the presence of this region alone yields a stable but enzymatically inactive protease (7, 15), highlighting the importance of the C terminus of NS2Bc (residues 67 to 95). Nuclear magnetic resonance (NMR) studies have revealed that the C terminus of NS2Bc is disordered in solution, and constructs lacking this segment are defective in substrate binding (7).
In the absence of structures of DENV protease-substrate complexes, our current understanding of the role of NS2Bc is derived from the related WNVpro. Two different studies of the crystal structure of WNVpro bound to a substrate-based peptide inhibitor or aprotinin showed that NS2B forms a belt around NS3pro, with the loop between β strands 2 and 3 (Leu79 to Phe85) interacting directly with the active site (2, 7). In contrast, NS2Bc adopts a distinctly different conformation in the apo form. While β strand 1 is identical to the inhibitor-bound structures, forming part of the N-terminal β barrel, the conformation differs significantly beyond Trp61 (Trp62 in WNV). The segment between Glu63 and Ser71 forms a helix, followed by a β sheet that runs antiparallel to the B2A β strand of NS3pro. This conformation is partly conserved in WNVpro, as well, giving rise to the hypothesis that NS2Bc in flaviviruses in general may adopt these two distinct conformations in the apo- and inhibitor-bound forms (2).
Novel conformation of DEN1 NS2Bc.
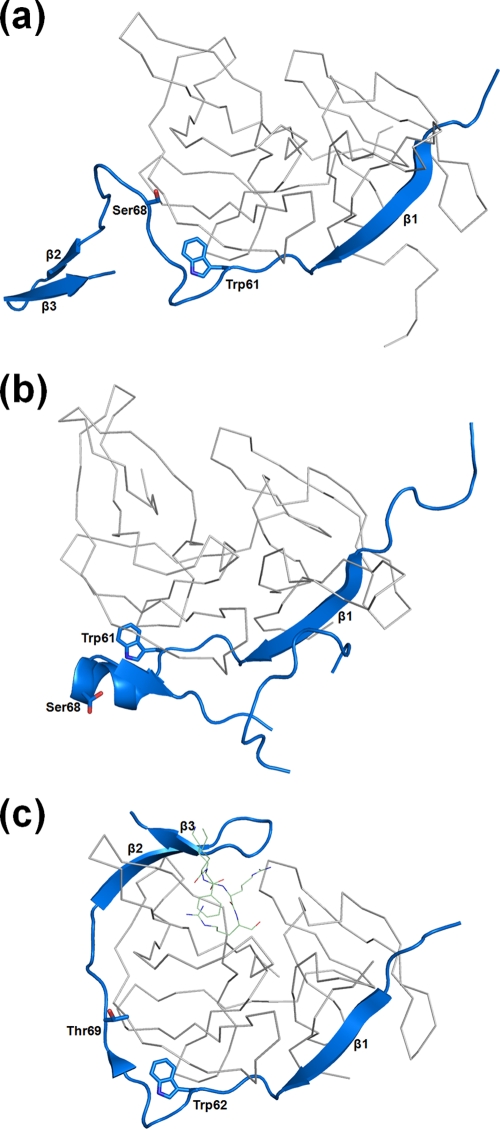
Interestingly, the NS2Bc in DEN1proΔ11-20 adopts a unique conformation hitherto not seen in the other known apoprotease structures (Fig. 4a). While β strand 1 is positioned similarly to the DEN2pro and WNVpro structures, the similarity does not extend beyond Trp61. This explains our initial failed attempts at molecular replacement when the NS2B of DEN2pro was included in the search model and the need to manually build this region of the structure. Where the apo DEN2pro NS2Bc (similar to WNVpro) takes a helical turn at Glu63 (Fig. 4b), DEN1proΔ11-20 NS2Bc adopts an extended conformation with a well-defined β2-β3 hairpin.
FIG. 4.
