Abstract
We have continued studies to further understand the role of the ubiquitin-proteasome system (UPS) in human cytomegalovirus (HCMV) infection. With specific inhibitors of the proteasome, we show that ongoing proteasome activity is necessary for facilitating the various stages of the infection. Immediate-early protein 2 expression is modestly reduced with addition of proteasome inhibitors at the onset of infection; however, both early and late gene expression are significantly delayed, even if the inhibitor is removed at 12 h postinfection. Adding the inhibitor at later times during the infection blocks the further accumulation of viral early and late gene products, the severity of which is dependent on when the proteasome is inhibited. This can be attributed primarily to a block in viral RNA transcription, although DNA synthesis is also partially inhibited. Proteasome activity and expression increase as the infection progresses, and this coincides with the relocalization of active proteasomes to the periphery of the viral DNA replication center, where there is active RNA transcription. Interestingly, one 19S subunit, Rpn2, is specifically recruited into the viral DNA replication center. The relocalization of the subunits requires viral DNA replication, but their maintenance around or within the replication center is not dependent on continued viral DNA synthesis or the proteolytic activity of the proteasome. These studies highlight the importance of the UPS at all stages of the HCMV infection and support further studies into this pathway as a potential antiviral target.
The fundamental role of the ubiquitin-proteasome system (UPS), not only in general proteolysis but also in the regulation of several different cellular systems, has gained increasing attention in recent years. These processes include cell cycle regulation, signal transduction, apoptosis, and antigen presentation, among others (11, 14). Numerous studies have also linked the UPS to transcription regulation, DNA repair, and chromatin remodeling, at both a proteolytic and nonproteolytic level (9, 13, 32, 34, 40, 49). Thus, its potential role in disease pathogenesis has also been an area of great interest. Different viral strategies have evolved that either utilize or subvert the UPS in facilitating a productive infection (3, 5, 18, 54). Among these is human cytomegalovirus (HCMV), which is a betaherpesvirus endemic within the human population that can cause serious disease in immunocompromised individuals and is also the leading infectious cause of birth defects.
In brief, the UPS utilizes a highly regulated process in which the proteasome selectively degrades proteins that have become ubiquitinated through a multistep mechanism involving E1 (ubiquitin-activating enzyme), E2 (ubiquitin-conjugating enzyme), and E3 (ubiquitin ligase enzyme) (19). The mammalian 26S proteasome usually comprises one or two 19S regulatory subcomplexes on either end of the 20S catalytic core complex (45). The 19S is further subdivided into the base and lid. The base is composed of six AAA (ATPases associated with different cellular activities) ATPase subunits (i.e., Rpt1 to -6), forming a hexameric base ring, plus three non-ATPase subunits (i.e., Rpn1, Rpn2, and Rpn10/S5a). The ATPase subunits are also collectively known as the APIS (19S ATPase proteins independent of 20S). The 19S lid is composed of nine non-ATPases (i.e., Rpn3, Rpn5, Rpn6, Rpn7, Rpn8, Rpn9, Rpn11, Rpn12, and Rpn15). Regulatory functions of the 19S include polyubiquitin recognition, substrate binding, facilitation of deubiquitination, protein unfolding, and translocation into the 20S for degradation (2). The 20S catalytic core is formed by four stacked rings of α(1-7), β(1-7), β(1-7), and α(1-7) subunits and primarily functions in protein degradation via the catalytic β1, β2, and β5 subunits, which contain caspase-like, trypsin-like, and chymotrypsin-like peptidase activity, respectively. The β1, β2, and β5 subunits can also be substituted by the β1i, β2i, and β5i subunits to form the immunoproteasomes upon gamma interferon stimulation. Other than the 19S, several non-ATPase complexes (i.e., PA28/11S REG and Blm10/PA200) can associate with the 20S unit and differentially alter its activity (48). Since these complexes do not bind polyubiquitin chains, they likely regulate ubiquitin-independent proteolytic activity.
One of the first indications that HCMV exploits the UPS was the discovery that the virus expresses two proteins, US2 and US11, that facilitate evasion of host immune surveillance by relocalizing major histocompatibility complex class I molecules from the endoplasmic reticulum to the cytoplasm for proteasome-mediated degradation (for a review, see reference 58). Subsequently it was found that the input viral tegument protein pp71 interacts with ND10-associated Daxx and promotes its ubiquitin-independent and proteasome-mediated degradation, thus facilitating viral transcription (8, 22, 23, 26, 39, 51). pp71 can also induce ubiquitin-independent proteasome-mediated degradation of unphosphorylated Rb, p107, and p130 (27, 28). HCMV has also been shown to inhibit the degradation of several cell cycle proteins by inactivating the anaphase-promoting complex, an E3 ubiquitin ligase (4, 59, 61, 62). In general, inhibition of the proteasome appears to impact negatively on viral replication (31, 46, 50).
