Abstract
Burkitt lymphoma (BL) is etiologically associated with Epstein-Barr virus (EBV). EBV-positive BL tumors display two latent forms of infection. One is referred to as latency I infection, in which EBV expresses the virus genome maintenance protein EBNA1 as the only viral protein. The other is referred to as Wp-restricted latency and was recently identified in a subset of BL tumors. In these tumors, EBV expresses EBNA1, EBNA3A, EBNA3B, EBNA3C, a truncated form of EBNA-LP, and the viral Bcl-2 homologue BHRF1, all of which are driven by the BamHI W promoter (Wp). To investigate the role of EBV in Wp-restricted BL, we conditionally expressed a dominant-negative EBNA1 (dnEBNA1) mutant which interrupts the virus genome maintenance function of EBNA1 in the P3HR-1 BL cell line. Induction of dnEBNA1 expression caused loss of the EBV genome and resulted in apoptosis of P3HR-1 cells in the absence of exogenous apoptosis inducers, indicating that P3HR-1 cells cannot survive without EBV. Stable transfection of the BHRF1 gene into P3HR-1 cells rescued the cells from the apoptosis induced by dnEBNA1 expression, whereas stable transfection of truncated EBNA-LP, EBNA3A, or EBNA3C did not. Moreover, knockdown of BHRF1 expression in P3HR-1 cells resulted in increased cell death. These results indicate that EBV is essential for the survival of P3HR-1 cells and that BHRF1 functions as a survival factor. Our finding implies a critical contribution of BHRF1 to the pathogenesis of Wp-restricted BLs.
Epstein-Barr virus (EBV), a gammaherpesvirus with B-cell growth-transforming ability, is causally implicated in Burkitt lymphoma (BL) (see reference 40 for a review). In vitro, EBV infection efficiently transforms human B cells into lymphoblastoid cell lines (LCLs), which display a transcription program called latency III, during which the virus expresses 6 nuclear proteins (EBNA1, -2, -3A, -3B, -3C, and -LP), 3 integral membrane proteins (LMP1, -2A, and -2B), 2 small nonpolyadenylated EBV-encoded RNAs (EBERs) (EBER1 and EBER2), BamHI A rightward transcripts (BARTs), and microRNAs (see reference 24 for a review). However, EBV-positive BL tumors express a limited set of viral genes and do not express EBNA2 or LMP1, which play essential roles in B-cell growth transformation. Two latency forms of infection are known in EBV-positive BLs. One is referred to as latency I infection, in which EBV expresses only the virus genome maintenance protein EBNA1 (whose expression is driven by the Qp promoter), EBERs, and BARTs. In addition to the classical latency I BLs, another subset of BLs that express EBNA1, EBNA3A, EBNA3B, EBNA3C, truncated EBNA-LP, EBERs, and BARTs was recently identified, and this form of infection is referred to as Wp-restricted latency because the expression of the EBNAs is driven by the Wp promoter (19). These Wp-restricted BLs are characterized by infection with an EBV that carries a deletion of the EBNA2 gene and its surrounding sequences (19). A more recent report has shown that Wp-restricted BLs express the EBV-encoded Bcl-2 homologue BHRF1 as a latent gene (21). BHRF1 was primarily identified as a member of the early antigen complex (38). However, it was also reported that BHRF1 could be expressed as a latent gene (3).
The hallmark of BL tumors is reciprocal translocation between the c-myc gene and one of the immunoglobulin (Ig) loci, which leads to constitutive and deregulated expression of c-myc (26). The c-myc/Ig translocation appears to play a critical role in the pathogenesis of BL (30, 49). Activation of c-myc not only stimulates cell proliferation, but also increases susceptibility to apoptosis (6). In BLs, EBV is postulated to counteract the proapoptotic effects that are probably induced by c-myc activation, because several lines of evidence have demonstrated that EBV provides a survival advantage to BL cells. It has been shown that EBV-positive BL lines are more resistant to apoptotic triggers than EBV-negative BL lines (22, 28, 31, 36, 41). EBERs confer resistance to apoptosis induced by various stimuli in BL lines (27, 36). EBNA1 is also reported to have an antiapoptotic function. Inhibition of EBNA1 by a retrovirus vector expressing a dominant-negative derivative of EBNA1 (dnEBNA1) decreased survival and cloning efficiency in BL lines (23). Enforced expression of EBNA1 in EBV-negative cells inhibits apoptosis induced by p53 (23, 42). Furthermore, Wp-restricted BL lines are more resistant to apoptosis than latency I BL lines (20, 22). Some viral proteins that are expressed in Wp-restricted but not in latency I BLs are reported to have antiapoptotic functions. For example, the truncated form of EBNA-LP that is specifically expressed in Wp-restricted BLs confers resistance to apoptosis induced by verotoxin 1 or staurosporine (10). EBNA3A and EBNA3C cooperate to repress expression of the proapoptotic tumor suppressor Bim and contribute to resistance to apoptosis induced by cytotoxic agents, such as nocadazole and cisplatin (1). Enforced BHRF1 expression protects latency I BL cells from ionomycin-induced apoptosis (21). These previous reports demonstrate that various EBV gene products have the ability to confer resistance to apoptosis induced by exogenous apoptotic triggers. However, it is still unclear whether EBV is essential for the survival of EBV-positive BL cells in the absence of exogenous apoptotic stimuli and, if so, which viral gene plays a central role in the survival.
The experiments reported here focus on the role of EBV in the growth and survival of Wp-restricted BL cells. We established a Wp-restricted P3HR-1 BL cell line in which dnEBNA1 was conditionally expressed in a tetracycline-regulated manner. The EBV genome disappeared from the cells after induction of dnEBNA1 expression because dnEBNA1 interrupted the virus genome maintenance function of EBNA1 (23, 25, 32, 37). The growth and survival of P3HR-1 cells in the absence and presence of dnEBNA1 expression were compared. Our data presented here clearly show that EBV is essential for the survival of P3HR-1 cells and that BHRF1 functions as a survival factor.
MATERIALS AND METHODS
Cell lines.
P3HR-1 (HH514 cl. 16), a Wp-restricted BL cell line carrying an EBV genome in which the EBNA2 gene and part of the EBNA-LP gene are deleted (5, 14, 15, 39); an EBV-negative subline of Akata BL (44); and an EBV-negative subline of Daudi BL (36) were grown in RPMI 1640 medium supplemented with 10% fetal bovine serum. P3-dnEBNA1 and Ak(−)-dnEBNA1 cells that conditionally express a dnEBNA1 in a tetracycline-regulated (Tet-off) manner were maintained in complete medium (RPMI 1640 medium supplemented with 10% fetal bovine serum) supplemented with G418 (400 μg/ml) and doxycycline (Dox) (100 ng/ml). P3-dnEBNA1 cell lines that were stably transfected with Bcl-2 (P3-dnEBNA1-Bcl2), BHRF1 (P3-dnEBNA1-BHRF1), truncated EBNA-LP (P3-dnEBNA1-LPd45), EBNA3A (P3-dnEBNA1-E3A), EBNA3C (P3-dnEBNA1-E3C), or EBERs (P3-dnEBNA1-EBER) were maintained in complete medium supplemented with G418, hygromycin B (300 μg/ml), and doxycycline.