Conformation of DEN1proΔ11-20 NS2Bc. Shown is NS2Bc (blue) of DEN1proΔ11-20 (a), DEN2pro (b), and WNVpro (c) in complex with a peptidic inhibitor. The positions of Trp61 and Ser68 and the corresponding residues in WNVpro, Trp62, and Thr69 are shown as sticks. The corresponding NS3pro structures are represented as ribbons (gray).
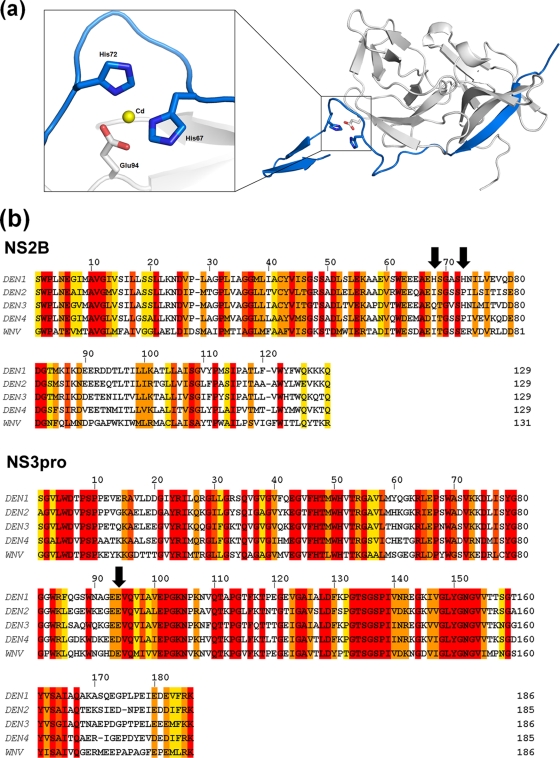
Although it is a valid concern that this alternate conformation may be an artifact resulting from the deletion of residues 11 to 20 in this construct, several observations argue against this. First, this path of NS2Bc closely follows that of the cofactor in the inhibitor-bound form of WNVpro until Ser68 (Fig. 4c). Beyond this point, the lack of a bound substrate or inhibitor precludes its forming a belt around NS3pro to interact with the substrate-binding pocket. Instead, it adopts an extended conformation, with two unique features stabilizing this novel structural feature of the NS2Bc C terminus. The first is a metal-coordinated site composed of His67 and His72 (of NS2B) and Glu94 (of NS3), shown in Fig. 5a. It is interesting that this is possibly an exclusive structural feature of DEN1, as the two His residues are not conserved in the other flaviviral NS2B sequences (Fig. 5b). It is likely that an ion(s) that occupies this site in the native protein has been replaced by the divalent cations present under the crystallization conditions. Of the four cations under the crystallization conditions (Ni, Co, Cd, and Mg), Cd fit the electron density map most appropriately. Second, the extended segment of NS2Bc is also stabilized by strong crystal contacts with the NS2Bcs of neighboring molecules.
FIG. 5.
A unique metal-coordinated site in DEN1proΔ11-20. (a) A metal-coordinated site composed of His67 and His72 of NS2Bc (blue) with Glu94 (gray) of NS3. The central cadmium atom is represented in yellow. (b) Sequence alignment of the NS2B and NS3pro sequences of DEN1 to -4 and WNV. The arrows indicate His67, His72, and Glu94 of DEN1. The alignment was generated using ClustalW2 (European Bioinformatics Institute [http://www.ebi.ac.uk/]). The shaded residues are colored according to the degree of conservation, ranging from yellow (least) to red (most conserved).
Given that the NS2Bc C terminus is disordered in solution prior to substrate binding (2), it has recently been proposed that this segment may adopt multiple conformations, with those seen in crystal structures being dictated by crystal-packing forces (13). While the structure of DEN1proΔ11-20 partially supports this hypothesis, it also brings into focus for the first time the influence of nonconserved sequence elements encoded by the genomes of the different serotypes. This could potentially have far-reaching implications in the context of structure-based drug design, specifically in the case of dengue, where simultaneous targeting of all four serotypes is viewed as essential for effective treatment.
Proteolytic activity of Den1proΔ11-20.
