Abstract
We previously reported that human immunodeficiency virus type 1 (HIV-1) develops resistance to the cholesterol-binding compound amphotericin B methyl ester (AME) by acquiring mutations (P203L and S205L) in the cytoplasmic tail of the transmembrane envelope glycoprotein gp41 that create cleavage sites for the viral protease (PR). In the present study, we observed that a PR inhibitor-resistant (PIR) HIV-1 mutant is unable to efficiently cleave the gp41 cytoplasmic tail in P203L and S205L virions, resulting in loss of AME resistance. To define the pathway to AME resistance in the context of the PIR PR, we selected for resistance with an HIV-1 isolate expressing the mutant enzyme. We identified a new gp41 mutation, R236L, that results in cleavage of the gp41 tail by the PIR PR. These results highlight the central role of gp41 cleavage as the primary mechanism of AME resistance.
Cholesterol-enriched membrane microdomains, often referred to as lipid rafts (4, 18, 24), play an important role in the replication of many enveloped viruses, including human immunodeficiency virus type 1 (HIV-1) (22, 30). Lipid rafts are involved in both HIV-1 entry and egress (reviewed in references 6, 22, and 30), and the lipid bilayer of HIV-1 virions is significantly enriched in cholesterol and highly saturated lipids characteristic of lipid rafts (3, 5, 8). We recently demonstrated that the cholesterol-binding polyene fungal antibiotic amphotericin B methyl ester (AME) potently inhibits HIV-1 replication. The antiviral activity of AME is due to a profound inhibition of viral entry (27, 28) and impairment of virus particle production (29).
In our previous studies, we showed that the propagation of HIV-1 in the presence of AME leads to viral escape from this compound. The mutations that confer resistance map to the cytoplasmic tail (CT) of the gp41 transmembrane envelope (Env) glycoprotein (27, 28). AME-resistant mutants (P203L and S205L) overcome the defect in viral entry imposed by AME by a novel mechanism of resistance whereby the gp41 CT is cleaved by the viral protease (PR) after incorporation of Env into virions (28). The introduction of stop codons into the gp41-coding region that prematurely truncate the CT also renders virions AME resistant. In the present study, we evaluated the interplay between protease inhibitor resistance (PIR) mutations and AME resistance.
PIR mutations in PR abrogate the ability of the P203L and S205L gp41 mutations to confer AME resistance.
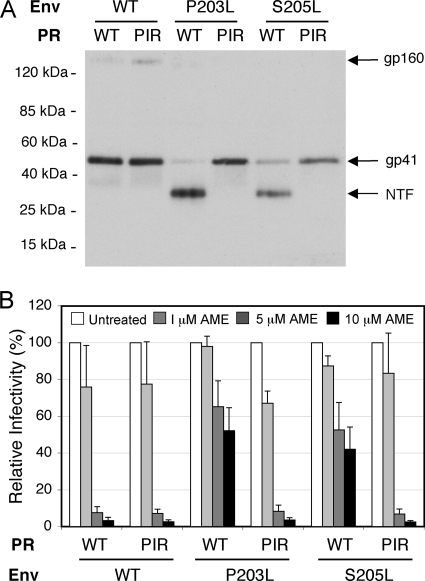
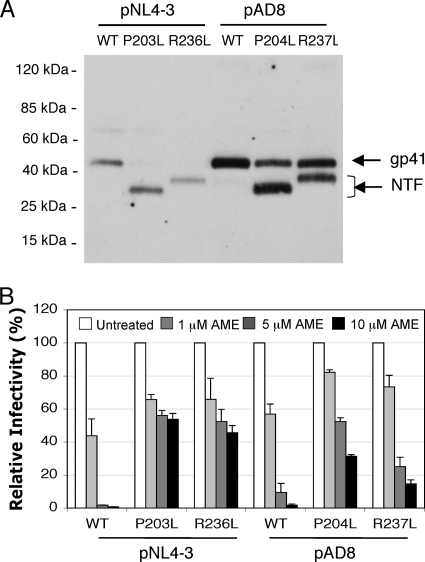
Because the AME-resistant phenotype of the P203L and S205L mutants results from cleavage of the gp41 CT by the viral PR, we examined the interplay between the emergence of AME resistance and mutations in PR that confer resistance to clinically approved PR inhibitors. For this purpose, we used a PIR mutant bearing five substitutions in PR (L10R/M46I/L63P/V82T/I84V) in the context of the pNL4-3 molecular clone (10). We prepared NL4L4-3/WT, NL4-3/P203L, and NL4-3/S205L virions containing either wild-type (WT) or PIR PR by transfecting the respective molecular clones into 293T cells (16). Virion lysates were immunoblotted with the 2F5 anti-gp41 monoclonal antibody, which recognizes an epitope in the ectodomain of gp41 (11, 21). As we reported previously (28), the WT PR cleaved the AME-resistant P203L and S205L gp41 mutants but not WT gp41 (Fig. 1A). Unexpectedly, we observed that the PIR PR was unable to cleave either of the gp41 mutants. These results suggest that the PIR mutations might reverse the AME-resistant phenotype of the P203L and S205L gp41 mutants. To test this possibility, we measured the infectivity of P203L and S205L mutant virions bearing WT or PIR PR in the TZM-bl indicator cell line (23, 31). We observed that as reported earlier (28), WT HIV-1 was highly sensitive to AME (with a reduction in infectivity of >50-fold at 10 μM) while the P203L and S205L mutants were largely resistant (with infectivity reduced by only approximately twofold at 10 μM) (Fig. 1B). Interestingly, and consistent with the gp41-processing data in Fig. 1A, both the P203L and S205L mutants became fully sensitive to AME in the context of the PIR PR (Fig. 1B). These data demonstrate that the PIR PR, unlike the WT enzyme, is unable to render the P203L and S205L mutants AME resistant by cleaving their gp41 CTs.
FIG. 1.
PIR mutations abrogate the ability of the P203L and S205L gp41 mutations to confer AME resistance. (A) 293T cells were transfected with WT pNL4-3 (WT) (1) or AME-resistant Env mutants (P203L and S205L) (27, 28) encoding WT or PIR PR. The PIR substitutions (L10R/M46I/L63P/V82T/I84V) (10) were introduced into the pNL4-3/P203L and pNL4-3/S205L mutants by subcloning the 3,737-bp ApaI-EcoRI fragment containing the PIR changes as described previously (2). Virus-containing supernatants were harvested and virions were concentrated by ultracentrifugation (14). Viral lysates were subjected to immunoblotting with the 2F5 monoclonal anti-gp41 antibody (11). Molecular mass markers are shown on the left. NTF, gp41 N-terminal fragment. (B) TZM-bl cells were infected with virus stocks produced by transfecting 293T cells with molecular clones encoding WT or AME-resistant Env mutants (P203L, S205L) and WT or PIR PR. Infections were carried out for 2 h in the absence or presence of 1, 5, or 10 μM AME. Infected cells were washed and cultured in the absence of AME; luciferase activity was measured 2 days postinfection. The infectivity of the PIR mutant was similar to that of the WT. Data shown are means ± standard errors (SEs) (n = 4).
Selection for AME resistance in the context of PIR PR.
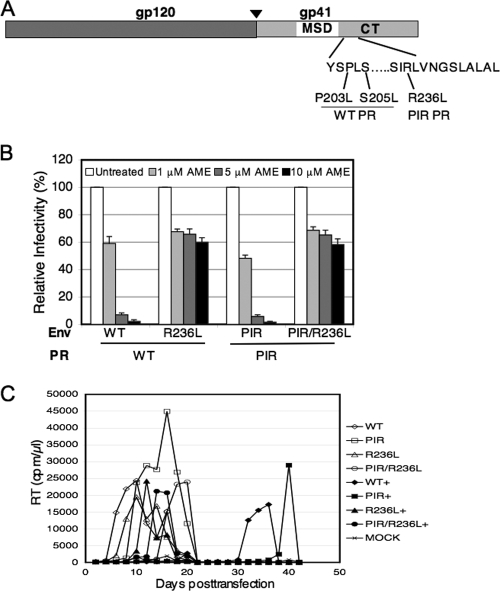
To determine whether NL4-3 encoding the PIR PR is able to acquire AME resistance and, if so, by what mechanism, we attempted to select for AME resistance in the context of the PIR PR. The replication of NL4-3/PIR was carried out in parallel with that of WT NL4-3 in Jurkat T cells in the absence of AME or in the continual presence of 5 or 10 μM compound. In the absence of AME, replication of both NL4-3 and NL4-3/PIR peaked on day 8. In the presence of AME, replication was significantly delayed: replication of the WT peaked on day 14 and that of NL4-3/PIR on day 24 (data not shown). To determine whether mutations were acquired during the course of virus propagation in the presence of AME, genomic DNA was extracted from cells harvested at peak reverse transcriptase (RT) activity from both WT NL4-3- and NL4-3/PIR-infected cultures. The Env-coding region was amplified by PCR, using the HIV-1 genomic DNA as a template (27, 28). In the WT-infected cultures, we observed the S205L mutant, confirming our previous result (28). In contrast, in the PIR-infected cultures, we identified a new mutation, R236L (Fig. 2A).