Plasmids.
dnEBNA1, which encodes amino acids (aa) 379 to 386 fused in frame to aa 451 to 641 of EBNA1 (Fig. 1A) (25, 37), was generated by PCR using EBNA1 cDNA of EBV strain B95-8 as a template and 5′-GAATTCGGATCCATCATGAAGAGGCCCAGGAGTCCCAGTAGTCGGGGTCAGGGTGATGGAGG-3′ and 5′-GAATTCGGATCCTCACTCCTGCCCTTCCTCAC-3′ as primers. The PCR product was digested with BamHI and cloned into the BamHI site of pSG5 (Stratagene) to make pSG5-dnEBNA1. The simian virus 40 (SV40) promoter-driven neomycin resistance gene derived from pcDNA3 (Invitrogen) was cloned into XbaI- and XhoI-digested pBluescript II KS(+) (Stratagene), referred to as pBS-Neo. The tetracycline-responsive gene expression cassette derived from pUHD10-3 (11) was inserted into XhoI- and HindIII-digested pBS-Neo, referred to as pBSN-tet. The dnEBNA1 gene derived from pSG5-dnEBNA1 was inserted into the BamHI site of pBSN-tet to make pBSN-tet-dnEBNA1. The tetracycline-regulatable transactivator (tTA) derived from pUHD15-1 (11) was inserted into the NheI site of pBSN-tet-dnEBNA1, and the resulting vector was named pBSN-tetOff-dnEBNA1. The human Bcl-2α cDNA derived from pBcl-2 (29) was cloned into the EcoRI site of pSG5 to make pSG5-Bcl2, and the hygromycin resistance gene derived from pSilencer 2.1-U6 hygro (Ambion) was inserted into the XbaI site of pSG5-Bcl2 to make pSG5-Bcl2-hyg. The BHRF1 gene was PCR amplified using P3HR-1 genomic DNA as a template and 5′-TTAAGAATTCAAGATGGCCTATTCAACAAG-3′ and 5′-AATTGAATTCTTAGTGTCTTCCTCTGGAGA-3′ as primers. The PCR product was digested with EcoRI and cloned into the EcoRI site of pSG5. The hygromycin resistance gene was inserted into the XbaI site of the vector to make pSG5-BHRF1-hyg. pSG5-flagEBNA3A and pSG5-flagEBNA3C have been described previously (35). The hygromycin resistance gene was inserted into the XbaI site of pSG5-flagEBNA3A or pSG5-flagEBNA3C, referred to as pSG5-E3A-hyg or pSG5-E3C-hyg, respectively. The SspI-XbaI fragment derived from pEKS10 (27) was cloned into HincII- and XbaI-digested pBluescript II KS(+) to make pBS-EKS10, and the hygromycin resistance gene was inserted into the XbaI site of pBS-EKS10 to make pBS-EKS10-hyg. EBNA-LP cDNA with four W1W2 repeats and the unique domain encoded by Y1Y2, derived from B95-8 virus-transformed IB4 LCL, was cloned into EcoRI- and BamHI-digested pBluescript II KS(+) to make pBS-EBNA-LP. The EBNA-LP stop codon mutant encoding the form of the protein with Y1Y2 deleted, referred to as LPd45 (12), was generated by inserting the sequence 5′-GTCTAGACTAG-3′ into SfiI- and BamHI-digested pBS-EBNA-LP by linker ligation. The LPd45 cDNA fragment was then excised and cloned into EcoRI- and BamHI-digested pSG5, followed by insertion of the hygromycin resistance gene to make pSG5-LPd45-hyg. To make an oriP plasmid expressing short hairpin RNA (shRNA) and enhanced green fluorescent protein (EGFP), a human U6 RNA pol III promoter-driven shRNA targeting BHRF1 (or control shRNA) and an SV40 promoter-driven EGFP gene cassette were subcloned into an XbaI-NruI-digested pCEP4 vector (Invitrogen). The shRNA-targeted sequences were as follows: BHRF1 (5′-GCGTTATCATGTGTTGCTTGA-3′) and firefly luciferase (control) (5′-GGATTTCAGTCGATGTACACGTTCGTC-3′).
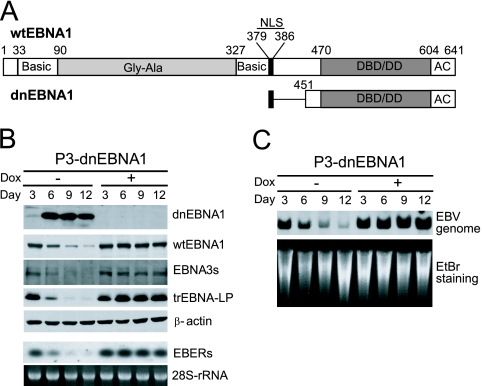
FIG. 1.
Enforced expression of dnEBNA1 in P3HR-1 cells results in loss of the EBV genome and viral gene products. (A) Schematic representations of wt EBNA1 and dnEBNA1. The basic amino acid-rich chromosome association domains (Basic), the Gly-Gly-Ala repeats (Gly-Ala), the nuclear localization sequence (NLS), the DNA-binding and dimerization domain (DBD/DD), and the acidic domain (AC) are shown. The numbers indicate the positions of amino acid residues. (B) P3-dnEBNA1 cells were cultured in the absence (−) or presence (+) of Dox. Every 3 days, the cultures were split. Total cell lysates were prepared at the indicated time points and subjected to Western blot analysis with EBV-immune human sera (reactive to dnEBNA1, wt EBNA1, or EBNA3s), anti-EBNA-LP (reactive to truncated [tr] EBNA-LP), or anti-β-actin antibody (β-actin). Total RNAs were prepared at the indicated time points and subjected to Northern blot analysis to detect EBERs. Ethidium bromide-stained 28S rRNA as a loading control is shown below. (C) P3-dnEBNA1 cells were cultured in the absence (−) or presence (+) of Dox. Every 3 days, the cultures were split. Genomic DNAs were prepared at the indicated time points, digested with EcoRI, and subjected to Southern blot analysis to detect the EBV genome. Ethidium bromide-stained DNA as a loading control is shown below (EtBr staining).
Establishment of stable cell lines.
P3-dnEBNA1 cells and Ak(−)-dnEBNA1 cells were made by electroporating P3HR-1 cells and EBV-negative Akata cells with linearized pBSN-tetOff-dnEBNA1, followed by selection with 700 μg/ml of G418 in the presence of doxycycline (100 ng/ml). Clones that expressed high levels of dnEBNA1 in response to doxycycline withdrawal and little or no detectable dnEBNA1 in the presence of doxycycline were chosen by immunoblotting. P3-dnEBNA1 cell lines that stably expressed the transfected, truncated EBNA-LP (P3-dnEBNA1-LPd45), EBNA3A (P3-dnEBNA1-E3A), EBNA3C (P3-dnEBNA1-E3C), EBERs (P3-dnEBNA1-EBER), Bcl-2 (P3-dnEBNA1-Bcl2), or BHRF1 (P3-dnEBNA1-BHRF1) were made by electroporating P3-dnEBNA1 cells with a linearized pSG5-LPd45-hyg, pSG5-E3A-hyg, pSG5-E3C-hyg, pBS-EKS10-hyg, pSG5-Bcl2-hyg, or pSG5-BHRF1-hyg, respectively, followed by selection with G418 (400 μg/ml) and hygromycin B (300 μg/ml) in the presence of doxycycline (100 ng/ml).
Cell growth.
Cells (1 × 106) were extensively washed with medium without doxycycline and cultured in 25-cm2 culture flasks in 10 ml of complete medium (1 × 105 cells per milliliter) with or without doxycycline. Every 3 days, the viable-cell number was determined with a hemocytometer based on trypan blue exclusion, and the cultures were diluted to 1 × 105 cells per milliliter by adding similar amounts of fresh medium to maintain optimum growth. Total viable-cell numbers were calculated based on the expansion from the initial 1 × 106 cells.
Cell cycle analysis.
Approximately 106 cells were fixed and stained with phosphate-buffered saline (PBS) containing propidium iodide and RNase A. The cells were analyzed by using a FACSCalibur and Cell Quest software (BD Bioscience). Apoptotic cells were measured by the appearance of cells with sub-G1 DNA content (a DNA content of less than 2N).
Western blotting.
Whole-cell lysates were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and the proteins were blotted onto nitrocellulose membranes. The membranes were incubated with EBV-immune human sera (reactive to EBNAs and dnEBNA1), anti-EBNA-LP (4D3) (43), anti-BHRF1 (5B11; Millipore), anti-LMP1 (S12), anti-LMP2A (14B7; Santa Cruz), anti-Bcl-2 (clone 7; BD), anti-PARP (4C10-5; BD), anti-caspase 9 (5B4; MBL), or anti-β-actin (AC-15; Sigma) antibody. The membranes were reacted with horseradish peroxidase-conjugated species-specific secondary antibodies and developed with enhanced-chemiluminescence (ECL) detection reagents (GE Healthcare).
