Abstract
The worst known H1N1 influenza pandemic in history resulted in more than 20 million deaths in 1918 and 1919. Although the underlying mechanism causing the extreme virulence of the 1918 influenza virus is still obscure, our previous functional genomics analyses revealed a correlation between the lethality of the reconstructed 1918 influenza virus (r1918) in mice and a unique gene expression pattern associated with severe immune responses in the lungs. Lately, microRNAs have emerged as a class of crucial regulators for gene expression. To determine whether differential expression of cellular microRNAs plays a role in the host response to r1918 infection, we compared the lung cellular “microRNAome” of mice infected by r1918 virus with that of mice infected by a nonlethal seasonal influenza virus, A/Texas/36/91. We found that a group of microRNAs, including miR-200a and miR-223, were differentially expressed in response to influenza virus infection and that r1918 and A/Texas/36/91 infection induced distinct microRNA expression profiles. Moreover, we observed significant enrichment in the number of predicted cellular target mRNAs whose expression was inversely correlated with the expression of these microRNAs. Intriguingly, gene ontology analysis revealed that many of these mRNAs play roles in immune response and cell death pathways, which are known to be associated with the extreme virulence of r1918. This is the first demonstration that cellular gene expression patterns in influenza virus-infected mice may be attributed in part to microRNA regulation and that such regulation may be a contributing factor to the extreme virulence of the r1918.
H1N1 influenza A viruses continue to pose serious threats to public health, as exemplified by the ongoing 2009 H1N1 influenza pandemic. The 1918-1919 H1N1 influenza pandemic was even deadlier in comparison, causing more than 20 million deaths worldwide. The keys to unlocking the mystery of the extreme virulence of the 1918 virus were provided with the reconstruction of the virus (reconstructed 1918 influenza virus [r1918]) by reverse genetics (37). The lethality of r1918 has since been examined in both mouse and macaque models (17, 18). Unlike the nonlethal infections of some other H1N1 influenza virus strains, such as A/Texas/36/91 (Tx/91) or A/Kawasaki/173/01 (K173), the r1918 causes severe and lethal pulmonary disease. We subsequently conducted functional genomics analyses that revealed that the extreme virulence of r1918 was correlated with atypical expression of immune response-related genes, including massive induction of cellular genes related to inflammatory response and cell death pathways (17, 18). In spite of these findings, the mechanistic basis for these atypical gene expression patterns remains unknown.
Cellular gene expression is a complicated process and is subject to regulation by many cellular factors. As a group of newly identified cellular regulators, microRNAs are known to regulate the expression of a large number of targets, mainly cellular genes. Through mRNA degradation or translational repression of their targets, microRNAs regulate a wide range of crucial physiologic and pathological processes. For example, miR-34a acts as a tumor suppressor by inhibiting the expression of sirt1 (40), whereas miR-21 contributes to myocardial disease by inhibiting the expression of spry1 (36). By targeting zeb1/2, the miR-200 family members play roles in maintaining the epithelial phenotype of cancer cells (27). Furthermore, Let-7s regulates the expression of hbl-1, which drives the developmental progression of epidermal stem cells (5). Cellular microRNAs also play critical roles in virus-host interactions. The cellular microRNA miR-122 is an indispensable factor in supporting hepatitis C virus (HCV) replication (16), whereas miR-196 and miR-296 substantially attenuate viral replication through type I interferon (IFN)-associated pathways in liver cells (28). Furthermore, miR-125b and miR-223 directly target human immunodeficiency virus type 1 (HIV-1) mRNA, thereby attenuating viral gene expression in resting CD4+ T cells (14), and miR-198 modulates HIV-1 replication indirectly by repressing the expression of ccnt1 (34), a cellular factor necessary for HIV-1 replication. More importantly, viruses may promote their life cycles by modulating the intracellular environment through actively regulating the expression of multiple cellular microRNAs. For example, human T-cell lymphotropic virus type 1 (HTLV-1) modulates the expression of a number of cellular microRNAs in order to control T-cell differentiation (3). Similarly, human cytomegalovirus (HCMV) selectively manipulates the expression of miR-100 and miR-101 to facilitate its own replication (38). In contrast, the involvement of microRNAs during influenza A virus infection or pathogenesis is largely unknown.
To determine whether cellular microRNAs play a role in the host response to influenza virus infection, we performed a systematic profiling of cellular microRNAs in lung tissues from mice infected with r1918 or a nonlethal seasonal influenza virus, Tx/91 (17). We identified a group of microRNAs whose expression patterns differentiated the host response to r1918 and Tx/91 infection. We assessed the potential functions of differentially expressed microRNAs by analyzing the predicted target genes whose expression was inversely correlated with the expression of these microRNAs. Our report provides a new perspective on the contribution of microRNAs to the pathogenesis of lethal 1918 influenza virus infection.
MATERIALS AND METHODS
Viruses.
Recombinant influenza A H1N1 virus strains r1918 and Tx/91 were generated with a reverse genetics system (10). The virus stocks were grown in Madin-Darby canine kidney (MDCK) cells. The Tx/91 virus (H1N1) has been characterized previously (37). The sequences of the r1918 genes corresponded to the published sequence of the influenza A/South Carolina/1/18 (H1N1) virus hemagglutinin (HA) open reading frame (29) and the influenza A/Brevig Mission/1/18 (H1N1) virus NA, M, NS, NP, PA, PB1, and PB2 open reading frames (2, 30, 31, 35). The r1918 contains the 5′ and 3′ noncoding regions corresponding to those of influenza virus A/WSN/33 (H1N1).
Mouse experiments.
Influenza virus infections in mice were conducted as described previously (17). All animal work was performed in a specially separated negative-pressure HEPA-filtered biosafety level 3 (BSL3) laboratory, with enhancements. All personnel wore half-body Racal hoods with backpack HEPA-filtered air supplies (37).
RNA isolation.
For total RNA extraction, entire lungs from 3 mice infected by r1918 or Tx/91 were harvested on days 1, 3, and 5 postinfection (p.i.); entire lungs from 5 uninfected mice were harvested on days 3 and 5 p.i. Whole mouse lung tissues were homogenized in buffer RLT (Invitrogen, CA). Total RNAs were isolated by use of the RNeasy kit (Invitrogen, CA), according to the modified manufacturer protocol in which buffer RW1 was omitted to ensure the retention of small RNAs. Small RNA integrity was confirmed by a Bioanalyzer (Agilent small RNA kits; Agilent Technologies, CA). Ten mock samples from days 3 and 5 were pooled as a common reference.
MicroRNA microarray analysis.