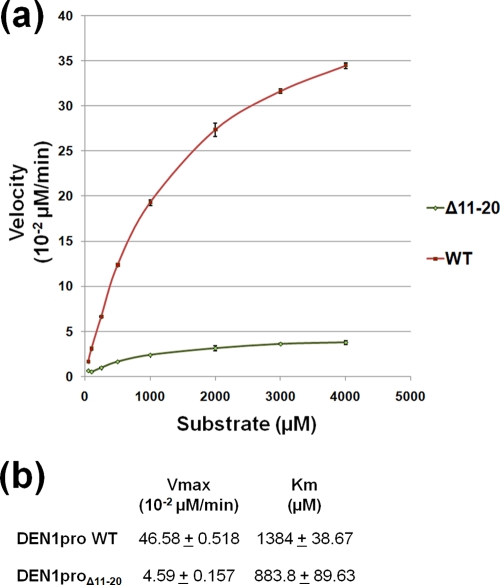
In in vitro protease assays using a chromogenic substrate corresponding to the NS3/4A junction, the DEN1proΔ11-20 mutant has a substrate affinity similar to that of wild-type (WT) DEN1pro (Fig. 6). This stands to reason, as this mutant possesses the necessary sequences of NS2B and NS3pro implicated in substrate binding. However, the mutant does separate the substrate-binding activity of the DEN1 protease from its catalytic efficiency. The catalytic turnover rate of the mutant is an order of magnitude lower than that of the WT protein. How the 10 deleted residues, from 11 to 20, in the NS3pro sequence influence its enzymatic activity is not apparent from the known structures. In the available NS2Bc-fused flaviviral protease structures, this region is either disordered or forms a loop in close proximity to the D1-E1A β hairpin. This hairpin is essentially identical in the DEN1proΔ11-20 structure, so the deletion does not seem to cause gross structural changes in the region. One likely explanation is that the shortening of the sequence promotes a certain degree of inflexibility, deterring conformational changes during the catalytic cycle. This is supported by the finding that mutating charged residues in the 16-to-20 region did not have a significant effect on the catalytic activity of the protease (16), whereas deleting the same region had a detrimental effect (data not shown).
FIG. 6.
Enzyme kinetics of DEN1proΔ11-20. (a) Enzyme reaction kinetics for DEN1proΔ11-20 (green) compared to WT DEN1pro (red). The reactions were carried out with the peptide substrate FAAGRK-pNA, corresponding to the NS3/4A junction. Free pNA was measured as absorbance at 405 nm and converted to the concentration released using a pNA standard curve. At least triplicates of reactions, with substrate concentrations ranging from 50 μM to 4 mM and the enzyme at 0.4 μM, were tested. (b) The Vmax and Km for WT DEN1pro and DEN1proΔ11-20 were calculated based on Michaelis-Menten kinetics using GraphPad Prism software.
Conclusion.
In this paper, we report the crystal structure of an optimal construct of the two-component protease NS2Bc/NS3pro from DEN1. The novel, potentially DEN1-specific extended conformation of NS2Bc seen in this structure reiterates the flexibility and multiple conformations adopted by this region prior to substrate binding. The observation that it partially follows the path of the cofactor in the inhibitor-bound form of the related WNVpro and the presence of a stabilizing metal-coordinated site within the DEN1 sequence provide evidence against this being a mutational or crystallization artifact. In conjunction with the available DEN2 and DEN4 structures, this provides a comprehensive view of the protease, along with some key areas of difference between the serotypes. This knowledge of the similarities and differences among the DENVs themselves and compared to other closely related flaviviruses forms a fundamental prerequisite for future drug design efforts to manipulate the viral proteolytic function by either directly targeting the active site or modifying the essential interaction between NS2B and NS3.