Southern blotting.
Genomic DNA was extracted and digested with EcoRI, separated by electrophoresis in a 0.8% agarose gel, and transferred to a nylon membrane. The EcoRI K fragment of Akata EBV (corresponding to the EcoRI J fragment of B95-8 EBV) was used as a probe. Probe labeling was carried out using the AlkPhos direct-labeling kit, and signals were detected with the CDP-Star detection reagent (GE Healthcare).
Northern blotting.
Total RNA was isolated and electrophoresed on a 1% agarose gel containing 0.66 M formaldehyde and then transferred to a nylon membrane. The EcoRI K fragment of Akata EBV was used as a probe to detect EBERs. Probe labeling and signal detection were carried out with the AlkPhos direct-labeling kit and the CDP-Star detection reagent.
Immunofluorescence.
Cells were smeared on glass slides and fixed. Indirect immunofluorescence was performed with C30-1 (specific to gp110) and C844-1 (specific to BHRF1) (18) mouse monoclonal antibodies, anti-BZLF1 rabbit polyclonal antibody, or EBV-immune human sera as primary antibodies. A Cy3-conjugated anti-mouse IgG, Cy3-conjugated anti-rabbit IgG, or fluorescein isothiocyanate (FITC)-conjugated anti-C3C antibody was used as a secondary antibody. The slides were counterstained with 4′,6-diamidino-2-phenylindole (DAPI).
Knockdown experiment.
P3HR-1 cells were transfected with oriP plasmids expressing shRNA and EGFP by using an electroporator (CUY-21; NEPA Gene). Square electric pulses were applied at 275 V (pulse length, 0.5 ms; 1 pulse; interval, 50 ms), followed by 20 V (pulse length, 50 ms; 10 pulses; interval, 50 ms). Four days after transfection, the cells were harvested and stained with annexin V-phycoerythrin (PE) (MBL) according to the manufacturer's instructions and were analyzed by using FACSCalibur and Cell Quest software. Sorting of EGFP-positive cells was performed on a FACS Vantage cell sorter (BD).
RESULTS
Enforced expression of dnEBNA1 in P3HR-1 cells results in loss of the EBV genome and viral gene products.
dnEBNA1, which contains the nuclear localization sequence, the DNA-binding and dimerization domain, and the acidic domain of EBNA1 (Fig. 1A), has been shown to inhibit the function of EBNA1 (16, 17, 23, 25, 32, 37). We established P3-dnEBNA1, a cell line derived from a Wp-restricted P3HR-1 BL cell line that conditionally expresses dnEBNA1 in a tetracycline-regulated manner. Western blot analysis revealed that dnEBNA1 was not expressed when P3-dnEBNA1 cells were cultured in the presence of Dox, whereas dnEBNA1 expression was strongly induced after the withdrawal of Dox from the medium (Fig. 1B). Next, we examined the level of the EBV genome in P3-dnEBNA1 cells by Southern blot analysis. The level of the EBV genome did not change in the presence of Dox but progressively decreased in the absence of Dox, indicating that the EBV genome was lost from the cells after dnEBNA1 induction (Fig. 1C). Consistent with the loss of the EBV genome, the expression levels of viral gene products, such as wild-type (wt) EBNA1, EBNA3s, and truncated EBNA-LP proteins, and EBERs also decreased in the absence of Dox (Fig. 1B). Therefore, as expected, these results indicate that enforced dnEBNA1 expression caused the loss of the EBV genome from P3HR-1 cells, as well as the loss of viral gene products.
Loss of the EBV genome from P3HR-1 cells coincides with the suppression of cell growth and the induction of apoptosis.
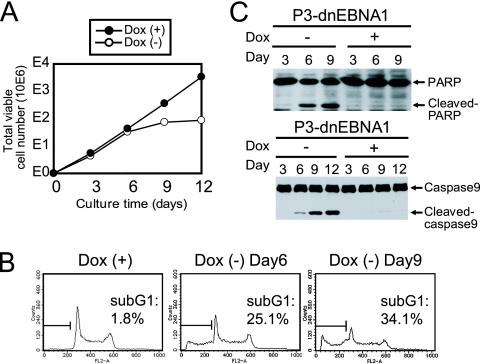
We evaluated the contribution dnEBNA1 expression makes to the growth and survival of P3HR-1 cells. The number of viable P3-dnEBNA1 cells exponentially increased in the presence of Dox (Fig. 2A). However, in the absence of Dox, viable-cell numbers dropped dramatically compared to those seen in the presence of Dox (Fig. 2A). To examine whether apoptosis occurs in P3HR-1 cells after dnEBNA1 induction, cell cycle analyses were performed (Fig. 2B). The percentage of the sub-G1 fraction was only 1.8% in P3-dnEBNA1 cells in the presence of Dox but was 25.1% and 34.1% 6 and 9 days, respectively, after the withdrawal of Dox. Moreover, Western blot analysis revealed an increase in cleaved PARP and cleaved caspase 9 in the absence of Dox (Fig. 2C). Therefore, these data indicate that enforced expression of dnEBNA1 results in apoptosis and reduced P3HR-1 cell growth.
FIG. 2.
Enforced expression of dnEBNA1 in P3HR-1 cells results in the suppression of cell growth and the induction of apoptosis. (A) P3-dnEBNA1 cells were cultured in the absence (−) or presence (+) of Dox. Every 3 days, viable cells were counted, and the cells were split as described in Materials and Methods. The total numbers of viable cells derived from the initial cultures were calculated and plotted at each time point. The number “10E6” along the y axis indicates that the cell number plotted should be multiplied by 1,000,000. (B) P3-dnEBNA1 cells were cultured in the absence (−) or presence (+) of Dox for the indicated number of days. The cells were fixed and stained with propidium iodide, and cell cycle analysis was performed with a FACSCalibur. The percentages of apoptotic cells were determined and are shown (sub-G1). (C) P3-dnEBNA1 cells were cultured in the absence (−) or presence (+) of Dox. Total cell lysates were prepared at the indicated time points and subjected to Western blot analysis with anti-PARP (top) or anti-caspase 9 (bottom) antibody.
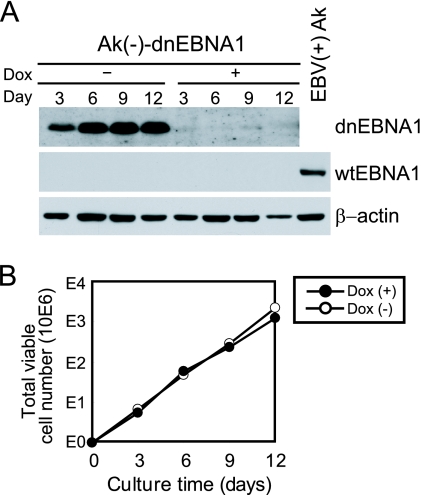
To investigate the possibility that dnEBNA1 nonspecifically attenuates cell growth, we established Ak(−)-dnEBNA1, a cell line derived from an EBV-negative subline of Akata BL cells that conditionally expresses dnEBNA1 in a tetracycline-regulated manner. Similar to P3-dnEBNA1 cells, Ak(−)-dnEBNA1 cells expressed dnEBNA1 in the absence of Dox, but not in its presence (Fig. 3A). However, the numbers of viable Ak(−)-dnEBNA1 cells increased similarly in the absence and presence of Dox (Fig. 3B). Namely, the negative effect of dnEBNA1 on cell growth was not observed in EBV-negative cells, indicating that dnEBNA1 is not nonspecifically toxic.
FIG. 3.