MicroRNA expression profiling was carried out using the Agilent mouse microRNA microarray (Agilent, CA). The microRNA microarray was designed based on the Sanger miRBase release 10.1 and contained probes for 567 mouse microRNAs (Agilent design ID 019119). Total RNA (100 ng) was used for making microRNA probes, according to Agilent's protocol. Probes were hybridized at 55°C for 22 h. The slides were then washed using gene expression wash buffer 1 at 16°C for 5 min and using gene expression wash buffer 2 at 37°C for 5 min. After being washed, the slides were scanned using an Agilent slide scanner. Microarray results were extracted using Agilent Feature Extraction software. The total microRNA signal from the GeneView result files, which summarized the measurements of all probes for each microRNA on an array, was used in the analyses. Detection of a microRNA was based on the “gIsGeneDetected” column. Expression data were normalized across arrays using a median-centered approach. Differential expression of microRNAs between two groups of samples was assessed by ANOVA (analysis of variance). The expression change (fold change) of a microRNA in an r1918-infected sample compared to that in the time-matched Tx/91-infected sample was calculated; the fold change of a microRNA in an r1918-infected sample or a Tx/91-infected sample relative to the time-matched mock sample was also calculated. A cutoff ANOVA (P ≤ 0.01) and an absolute fold change of ≥1.5 were used to select the differentially expressed microRNAs.
Quantitative real-time PCR.
Quantitative real-time PCR (qRT-PCR) was used to validate microRNA expression changes. For each microRNA qRT-PCR assay, the total RNA input used was 10 ng per sample. Reverse transcription was performed using TaqMan reverse transcription reagents (Applied Biosystems, CA). The primer and probe sets for qRT-PCR were selected from the Assays-on-Demand product list (Applied Biosystems, CA). qRT-PCR was performed using an ABI 7900 real-time PCR system and TaqMan chemistry. Each assay was run in quadruplicates with TaqMan 2× PCR universal master mix NO AmpErase UNG (Applied Biosystems, CA) for microRNA detections in a 20-μl total reaction volume. The relative quantification of a microRNA in the r1918 samples relative to the time-matched Tx/91 samples was calculated using the equation 2−ΔΔCT (threshold cycle). Probes used for analysis included mmu-miR-1, mmu-miR-223, mmu-miR-21, mmu-miR-34a, mmu-miR-30a, mmu-miR-200a, and mmu-miR-200b (Applied Biosystems).
mRNA microarray analysis.
The mRNA microarray experiments were conducted in a previous study (17). The mRNA microarray raw data from the previous study were reprocessed for a direct comparison of the cellular gene expression profiles between r1918- and Tx/91-infected lungs using Resolver 7.2 (Rosetta Biosoftware, WA). The gene expression profiles of Tx/91-infected lungs from 3 animals for each time point were in silico pooled and used as time-matched references. The cellular gene expressions in lungs from three r1918-infected animals at a particular time point were compared to those from the time-matched references to assess relative expression changes. Fold change was used to indicate the gene expression change upon r1918 infection relative to the time-matched Tx/91 infection. All primary data are published in accordance with the proposed microarray experiment (MIAME) standards on the Katze Lab website (http://viromics.washington.edu).
MicroRNA target database.
The identifiers of predicted mouse microRNA targets were downloaded from the Sanger Institute (http://microrna.sanger.ac.uk/targets/v5/). To make the gene identifiers in the target database consistent with the gene identifiers on the Agilent microarray platform, the Ensembl gene IDs in the Sanger mouse target database were translated into Entrez Gene IDs by BioMart (http://www.ensembl.org/biomart).
Statistical analysis.
To assess the expression patterns of studied microRNAs and their target genes, the correlation coefficients among the mean values of 310 differentially expressed microRNAs and their target genes at days 1, 3, and 5 were calculated in the R environment (http://www.r-project.org/). The differentially expressed microRNAs between the r1918- and Tx/91-infected samples were selected by the cutoff ANOVA (P ≤ 0.01) and the absolute fold change of ≥1.5. The differentially expressed microRNA target genes between the r1918- and Tx/91-infected samples were selected by having P values of ≤0.01 alone. Then, only the microRNA-target pairs with inversely correlated expression patterns were selected for further analyses. The probabilities of enrichment of inversely correlated target by chance were assessed by hypergeometric (HG) tests by considering the following four factors: the number of genes on the arrays, the number of targets on the arrays, the number of differentially expressed genes on the arrays, and the number of inversely correlated targets on the arrays. The calculation for HG tests was performed in the R environment. An HG test, with P values of ≤0.05, was used as the cutoff for statistically significant enrichment.
Bioinformatics analysis.
The enrichment of functional categories was assessed using GeneGo (http://www.GeneGo.com). RNA expression data were analyzed in the context of known biological functions, regulatory networks, and other higher-order pathways. GeneGo was used to identify biological functions or diseases that were the most significant. For all analyses, Fisher's exact test was used to determine the probability that each biological function assigned to that data set was due to chance alone. The Entrez Gene IDs of selected genes at days 1, 3, and 5 are used for the pathway analysis, respectively, with P set at 0.01.
RESULTS
r1918 and Tx/91 induce distinct cellular microRNA expression patterns in mouse lungs.
In a study published in 2006 (17), we reported a correlation between the extreme virulence of r1918 and an aberrant cellular gene expression profile in the lungs of infected mice. Compared with pathology observed in mice infected by the nonlethal influenza virus Tx/91, mice infected by r1918 exhibited more severe pulmonary lesions accompanied by excessive lung inflammation. However, we did not suggest a potential mechanism causing the aberrant gene expression. During the past few years, microRNAs have emerged as a group of key regulators of cellular gene expression. We therefore sought to determine whether gene expression changes might be influenced by the differential expression of cellular microRNAs during influenza virus infection. To test this hypothesis, we made use of archived lung samples from the previous study to assess cellular microRNA expression. We also utilized the gene expression data from the same study to assess the functional associations of the differentially expressed cellular microRNAs from the perspective of their differentially expressed targets.
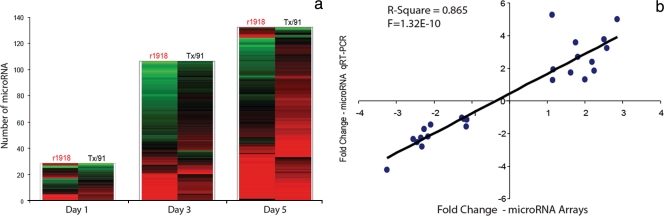
We started by directly comparing the cellular microRNA expression profiles present in mouse lung tissues obtained from r1918- and Tx/91-infected animals on days 1, 3, and 5 p.i. Among 567 microRNAs present on the arrays, 310 were detected in mouse lung tissues and were included into our analyses (see Table S1 in the supplemental material). In order to select the microRNAs with differences in abundance upon r1918 and Tx/91 infections, we adopted a cutoff 1.5-fold change (P < 0.01) at one or more of three time points. A 1.5-fold cutoff on microRNA expression changes was used based on previous microRNA profiling studies, confirming that this difference can have a significant impact on the biology of cells (13, 23). The expression patterns of differentially expressed microRNAs between r1918- and Tx/91-infected lungs relative to a common reference, a pooled mock sample, are shown in Fig. 1. Notably, the number of differentially expressed microRNAs between the two virus-infected samples increased over time. Twenty-nine microRNAs exhibited differential expression on day 1 p.i. The number increased to 106 on day 3 and increased further to 132 on day 5. Thus, our data indicate that the lethal r1918 and nonlethal Tx/91 infections induce distinct cellular microRNA expression patterns in mouse lungs.