Viral gene expression is temporally regulated into three different kinetic classes: immediate early (IE), early, and late (for a review, see reference 41). Expression of early genes is dependent on IE gene expression and transactivation. Early genes encode proteins for viral DNA replication, which is necessary for subsequent late gene expression. Late genes encode more structural proteins that function in virion maturation and egress. Productive viral infection requires that cells be infected in G0/G1 phase of the cell cycle such that IE genes can be expressed immediately after infection. We previously showed that if cells are infected in S phase, there is no viral IE gene expression until cells return to G1. Interestingly, this block to IE gene expression could be relieved by addition of a proteasome inhibitor (15).
We have continued our studies on the interplay between HCMV and the UPS, further delineating the molecular mechanisms involved in the suppression of HCMV replication by proteasome inhibitors. Viral early and late protein expression levels are greatly reduced upon proteasome inhibitor treatment. This appears to be due primarily to a defect in continued viral gene transcription as well as a decrease in viral DNA synthesis, both of which are facilitated by the early replication proteins. Not only is proteasome activity required for efficient viral gene expression, but also the levels and activity of the proteasome increase during the course of infection, correlating with times of viral early and late gene expression. Based on immunofluorescence assays (IFA), specific 19S subunits appear to be recruited to sites of viral replication. Interestingly, the 19S non-ATPase subunit Rpn2 accumulates in the viral DNA replication centers, whereas the other proteasome subunits analyzed are relocalized to the periphery of the replication centers, where there is active RNA transcription.
MATERIALS AND METHODS
Cells and virus.
Human foreskin fibroblasts (HFFs), obtained from the University of California, San Diego, Medical Center, were cultured in minimum essential medium with Earle's salts supplemented with 10% heat-inactivated fetal bovine serum, 1.5 μg/ml amphotericin B, 2 mM l-glutamine, 200 U/ml penicillin, and 200 μg/ml streptomycin. All cell culture media were from Gibco-BRL. Cells were kept in incubators maintained at 37°C and 7% CO2. The Towne strain of HCMV was obtained from the American Type Culture Collection (VR 977) and propagated as previously described (56).
Cell synchronization and infection.
All experiments were performed under G0 synchronization conditions (52). Cells were trypsinized 3 days after the monolayer reached confluence and then replated at a lower density to induce cell cycle progression. At the time of replating, cells were either infected with HCMV at the indicated multiplicity of infection (MOI) or mock infected with tissue culture supernatants as previously described (52). For proteasome inhibition assays, cell cultures were incubated with 2.5 μM MG132, 10 μM lactacystin, 100 nM salinosporamide A (Sal A), or dimethyl sulfoxide (DMSO) as the vehicle control at the times indicated. MG132 and lactacystin were obtained from Calbiochem, while Sal A was a gift from Bradley Moore (Scripps Institute of Oceanography, University of California, San Diego). The viral DNA inhibitors ganciclovir (GCV) and phosphonoacetic acid (PAA) were obtained from Sigma and used at 20 μM and 360 μM, respectively. Cells were harvested at the indicated times postinfection (p.i.) and processed as described for each experiment. All experiments were performed at least twice.
Western blot analysis.
Cells were lysed in Laemmli reducing sample buffer (62.5 mM Tris [pH 6.8], 2% SDS, 10% glycerol, 5% β-mercaptoethanol) supplemented with a protease inhibitor cocktail (Roche) and phosphatase inhibitors (50 mM sodium fluoride, 10 mM β-glycerophosphate, and 1 mM sodium orthovanadate), and the lysates were sonicated and boiled. Equal amounts of lysate (i.e., by cell number) were loaded onto SDS-PAGE gels unless otherwise stated. Following electrophoresis, proteins were transferred to nitrocellulose (Schleicher & Schuell), and Western blot analyses were performed using appropriate antibodies. The Supersignal West pico and West femto chemiluminescent detection methods (Pierce) were used to visualize the proteins according to the manufacturer's instructions.
Quantitative real-time PCR.