FIG. 2.
The R236L mutation confers resistance to AME in the context of WT or PIR PR. (A) Identification of the R236L mutation. The env region was PCR amplified from genomic DNA purified at the peak of viral replication from WT- and PIR-infected Jurkat cultures treated with AME, and the PCR product was sequenced as described previously (27). The organization of HIV-1 Env is indicated, with an arrowhead showing the cellular protease cleavage site between gp120 and gp41. Locations of the R236L mutation and the previously described AME-resistant mutants (P203L, S205L) selected in the context of WT PR (27, 28) are shown. MSD, membrane-spanning domain. (B) TZM-bl cells were infected with virions bearing WT or R236L Env and WT or PIR PR. The R236L mutant was constructed by QuikChange site-directed mutagenesis (Stratagene), and the PIR mutations were introduced into the pNL4-3/R236L clone as described in the legend to Fig. 1. Infections were carried out in the absence or presence of 1, 5, or 10 μM AME as described in the legend to Fig. 1. Data shown are means ± SEs of the results from three independent experiments performed in duplicate. The relative infectivities of the NL4-3/PIR, NL4-3/R236L, and NL4-3/PIR/R236L mutants, compared to that of the WT, were 97%, 87%, and 72%, respectively. (C) Five million Jurkat cells were transfected with 5 μg of the indicated molecular clones (5 μg) by using the DEAE-dextran transfection procedure and were cultured in the absence or continual presence (+) of 5 μM AME. Cultures were split 1:3 every 2 days, and supernatants were reserved for RT assay at each time point.
The R236L mutation confers AME resistance in the context of either WT or PIR PR.
To determine whether the R236L mutation provides AME resistance, we introduced this gp41 substitution into pNL4-3 molecular clones encoding either WT or the PIR PR. Single-cycle infectivity assays were performed in TZM-bl cells (23, 31) in the presence or absence of AME. The infectivity of the R236L mutant was reduced less than twofold in the presence of 10 μM AME, whereas WT infectivity was reduced more than 50-fold (Fig. 2B). Interestingly, the R236L mutant was AME resistant in the context of either WT or PIR PR. To confirm that the R236L mutant was AME resistant, we analyzed the replication kinetics of R236L and PIR/R236L mutants in the presence and absence of AME in Jurkat T cells. Cells were transfected with pNL4-3, pNL4-3/PIR, pNL4-3/R236L, and pNL4-3/PIR/R236L molecular clones by using the DEAE-dextran method (17) and were cultured continuously in the absence of AME or in the presence of 5 μM compound (Fig. 2C). AME delayed the replication of WT virus and the PIR mutant virus by approximately 2 weeks. In contrast, replication of the R236L mutant was delayed by only 2 days in the presence of AME, and replication of the PIR/R236L mutant actually peaked several days earlier in the presence of AME than in its absence. These results indicate that the R236L mutation confers resistance in the context of either WT or PIR PR both in single-cycle infectivity assays and in spreading infections.
R236L gp41 is cleaved by both WT and PIR PR.