FIG. 1.
The infections of H1N1 influenza viruses r1918 and Tx/91 induce distinct cellular microRNA expression patterns in mouse lungs. (a) Distinct cellular microRNA expression patterns in r1918- and Tx/91-infected mouse lungs. The columns correspond to the expression patterns of differentially expressed microRNAs between r1918- and Tx/91-infected mouse lungs on days 1, 3, and 5. The y axis indicates the number of differentially expressed microRNAs. The microRNAs satisfied a cutoff ANOVA P value of ≤0.01 of direct comparison and the absolute fold change between r1918 and Tx/91 of ≥1.5. Red represents microRNA with increased expression in r1918- or Tx/91-infected samples, relative to mock-infected samples. Green represents microRNA with decreased expression in r1918- or Tx/91-infected samples, relative to mock-infected samples. (b) Correlation coefficient of microRNA expression changes derived from microRNAs arrays and TaqMan qRT-PCR assays. Each spot represents one microRNA per time point measurement. The x axis represents the fold change in r1918-infected samples relative to Tx/91-infected samples derived from microRNA arrays. The y axis represents the fold change in r1918-infected samples relative to Tx/91-infected samples derived from TaqMan qRT-PCR assays. The overall correlation coefficient is represented by the R2 value. A total of 8 microRNAs from the three days were tested. One spot represents three biological duplicates.
To confirm these results, we measured the relative abundance of a few select microRNAs using TaqMan qRT-PCR assays. Eight cellular microRNAs with known functions, including mmu-miR-34a, mmu-miR-30a, mmu-miR-200a, mmu-miR-200b, mmu-miR-133a, mmu-miR-21, mmu-miR-1, and mmu-miR-223, were tested by qRT-PCR. The fold change of a particular microRNA in the r1918 sample relative to the time-matched Tx/91 sample was calculated (see Materials and Methods). The value of the coefficient of determination, R2, was used to indicate the correlation coefficient among the fold changes derived from microarrays and from qRT-PCR assays on the same pair of samples. The calculated R2 value of 0.865 indicates a high correlation coefficient between the results obtained from microRNA microarrays and those obtained from qRT-PCR (Fig. 1b).
Differences in microRNA expression during r1918 and Tx/91 infections.
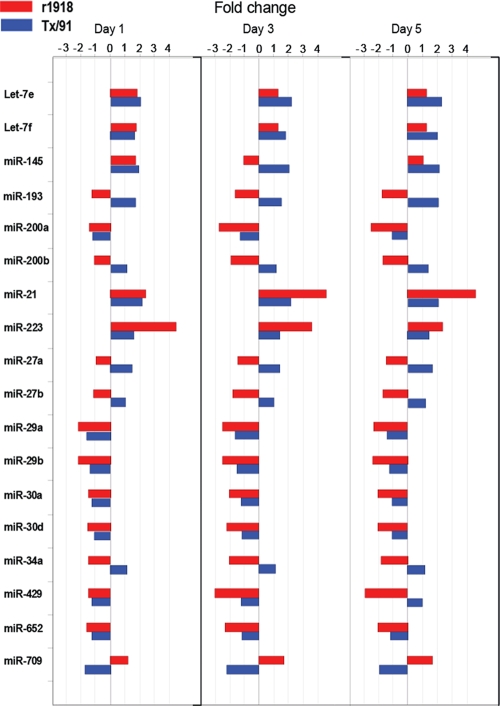
Among more than 130 cellular microRNAs showing distinct expression patterns between the lethal r1918 and the nonlethal Tx/91 infections, we focused on 18 microRNAs for further analyses because of their high abundance in lung tissues (see Table S1 in the supplemental material). These microRNAs had different expression patterns between the r1918- and Tx/91-infected lungs, including directly opposite regulation (up in one and down in the other), regulation in the same direction but to differing degrees, or regulation during one infection but not the other (Fig. 2). For example, miR-193 was strongly downregulated during r1918 infection, while it was upregulated during Tx/91 infection. In contrast, miR-709 was strongly upregulated during r1918 infection, while it was strongly downregulated during Tx/91 infection. miR-223 and miR-21, which were strongly upregulated in r1918 infection, were moderately upregulated only upon Tx/91 infection. On the other hand, while strongly downregulated in r1918 infection, miR-29a and miR-29b were moderately downregulated only upon Tx/91 infection. Finally, the expression levels of miR-200a, miT-34a, and miR-30a were downregulated in r1918 infection but were below the cutoff in Tx/91 infection. Taken together, our data indicated that some of the most abundantly expressed cellular microRNAs demonstrated distinct expression patterns between the infections of r1918 and Tx/91 in mouse lungs.
FIG. 2.
A total of 18 microRNAs demonstrate various expression patterns between r1918-infected and Tx/91-infected lungs. All fold changes shown here are relative to mock-infected lungs. A red bar indicates an r1918 infection, and a blue bar indicates a Tx/91 infection.
Interestingly, these microRNAs have been implicated in multiple key functions. MiR-223 and Let-7 have been shown to be involved in immune responses, and miR-223 is a negative modulator of neutrophil activation and neutrophil-mediated killing (16). A decreased expression level of Let-7 is associated with the activation of NF-κB in response to microbial challenge (12). Upregulation of miR-21 is closely related to airway inflammation (21), a symptom of lethal r1918 infection, and miR-34a is associated with tumorigenesis, as the mutual activation of MiR-34a and p53 has been shown both in a human cell line (40) and in patients (24). In addition, a stable expression of miR-200a is critical in maintaining the phenotype of epithelial cells (11). Taken together, because these differentially expressed microRNAs are known to be related to important functions, their differential expression is likely to contribute to physiological changes in lung tissue during r1918 infection.
Expression changes of predicted mRNA targets are inversely correlated with the expression changes of their corresponding microRNAs.
To better understand the roles of these 18 microRNAs which were differentially expressed between r1918 and Tx/91 infections, we investigated the functional associations of the predicted cellular targets potentially regulated by these microRNAs. As microRNAs predominately function as repressors of target gene expression, we therefore focused only on the targets whose expression was inversely correlated with the expression of their corresponding microRNAs. The strategy for selection of inversely correlated targets is represented in Fig. 3. To make the analysis more straightforward, we directly compared microRNA and cellular gene expression between r1918 and Tx/91 infections to assess expression changes. In the following analyses, an upregulation means that a microRNA or a cellular gene was expressed more abundantly during r1918 infection relative to Tx/91 infection, while a downregulation means that a microRNA or a cellular gene was expressed less abundantly during r1918 infection relative to Tx/91 infection.