DNA from mock- or HCMV-infected cells was isolated using a DNA blood minikit (Qiagen), and the concentration was determined by UV spectrophotometry. Quantitative real-time PCR (qPCR) was performed with the ABI Prism 7000 sequence detection system (Applied Biosystems) using the TaqMan Universal PCR master mix (Applied Biosystems) along with oligonucleotide primers and TaqMan dually labeled (5′-6-carboxyfluorescein and 3′-black hole quencher-1) probes (Integrated DNA Technologies) with 40 ng DNA per reaction mixture. Probes were targeted to an unspliced region of HCMV UL77, IE2 (UL122), and the glyceraldehyde 3-phosphate dehydrogenase (GAPDH) promoter, which was used for normalization of input DNA, as previously described (60). Standard curves were generated using DNA isolated from cells harvested at 48 h p.i. The primers and probes used were as follows: UL77 forward (5′-CGTTGCCCGGGAACG-3′), UL77 reverse (5′-GGTGTGAAAGCGGATAAAGGG-3′), and UL77 probe (5′-ACCTAGCTACTTTGGAATCACGCAGAACGA-3′); IE2 forward (5′-GCGCAATATCATGAACGA-3′), IE2 reverse (5′-GATTGGTGTTGCGGAACATG-3′), and IE2 probe (5′-TCGGCGGGGTCGC-3′); GAPDH promoter forward (5′-TTTCATCCAAGCGTGTAAGGG-3′), reverse (5′-CAGGACTGGACTGTGGGCA-3′), and probe (5′-CCCCGTCCTTGACTCCCTAGTGTC-3′).
Northern blot analysis.
All reagents and kits were obtained from Ambion. Total RNA from mock- or HCMV-infected cells was isolated using the RNAqueous kit with concentrations measured by UV spectrophotometry. Northern blot assays were done using the NorthernMax kit per the manufacturer's instructions. Briefly, 10 μg RNA was resolved in a 1% agarose formaldehyde gel and transferred to a BrightStar-Plus membrane. Biotinylated probes were generated using the BrightStar psoralen-biotin kit with the following DNA fragments: UL44 (Afl II/SacII), UL99 (KpnI/SacII), UL83 (SacII), UL82 (PvuI/SacII), and GAPDH (582-bp Afl III). Membranes were hybridized to probes at 42°C overnight, washed, developed using the BrightStar BioDetect kit, and exposed to film for autoradiography.
Immunofluorescence assays.
Cells were seeded onto glass coverslips at the time of infection and fixed with 2 to 4% paraformaldehyde for 20 min at the indicated times p.i. Cells were permeabilized with 0.2% Triton X-100 for 5 min and washed in phosphate-buffered saline (PBS) prior to immunofluorescence staining. Normal goat serum (10% in PBS; Jackson ImmunoResearch) was used as a blocking solution and antibody diluent. Mouse or rabbit IgG (Jackson ImmunoResearch) served as negative controls. Following primary antibody incubation and subsequent washes in PBS, coverslips were incubated with appropriate fluorescein isothiocyanate- or tetramethyl rhodamine isothiocyanate-conjugated secondary antibody (Jackson ImmunoResearch) plus Hoescht stain. Coverslips were treated with SlowFade Gold (Molecular Probes), an antiphotobleaching reagent, and mounted onto a slide for imaging. Costained samples were analyzed by a DeltaVision deconvolution microscopy system (Applied Precision) with SoftWoRx software (Applied Precision) on a Silicon Graphics O2 workstation. Images were taken at 0.2-μm increments along the z axis with a Photometrics charge-coupled-device camera mounted on a fluorescence/differential interference contrast microscope. The fluorescence data sets were deconvolved and analyzed by DeltaVision SoftWoRx programs. Adobe Photoshop was used to prepare images for the figures.
BrU and BrdU pulse-labeling assays.
HFFs were infected with HCMV at an MOI of 2 or mock infected and seeded onto coverslips. At 36 h p.i., cells were rinsed in PBS and incubated with fresh medium containing 1 mM bromouridine (BrU; Sigma) or 10 μM bromodeoxyuridine (BrdU; Sigma). DMSO was used as a negative control. Cells were fixed at 20, 30, and 60 min postlabeling and processed by IFA using an anti-BrdU antibody (Sigma), which detects both BrU and BrdU.
Microinjections.
Nuclear microinjections (MI) of 250 μg/ml DQ-ovalbumin (DQ-ova; Molecular Probes, Invitrogen) and blue dextran (injection control) were done on HFFs infected with HCMV at an MOI of 2 or mock infected at 40 h p.i. Coinjections with 150 nM Sal A were used to inhibit proteasome activity as a negative control. The cells, which were seeded onto coverslips, were transferred into serum-free medium 30 to 60 min prior to MI and further incubated for an additional 30 min after microinjection to allow for proteolytic degradation of the DQ-ova. Cells were then fixed with 3.7% formaldehyde and processed for IFA.
Antibodies.
All antibodies to proteasome subunits were obtained from Biomol, except for 19S subunit Rpn1 (Calbiochem). Antibodies to HCMV proteins UL57, UL44, UL83 (CH12), UL99 (CH19), and IE1/IE2 (CH16.0) were from Virusys, while those to UL97, UL85, and UL86 were gifts from William Britt (University of Alabama, Birmingham). Other antibodies used included those to actin (AC-15; Sigma), GAPDH (6c5; Fitzgerald), BrdU (Sigma), H3K4 (Upstate), ARNA3 (Chemicon), H14 and H5 (Covance), and p53 and wee1 (Santa Cruz Biotechnology).