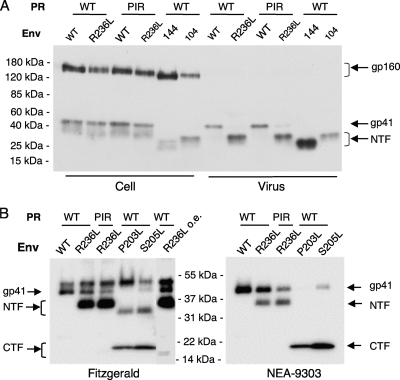
As mentioned above, the AME-resistant mutants P203L and S205L undergo PR-mediated cleavage in the gp41 CT (28) (Fig. 1A). The observation that the R236L virus is AME resistant (Fig. 2B and data not shown) raised the possibility that this mutant also undergoes PR-mediated gp41 cleavage. To examine this question, we transfected HeLa or 293T cells with molecular clones pNL4-3, pNL4-3/PIR, pNL4-3/R236L, and pNL4-3/PIR/R236L. As controls, we included the gp41 truncation mutants pNL4-3/CTdel-144 and pNL4-3/CTdel-104 (15, 16, 20), which encode gp41 glycoproteins truncated by 144 and 104 amino acids, respectively, as a result of engineered premature-termination codons. Cell and viral lysates were prepared as described previously (28) and immunoblotted with the 2F5 anti-gp41 monoclonal antibody (11) (Fig. 3A). In virion lysates, WT gp41 was 41 kDa, and the CTdel-144 and CTdel-104 gp41 bands were of the expected (smaller) sizes. Interestingly, in the context of either WT or PIR PR, the R236L mutant virions showed a gp41 band of ∼32 kDa, intermediate in size between that of the CTdel-144 and CTdel-104 mutants. This band is designated the N-terminal fragment (NTF) because it is detected with an antibody specific for the ectodomain of gp41 (Fig. 3A). Viral lysates were also immunoblotted with two additional gp41 antibodies: the Fitzgerald polyclonal antibody and the NEA-9303 monoclonal antibody (Fig. 3B). We included our previously characterized AME-resistant mutants, the P203L and S205L mutants (28), for comparison. The Fitzgerald anti-gp41 polyclonal antiserum detected the full-length gp41 and the NTF in R236L viral lysates but poorly detected the cleaved C-terminal fragment (CTF) for this mutant. In contrast, this antibody efficiently detected the NTF and CTF for the P203L and S205L mutants (Fig. 3B). The molecular mass of the smaller gp41 CTF from R236L is ≈13 kDa (Fig. 3B, overexposed lane [R236L o.e.]). The NEA-9303 anti-gp41 monoclonal antibody detected the CTF of the P203L and S205L mutants, but this antibody detected the NTF but not the CTF of the R236L mutant. These results demonstrate that the epitope recognized by the NEA-9303 antibody lies between the sites of PR-mediated cleavage of the S205L and R236L mutants. Based on the size of the NTF and CTF observed with the R236L mutant, the data in Fig. 3 suggest that the R236L gp41 CT is cleaved by PR in the vicinity of the R236L mutation.
FIG. 3.
Cleavage of the R236L gp41 CT by WT and PIR PR. (A) 293T cells were transfected with WT or the R236L mutant molecular clones encoding WT or PIR PR. Cell and viral lysates were prepared as described previously (28) and subjected to immunoblotting with the 2F5 anti-gp41 monoclonal antibody. To evaluate the size of the truncated gp41 from the R236L Env mutant, gp41 CT truncation mutants CTdel-144 (144) and CTdel-104 (104) (13, 16, 20) were included for comparison. (B) 293T cells were transfected with WT or AME-resistant mutants (P203L, S205L, R236L, PIR/R236L), and viral lysates were subjected to immunoblotting with the Fitzgerald polyclonal anti-gp41 antibody or the anti-gp41 monoclonal antibody NEA-9303 (19) as described previously (28). Positions of gp160, gp41, and the gp41 N-terminal fragment (NTF) and C-terminal fragment (CTF) are shown. On the right side of the Fitzgerald blot is an overexposed lane of the R236L mutant (R236L o.e.) that shows the ≈13-kDa CTF for this mutant. Molecular mass markers are indicated.
Identification of the cleavage site in the CT of R236L gp41.
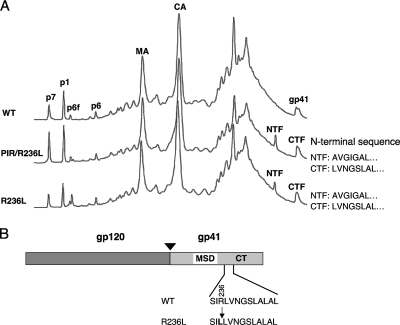
To identify the site of PR-mediated cleavage in the CT of R236L gp41, we concentrated the virions after transfecting HeLa or 293T cells and purified them by high-performance liquid chromatography (HPLC) as described previously (9, 28). Protein elution profiles of HPLC fractions detected at 280 nm showed peaks in the R236L and R236L/PIR virions that were not detected in WT virions (Fig. 4A, NTF and CTF). To determine which of the HPLC fractions contained gp41 NTF and CTF, we performed Western blotting analysis of these fractions by using Fitzgerald anti-gp41 polyclonal antiserum. Our analysis indicated that fractions 60 to 62 contained the NTF and fraction 66 contained the CTF (data not shown). We performed N-terminal protein sequencing of both NTF and CTF to map the site of gp41 cleavage. In the context of both WT and PIR PR, the N-terminal sequences obtained for the NTF and CTF were AVGIGAL and LVNGSLAL, respectively (Fig. 4B). These results demonstrate that the R236L mutant is cleaved by PR between Leu-236 and Leu-237 (Fig. 4B), indicating that the R236L mutation in the gp41 CT creates a cleavage site that can be processed by either WT or the PIR PR, resulting in truncation of the gp41 CT by 109 amino acids.