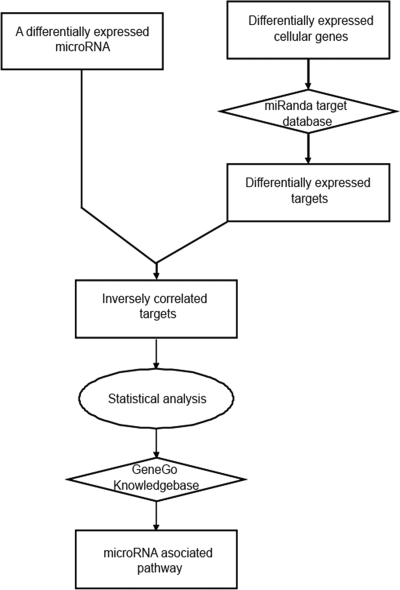
FIG. 3.
Schematic representation of the strategy for assessing the functional associations of the differentially expressed microRNAs. A rectangular shape represents the type of microRNA, gene, or pathway. A diamond shape represents the type of database. An oval shape represents the type of analysis.
First, to select the differentially expressed microRNA targets, we overlaid the miRanda microRNA target database onto the cellular genes differentially expressed between r1918 and Tx/91 infections. We chose the miRanda database because the miRanda algorithm predicts more microRNA targets by scanning the microRNA binding sites in both the 3′ untranslated region (UTR) and the protein-coding region. Then, we selected the predicted targets, whose expression was inversely correlated with their corresponding microRNAs. Finally, we evaluated the enrichment of inversely correlated targets using a hypergeometric (HG) test. If the enrichment of inversely correlated targets of a particular microRNA was statistically significant, we analyzed the functional associations of these targets using GeneGo, a commercial tool for functional analysis.
Importantly, the enrichment of inversely correlated targets of all 18 microRNAs was statistically significant (P ≤ 0.05) at one time point at the least (Table 1). This suggests that the inverse correlation between a particular microRNA shown in Table 1 and its targets was not due to chance but was more likely due to microRNA-mediated regulation. The results obtained from the HG test also indicate an increasing impact mediated by microRNAs on cellular gene expression while the infection progressed. On day 1 p.i, only 3 microRNAs had P values of ≤0.05 among the 18 microRNAs. The number of microRNAs increased to 13 and 12 on days 3 and 5, respectively. Moreover, the HG test results suggested that microRNA-mediated regulation may be time specific. For example, although miR-223 was upregulated during r1918 infection at all three time points, only the HG test results from day 1 had a P of ≤0.05. These results suggest that miR-223-mediated regulation of gene expression was predominately elicited on day 1, an early stage of infection. In contrast, although miR-29a was significantly downregulated in r1918 infection at all three time points, the HG test results from days 3 and 5, but not day 1, had P values of ≤0.05; by inference, miR-29a-mediated regulation on target gene expression occurred predominately at a later stage of infection. Furthermore, although miR-34a was downregulated in r1918 infection at all three time points, the HG test results from only day 3 had a P of ≤0.05, suggesting that miR-34a-mediated regulation on gene expression may be transient. Taken together, our statistical analysis indicates a significant enrichment of the inversely related targets of 18 highly abundant microRNAs during r1918 infection; these microRNAs may regulate target gene expression at different stages of infections.
TABLE 1.
Summary of 18 microRNAs with enrichment of inversely correlated targets
| microRNA | Results obtained ona: |
|||||
|---|---|---|---|---|---|---|
| Day 1 |
Day 3 |
Day 5 |
||||
| Regulation | P valueb | Regulation | P valueb | Regulation | P valueb | |
| Let-7e | * | ** | D | 0.006 | D | 0.009 |
| Let-7f | * | ** | D | ** | D | 0.012 |
| miR-145 | * | ** | D | 0.047 | D | ** |
| miR-193 | D | 0.029 | D | 3.0E-04 | D | 0.028 |
| miR-200a | * | ** | D | 0.017 | D | 2.0E-04 |
| miR-200b | * | ** | D | ** | D | 0.001 |
| miR-21 | * | ** | U | 0.012 | U | ** |
| miR-223 | U | 0.028 | U | ** | U | ** |
| miR-27a | D | ** | D | 0.003 | D | 0.010 |
| miR-27b | D | ** | D | 0.002 | D | 0.020 |
| miR-29a | D | ** | D | 0.010 | D | 0.002 |
| miR-29b | D | ** | D | 0.010 | D | 8.0E-04 |
| miR-30a | * | ** | D | ** | D | 0.015 |
| miR-30d | D | ** | D | 0.032 | D | 6.0E-04 |
| miR-34a | D | ** | D | 0.003 | D | ** |
| miR-429 | * | ** | D | 0.028 | D | 7.0E-04 |
| miR-652 | D | ** | D | 0.009 | D | ** |
| miR-709 | U | 0.018 | U | ** | U | ** |
U, upregulation; D, downregulation; *, Unchanged; **, P ≥ 0.05.
Hypergeometric test P values.
Inversely correlated microRNA targets are involved in key functions.
To further characterize the roles of these 18 differentially expressed microRNAs during r1918 infection, we interrogated their inversely correlated targets for functional associations. By using the functional analysis tool GeneGo, we were able to assign top gene ontology (GO) terms to the selected microRNAs (Table 2).
TABLE 2.
Summary of functional associations of 18 microRNAs
| microRNA | No. of inversely correlated targetsa |
No. of profiled targets | GO pathway(s) | ||
|---|---|---|---|---|---|
| Day 1 | Day 3 | Day 5 | |||
| let-7e | * | 167 | 143 | 584 | Response to long exposure to lithium ion |
| Skeletal muscle fiber adaptation | |||||
| let-7f | * | * | 138 | 557 | Immune response |
| Lymphocyte-mediated immunity | |||||
| miR-145 | * | 152 | * | 561 | Cardiac muscle contraction |
| Mitochondrial transport | |||||
| miR-193 | 104 | 145 | 121 | 485 | Establishment of localization in cell |
| Blood circulation | |||||
| miR-200a | * | 159 | 162 | 596 | JAK-STAT cascade |
| Viral genome replication | |||||
| miR-200b | * | * | 161 | 612 | Regulation of fever |
| Positive regulation of prostaglandin biosynthetic process | |||||
| miR-21 | * | 139 | * | 467 | Protein homotetramerization |
| miR-223 | 124 | * | * | 470 | Cell division |
| miR-27a | * | 154 | 137 | 546 | Cell death |
| Vascular process in circulatory system | |||||
| Regulation of immune response | |||||
| miR-27b | * | 154 | 136 | 542 | Intracellular signaling cascade |
| Cell death | |||||
| Vascular process in circulatory system | |||||
| miR-29a | * | 184 | 173 | 676 | Cell cycle |
| DNA recombination | |||||
| DNA damage response | |||||
| Signal transduction | |||||
| miR-29b | * | 170 | 163 | 628 | Cell cycle |
| DNA recombination | |||||
| Extracellular matrix organization | |||||
| miR-30a | * | * | 142 | 598 | Positive regulation of fever response to lithium ion |
| Muscle thin filament assembly | |||||
| miR-30d | * | 150 | 144 | 569 | Muscle thin filament assembly |
| Positive regulation of fever | |||||
| Response to lithium ion | |||||
| miR-34a | * | 181 | * | 653 | Calcium ion homeostasis |
| miR-429 | * | 142 | 142 | 535 | Regulation of fever |
| Regulation of cellular component organization | |||||
| Positive regulation of prostaglandin biosynthetic process | |||||
| miR-652 | * | 140 | * | 504 | Induction of apoptosis by intracellular signals |
| Regulation of molecular function | |||||
| miR-709 | 177 | * | * | 688 | Spindle organization |
| Cell cycle phase | |||||
*, hypergeometric test P value > 0.05.