RESULTS
Viral protein expression is delayed upon inhibition of proteasome activity.
Previous studies assessing the impact of proteasome inhibitors on HCMV infection showed a decrease in viral titer and an apparent defect at all stages of the infection, with a significant reduction in viral protein expression (31, 46). These studies were done with the proteasome inhibitor added after viral adsorption and maintained in culture through the time course. Given the temporal kinetics of HCMV gene expression, it was not possible to determine from these studies whether the early and late stages of the infection were blocked differentially, independent of the inhibitory effects on the IE proteins. It was unclear whether the decrease in early or late gene expression could be attributed to a specific defect in viral DNA synthesis and/or gene transcription or to the decrease in IE expression, which would subsequently affect transactivation of the early genes required for viral DNA synthesis. The block in IE protein expression was later shown to be MOI dependent (31, 50), while early and late protein expression remained significantly impaired at all MOIs tested (31), further indicating that the stages of viral gene expression may be differentially affected by proteasome inhibition. Another potential complication that needs to be considered when studying the effects of proteasome inhibitors on HCMV gene expression is the time at which the virus and inhibitor are added in relation to the cell cycle. Expression of IE proteins can be detected in cells infected in G0/G1 by 1 to 2 h p.i. (15, 25). Cells infected in S phase fail to express IE proteins until cells divide and cycle to the next G1 phase; however, addition of a proteasome inhibitor was able to relieve this block in IE gene expression (15, 52). Thus, infection of an asynchronous cell population would likely result in asynchronous viral gene expression. To more accurately assess the role of the proteasome in each stage of viral replication, it was important to use cells that had been synchronized to enter the cell cycle at the time of infection. For the experiments presented here, cells were synchronized in G0 and released into G1 at the time of infection as described in Materials and Methods.
To further delineate the stage(s) of HCMV replication that is proteasome dependent, G0-synchronized HFFs were released into G1 by trypsinization, replating at a lower density, and infection at an MOI of 2 at the time of replating. Proteasome inhibitors were added at different times p.i., and viral protein expression, correlating to the different kinetic classes, was examined by Western blotting. Three different proteasome inhibitors (i.e., MG132, lactacystin, or Sal A) were used to minimize the risk that the results were due to off-target effects. MG132 is a reversible inhibitor of the chymotrypsin-like activity of the proteasome, lactacystin blocks both the chymotrypsin-like and trypsin-like activities, and Sal A inhibits all three chymotrypsin-like, trypsin-like, and caspase-like activities. The effects of all three inhibitors on the infection were essentially the same.
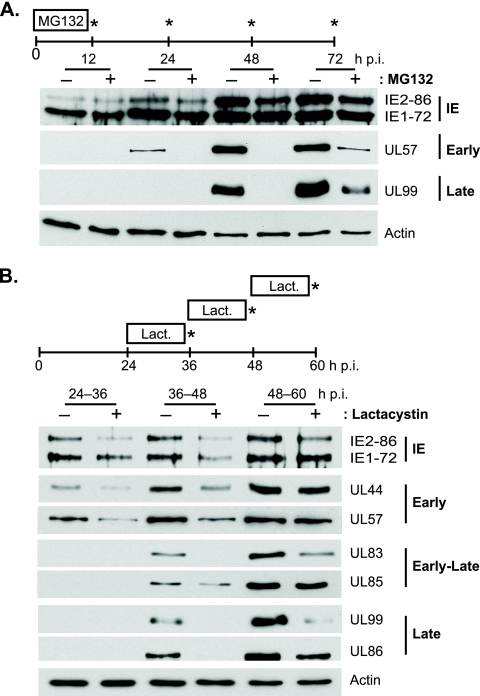
In Fig. 1A, MG132 was added with the virus inoculum at the beginning of the infection and then washed out at 12 h p.i. Fresh medium was added and samples were harvested at 12, 24, 48, and 72 h p.i. Although the drug was added with the virus, no apparent constraints on viral entry were observed, as viral IE1-72 and IE2-86 protein expression at 12 h p.i. were comparable in the presence and absence of the proteasome inhibitor; however, IE2-86 expression appeared slightly delayed thereafter, with decreased levels observed in the inhibitor-treated samples at 24, 48, and 72 h p.i. A significant delay in early (i.e., UL57) and late (i.e., UL99) protein expression was also observed. These results are consistent with previous studies (31, 46, 50) that showed that the addition of proteasome inhibitors at the time of infection negatively impacts viral protein expression. The lack of effect on IE1-72 expression was likely due to the MOI used in this assay (31). Interestingly, both UL57 and UL99 remained significantly suppressed even after the 60-h release from the drug, further underscoring the importance of proteasome activity at the early stages of the infection in facilitating viral replication and the switch to early and late gene expression. This may in part be attributed to the decrease observed in IE2-86 expression.