FIG. 4.
Identification of the PR cleavage site in R236L gp41. (A) Separation of viral proteins by HPLC. Virions were prepared in 293T cells by transfection with pNL4-3 derivatives encoding WT (WT) or R236L Env (R236L) mutants with WT or PIR PR (PIR/R236L). Virions were concentrated by ultracentrifugation, and viral proteins were separated by HPLC as described previously (28); the A280 chromatograms are shown. Major viral protein peaks identified by immunoblot or protein sequence analysis are labeled. Peaks representing matrix (MA), capsid (CA), full-length gp41, gp41 NTF, and gp41 CTF are shown. The NTF and CTF derived from R236L and PIR/R236L virions were subjected to Edman degradation, and the N-terminal sequences were analyzed by using an automated 477 protein sequencer (Applied Biosystems, Foster City, CA). (B) Cleavage site in the R236L gp41. The organization of HIV-1 Env is as shown in Fig. 2A, and the position of the residue 236 mutation is indicated in bold. The PR-mediated cleavage site between gp41 residues 236 and 237 is indicated by an arrow.
The P204L and R237L mutations in the AD8 strain of HIV-1 confer partial gp41 cleavage and resistance to AME.
To determine whether the P203L and R236L mutations identified here and in our earlier study also induced AME resistance in another strain of HIV-1, we introduced the analogous mutations into the AD8 molecular clone (26). Site-directed mutagenesis was used to change the gp41 coding region at positions 204 and 237 from YSPLS to YSLLS and SVRLV to SVLLV, respectively. As is the case for the analogous mutations in pNL4-3, these substitutions create new di-Leu motifs in gp41. We then examined whether these mutations induce the cleavage of gp41 by PR. 293T cells were transfected with WT, P204L, or R237L mutant pAD8 molecular clones, and virions were collected 24 h after transfection. As controls, we included WT and AME-resistant (P203L and S205L) pNL4-3 molecular clones in the analysis. Viral lysates were prepared and immunoblotted with antibody (Ab) specific to the N terminus of gp41. The results indicated that both P204L and R237L mutants in the context of the AD8 strain undergo gp41 CT cleavage, although the extent of cleavage is reproducibly less than observed with the AME-resistant NL4-3 mutants (Fig. 5A). We also tested the sensitivity of the P204L and R237L AD8 mutants to AME relative to that of the WT AD8 parental virus. We again included WT and AME-resistant pNL4-3 mutants as controls. Virus stocks generated in 293T cells were used to infect TZM-bl cells in the presence or absence of AME. We observed that WT AD8 virions were highly sensitive to AME, with a >50-fold reduction in infectivity at 10 μM AME. In contrast, the P204L and R237L mutants were partially resistant; virus infectivity was reduced by only three- and sixfold for P204L and R237L, respectively, at 10 μM AME (Fig. 5B). These results demonstrate that the AME resistance mutations selected in the context of NL4-3 also confer a significant level of PR-mediated CT cleavage and AME resistance when present in the AD8 strain of HIV-1.
FIG. 5.
The P204L and R237L mutations in the AD8 strain of HIV-1 confer resistance to AME. (A) 293T cells were transfected with WT pAD8 or the P204L and R237L AD8 mutants constructed by QuikChange site-directed mutagenesis (Stratagene). One day posttransfection, viral lysates were prepared as described in the legends to Fig. 1 and 3 and were subjected to immunoblotting with the 2F5 anti-gp41 monoclonal antibody. In this experiment, NL4-3 and its AME-resistant derivatives (P203L and R236L) were included as controls. (B) TZM-bl cells were infected with virus stocks derived from WT NL4-3 or AD8 or the indicated mutants. Infections were carried out in the absence or presence of 1, 5, or 10 μM AME as described in the legend to Fig. 1. Data shown are means ± SEs of the results from three independent experiments performed in duplicate.