Enforced expression of dnEBNA1 does not affect the growth of EBV-negative Akata cells. (A) Ak(−)-dnEBNA1 cells were cultured in the absence (−) or presence (+) of Dox. Total cell lysates were prepared and subjected to Western blot analysis with EBV-immune human serum (reactive to dnEBNA1 and wt EBNA1) and anti-β-actin antibody. EBV-positive Akata cells served as a positive control for wt EBNA1 detection [EBV(+)Ak]. (B) Ak(−)-dnEBNA1 cells were cultured in the absence (−) or presence (+) of Dox. Viable cells were counted every 3 days, and the cells were split. The total numbers of viable cells derived from the initial cultures were calculated and plotted.
Stable overexpression of Bcl-2 rescues P3HR-1 cells from dnEBNA1-induced cell growth inhibition and apoptotic cell death.
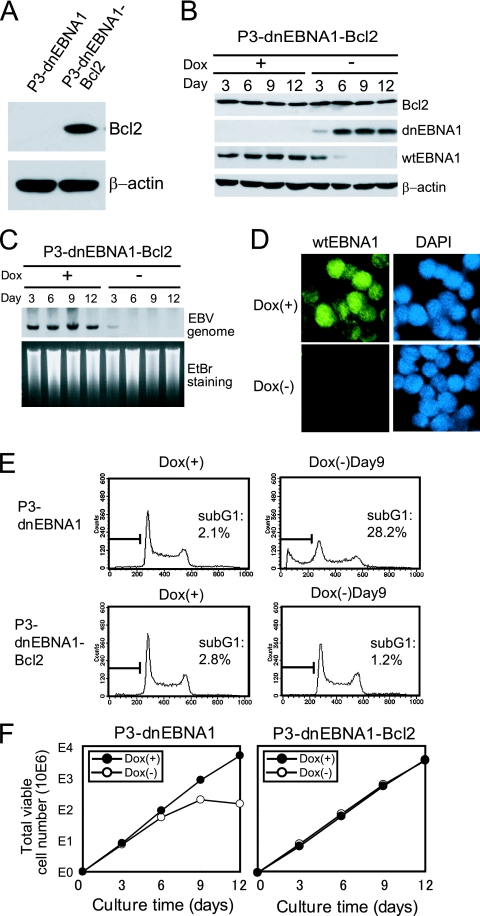
We next asked whether apoptosis is the main cause of the loss of P3HR-1 cell growth under dnEBNA1-induced conditions. We established a P3-dnEBNA1-Bcl2 cell line by stably transfecting an antiapoptotic Bcl-2 gene into P3-dnEBNA1 cells. As expected, P3-dnEBNA1-Bcl2 cells, but not P3-dnEBNA1 cells, had an increased level of Bcl-2 protein (Fig. 4A). When P3-dnEBNA1-Bcl2 cells were cultured in medium without Dox, dnEBNA1 expression was induced and the EBV genome was lost (Fig. 4B and C), as observed with parental P3-dnEBNA1 cells (Fig. 1B and C). Western blot and immunofluorescence analyses also revealed the disappearance of the wt EBNA1 protein after dnEBNA1 induction (Fig. 4B and D). In contrast to parental P3-dnEBNA1 cells, the sub-G1 fraction of P3-dnEBNA1-Bcl2 cells did not increase after dnEBNA1 induction, showing that Bcl-2 overexpression blocked dnEBNA1-induced apoptosis, as expected (Fig. 4E). Importantly, the growth rate of P3-dnEBNA1-Bcl2 cells in the absence of Dox was almost the same as in its presence (Fig. 4F). These data demonstrate that stable overexpression of Bcl-2 almost completely protected P3HR-1 cells, not only from dnEBNA1-induced apoptosis, but also from growth inhibition. Hence, apoptosis is likely to be the main cause of the loss of P3HR-1 cell growth under dnEBNA1-induced conditions.
FIG. 4.
Stable overexpression of Bcl-2 rescues P3HR-1 cells from dnEBNA1-induced growth inhibition and apoptosis. (A) P3-dnEBNA1 and P3-dnEBNA1-Bcl2 cells were subjected to Western blot analysis with anti-Bcl-2 and anti-β-actin antibodies. (B) P3-dnEBNA1-Bcl2 cells cultured in the absence (−) or presence (+) of Dox were subjected to Western blot analysis with EBV-immune human serum (reactive to dnEBNA1 and wt EBNA1), anti-Bcl-2, and anti-β-actin antibodies. (C) P3-dnEBNA1-Bcl2 cells cultured in the absence (−) or presence (+) of Dox were subjected to Southern blot analysis to detect the EBV genome. (D) P3-dnEBNA1-Bcl2 cells were cultured in the absence (−) or presence (+) of Dox for 12 days. The cells were processed for immunofluorescence analyses with EBV-immune human serum (reactive to wt EBNA1, but not to dnEBNA1) as a primary antibody and FITC-conjugated anti-C3C antibody as a secondary antibody. (E) P3-dnEBNA1 cells and P3-dnEBNA1-Bcl2 cells were cultured in the absence (−) or presence (+) of Dox for 9 days. Cell cycle analysis was performed, and the percentages of cells in the sub-G1 fraction were determined. (F) P3-dnEBNA1 and P3-dnEBNA1-Bcl2 cells were cultured in the absence (−) or presence (+) of Dox. Viable cells were counted every 3 days, and the cells were split. The total numbers of viable cells derived from the initial cultures were calculated and plotted.
Stable transfection with EBERs, truncated EBNA-LP, EBNA3A, or EBNA3C does not protect P3HR-1 cells from dnEBNA1-induced growth inhibition.
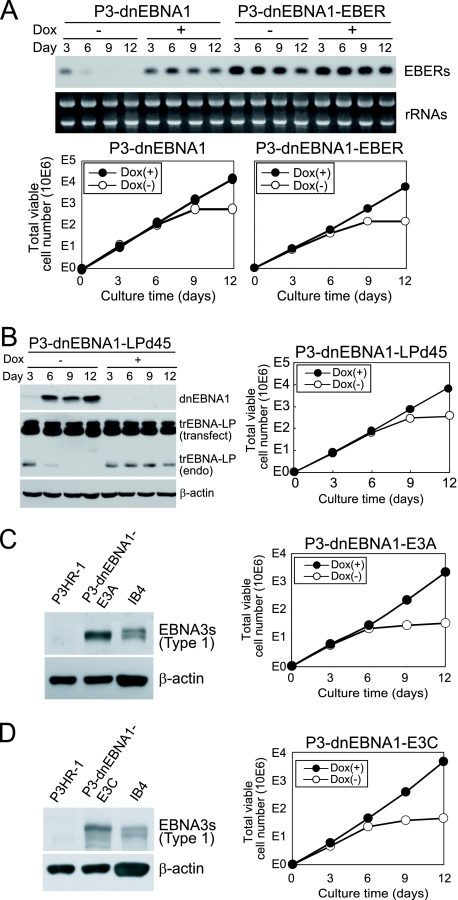
The correlation between the loss of the EBV genome and the growth inhibition of P3HR-1 cells suggests that one or more viral gene products are responsible for the survival of P3HR-1 cells. In P3HR-1 cells, EBV expresses viral genes, including nuclear proteins (EBNA1, EBNA3s, and truncated EBNA-LP) and EBERs (Fig. 1B). We transfected a vector expressing EBERs, truncated EBNA-LP, EBNA3A, or EBNA3C into P3-dnEBNA1 cells and established stable cell lines expressing the respective EBV gene products. A P3-dnEBNA1 cell line stably transfected with the EBER gene (P3-dnEBNA1-EBER) clearly expressed more EBERs than the parental cells in the presence of Dox (Fig. 5A). In the absence of Dox, EBER expression in P3-dnEBNA1-EBER cells decreased but was still present on day 12, indicating that EBER expression provided by the transfected vector remained after dnEBNA1 induction. However, the growth of P3-dnEBNA1-EBER cells was clearly more inhibited by the absence of Dox than by its presence (Fig. 5A). We also confirmed that P3-dnEBNA1 cell lines stably transfected with a truncated EBNA-LP (P3-dnEBNA1-LPd45), EBNA3A (P3-dnEBNA1-E3A), or EBNA3C (P3-dnEBNA1-E3C) gene expressed the transfected genes (Fig. 5B, C, and D). The growth of these stable cell lines was also inhibited when they were cultured in media without Dox (Fig. 5B, C, and D). These results demonstrate that exogenous expression of EBERs, truncated EBNA-LP, EBNA3A, or EBNA3C alone does not protect P3HR-1 cells from dnEBNA1-induced growth inhibition. The expression of LMP1 or LMP2A was barely detectable or marginal in P3HR-1 cells by Western blot analysis (data not shown), which is consistent with the previous report that LMP1 and LMP2A proteins were undetectable in Wp-restricted BL lines (19).