FIG. 1.
HCMV protein expression is delayed by proteasome inhibitor treatment. HFFs infected with HCMV at an MOI of 2 were treated with MG132 (2.5 μM) from 0- to 12 h p.i. with cells harvested at the times indicated (A) or lactacystin (Lact; 10 μM) from 24 to 36, 36 to 48, or 48 to 60 h p.i. with cells harvested at the end of treatment (B) and processed for Western blot analyses with antibodies to HCMV proteins IE2, IE1, UL44, UL57, UL99, UL83, UL85, and UL86. Actin is shown as a loading control.
To further assess the impact of proteasome inhibition on the later stages of the infection, lactacystin was added to the infected cells at 24, 36, or 48 h p.i. After 12 h in the presence or absence of the drug, the cells were harvested and processed for Western blot analyses (Fig. 1B). Decreased IE2-86 protein expression was still observed with the proteasome inhibitor added at these later times of infection. IE1-72 expression was slightly lower with the drug present between 36 and 48 h p.i., and early protein expression (i.e., UL44 and UL57) was decreased when proteasome activity was inhibited from 24 to 36 h p.i or from 36 to 48 h p.i., but little to no effect was seen with the 48- to 60-h p.i. treatment. Late protein expression (i.e., UL83, UL99, and UL86) was also significantly reduced when the drug was present from 36 to 48 h p.i. or 48 to 60 h p.i., except for UL85 expression, which showed only a slight decrease with drug treatment at the different time periods. Interestingly, UL44 and UL57 levels after proteasome inhibition from 36 to 48 h p.i. were similar to that observed at 36 h p.i. with no treatment. Likewise, UL83 and UL99 levels after the 48- to 60-h p.i. drug treatment were comparable to those seen at 48 h p.i. with no drug. Similar results were also obtained with PYR-41, an inhibitor of the ubiquitin-activating enzyme E1 (data not shown). These results suggested that proteasome inhibition prevented further accumulation of the viral proteins, but we could not distinguish whether protein processing is stalled at the translational level or if an earlier step (i.e., transcription or DNA replication) is inhibited. However, given that the protein expression is less affected after the prime expression period for each kinetic class (i.e., early proteins are not as affected when the inhibitor is added at late times of infection), the latter possibility seemed more likely.
Proteasome inhibitors prevent further accumulation of viral transcripts.
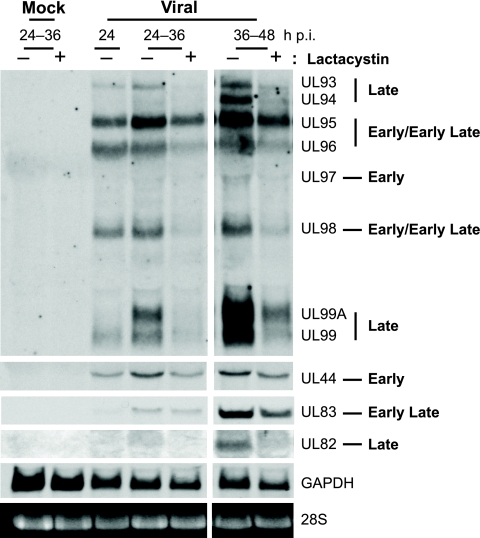
To differentiate whether protein expression was affected at the RNA or protein level, Northern blot assays were used to assess RNA levels of representative early and late transcripts. One of the probes used in this study is directed to the coding region of UL99, which allowed for the simultaneous detection and analysis of the 3′ coterminal transcripts from the UL93 to UL99 coding region that are expressed with different temporal kinetics during the infection (63). As defined in the study by Wing et al. (63), kinetic class determination was based on whether transcript expression was dependent on viral DNA replication. UL97 transcript expression was independent of viral DNA replication and defined as early, whereas UL93 and UL94 were dependent and designated as late transcripts. UL95, UL96, and UL98 RNA levels were somewhat reduced after GCV (a viral DNA replication inhibitor) treatment and were classified as early-late transcripts. Interestingly, UL96 and UL98 expression was also observed to be higher at 5 h p.i. than at 72 h p.i. with GCV treatment, indicating that these genes may be under the control of multiple promoter elements and expressed in more than one kinetic class.