Conclusions.
In this study, we examined the effect of mutations in the HIV-1 PR on the development of resistance to the cholesterol-binding entry inhibitor AME. Interestingly, we observed that the AME-resistant mutants selected in the presence of WT PR (P203L and S205L) (27, 28) are no longer AME resistant in the context of the PIR PR. This observation suggests that the PIR PR differs in its substrate preference relative to the WT enzyme such that it is unable to cleave the gp41 CT of the P203L and S205L mutants. To identify the AME resistance pathway followed by the virus in the context of PIR PR, we performed de novo selections with the NL4-3/PIR clone. We identified a mutation, R236L, that like P203L and S205L creates a PR cleavage site in the gp41 CT. Unlike the P203L and S205L mutants, however, the R236L mutant gp41 serves as a substrate for both WT and PIR PR. Each of the three independent AME-resistant mutants that we identified (P203L, S205L, and R236L) confers AME resistance by a PR-mediated gp41 cleavage mechanism, and as we reported previously (27, 28), artificial truncation of gp41 also relieves the AME-imposed block to infectivity in single-round assays. These data highlight the importance of gp41 truncation in AME resistance. We also observed that the phenomenon of AME resistance mediated by PR-mediated gp41 cleavage is not limited to the NL4-3 strain of HIV-1. Mutations analogous to NL4-3 P203L and R236L (P204L and R237L, respectively) also induce gp41 cleavage and partial AME resistance in the context of the AD8 strain of HIV-1. The more-limited gp41 cleavage and AME resistance in the AD8 context suggest that strain-specific sequence variation or differences in gp41 CT conformation between NL4-3 and AD8 may render the AD8 sequences less optimal for PR-mediated cleavage.
We note that thus far all AME resistance mutations that we have identified (P203L, S205L, and R236L) generate di-Leu motifs (this study and references 27 and 28). This suggests that such motifs are favored as targets for PR-mediated gp41 CT cleavage. It is intriguing that the WT and PIR PR display different sequence preferences for the di-Leu motifs at residues 203 and 205 versus residue 236. Also worth noting is that the WT NL4-3 gp41 CT contains three di-Leu motifs (gp41 residues 273/274, 288/289, and 303/304) and one tri-Leu motif (gp41 residues 263-265) (GenBank accession no. AF324493). These naturally occurring di- and tri-Leu motifs are more than 60 amino acids downstream from the gp41 membrane-spanning domain, and we have no evidence that they are naturally cleaved by PR. These observations suggest that di-Leu motifs must be located within 30 or so residues of the membrane-spanning domain in order to be targeted by PR. Furthermore, we previously showed that truncation of the gp41 CT by 30 or 63 residues from the C terminus did not confer resistance to AME; the acquisition of PR cleavage sites at these positions would thus not be selected for during the development of AME resistance. It is noteworthy that these naturally occurring di-Leu motifs also exist in the AD8 strain (GenBank accession no. AF004394) and in many other HIV-1 isolates (www.hiv.lan1.gov/content/index).
Additional work will be required to determine why gp41 truncation bypasses the infectivity block imposed by AME. We speculate that gp41 truncation may induce a conformational change in the ectodomain of gp41 or in gp120 (7, 12, 25, 32) that enables virion-cell fusion to occur even in the presence of AME or increases the flexibility or diffusion of the Env complex in the virion lipid bilayer such that the AME-imposed restriction to infectivity is eliminated.
Acknowledgments
We thank members of the Freed laboratory for helpful discussions and critical reviews of the manuscript and K. Felton and K. Waki for assistance with DNA cloning. The HIV-1 gp41 monoclonal antibody 2F5 and TZM-bl cells were obtained from the NIH AIDS Research and Reference Reagent Program. AME was generously supplied by Karykion Corporation, Princeton, NJ.
This research was supported by the Intramural Research Program of the Center for Cancer Research, National Cancer Institute, NIH, and by the Intramural AIDS Targeted Antiviral Program.
Footnotes
Published ahead of print on 30 December 2009.