FIG. 5.
Stable transfection with EBERs, truncated EBNA-LP, EBNA3A, or EBNA3C does not protect P3HR-1 cells from dnEBNA1-induced growth inhibition. (A) P3-dnEBNA1 and P3-dnEBNA1-EBER cells cultured in the absence (−) and presence (+) of Dox were subjected to Northern blot analysis to detect EBERs. An ethidium bromide-stained rRNA loading control is also shown. Viable cells were counted, and the total numbers of viable cells derived from the initial cultures were calculated and plotted. (B) P3-dnEBNA1-LPd45 cells were subjected to Western blot analysis with EBV-immune human serum (reactive to dnEBNA1), anti-EBNA-LP, and anti-β-actin antibodies (left). The bands representing transfected truncated (tr) EBNA-LP (transfect) and endogenous trEBNA-LP (endo) are shown. Viable cells were counted, and the total numbers of viable cells derived from the initial cultures were calculated and plotted (right). (C and D) P3HR-1, P3-dnEBNA1-E3A, and P3-dnEBNA1-E3C cells were subjected to Western blot analysis with human serum (reactive to type 1 EBNA3s, but not to type 2 EBNA3s) and anti-β-actin antibody. Note that transfected EBNA3s are type 1 and P3HR-1s endogenous EBNA3s are type 2. An IB4 lysate was used as a positive control for type 1 EBNA3A and type 1 EBNA3C (left). Viable cells were counted, and the total numbers of viable cells derived from the initial cultures were calculated and plotted (right).
BHRF1 is constitutively expressed in P3HR-1 cells as a latent protein.
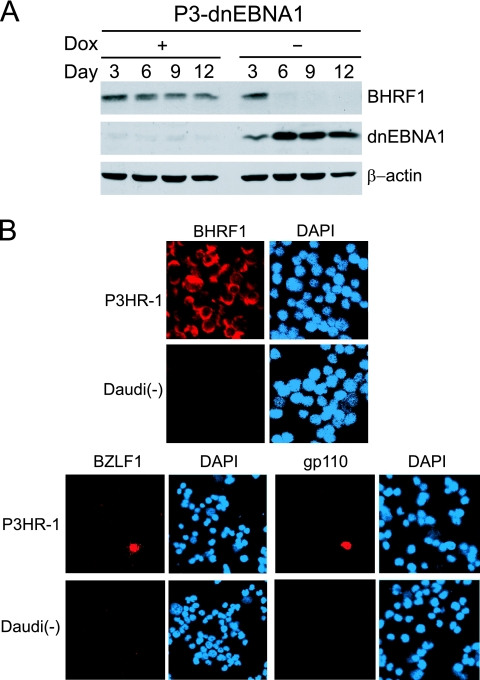
BHRF1, an EBV-encoded Bcl-2 homologue, has been classified as an early lytic gene (38). In addition to its expression in lytic infection, it has recently been reported that BHRF1 is also expressed as a latent gene in Wp-restricted BLs (21), which prompted us to examine BHRF1 expression in P3HR-1 cells. Western blot analysis revealed that P3-dnEBNA1 cells clearly expressed BHRF1 in the presence of Dox, and the expression disappeared after inducing dnEBNA1 by Dox withdrawal (Fig. 6A). To determine whether BHRF1 is expressed as a latent protein in P3HR-1 cells, we performed immunofluorescence analyses of BHRF1 expression, as well as the expression of the immediate-early protein BZLF1 and the late protein gp110. Less than 1% of P3HR-1 cells were positive for BZLF1 or gp110, indicating that spontaneous lytic replication had occurred only in a small population of P3HR-1 cells (Fig. 6B). In contrast, almost all P3HR-1 cells were positive for BHRF1 expression (Fig. 6B). These results signify that BHRF1 is constitutively expressed in latently infected P3HR-1 cells and that dnEBNA1 expression results in the loss of BHRF1 expression in these cells.
FIG. 6.
Expression of BHRF1 as a latent protein in P3HR-1 cells. (A) P3-dnEBNA1 cells cultured in the absence (−) or presence (+) of Dox were subjected to Western blot analysis with EBV-immune human serum (reactive to dnEBNA1), anti-BHRF1, and anti-β-actin antibodies. (B) P3HR-1 cells and EBV-negative Daudi cells [Daudi(−)] were processed for immunofluorescence analyses with mouse anti-BHRF1, rabbit anti-BZLF1, or mouse anti-gp110 antibody as a primary antibody and Cy3-conjugated anti-mouse IgG or Cy3-conjugated anti-rabbit IgG as a secondary antibody.
Stable transfection with the BHRF1 gene rescues P3HR-1 cells from dnEBNA1-induced apoptosis and cell growth inhibition.
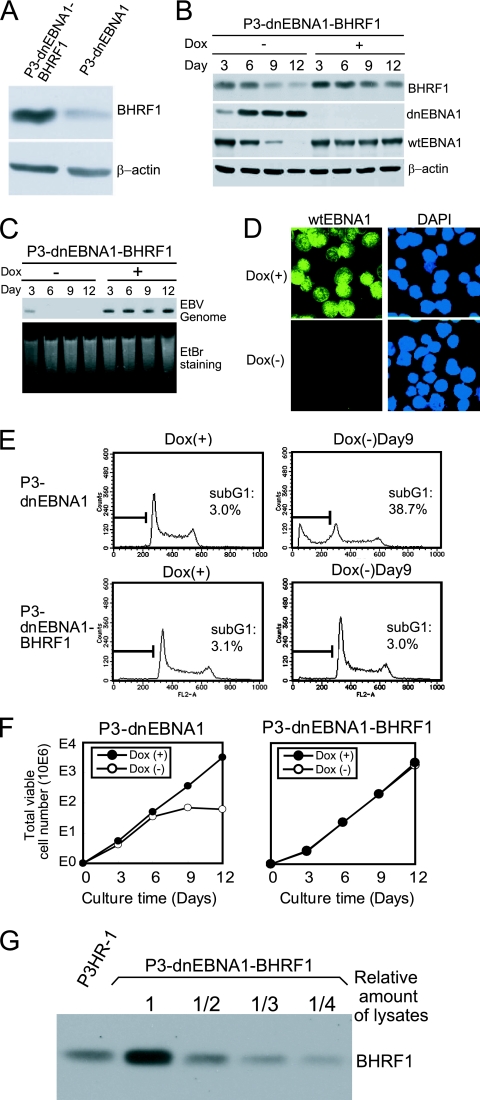
Next, we transfected a BHRF1 expression vector into P3-dnEBNA1 cells and established a stable cell line, P3-dnEBNA1-BHRF1. P3-dnEBNA1-BHRF1 cells expressed more BHRF1 than the parental P3-dnEBNA1 cells in the presence of Dox (Fig. 7A). When P3-dnEBNA1-BHRF1 cells were cultured in medium without Dox, dnEBNA1 expression was induced, the EBV genome was lost, and the wt EBNA1 protein disappeared (Fig. 7B, C, and D). BHRF1 expression in P3-dnEBNA1-BHRF1 cells also decreased in the absence of Dox but was still present on day 12 (Fig. 7B). This result indicates that BHRF1 expression provided by the transfected vector remained after loss of the EBV genome. Notably, in contrast to P3-dnEBNA1 cells, neither apoptosis nor cell growth inhibition was observed when P3-dnEBNA1-BHRF1 cells were cultured in the absence of Dox (Fig. 7E and F). These results demonstrate that stable transfection of the BHRF1 gene alone can rescue P3HR-1 cells from dnEBNA1-induced apoptosis and growth inhibition. We compared the level of BHRF1 expression in P3-dnEBNA1-BHRF1 cells to that in P3HR-1 cells. Western blot analysis using serially diluted cell lysates revealed that the level of BHRF1 expression in P3-dnEBNA1-BHRF1 cells in the presence of Dox was almost twice that in P3HR-1 cells (Fig. 7G). This means that the level of BHRF1 expression provided by the transfected vector was almost equivalent to that of endogenous BHRF1 expression in P3HR-1 cells. Accordingly, these data indicate that the expression level of BHRF1 in P3HR-1 cells is enough to prevent P3HR-1 cells from undergoing apoptosis following eradication of the EBV genome.