In our study, HFFs were synchronized in G0 and infected with HCMV at an MOI of 2 or mock infected at the time of release into G1. Lactacystin (10 μM) was added from 24 to 36 h p.i. or 36 to 48 h p.i., the times during which the greatest effects on protein accumulation were observed, with total RNA isolated from cells at the end of treatment. GAPDH and 28S RNA levels were used as loading controls. Using the probe directed to UL99, we found that the UL95, UL96, and UL98 transcripts were expressed at high levels by 24 h p.i. (Fig. 2). Addition of the proteasome inhibitor at 24 to 36 h p.i. or 36 to 48 h p.i. appeared to prevent further accumulation of UL95, as expression levels remained similar to when the drug was added, while levels continued to increase with no drug. Interestingly, both UL96 and UL98 RNA levels dropped below that seen at 24 h p.i. after each drug addition period, which may reflect a shorter half-life of these RNAs. The expression of the UL93, UL94, and UL99 transcripts was not readily detected until 36 or 48 h p.i., consistent with late kinetics, and was greatly reduced upon treatment with the proteasome inhibitor. The early UL97 transcript was not as readily detectable but showed no significant changes with the 36- to 48-h p.i. drug addition upon longer exposures of the blot (data not shown).
FIG. 2.
Proteasome inhibition prevents further accumulation of viral transcripts. HFFs infected with HCMV at an MOI of 2 or mock infected were treated with lactacystin (10 μM) at the times indicated and harvested at the end of treatment. Total RNA was isolated and processed for Northern blot analyses with probes to the 3′ coterminal regions of UL92 to UL99, UL44, UL83, and UL82. GAPDH mRNA and 28S rRNA are shown as loading controls.
To ensure that the observed downregulation was not specific to the UL99 transcription unit, we also tested UL44 (early), UL83 (early-late), and UL82 (late) RNA expression levels. As shown in Fig. 2, UL44 expression was reduced in both treatment periods, although less of an effect was seen with the 36- to 48-h p.i. lactacystin addition. UL82 expression was also significantly decreased with the proteasome inhibitor at 36 to 48 h p.i. The effects on UL83 were more variable, as expression levels with the inhibitor added at 24 to 36 h p.i. showed little to no difference, while addition of the inhibitor at 36 to 48 h p.i. showed a modest decrease. In general, inhibition of the proteasome appears to prevent further expression or accumulation of the early transcripts (i.e., UL44 and UL95), while the early-late transcripts of UL96 and UL98 may have shorter half-lives. The significant reduction in the late transcripts (i.e., UL93, UL94, UL99, and UL82) may be due to a block in switching to late gene expression in the presence of the inhibitor.
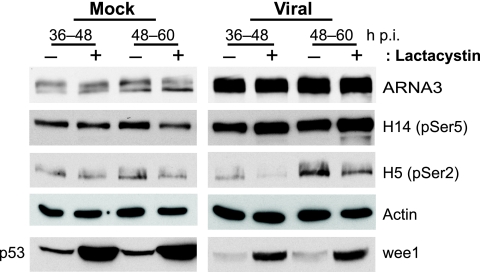
Decreased viral late gene expression and viral yield have also been observed in herpes simplex virus type 1 (HSV-1)-infected cells treated with MG132 or lactacystin (10). The associated recovery of overall RNA polymerase II (RNAP II) levels, especially that of pSer2-RNAP II, which normally decreases during HSV-1 infection, led to the hypothesis that proteasome inhibitor treatment prevents the degradation of stalled RNAP II complexes at later times of infection (10, 17, 35). To determine whether this may also be the case with HCMV, RNAP II levels during HCMV infection were assessed with or without proteasome inhibition. Mock- or HCMV-infected cells were treated with lactacystin at 36 to 48 h p.i. or 48 to 60 h p.i. and harvested at the end of treatment for Western blot analyses using RNAP II phospho-specific antibodies. As reported previously (57), unlike HSV-1, an overall increase in RNAP II expression was observed during HCMV infection, as shown by both the ARNA3 (RNAP IIo/a) and H14 (pSer5-RNAP IIo) antibodies, while pSer2-RNAP II (H5) levels did not accumulate in the HCMV-infected cells until later during the infection (Fig. 3). Furthermore, no significant effect on the various forms of RNAP II was observed with lactacystin treatment in either mock- or HCMV-infected cells. Actin is shown as a loading control, while p53 and wee1 (53) served as positive controls for proteasome inhibition in mock- or virus-infected cells, respectively.
FIG. 3.
RNAP II expression is increased during HCMV infection but not affected upon proteasome inhibition. HFFs infected with HCMV at an MOI of 2 or mock infected were treated with lactacystin (10 μM) at the times indicated and harvested at the end of treatment for Western blot analyses with antibodies to RNAP IIo/a (ARNA3), pSer5-RNAP IIo (H14), pSer2-RNAP IIo (H5), p53, wee1, and actin.
Viral DNA synthesis is modestly decreased upon proteasome inhibitor treatment.