GO analysis indicated that many of the inversely correlated targets could be related to influenza virus pathogenesis. For example, the inversely correlated targets of four microRNAs are associated with the immune response. The targets of Let-7f are associated with lymphocyte-mediated immune response, while the targets of miR-200a are associated with viral gene replication and the JAK-STAT signaling pathway, which is closely related to the type I IFN-mediated innate immune response. The inversely correlated miR-34a targets are associated with calcium ion homeostasis, which is critical for immune cell activation. In addition, the targets of miR-27a are associated with regulation of the immune response. The inversely correlated targets of three microRNAs, including miR-652, miR-27a, and miR-27b, are associated with apoptosis and cell death. Intriguingly, atypical expression of immune response-related and cell death-related genes was previously shown to be related to the extreme virulence of r1918 in mouse and macaque models of infection (17, 18). Furthermore, the inversely correlated targets of miR-223, miR-29a, miR-29b, and miR-709 are related to cell division and the cell cycle. Finally, the inversely correlated targets of four microRNAs, including miR-200b, -30a, -30d, and -429, are associated with the regulation of fever, a key sign of illness during influenza virus infection. To better understand how specific microRNAs affected these biological functions, we analyzed two related pathways comprising their inversely correlated targets.
Type I interferon pathway.
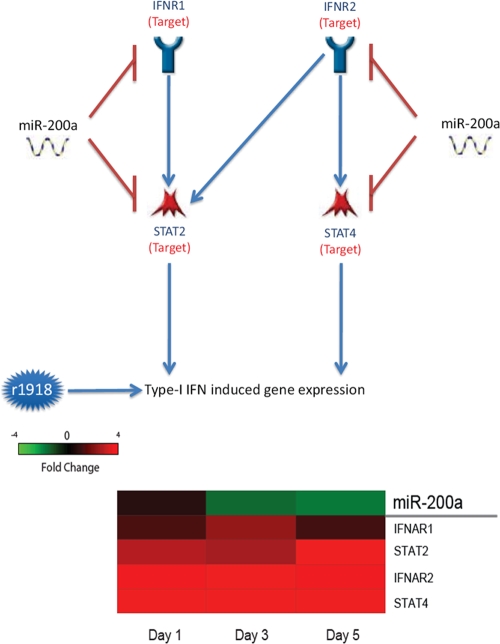
Type I IFN plays a key role in the host immune response to virus infection. Influenza virus triggers the activation of the type I IFN signaling pathway, which in turn represses virus replication. However, the activation of IFN also activates the damaging inflammatory response, a contributing factor to the lethal infection of r1918 in vivo. Our analysis of microRNA targets indicated that the type I IFN pathway was subject to microRNA-mediated regulation, since key genes in this pathway, such as IFNAR1 and STAT2, are direct targets of miR-200a, and their expression was inversely correlated with the expression of this microRNA (Fig. 4). Our data suggest that the downregulation of miR-200a in r1918-infected lungs may induce the upregulation of key genes in the type I IFN signaling pathway. Indeed, many IFN-stimulated genes demonstrated increased expression levels in r1918-infected lungs compared with those in the Tx/91-infected lungs (17, 18).
FIG. 4.
miR-200a is implicated in the type I interferon signaling pathway. (Top) Type I interferon signaling pathway comprised of miR-200a targets; (bottom) expression changes of miR-200a and its targets associated with the type I interferon pathway in r1918-infected samples. Red represents microRNA or targets that have increased expression in r1918-infected samples, relative to Tx/91-infected samples. Green represents microRNA or targets that have decreased expression in r1918-infected samples, relative to Tx/91-infected samples.
CREB pathway.
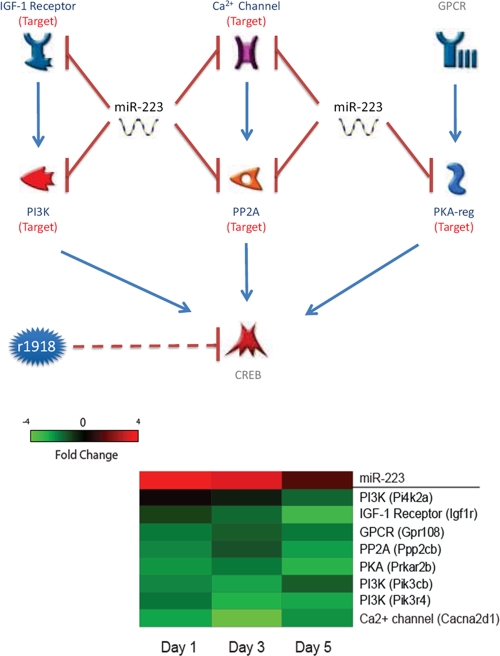
The cyclic AMP (cAMP) responsive element binding protein, CREB, is a transcription factor that regulates the expression of hundreds of genes. CREB-null mice die immediately after birth from respiratory distress (33), as CREB is involved in critical functions, including T-cell development (33) and cell survival (6). The activity of CREB is regulated by multiple upstream pathways, including insulin-like growth factor, Ca2+, and G protein-coupled receptor signaling pathways. Many key genes in the CREB upstream pathways are miR-233 targets, and concomitant with the strongly increased expression of miR-233 in r1918-infected lungs, the miR-223 targets in the CREB upstream pathways were significantly downregulated (Fig. 5). Our data therefore suggest that upregulation of miR-223 may repress the activity of CREB.
FIG. 5.
miR-223 is implicated in the CREB signaling pathway. (Top) CREB pathway comprised of miR-223 targets; (bottom) expression changes of miR-223 and its targets associated with the CREB pathway in r1918-infected samples. Red represents microRNA or targets that have increased expression in r1918-infected samples, relative to Tx/91-infected samples. Green represents microRNA or targets that have decreased expression in r1918-infected samples, relative to Tx/91-infected samples.