The flexibility and serotype-specific conformations of the C terminus of NS2Bc argue against the likelihood of the discovery of a single broad-spectrum flaviviral inhibitor that modulates the protease activity by trapping the cofactor in a conformation unfavorable for substrate binding. Drug discovery efforts against flaviviruses are further challenged by the huge conformational change undergone by NS2B upon substrate binding itself, as is known for the WNV protease so far. This gives rise to an immediate need in the field for structures of the DENV proteases in complex with substrate/inhibitors to further distill our knowledge of the commonalities within this viral family to focus drug design efforts on the regions with higher probability for efficient targeting across the board. Unfortunately, as is the case with DEN2pro, the active site in this DEN1proΔ11-20 structure is also buried in crystal contacts, as is the C terminus of NS2Bc, precluding the diffusion of substrates and inhibitors into the active site by soaking of the crystals. This necessitates the crystallization of these proteases under other conditions with altered crystal packing to be useful in small-molecule- and fragment-based screens. Interestingly, the deletion of residues 11 to 20 within DEN1 NS3pro results in a construct that retains the substrate-binding capacity of the WT enzyme, but with a significant reduction in enzymatic efficiency, allowing the separation of substrate binding from subsequent catalysis, another potentially useful tool in drug design efforts that target either one or both of these functions. This apparent requirement for the enzyme's flexibility in order to achieve full catalytic efficiency further gives rise to an additional avenue for small-molecule inhibitor discovery: those that act by rigidifying the protease rather than by targeting the active site. This has the added potential to lead to broad-spectrum inhibitors if the flexibility of the protease is a conserved feature of flaviviruses in general, a notion that warrants further research.
Acknowledgments
We thank Anand Kolatkar and Joshua Kunken for computational support. The support of beamline scientists at APS GM/CA-CAT and SSRL Beamline 11-1 during crystal screening and data collection is gratefully acknowledged.
The project described was supported by grant number U54AI065359 from the National Institute of Allergy and Infectious Diseases (NIAID). The work was also supported by the Accelerated Technologies Center for Gene to 3D Structure (ATCG3D), a specialized center of the Protein Structure Initiative Research Network funded by National Institutes of Health (NIH) and National Institute of General Medical Science (NIGMS) grant number U54 GM074961. GM/CA CAT has been funded in whole or in part with Federal funds from the National Cancer Institute (Y1-CO-1020) and the NIGMS (Y1-GM-1104). Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under contract no. DE-AC02-06CH11357. Portions of this research were carried out at the Stanford Synchrotron Radiation Lightsource, a national user facility operated by Stanford University on behalf of the U.S. Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, and by the NIH, National Center for Research Resources (NCRR), Biomedical Technology Program, and the NIGMS.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIAID, NIGMS, or NIH.
This is TSRI manuscript number 20394.
Footnotes
Published ahead of print on 30 December 2009.
REFERENCES
- 1.Adams, P. D., R. W. Grosse-Kunstleve, L. W. Hung, T. R. Ioerger, A. J. McCoy, N. W. Moriarty, R. J. Read, J. C. Sacchettini, N. K. Sauter, and T. C. Terwilliger. 2002. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58:1948-1954. [DOI] [PubMed] [Google Scholar]