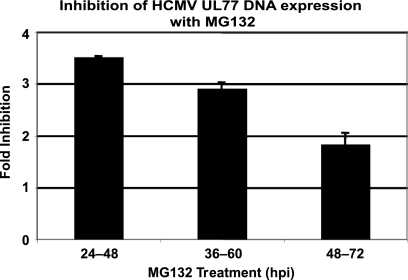
Previously, it was shown that the presence of the proteasome inhibitor from the beginning of the infection inhibited viral DNA replication (31, 46), but this could have been the result of reduced expression of the viral early proteins necessary for viral DNA synthesis. To address this question more directly, HFFs were infected with HCMV at an MOI of 2 and were treated with MG132 during the intervals of 24 to 48 h p.i., 36 to 60 h p.i., or 48 to 72 h p.i. Cells were harvested at the end of each treatment period and analyzed for viral DNA accumulation by qPCR with primers and probe to the UL77 gene. A probe to the GAPDH promoter was used as a control for input DNA and normalization. Values are expressed as the fold inhibition of UL77 DNA accumulation in the MG132 treated compared to DMSO during each treatment period (Fig. 4). The accumulation of viral DNA decreased by ∼3.5-fold with the 24- to 48-h p.i. treatment. Similar decreases were seen with the later treatments, with ∼3-fold for 36 to 60 h p.i. and ∼2-fold for 48 to 72 h p.i. Since UL77 is relatively close to the origin of viral DNA replication, we also used primers and probe to the IE2 (UL122) region of the genome, which is further from the origin, to determine whether there might be a greater decrease in apparent viral DNA synthesis due to preferential effects on the elongation step of DNA replication. No differences were observed, however, indicating that the inhibition is likely at the initiation step or just after conversion to the elongation step (data not shown).
FIG. 4.
DNA synthesis is downregulated upon proteasome inhibitor treatment. Viral DNA accumulation was measured by qPCR with primers and probe to the UL77 gene using total DNA isolated from HCMV-infected HFFs (MOI of 2) treated with DMSO or MG132 (2.5 μM) for 24 h at the postinfection times indicated. Values were normalized to that for the GAPDH promoter and are expressed as the fold inhibition of DNA accumulation in MG132-treated samples compared to DMSO control samples during the 24-h period. Mean values from two experiments are shown.
Proteasome activity and expression increase at later times of the infection.
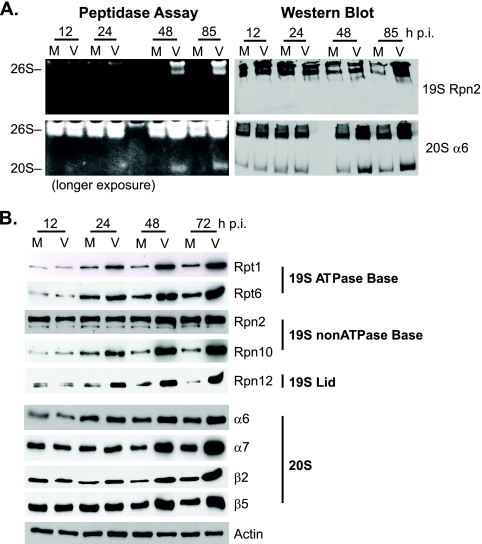
Given the decrease in viral RNA expression upon proteasome inhibition and the many reports indicating a functional role of proteasomes in transcription, we hypothesized that the proteasome facilitates HCMV transcription. If this were the case, then proteasome activity or abundance might increase as the infection progresses. The 26S proteasome is composed of two subcomplexes, the 19S regulatory complex and the 20S catalytic core. Although free 20S maintains some catalytic activity, the 26S complex is responsible for the major proteolytic activity in the cell. In-gel peptidase assays using a fluorogenic substrate were performed to measure the proteolytic activity of mock- or HCMV-infected cells harvested at 12, 24, 48, and 85 h p.i. Increased peptidase activity was observed in the virus-infected samples beginning 24 to 48 h p.i. and remained at elevated levels throughout the time course, while levels were unchanged in the uninfected cells (Fig. 5A). Separate gels were run for Western blot analyses. Alignment of blots probed for 19S subunit Rpn2 and 20S subunit α6 showed that the slower-migrating bands corresponded to the 26S proteasome complex (i.e., 19S and 20S), whereas the lower-molecular-weight band represented free 20S activity (Fig. 5A).
FIG. 5.
Proteasome activity and subunit expression increases during HCMV infection. (A) Mock-infected (M) or virus-infected (V) HFFs were harvested at the times indicated. Cell lysates were processed for an in-gel peptidase assay by using a fluorigenic peptide to measure proteasome activity. Western blot assays for 19S Rpn2 and 20S α6 were done in parallel on separate gels to identify the 26S and 20S proteasome fractions. (B) Western blot analyses of proteasome subunit expression in mock- or virus-infected HFFs over an infection time course. Representative subunits from each subcomplex are shown. Actin was used as a loading control.