FIG. 7.
Stable transfection of the BHRF1 gene protects P3HR-1 cells from dnEBNA1-induced apoptosis and growth inhibition. (A) P3-dnEBNA1 and P3-dnEBNA1-BHRF1 cells cultured in medium with Dox were subjected to Western blot analysis with anti-BHRF1 and anti-β-actin antibodies. (B) P3-dnEBNA1-BHRF1 cells cultured in the absence (−) or presence (+) of Dox were subjected to Western blot analysis with EBV-immune human serum (reactive to dnEBNA1 and wt EBNA1), anti-BHRF1, and anti-β-actin antibodies. (C) P3-dnEBNA1-BHRF1 cells cultured in the absence (−) or presence (+) of Dox were subjected to Southern blot analysis to detect the EBV genome. (D) P3-dnEBNA1-BHRF1 cells were cultured in the absence (−) or presence (+) of Dox for 12 days. The cells were processed for immunofluorescence analyses with EBV-immune human serum (reactive to wt EBNA1, but not to dnEBNA1) as a primary antibody and FITC-conjugated anti-C3C antibody as a secondary antibody. (E) P3-dnEBNA1 and P3-dnEBNA1-BHRF1 cells were cultured in the absence (−) or presence (+) of Dox for 9 days. Cell cycle analysis was performed, and the percentages of cells in the sub-G1 fraction were determined. (F) P3-dnEBNA1 and P3-dnEBNA1-BHRF1 cells were cultured in the absence (−) or presence (+) of Dox. Viable cells were counted every 3 days, and the cells were split. The total numbers of viable cells derived from the initial cultures were calculated and plotted. (G) Cell lysates of P3HR-1 and serially diluted cell lysates of P3-dnEBNA1-BHRF1 were subjected to Western blot analysis with anti-BHRF1 antibody.
Knockdown of BHRF1 expression in P3HR-1 cells results in increased cell death.
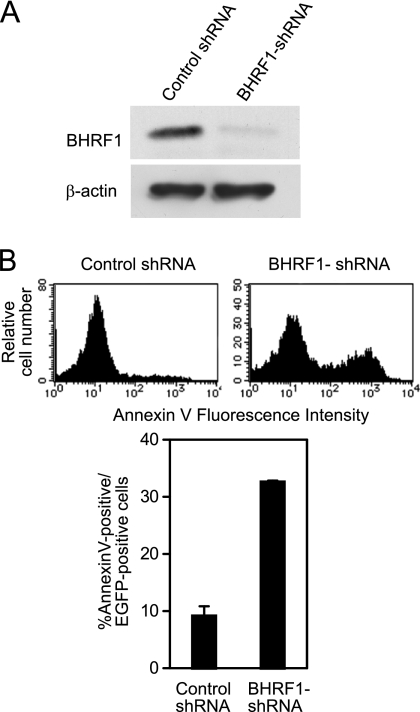
To examine whether knockdown of BHRF1 hampers P3HR-1 cell survival, we transfected an oriP plasmid expressing shRNA against BHRF1 (BHRF1-shRNA) and EGFP as a transfection marker into P3HR-1 cells. Western blot analysis revealed that BHRF1-shRNA could significantly knock down BHRF1 expression in P3HR-1 cells (Fig. 8A). The percentage of cells with positive annexin V staining among BHRF1 shRNA-transfected cells was clearly higher than that among control shRNA-transfected cells (Fig. 8B). The data indicate that knockdown of BHRF1 causes an increase in cell death, further supporting the importance of BHRF1 in P3HR-1 cell survival.
FIG. 8.
Knockdown of BHRF1 expression in P3HR-1 cells results in increased cell death. (A) P3HR-1 cells were transfected with oriP plasmid expressing BHRF1-shRNA (or control shRNA) and EGFP. Three days after transfection, EGFP-positive cells were sorted and subjected to Western blot analysis with anti-BHRF1 and anti-β-actin antibodies. (B) P3HR-1 cells transfected with BHRF1-shRNA (or control shRNA) and EGFP were stained with annexin V-PE 4 days after transfection. The histograms represent annexin V staining after gating on EGFP-positive cells (top). The percentages of annexin V-positive cells among EGFP-positive cells are shown (bottom). Each bar represents the average of triplicate transfections, with the error bar representing the standard error of the mean.
DISCUSSION
The present study was performed to clarify the role of EBV in the growth and survival of Wp-restricted BL cells. Our data show that dnEBNA1 expression in Wp-restricted P3HR-1 BL cells resulted in the loss of the EBV genome, which was accompanied by cell growth inhibition, as evidenced by a drop in the cell number increase and by an increase in apoptosis in the absence of exogenous apoptosis inducers. Moreover, dnEBNA1-induced cell growth inhibition was almost completely counteracted by exogenous overexpression of the antiapoptotic protein Bcl-2. Thus, rather than promoting cell cycle progression in P3HR-1 cells, EBV appears to play an essential role in inhibiting apoptosis.
We demonstrated that dnEBNA1-induced apoptosis of P3HR-1 cells was almost completely inhibited by the exogenous expression of BHRF1. The expression level of BHRF1 observed in parental P3HR-1 cells was enough to prevent P3HR-1 cells from undergoing apoptosis. BHRF1-transfected P3HR-1 cells could survive and grow without the EBV genome. Moreover, knockdown of BHRF1 expression in P3HR-1 cells resulted in increased cell death. These results indicate that dnEBNA1-induced apoptosis in P3HR-1 cells was caused by a loss of BHRF1 expression following the elimination of the EBV genome. BHRF1 has a high degree of homology to Bcl-2 and inhibits apoptosis induced by various stimuli (8, 13, 21). Austin et al. first demonstrated that BHRF1 could be expressed as a latent gene (3). Kelly et al. have recently reported the existence of Wp promoter-driven BHRF1 transcripts in Wp-restricted BLs, indicating that BHRF1 is expressed as a latent gene in these BLs (21). Our immunofluorescence data presented here clearly reveal that BHRF1 protein is expressed in latently infected P3HR-1 cells, which provides further evidence for BHRF1 expression as a latent gene in Wp-restricted BL cells. It has also been demonstrated that stable BHRF1 transfection protects latency I BL cells from ionomycin-induced apoptosis (21). However, the role of BHRF1 in BL cells in the absence of exogenous apoptotic triggers has never been examined. Thus, our present study is the first to show that BHRF1 functions as a survival factor in Wp-restricted BL cells under normal culture conditions and in the absence of exogenous apoptotic stimuli.
In contrast to BHRF1, we could not find any role for EBNA3A, EBNA3C, truncated EBNA-LP, or EBERs in the survival of P3HR-1 cells when the cells were cultured in the absence of exogenous apoptosis inducers. Exogenous expression of EBNA3A, EBNA3C, truncated EBNA-LP, or EBERs did not protect P3HR-1 cells from dnEBNA1-induced growth inhibition. This was somewhat surprising, as EBNA3A, EBNA3C, EBNA-LP, and EBERs play important roles in B-cell growth transformation and/or in LCL growth (33, 34, 35, 46, 47, 48). As EBNA3A and EBNA3C, truncated EBNA-LP, and EBERs have been reported to repress apoptosis induced by exogenous apoptotic triggers (1, 10, 27, 36), these viral genes may have roles to play under more severe conditions where various exogenous apoptotic stimuli may threaten the survival of BL cells.