- 2.Aleshin, A. E., S. A. Shiryaev, A. Y. Strongin, and R. C. Liddington. 2007. Structural evidence for regulation and specificity of flaviviral proteases and evolution of the Flaviviridae fold. Protein Sci. 16:795-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Collaborative Computational Project Number 4. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50:760-763. [DOI] [PubMed] [Google Scholar]
- 4.De Clercq, E. 2007. The design of drugs for HIV and HCV. Nat. Rev. Drug Discov. 6:1001-1018. [DOI] [PubMed] [Google Scholar]
- 5.Ekonomiuk, D., X. C. Su, K. Ozawa, C. Bodenreider, S. P. Lim, Z. Yin, T. H. Keller, D. Beer, V. Patel, G. Otting, A. Caflisch, and D. Huang. 2009. Discovery of a non-peptidic inhibitor of West Nile virus NS3 protease by high-throughput docking. PLoS Negl. Trop. Dis. 3:e356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Emsley, P., and K. Cowtan. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126-2132. [DOI] [PubMed] [Google Scholar]
- 7.Erbel, P., N. Schiering, A. D'Arcy, M. Renatus, M. Kroemer, S. P. Lim, Z. Yin, T. H. Keller, S. G. Vasudevan, and U. Hommel. 2006. Structural basis for the activation of flaviviral NS3 proteases from dengue and West Nile virus. Nat. Struct. Mol. Biol. 13:372-373. [DOI] [PubMed] [Google Scholar]
- 8.Ericsson, U. B., B. M. Hallberg, G. T. Detitta, N. Dekker, and P. Nordlund. 2006. Thermofluor-based high-throughput stability optimization of proteins for structural studies. Anal. Biochem. 357:289-298. [DOI] [PubMed] [Google Scholar]
- 9.Falgout, B., M. Pethel, Y. M. Zhang, and C. J. Lai. 1991. Both nonstructural proteins NS2B and NS3 are required for the proteolytic processing of dengue virus nonstructural proteins. J. Virol. 65:2467-2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnston, P. A., J. Phillips, T. Y. Shun, S. Shinde, J. S. Lazo, D. M. Huryn, M. C. Myers, B. I. Ratnikov, J. W. Smith, Y. Su, R. Dahl, N. D. Cosford, S. A. Shiryaev, and A. Y. Strongin. 2007. HTS identifies novel and specific uncompetitive inhibitors of the two-component NS2B-NS3 proteinase of West Nile virus. Assay Drug Dev. Technol. 5:737-750. [DOI] [PubMed] [Google Scholar]
- 11.Khumthong, R., C. Angsuthanasombat, S. Panyim, and G. Katzenmeier. 2002. In vitro determination of dengue virus type 2 NS2B-NS3 protease activity with fluorescent peptide substrates. J. Biochem. Mol. Biol. 35:206-212. [DOI] [PubMed] [Google Scholar]
- 12.Kim, J. L., K. A. Morgenstern, C. Lin, T. Fox, M. D. Dwyer, J. A. Landro, S. P. Chambers, W. Markland, C. A. Lepre, E. T. O'Malley, S. L. Harbeson, C. M. Rice, M. A. Murcko, P. R. Caron, and J. A. Thomson. 1996. Crystal structure of the hepatitis C virus NS3 protease domain complexed with a synthetic NS4A cofactor peptide. Cell 87:343-355. [DOI] [PubMed] [Google Scholar]
- 13.Lescar, J., D. Luo, T. Xu, A. Sampath, S. P. Lim, B. Canard, and S. G. Vasudevan. 2008. Towards the design of antiviral inhibitors against flaviviruses: the case for the multifunctional NS3 protein from dengue virus as a target. Antiviral Res. 80:94-101. [DOI] [PubMed] [Google Scholar]
- 14.Leung, D., K. Schroder, H. White, N. X. Fang, M. J. Stoermer, G. Abbenante, J. L. Martin, P. R. Young, and D. P. Fairlie. 2001. Activity of recombinant dengue 2 virus NS3 protease in the presence of a truncated NS2B co-factor, small peptide substrates, and inhibitors. J. Biol. Chem. 276:45762-45771. [DOI] [PubMed] [Google Scholar]