Taken together, our study reveals that influenza virus infection induces changes in the cellular microRNAome and that unique patterns of differential expression of microRNAs may contribute to the extreme virulence of r1918 influenza virus infection by regulating the expression of cellular targets involving immune response and other critical cellular functions.
DISCUSSION
Potential mechanisms utilized by r1918 to regulate cellular microRNA expression.
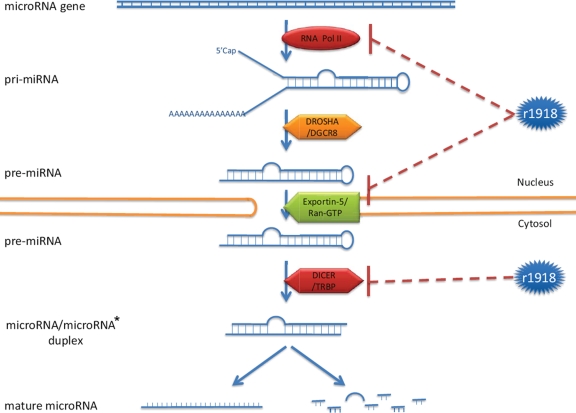
In this study, we identified microRNAs that were differentially expressed during lethal r1918 and nonlethal Tx/91 virus infections. Interestingly, 15 out of 18 abundantly expressed microRNAs were expressed at significantly lower levels during r1918 infection relative to infection with Tx/91. Because microRNA biogenesis is a complicated process and involves many cellular molecules, the abundance of cellular microRNAs can be regulated at multiple levels during influenza virus infection (Fig. 6). In particular, cellular microRNAs are predominately transcribed by RNA polymerase II (19), whose stability is compromised by the polymerase complex of influenza A viruses (32). In this regard, the viral polymerase complex is a major determinant of the pathogenicity of the 1918 pandemic virus (39). An elevated degradation of RNA polymerase II by the r1918 polymerase complex may in turn result in a decreased transcription of cellular microRNA genes, accounting for the overall reduced expression levels in the cellular microRNAome we observed in this study. Notably, although the titer of r1918 was higher than that of Tx/91 at each time point (17), the difference in viral titer remained constant, whereas the number of microRNAs that were differentially expressed between the two infections increased steadily over time (Fig. 1a). This finding suggests that differences in virus replication levels are insufficient to account for the distinct microRNA expression patterns elicited by the two viruses. Nonetheless, we cannot completely rule out that virus replication rates may, in small part, play a role in the regulation we observed. The r1918 may also decrease the stability of pre-microRNA, the microRNA precursor, by the cap-snatching mechanism (4). Furthermore, the export of pre-microRNA may be repressed due to competition for Ran-GTP from the NS2 protein of influenza virus (25). Influenza virus may also downregulate cellular microRNA expression by repressing the expression of Dicer (22), one of the most important components in the microRNA biogenesis machinery (Fig. 6, bottom).
FIG. 6.
The r1918 might potentially regulate the cellular microRNA expression at multiple levels.
Relationship between cellular microRNAs and viral pathogenesis.
MicroRNAs are potent gene expression regulators and are expressed in a variety of organisms. Although viral microRNA genes have been discovered in some viruses, including herpesviruses and human immunodeficiency virus type 1 (HIV-1) (7, 26), no influenza virus-encoded microRNA has been identified yet. From the perspective of influenza virus, alteration of cellular microRNA expression may be a critically important component of the virus-host interaction. Indeed, as a single microRNA may regulate the expression of hundreds of cellular genes, it may be more efficient for viruses to regulate the expression of a large number of cellular genes through a single microRNA rather than to regulate cellular gene expression directly. Also, influenza A virus is known for its ability to repress cellular gene expression at the posttranscriptional level by the cap-snatching mechanism (8). Alteration of the expression of cellular microRNAs may therefore provide a supplementary mechanism for influenza virus to regulate cellular gene expression. Compared to the cap-snatching mechanism, the microRNA mechanism is more selective, allowing for more directed regulation of cellular gene expression.
In this study, we observed three distinct microRNA expression patterns between the r1918- and the Tx/91-infected lungs compared with those of the mock-infected lungs. For example, miR-21 was upregulated in both infections but more strongly during r1918 infection. This indicates that miR-21 expression can be altered during influenza virus infection, irrespective of the lethality of the virus. It also suggests that regulating cellular microRNA expression may be a common activity among different influenza viruses; however, the pathogenic capacity of the viruses may eventually be determined by the expression pattern of a specific microRNA or a group of microRNAs.
Role of cellular microRNAs in the host immune response.
From the perspective of the host, thwarting influenza virus infection requires a tightly controlled antiviral immune response, as the response can have inherently damaging side effects. On one hand, the host antiviral immune response needs to be activated in order to abate virus infection. On the other hand, the immune response cannot be overly activated, which otherwise may result in a cytokine storm, severe tissue damage, and even death. Thus, the host antiviral immune response needs to be precisely regulated (1, 17), as an aberrant induction will lead to devastating consequences. A precise activation of antiviral immunity requires delicate regulation from the host, which may involve a number of cellular microRNAs. As the expression of a cellular mRNA is usually regulated by multiple microRNAs, it can be fine-tuned to the optimal level by various combinations of microRNAs (9). Changes in cellular microRNA expression during r1918 influenza virus infection will essentially disrupt the fine-tuning mechanism of the host in controlling antiviral immunity, resulting in a lethal infection.
In this study, we identified a potential role for miR-200a in regulating the immune response. Previously, miR-200a was known only for regulating the epithelial-to-mesenchymal transition by targeting zeb1 and sip1 (27). In r1918-infected mice, we not only found inversely correlated expression of miR-200a and sip1 but also found evidence implicating miR-200a in the type I IFN response. The aberrant activation of the type I IFN pathway during r1918 infection may contribute to an unconstrained inflammatory response (17, 18). Most significantly, we found increased expression of miR-200a target genes in the type I IFN pathway, including IFN-α receptors and STAT2/4. The upregulation of these cellular genes, owing to the downregulation of miR-200a, may induce profound effects during infection. As these genes are located at the upstream end of the IFN signaling cascade, a moderate expression change could be exponentially amplified at the downstream end of the cascade. As transcription factors, STAT2/4 are able to regulate the expression of a large number of IFN-stimulated genes (ISGs). An increase in the expression of STAT2/4 caused by the downregulation of miR-200a may induce an overwhelming increase of ISG expression. In line with our observations, massive induction of ISG expression has been observed in mice and macaques infected with the r1918 (17, 18).