The apoptotic pathway seems to be intrinsically activated in P3HR-1 cells, because the cells spontaneously undergo apoptosis in the absence of EBV. BL tumors carry a reciprocal chromosomal translocation between the c-myc gene and one of the Ig loci (26). The c-myc/Ig translocation results in constitutive and deregulated expression of the c-myc. Constitutive c-myc expression stimulates cell proliferation and increases susceptibility to apoptosis (2, 7). Accordingly, c-myc activation could be a driving force behind dnEBNA1-induced apoptosis in P3HR-1 cells. Our present data demonstrate that stable transfection with the cellular Bcl-2 or the viral Bcl-2 homologue makes the EBV genome dispensable in P3HR-1 cells, suggesting that the function of EBV can replace the functions of these antiapoptotic proteins. It has been reported that Bcl-2 and BHRF1 can inhibit apoptotic cell death induced by c-myc (4, 8, 9). Moreover, it is well known that bcl-2 collaborates with c-myc to accelerate lymphomagenesis in transgenic mice (45). Therefore, it seems possible that BHRF1 collaborates with c-myc in the development of Wp-restricted BLs.
The data presented here reinforce the idea that EBV contributes to BL pathogenesis by counteracting the proapoptotic effects that are probably induced by the c-myc/Ig translocation. Furthermore, our work indicates the critical requirement for BHRF1 in the maintenance of Wp-restricted BL cells. Therefore, removal of the EBV genome from cells by inhibiting EBNA1 function could be an effective and attractive therapeutic approach for the treatment of Wp-restricted BL tumors.
Acknowledgments
We thank Y. Kawaguchi for providing anti-EBNA-LP monoclonal antibody and K. Yamaguchi for cell sorting. We also thank T. Kanda, D. Iwakiri, M. Noguchi, and S. Imai for helpful suggestions.
This work was supported by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology, Japan (S.M. and K.T.).
Footnotes
Published ahead of print on 30 December 2009.
REFERENCES
- 1.Anderton, E., J. Yee, P. Smith, T. Crook, R. E. White, and M. J. Allday. 2008. Two Epstein-Barr virus (EBV) oncoproteins cooperate to repress expression of the proapoptotic tumour-suppressor Bim: clues to the pathogenesis of Burkitt's lymphoma. Oncogene 27:421-433. [DOI] [PubMed] [Google Scholar]
- 2.Askew, D. S., R. A. Ashmun, B. C. Simmons, and J. L. Cleveland. 1991. Constitutive c-myc expression in an IL-3-dependent myeloid cell line suppresses cell cycle arrest and accelerates apoptosis. Oncogene 6:1915-1922. [PubMed] [Google Scholar]
- 3.Austin, P. J., E. Flemington, C. N. Yandava, J. L. Strominger, and S. H. Speck. 1988. Complex transcription of the Epstein-Barr virus BamHI fragment H rightward open reading frame 1 (BHRF1) in latently and lytically infected B lymphocytes. Proc. Natl. Acad. Sci. U. S. A. 85:3678-3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bissonnette, R. P., F. Echeverri, A. Mahboubi, and D. R. Green. 1992. Apoptotic cell death induced by c-myc is inhibited by bcl-2. Nature 359:552-554. [DOI] [PubMed] [Google Scholar]
- 5.Bornkamm, G. W., J. Hudewentz, U. K. Freese, and U. Zimber. 1982. Deletion of the nontransforming Epstein-Barr virus strain P3HR-1 causes fusion of the large internal repeat to the DSL region. J. Virol. 43:952-968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dang, C. V., K. A. O'Donnell, K. I. Zeller, T. Nguyen, R. C. Osthus, and F. Li. 2006. The c-Myc target gene network. Semin. Cancer Biol. 16:253-264. [DOI] [PubMed] [Google Scholar]
- 7.Evan, G. I., A. H. Wyllie, C. S. Gilbert, T. D. Littlewood, H. Land, M. Brooks, C. M. Waters, L. Z. Penn, and D. C. Hancock. 1992. Induction of apoptosis in fibroblasts by c-myc protein. Cell 69:119-128. [DOI] [PubMed] [Google Scholar]
- 8.Fanidi, A., D. C. Hancock, and T. D. Littlewood. 1998. Suppression of c-Myc-induced apoptosis by the Epstein-Barr virus gene product BHRF1. J. Virol. 72:8392-8395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fanidi, A., E. A. Harrington, and G. I. Evan. 1992. Cooperative interaction between c-myc and bcl-2 proto-oncogenes. Nature 359:554-556. [DOI] [PubMed] [Google Scholar]
- 10.Garibal, J., E. Hollville, A. I. Bell, G. L. Kelly, B. Renouf, Y. Kawaguchi, A. B. Rickinson, and J. Wiels. 2007. Truncated form of the Epstein-Barr virus protein EBNA-LP protects against caspase-dependent apoptosis by inhibiting protein phosphatase 2A. J. Virol. 81:7598-7607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gossen, M., and H. Bujard. 1992. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl. Acad. Sci. U. S. A. 89:5547-5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harada, S., and E. Kieff. 1997. Epstein-Barr virus nuclear protein LP stimulates EBNA-2 acidic domain-mediated transcriptional activation. J. Virol. 71:6611-6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Henderson, S., D. Huen, M. Rowe, C. Dawson, G. Johnson, and A. Rickinson. 1993. Epstein-Barr virus-coded BHRF1 protein, a viral homologue of Bcl-2, protects human B cells from programmed cell death. Proc. Natl. Acad. Sci. U. S. A. 90:8479-8483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heston, L., M. Rabson, N. Brown, and G. Miller. 1982. New Epstein-Barr virus variants from cellular subclones of P3J-HR-1 Burkitt lymphoma. Nature 295:160-163. [DOI] [PubMed] [Google Scholar]
- 15.Hinuma, Y., and J. T. Grace. 1968. Cloning of Burkitt lymphoma cells cultured in vitro. Cancer 22:1089-1095. [DOI] [PubMed] [Google Scholar]
- 16.Hung, S. C., M. S. Kang, and E. Kieff. 2001. Maintenance of Epstein-Barr virus (EBV) oriP-based episomes requires EBV-encoded nuclear antigen-1 chromosome-binding domains, which can be replaced by high-mobility group-I or histone H1. Proc. Natl. Acad. Sci. U. S. A. 98:1865-1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kang, M. S., S. C. Hung, and E. Kieff. 2001. Epstein-Barr virus nuclear antigen 1 activates transcription from episomal but not integrated DNA and does not alter lymphocyte growth. Proc. Natl. Acad. Sci. U. S. A. 98:15233-15238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Katsumura, K. R., S. Maruo, Y. Wu, T. Kanda, and K. Takada. 2009. Quantitative evaluation of the role of Epstein-Barr virus immediate-early protein BZLF1 in B-cell transformation. J. Gen. Virol. 90:2331-2341. [DOI] [PubMed] [Google Scholar]
- 19.Kelly, G., A. Bell, and A. Rickinson. 2002. Epstein-Barr virus-associated Burkitt lymphomagenesis selects for downregulation of the nuclear antigen EBNA2. Nat. Med. 8:1098-1104. [DOI] [PubMed] [Google Scholar]
- 20.Kelly, G. L., A. E. Milner, R. J. Tierney, D. S. Croom-Carter, M. Altmann, W. Hammerschmidt, A. I. Bell, and A. B. Rickinson. 2005. Epstein-Barr virus nuclear antigen 2 (EBNA2) gene deletion is consistently linked with EBNA3A, -3B, and -3C expression in Burkitt's lymphoma cells and with increased resistance to apoptosis. J. Virol. 79:10709-10717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kelly, G. L., H. M. Long, J. Stylianou, W. A. Thomas, A. Leese, A. I. Bell, G. W. Bornkamm, J. Mautner, A. B. Rickinson, and M. Rowe. 2009. An Epstein-Barr virus anti-apoptotic protein constitutively expressed in transformed cells and implicated in Burkitt lymphomagenesis: the Wp/BHRF1 link. PLoS Pathog. 5:e1000341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kelly, G. L., A. E. Milner, G. S. Baldwin, A. I. Bell, and A. B. Rickinson. 2006. Three restricted forms of Epstein-Barr virus latency counteracting apoptosis in c-myc-expressing Burkitt lymphoma cells. Proc. Natl. Acad. Sci. U. S. A. 103:14935-14940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kennedy, G., J. Komano, and B. Sugden. 2003. Epstein-Barr virus provides a survival factor to Burkitt's lymphomas. Proc. Natl. Acad. Sci. U. S. A. 100:14269-14274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kieff, E., and A. B. Rickinson. 2007. Epstein-Barr virus and its replication, p. 2603-2654. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 5th ed. Lippincott Williams and Wilkins, Philadelphia, PA.