- 15.Luo, D., T. Xu, C. Hunke, G. Gruber, S. G. Vasudevan, and J. Lescar. 2008. Crystal structure of the NS3 protease-helicase from dengue virus. J. Virol. 82:173-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matusan, A. E., P. G. Kelley, M. J. Pryor, J. C. Whisstock, A. D. Davidson, and P. J. Wright. 2001. Mutagenesis of the dengue virus type 2 NS3 proteinase and the production of growth-restricted virus. J. Gen. Virol. 82:1647-1656. [DOI] [PubMed] [Google Scholar]
- 17.McArthur, J. H., A. P. Durbin, J. A. Marron, K. A. Wanionek, B. Thumar, D. J. Pierro, A. C. Schmidt, J. E. Blaney, Jr., B. R. Murphy, and S. S. Whitehead. 2008. Phase I clinical evaluation of rDEN4Delta30-200,201: a live attenuated dengue 4 vaccine candidate designed for decreased hepatotoxicity. Am. J. Trop. Med. Hyg. 79:678-684. [PMC free article] [PubMed] [Google Scholar]
- 18.McCoy, A. J., R. W. Grosse-Kunstleve, L. C. Storoni, and R. J. Read. 2005. Likelihood-enhanced fast translation functions. Acta Crystallogr. D Biol. Crystallogr. 61:458-464. [DOI] [PubMed] [Google Scholar]
- 19.Murshudov, G. N., A. A. Vagin, and E. J. Dodson. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53:240-255. [DOI] [PubMed] [Google Scholar]
- 20.Neuman, B. W., J. S. Joseph, K. S. Saikatendu, P. Serrano, A. Chatterjee, M. A. Johnson, L. Liao, J. P. Klaus, J. R. Yates III, K. Wuthrich, R. C. Stevens, M. J. Buchmeier, and P. Kuhn. 2008. Proteomics analysis unravels the functional repertoire of coronavirus nonstructural protein 3. J. Virol. 82:5279-5294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Othman, R., T. S. Kiat, N. Khalid, R. Yusof, E. I. Newhouse, J. S. Newhouse, M. Alam, and N. A. Rahman. 2008. Docking of noncompetitive inhibitors into dengue virus type 2 protease: understanding the interactions with allosteric binding sites. J. Chem. Infect. Model. 48:1582-1591. [DOI] [PubMed] [Google Scholar]
- 22.Otwinowski, Z., and W. Minor. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307-326. [DOI] [PubMed] [Google Scholar]
- 23.Rabablert, J., and S. Yoksan. 2009. Attenuated D2 16681-PDK53 vaccine: defining humoral and cell-mediated immunity. Curr. Pharm. Des. 15:1203-1211. [DOI] [PubMed] [Google Scholar]
- 24.Sampath, A., and R. Padmanabhan. 2009. Molecular targets for flavivirus drug discovery. Antiviral Res. 81:6-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun, W., D. Cunningham, S. S. Wasserman, J. Perry, J. R. Putnak, K. H. Eckels, D. W. Vaughn, S. J. Thomas, N. Kanesa-Thasan, B. L. Innis, and R. Edelman. 2009. Phase 2 clinical trial of three formulations of tetravalent live-attenuated dengue vaccine in flavivirus-naive adults. Hum. Vaccin. 5:33-40. [DOI] [PubMed] [Google Scholar]
- 26.Tomlinson, S. M., R. D. Malmstrom, A. Russo, N. Mueller, Y. P. Pang, and S. J. Watowich. 2009. Structure-based discovery of dengue virus protease inhibitors. Antiviral Res. 82:110-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yin, Z., S. J. Patel, W. L. Wang, W. L. Chan, K. R. Ranga Rao, G. Wang, X. Ngew, V. Patel, D. Beer, J. E. Knox, N. L. Ma, C. Ehrhardt, S. P. Lim, S. G. Vasudevan, and T. H. Keller. 2006. Peptide inhibitors of dengue virus NS3 protease. part 2: SAR study of tetrapeptide aldehyde inhibitors. Bioorg. Med. Chem. Lett. 16:40-43. [DOI] [PubMed] [Google Scholar]
- 28.Yin, Z., S. J. Patel, W. L. Wang, G. Wang, W. L. Chan, K. R. Rao, J. Alam, D. A. Jeyaraj, X. Ngew, V. Patel, D. Beer, S. P. Lim, S. G. Vasudevan, and T. H. Keller. 2006. Peptide inhibitors of dengue virus NS3 protease. Part 1: Warhead. Bioorg. Med. Chem. Lett. 16:36-39. [DOI] [PubMed] [Google Scholar]
- 29.Yusof, R., S. Clum, M. Wetzel, H. M. Murthy, and R. Padmanabhan. 2000. Purified NS2B/NS3 serine protease of dengue virus type 2 exhibits cofactor NS2B dependence for cleavage of substrates with dibasic amino acids in vitro. J. Biol. Chem. 275:9963-9969. [DOI] [PubMed] [Google Scholar]