In addition to directly regulating the expression of transcription factors, microRNAs may also impose indirect regulation. For instance, miR-223 may indirectly repress the activity of the transcription factor CREB by regulating its upstream pathways. In this study, we demonstrated that miR-223 may downregulate CREB activity by repressing three such upstream pathways, namely the IGF-1 receptor, Ca2+ channel, and GPCR pathways. In addition, miR-223 also repressed the expression of crucial intermediate molecules in these pathways, such as PI3K, PP2A, and PKA (Fig. 5). One critical function of CREB is maintaining cell survival and growth in vivo (20). Indirect repression of CREB activity by miR-223 would be expected to result in increased cell death, which is a signature of lethal r1918 infection in vivo (17, 18).
Future studies should focus on verifying the roles of microRNAs in contributing to the lethality of the r1918. As our data have for the first time laid out the correlation between expression of 18 microRNAs and r1918 pathogenesis, artificial manipulation of the expression of some cellular microRNAs by microRNA mimics or antagomirs would help identify the microRNAs responsible for the extreme virulence of the r1918. As the targets in the miRanda database were computationally identified, the microRNA-target regulatory relationships implicated in lethal r1918 infection in this study should be further validated experimentally in future studies. More importantly, the contribution of individual microRNAs to viral pathogenesis should be examined in vivo. For example, the recently produced miR-223-deficient mouse is a prime candidate to be used for evaluating the role of miR-223 during r1918 infection (15).
In this report, we implicated cellular microRNAs in the regulation of the host immune response during r1918 influenza virus infection. As the relationship between microRNA expression and r1918 pathogenicity has not been previously addressed, our findings provide a deeper understanding of the mechanisms underlying lethal influenza virus infection and may open new venues for in-depth studies of the influenza virus pathogenicity. As our data show, even without a virus-encoded microRNA, an RNA virus can utilize cellular microRNAs to manipulate cellular gene expression, a likely common scenario for RNA viruses. Therefore, microRNA profiling should be extended to studying other highly pathogenic RNA viruses. Finally, the results obtained from this study may eventually lead to the discovery of cellular targets for novel therapies against lethal influenza virus infection.
Supplementary Material
Acknowledgments
We thank Marcus Korth for critically reading the manuscript. We also thank Robert Palermo, Lynn Law, and Sean Proll for their helpful discussions.
This work was supported by the Public Health Service grant P01AI058113 from the National Institute of Allergy and Infectious Diseases.
The findings and conclusions in this report are those of the author(s) and do not necessarily represent the views of the funding agency.
Footnotes
Published ahead of print on 13 January 2010.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Aldridge, J. R., C. E. Moseley, D. A. Boltz, N. J. Negovetich, C. Reynolds, J. Franks, S. A. Brown, P. C. Doherty, R. G. Webster, and P. G. Thomas. 2009.TNF/iNOS-producing dendritic cells are the necessary evil of lethal influenza virus infection. Proc. Natl. Acad. Sci. U. S. A. 106:5306-5311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Basler, C. F., A. H. Reid, J. K. Dybing, T. A. Janczewski, T. G. Fanning, H. Zheng, M. Salvatore, M. L. Perdue, D. E. Swayne, A. García-Sastre, P. Palese, and J. K. Taubenberger. 2001. Sequence of the 1918 pandemic influenza virus nonstructural gene (NS) segment and characterization of recombinant viruses bearing the 1918 NS genes. Proc. Natl. Acad. Sci. U. S. A. 98:2746-2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bellon, M., Y. Lepelletier, O. Hermine, and C. Nicot. 2009. Deregulation of microRNA involved in hematopoiesis and the immune response in HTLV-I adult T-cell leukemia. Blood 113:4914-4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beloso, A., C. Martinez, J. Valcarcel, J. F. Santaren, and J. Ortin. 1992. Degradation of cellular mRNA during influenza virus infection: its possible role in protein synthesis shutoff. J. Gen. Virol. 73:575-581. [DOI] [PubMed] [Google Scholar]
- 5.Bethke, A., N. Fielenbach, Z. Wang, D. J. Mangelsdorf, and A. Antebi. 2009. Nuclear hormone receptor regulation of microRNAs controls developmental progression. Science 324:95-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonni, A., A. Brunet, A. E. West, S. R. Datta, M. A. Takasu, and M. E. Greenberg. 1999. Cell survival promoted by the Ras-MAPK signaling pathway by transcription-dependent and -independent mechanisms. Science 286:1358-1362. [DOI] [PubMed] [Google Scholar]
- 7.Cullen, B. R. 2006. Viruses and microRNAs. Nat. Genet. 38:S25-S30. [DOI] [PubMed] [Google Scholar]
- 8.Dias, A., D. Bouvier, T. Crepin, A. A. McCarthy, D. J. Hart, F. Baudin, S. Cusack, and R. W. H. Ruigrok. 2009. The cap-snatching endonuclease of influenza virus polymerase resides in the PA subunit. Nature 458:914-918. [DOI] [PubMed] [Google Scholar]
- 9.Flynt, A. S., and E. C. Lai. 2008. Biological principles of microRNA-mediated regulation: shared themes amid diversity. Nat. Rev. Genet. 9:831-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fodor, E., L. Devenish, O. G. Engelhardt, P. Palese, G. G. Brownlee, and A. García-Sastre. 1999. Rescue of influenza A virus from recombinant DNA. J. Virol. 73:9679-9682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gregory, P. A., A. G. Bert, E. L. Paterson, S. C. Barry, A. Tsykin, G. Farshid, M. A. Vadas, Y. Khew-Goodall, and G. J. Goodall. 2008. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell Biol. 10:593-601. [DOI] [PubMed] [Google Scholar]
- 12.Hu, G., R. Zhou, J. Liu, A. Y. Gong, A. N. Eischeid, J. W. Dittman, and X. M. Chen. 2009. MicroRNA-98 and let-7 confer cholangiocyte expression of cytokine-inducible Src homology 2-containing protein in response to microbial challenge. J. Immunol. 183:1617-1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu, S. J., G. Ren, J. L. Liu, Z. A. Zhao, Y. S. Yu, R. W. Su, X. H. Ma, H. Ni, W. Lei, and Z. M. Yang. 2008. MicroRNA expression and regulation in mouse uterus during embryo implantation. J. Biol. Chem. 283:23473-23484. [DOI] [PubMed] [Google Scholar]
- 14.Huang, J., F. Wang, E. Argyris, K. Chen, Z. Liang, H. Tian, W. Huang, K. Squires, G. Verlinghieri, and H. Zhang. 2007. Cellular microRNAs contribute to HIV-1 latency in resting primary CD4+ T lymphocytes. Nat. Med. 13:1241-1247. [DOI] [PubMed] [Google Scholar]