REFERENCES
- 1.Adachi, A., H. E. Gendelman, S. Koenig, T. Folks, R. Willey, A. Rabson, and M. A. Martin. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59:284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adamson, C. S., K. Waki, S. D. Ablan, K. Salzwedel, and E. O. Freed. 2009. Impact of human immunodeficiency virus type 1 resistance to protease inhibitors on evolution of resistance to the maturation inhibitor bevirimat (PA-457). J. Virol. 83:4884-4894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aloia, R. C., H. Tian, and F. C. Jensen. 1993. Lipid composition and fluidity of the human immunodeficiency virus envelope and host cell plasma membranes. Proc. Natl. Acad. Sci. U. S. A. 90:5181-5185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown, D. A., and E. London. 2000. Structure and function of sphingolipid- and cholesterol-rich membrane rafts. J. Biol. Chem. 275:17221-17224. [DOI] [PubMed] [Google Scholar]
- 5.Brugger, B., B. Glass, P. Haberkant, I. Leibrecht, F. T. Wieland, and H. G. Krausslich. 2006. The HIV lipidome: a raft with an unusual composition. Proc. Natl. Acad. Sci. U. S. A. 103:2641-2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campbell, S. M., S. M. Crowe, and J. Mak. 2001. Lipid rafts and HIV-1: from viral entry to assembly of progeny virions. J. Clin. Virol. 22:217-227. [DOI] [PubMed] [Google Scholar]
- 7.Chakrabarti, L., M. Emerman, P. Tiollais, and P. Sonigo. 1989. The cytoplasmic domain of simian immunodeficiency virus transmembrane protein modulates infectivity. J. Virol. 63:4395-4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan, R., P. D. Uchil, J. Jin, G. Shui, D. E. Ott, W. Mothes, and M. R. Wenk. 2008. Retroviruses human immunodeficiency virus and murine leukemia virus are enriched in phosphoinositides. J. Virol. 82:11228-11238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chertova, E., O. Chertov, L. V. Coren, J. D. Roser, C. M. Trubey, J. W. Bess, Jr., R. C. Sowder II, E. Barsov, B. L. Hood, R. J. Fisher, K. Nagashima, T. P. Conrads, T. D. Veenstra, J. D. Lifson, and D. E. Ott. 2006. Proteomic and biochemical analysis of purified human immunodeficiency virus type 1 produced from infected monocyte-derived macrophages. J. Virol. 80:9039-9052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Condra, J. H., W. A. Schleif, O. M. Blahy, L. J. Gabryelski, D. J. Graham, J. C. Quintero, A. Rhodes, H. L. Robbins, E. Roth, M. Shivaprakash, et al. 1995. In vivo emergence of HIV-1 variants resistant to multiple protease inhibitors. Nature 374:569-571. [DOI] [PubMed] [Google Scholar]
- 11.D'Souza, M. P., D. Livnat, J. A. Bradac, and S. H. Bridges. 1997. Evaluation of monoclonal antibodies to human immunodeficiency virus type 1 primary isolates by neutralization assays: performance criteria for selecting candidate antibodies for clinical trials. AIDS Clinical Trials Group Antibody Selection Working Group. J. Infect. Dis. 175:1056-1062. [DOI] [PubMed] [Google Scholar]
- 12.Edwards, T. G., S. Wyss, J. D. Reeves, S. Zolla-Pazner, J. A. Hoxie, R. W. Doms, and F. Baribaud. 2002. Truncation of the cytoplasmic domain induces exposure of conserved regions in the ectodomain of human immunodeficiency virus type 1 envelope protein. J. Virol. 76:2683-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Freed, E. O., and M. A. Martin. 1996. Domains of the human immunodeficiency virus type 1 matrix and gp41 cytoplasmic tail required for envelope incorporation into virions. J. Virol. 70:341-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Freed, E. O., and M. A. Martin. 1994. Evidence for a functional interaction between the V1/V2 and C4 domains of human immunodeficiency virus type 1 envelope glycoprotein gp120. J. Virol. 68:2503-2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Freed, E. O., and M. A. Martin. 1995. The role of human immunodeficiency virus type 1 envelope glycoproteins in virus infection. J. Biol. Chem. 270:23883-238836. [DOI] [PubMed] [Google Scholar]