- 25.Kirchmaier, A. L., and B. Sugden. 1997. Dominant-negative inhibitors of EBNA-1 of Epstein-Barr virus. J. Virol. 71:1766-1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klein, G. 1983. Specific chromosomal translocations and the genesis of B-cell-derived tumors in mice and men. Cell 32:311-315. [DOI] [PubMed] [Google Scholar]
- 27.Komano, J., S. Maruo, K. Kurozumi, T. Oda, and K. Takada. 1999. Oncogenic role of Epstein-Barr virus-encoded RNAs in Burkitt's lymphoma cell line Akata. J. Virol. 73:9827-9831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Komano, J., M. Sugiura, and K. Takada. 1998. Epstein-Barr virus contributes to the malignant phenotype and to apoptosis resistance in Burkitt's lymphoma cell line Akata. J. Virol. 72:9150-9156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Komano, J., and K. Takada. 2001. Role of bcl-2 in Epstein-Barr virus-induced malignant conversion of Burkitt's lymphoma cell line Akata. J. Virol. 75:1561-1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kovalchuk, A. L., C. F. Qi, T. A. Torrey, L. Taddesse-Heath, L. Feigenbaum, S. S. Park, A. Gerbitz, G. Klobeck, K. Hoertnagel, A. Polack, G. W. Bornkamm, S. Janz, and H. C. Morse III. 2000. Burkitt lymphoma in the mouse. J. Exp. Med. 192:1183-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leao, M., E. Anderton, M. Wade, K. Meekings, and M. J. Allday. 2007. Epstein-Barr virus-induced resistance to drugs that activate the mitotic spindle assembly checkpoint in Burkitt's lymphoma cells. J. Virol. 81:248-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mackey, D., and B. Sugden. 1999. The linking regions of EBNA1 are essential for its support of replication and transcription. Mol. Cell. Biol. 19:3349-3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mannick, J. B., J. I. Cohen, M. Birkenbach, A. Marchini, and E. Kieff. 1991. The Epstein-Barr virus nuclear protein encoded by the leader of the EBNA RNAs is important in B-lymphocyte transformation. J. Virol. 65:6826-6837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maruo, S., Y. Wu, S. Ishikawa, T. Kanda, D. Iwakiri, and K. Takada. 2006. Epstein-Barr virus nuclear protein EBNA3C is required for cell cycle progression and growth maintenance of lymphoblastoid cells. Proc. Natl. Acad. Sci. U. S. A. 103:19500-19505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maruo, S., E. Johannsen, D. Illanes, A. Cooper, and E. Kieff. 2003. Epstein-Barr virus nuclear protein EBNA3A is critical for maintaining lymphoblastoid cell line growth. J. Virol. 77:10437-10447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nanbo, A., K. Inoue, K. Adachi-Takasawa, and K. Takada. 2002. Epstein-Barr virus RNA confers resistance to interferon-alpha-induced apoptosis in Burkitt's lymphoma. EMBO J. 21:954-965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nasimuzzaman, M., M. Kuroda, S. Dohno, T. Yamamoto, K. Iwatsuki, S. Matsuzaki, R. Mohammad, W. Kumita, H. Mizuguchi, T. Hayakawa, H. Nakamura, T. Taguchi, H. Wakiguchi, and S. Imai. 2005. Eradication of Epstein-Barr virus episome and associated inhibition of infected tumor cell growth by adenovirus vector-mediated transduction of dominant-negative EBNA1. Mol. Ther. 11:578-590. [DOI] [PubMed] [Google Scholar]
- 38.Pearson, G. R., J. Luka, L. Petti, J. Sample, M. Birkenbach, D. Braun, and E. Kieff. 1987. Identification of an Epstein-Barr virus early gene encoding a second component of the restricted early antigen complex. Virology 160:151-161. [DOI] [PubMed] [Google Scholar]
- 39.Rabson, M., L. Gradoville, L. Heston, and G. Miller. 1982. Non-immortalizing P3J-HR-1 Epstein-Barr virus: a deletion mutant of its transforming parent, Jijoye. J. Virol. 44:834-844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rickinson, A. B., and E. Kieff. 2007. Epstein-Barr virus, p. 2655-2700. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 5th ed. Lippincott Williams and Wilkins, Philadelphia, PA.
- 41.Ruf, I. K., P. W. Rhyne, H. Yang, C. M. Borza, L. M. Hutt-Fletcher, J. L. Cleveland, and J. T. Sample. 1999. Epstein-Barr virus regulates c-MYC, apoptosis, and tumorigenicity in Burkitt lymphoma. Mol. Cell. Biol. 19:1651-1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saridakis, V., Y. Sheng, F. Sarkari, M. N. Holowaty, K. Shire, T. Nguyen, R. G. Zhang, J. Liao, W. Lee, A. M. Edwards, C. H. Arrowsmith, and L. Frappier. 2005. Structure of the p53 binding domain of HAUSP/USP7 bound to Epstein-Barr nuclear antigen 1: implications for EBV-mediated immortalization. Mol. Cell 18:25-36. [DOI] [PubMed] [Google Scholar]
- 43.Shaku, F., G. Matsuda, R. Furuya, C. Kamagata, M. Igarashi, M. Tanaka, M. Kanamori, Y. Nishiyama, N. Yamamoto, and Y. Kawaguchi. 2005. Development of a monoclonal antibody against Epstein-Barr virus nuclear antigen leader protein (EBNA-LP) that can detect EBNA-LP expressed in P3HR1 cells. Microbiol. Immunol. 49:477-483. [DOI] [PubMed] [Google Scholar]
- 44.Shimizu, N., A. Tanabe-Tochikura, Y. Kuroiwa, and K. Takada. 1994. Isolation of Epstein-Barr virus (EBV)-negative cell clones from the EBV-positive Burkitt's lymphoma (BL) line Akata: malignant phenotypes of BL cells are dependent on EBV. J. Virol. 68:6069-6073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Strasser, A., A. W. Harris, M. L. Bath, and S. Cory. 1990. Novel primitive lymphoid tumours induced in transgenic mice by cooperation between myc and bcl-2. Nature 348:331-333. [DOI] [PubMed] [Google Scholar]
- 46.Tomkinson, B., E. Robertson, and E. Kieff. 1993. Epstein-Barr virus nuclear proteins EBNA-3A and EBNA-3C are essential for B-lymphocyte growth transformation. J. Virol. 67:2014-2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu, Y., S. Maruo, M. Yajima, T. Kanda, and K. Takada. 2007. Epstein-Barr virus (EBV)-encoded RNA 2 (EBER2) but not EBER1 plays a critical role in EBV-induced B-cell growth transformation. J. Virol. 81:11236-11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yajima, M., T. Kanda, and K. Takada. 2005. Critical role of Epstein-Barr Virus (EBV)-encoded RNA in efficient EBV-induced B-lymphocyte growth transformation. J. Virol. 79:4298-4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhu, D., C. F. Qi, H. C. Morse III, S. Janz, and F. K. Stevenson. 2005. Deregulated expression of the Myc cellular oncogene drives development of mouse “Burkitt-like” lymphomas from naive B cells. Blood 105:2135-2137. [DOI] [PubMed] [Google Scholar]