- 15.Johnnidis, J. B., M. H. Harris, R. T. Wheeler, S. Stehling-Sun, M. H. Lam, O. Kirak, T. R. Brummelkamp, M. D. Fleming, and F. D. Camargo. 2008. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature 451:1125-1129. [DOI] [PubMed] [Google Scholar]
- 16.Jopling, C. L., M. Yi, A. M. Lancaster, S. M. Lemon, and P. Sarnow. 2005. Modulation of hepatitis C virus RNA abundance by a liver-specific microRNA. Science 309:1577-1581. [DOI] [PubMed] [Google Scholar]
- 17.Kash, J. C., T. M. Tumpey, S. C. Proll, V. Carter, O. Perwitasari, M. J. Thomas, C. F. Basler, P. Palese, J. K. Taubenberger, A. García-Sastre, D. E. Swayne, and M. G. Katze. 2006. Genomic analysis of increased host immune and cell death responses induced by 1918 influenza virus. Nature 443:578-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kobasa, D., S. M. Jones, K. Shinya, J. C. Kash, J. Copps, H. Ebihara, Y. Hatta, J. H. Kim, P. Halfmann, M. Hatta, F. Feldmann, J. B. Alimonti, L. Fernando, Y. Li, M. G. Katze, H. Feldmann, and Y. Kawaoka. 2007. Aberrant innate immune response in lethal infection of macaques with the 1918 influenza virus. Nature 445:319-323. [DOI] [PubMed] [Google Scholar]
- 19.Lee, Y., M. Kim, J. Han, K. H. Yeom, S. Lee, S. H. Baek, and V. N. Kim. 2004. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 23:4051-4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lonze, B. E., A. Riccio, S. Cohen, and D. D. Ginty. 2002. Apoptosis, axonal growth defects, and degeneration of peripheral neurons in mice lacking CREB. Neuron 34:371-385. [DOI] [PubMed] [Google Scholar]
- 21.Lu, T. X., A. Munitz, and M. E. Rothenberg. 2009. MicroRNA-21 is up-regulated in allergic airway inflammation and regulates IL-12p35 expression. J. Immunol. 182:4994-5002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matskevich, A. A., and K. Moelling. 2007. Dicer is involved in protection against influenza A virus infection. J. Gen. Virol. 88(Pt. 10):2627-2635. [DOI] [PubMed] [Google Scholar]
- 23.Mestdagh, P., P. Van Vlierberghe, A. De Weer, D. Muth, F. Westermann, F. Speleman, and J. Vandesompele. 2009. A novel and universal method for microRNA RT-qPCR data normalization. Genome Biol. 10:R64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mraz, M., K. Malinova, J. Kotaskova, S. Pavlova, B. Tichy, J. Malcikova, K. Stano Kozubik, J. Smardova, Y. Brychtova, M. Doubek, M. Trbusek, J. Mayer, and S. Pospisilova. 2009. miR-34a, miR-29c and miR-17-5p are downregulated in CLL patients with TP53 abnormalities. Leukemia 23:1159-1163. [DOI] [PubMed] [Google Scholar]
- 25.Neumann, G., M. T. Hughes, and Y. Kawaoka. 2000. Influenza A virus NS2 protein mediates vRNP nuclear export through NES-independent interaction with hCRM1. EMBO J. 19:6751-6758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Omoto, S., and Y. R. Fujii. 2005. Regulation of human immunodeficiency virus 1 transcription by nef microRNA. J. Gen. Virol. 86:751-755. [DOI] [PubMed] [Google Scholar]
- 27.Park, S. M., A. B. Gaur, E. Lengyel, and M. E. Peter. 2008. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 22:894-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pedersen, I. M., G. Cheng, S. Wieland, S. Volinia, C. M. Croce, F. V. Chisari, and M. David. 2007. Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature 449:919-922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reid, A. H., T. G. Fanning, J. V. Hultin, and J. K. Taubenberger. 1999. Origin and evolution of the 1918 “Spanish” influenza virus hemagglutinin gene. Proc. Natl. Acad. Sci. U. S. A. 96:1651-1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reid, A. H., T. G. Fanning, T. A. Janczewski, S. McCall, and J. K. Taubenberger. 2002. Characterization of the 1918 “Spanish” influenza virus matrix gene segment. J. Virol. 76:10717-10723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reid, A. H., T. G. Fanning, T. A. Janczewski, and J. K. Taubenberger. 2000. Characterization of the 1918 “Spanish” influenza virus neuraminidase gene. Proc. Natl. Acad. Sci. U. S. A. 97:6785-6790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rodriguez, A., A. Perez-Gonzalez, and A. Nieto. 2007. Influenza virus infection causes specific degradation of the largest subunit of cellular RNA polymerase II. J. Virol. 81:5315-5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rudolph, D., A. Tafuri, P. Gass, G. Hämmerling, B. Arnold, and G. Schütz. 1998. Impaired fetal T cell development and perinatal lethality in mice lacking the cAMP response element binding protein. Proc. Natl. Acad. Sci. U. S. A. 95:4481-4486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sung, T. L., and A. P. Rice. 2009. miR-198 inhibits HIV-1 gene expression and replication in monocytes and its mechanism of action appears to involve repression of cyclin T1. PLoS Pathog. 5:e1000263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taubenberger, J. K., A. H. Reid, R. M. Lourens, R. Wang, G. Jin, and T. G. Fanning. 2005. Characterization of the 1918 influenza virus polymerase genes. Nature 437:889-893. [DOI] [PubMed] [Google Scholar]
- 36.Thum, T., C. Gross, J. Fiedler, T. Fischer, S. Kissler, M. Bussen, P. Galuppo, S. Just, W. Rottbauer, S. Frantz, M. Castoldi, J. Soutschek, V. Koteliansky, A. Rosenwald, M. A. Basson, J. D. Licht, J. T. R. Pena, S. H. Rouhanifard, M. U. Muckenthaler, T. Tuschl, G. R. Martin, J. Bauersachs, and S. Engelhardt. 2008. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature 456:980-984. [DOI] [PubMed] [Google Scholar]
- 37.Tumpey, T. M., C. F. Basler, P. V. Aguilar, H. Zeng, A. Solorzano, D. E. Swayne, N. J. Cox, J. M. Katz, J. K. Taubenberger, P. Palese, and A. Garcia-Sastre. 2005. Characterization of the reconstructed 1918 Spanish influenza pandemic virus. Science 310:77-80. [DOI] [PubMed] [Google Scholar]
- 38.Wang, F. Z., F. Weber, C. Croce, C. G. Liu, X. Liao, and P. E. Pellett. 2008. Human cytomegalovirus infection alters the expression of cellular microRNA species that affect its replication. J. Virol. 82:9065-9074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Watanabe, T., S. Watanabe, K. Shinya, J. H. Kim, M. Hatta, and Y. Kawaoka. 2009. Viral RNA polymerase complex promotes optimal growth of 1918 virus in the lower respiratory tract of ferrets. Proc. Natl. Acad. Sci. U. S. A. 106:588-592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamakuchi, M., M. Ferlito, and C. J. Lowenstein. 2008. miR-34a repression of SIRT1 regulates apoptosis. Proc. Natl. Acad. Sci. U. S. A. 105:13421-13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.