- 16.Freed, E. O., and M. A. Martin. 1995. Virion incorporation of envelope glycoproteins with long but not short cytoplasmic tails is blocked by specific, single amino acid substitutions in the human immunodeficiency virus type 1 matrix. J. Virol. 69:1984-1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Freed, E. O., J. M. Orenstein, A. J. Buckler-White, and M. A. Martin. 1994. Single amino acid changes in the human immunodeficiency virus type 1 matrix protein block virus particle production. J. Virol. 68:5311-5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hancock, J. F. 2006. Lipid rafts: contentious only from simplistic standpoints. Nat. Rev. Mol. Cell Biol. 7:456-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kure, K., K. M. Weidenheim, W. D. Lyman, and D. W. Dickson. 1990. Morphology and distribution of HIV-1 gp41-positive microglia in subacute AIDS encephalitis. Acta Neuropathol. 80:393-400. [DOI] [PubMed] [Google Scholar]
- 20.Murakami, T., and E. O. Freed. 2000. The long cytoplasmic tail of gp41 is required in a cell type-dependent manner for HIV-1 envelope glycoprotein incorporation into virions. Proc. Natl. Acad. Sci. U. S. A. 97:343-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ofek, G., M. Tang, A. Sambor, H. Katinger, J. R. Mascola, R. Wyatt, and P. D. Kwong. 2004. Structure and mechanistic analysis of the anti-human immunodeficiency virus type 1 antibody 2F5 in complex with its gp41 epitope. J. Virol. 78:10724-10737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ono, A., and E. O. Freed. 2005. Role of lipid rafts in virus replication. Adv. Virus Res. 64:311-358. [DOI] [PubMed] [Google Scholar]
- 23.Platt, E. J., K. Wehrly, S. E. Kuhmann, B. Chesebro, and D. Kabat. 1998. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J. Virol. 72:2855-2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simons, K., and D. Toomre. 2000. Lipid rafts and signal transduction. Nature Rev. 1:31-39. [DOI] [PubMed] [Google Scholar]
- 25.Spies, C. P., G. D. Ritter, Jr., M. J. Mulligan, and R. W. Compans. 1994. Truncation of the cytoplasmic domain of the simian immunodeficiency virus envelope glycoprotein alters the conformation of the external domain. J. Virol. 68:585-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Theodore, T. S., G. Englund, A. Buckler-White, C. E. Buckler, M. A. Martin, and K. W. Peden. 1996. Construction and characterization of a stable full-length macrophage-tropic HIV type 1 molecular clone that directs the production of high titers of progeny virions. AIDS Res. Hum. Retroviruses 12:191-194. [DOI] [PubMed] [Google Scholar]
- 27.Waheed, A. A., S. D. Ablan, M. K. Mankowski, J. E. Cummins, R. G. Ptak, C. P. Schaffner, and E. O. Freed. 2006. Inhibition of HIV-1 replication by amphotericin B methyl ester: selection for resistant variants. J. Biol. Chem. 281:28699-28711. [DOI] [PubMed] [Google Scholar]
- 28.Waheed, A. A., S. D. Ablan, J. D. Roser, R. C. Sowder, C. P. Schaffner, E. Chertova, and E. O. Freed. 2007. HIV-1 escape from the entry-inhibiting effects of a cholesterol-binding compound via cleavage of gp41 by the viral protease. Proc. Natl. Acad. Sci. U. S. A. 104:8467-8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Waheed, A. A., S. D. Ablan, F. Soheilian, K. Nagashima, A. Ono, C. P. Schaffner, and E. O. Freed. 2008. Inhibition of human immunodeficiency virus type 1 assembly and release by the cholesterol-binding compound amphotericin B methyl ester: evidence for Vpu dependence. J. Virol. 82:9776-9781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Waheed, A. A., and E. O. Freed. 2009. Lipids and membrane microdomains in HIV-1 replication. Virus Res. 143:162-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wei, X., J. M. Decker, H. Liu, Z. Zhang, R. B. Arani, J. M. Kilby, M. S. Saag, X. Wu, G. M. Shaw, and J. C. Kappes. 2002. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob. Agents Chemother. 46:1896-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wyss, S., A. S. Dimitrov, F. Baribaud, T. G. Edwards, R. Blumenthal, and J. A. Hoxie. 2005. Regulation of human immunodeficiency virus type 1 envelope glycoprotein fusion by a membrane-interactive domain in the gp41 cytoplasmic tail. J. Virol. 79:12231-12241. [DOI] [PMC free article] [PubMed] [Google Scholar]