Western blot assays were also used to screen the various proteasome subunits to determine whether the elevated activity correlated with an increase in proteasome abundance during HCMV infection. HFFs that were infected with HCMV (MOI of 5) or mock infected were harvested over a 72-h time course. Expression levels of all 19S and 20S subunits were examined in these experiments, except for the 19S lid subunits Rpn3, Rpn5, Rpn6, Rpn9, Rpn15, and Rpn11, as well as the 20S subunit α1. Representative subunits from each subcomplex are shown with actin as a loading control (Fig. 5B). As with the peptidase assay, protein expression levels of most subunits were higher in the virus-infected cells beginning 24 to 48 h p.i. and continued to increase throughout the time course. A few subunits (i.e., Rpn2) showed little to no change in expression.
Proteasome subunits relocalize around viral replication centers.
IFA was used to determine whether proteasomes localize to sites of viral replication. Intracellular distribution studies regarding proteasome localization have led to conflicting results, but in general, proteasomes appear to be abundantly distributed throughout the cytosol and nucleus (but excluded from the nucleoli) and have been found associated with intermediate filaments, endoplasmic reticulum membrane, and centrosomes (21). Mock- or HCMV-infected (MOI of 5) HFFs were fixed onto coverslips and stained at various times p.i. using antibodies targeted against different proteasome subunits. We were limited in this analysis to the use of antibodies that did not show a high level of accumulation at the Golgi complex due to the presence of the virus-encoded Fc receptor. Subunits that have been tested include the 19S subunits Rpt1 to Rpt6, Rpn1, Rpn2, Rpn10, and Rpn12, as well as the 20S subunits α2, α4, α5, α6, and β1.
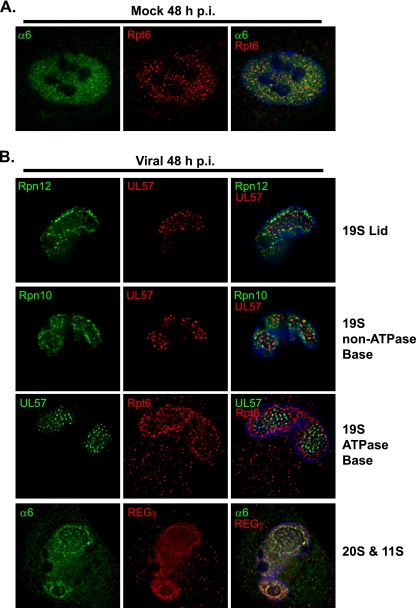
Mock-infected cells showed a faint diffuse staining of the various proteasome subunits throughout the cytosol with more prominent staining in the nucleus. This same localization pattern was maintained throughout the 72-h time course. The 19S subunit Rpt6 and 20S subunit α6 are shown costained in a mock-infected cell at 48 h p.i. as a representative example (Fig. 6A). The 20S α6 subunit appears to be more diffusely expressed than 19S Rpt6, which shows more punctate staining. This is consistent with 20S also being free or associated with different regulatory complexes. In the infected cells, a general increase in staining was observed for the majority of the subunits at 48 to 72 h p.i. (data are only shown for the 48-h time point in Fig. 6B), which corresponds to the increased protein expression seen by Western analysis. Cells were also costained with antibodies to viral replication proteins, UL57 or UL44, both of which localize throughout the viral replication center and are good markers for the size and localization of the replication center. The cells were subsequently analyzed by deconvolution microscopy to establish whether proteasomes localize to the viral replication centers. Several 19S subunits (i.e., Rpt6, Rpt1, Rpt5, Rpn10, and Rpn12), 11S subunit REGγ, and 20S α6 all exhibited some staining within the replication center and an enhanced accumulation at the rim of the replication center in infected cells beginning at ∼36 h p.i. (data are only shown for the 48-h time point in Fig. 6B). As representative examples, the 19S subunits Rpn12, Rpn10, and Rpt6 are shown costained with UL57, while 11S REGγ is shown costained with 20S α6 in a mid-cross-sectional plane at 48 h p.i. (Fig. 6B).
FIG. 6.
Proteasome subunits relocalize around the viral replication center during HCMV infection. Mock- or virus-infected HFFs were fixed onto coverslips at 48 h p.i., processed for IFA, and imaged by deconvolution microscopy of 0.2-μm sections at a magnification of 1,000× under oil immersion conditions. Midsectional planes of representative cells are shown. (A) Mock-infected HFFs were fixed at 48 h p.i. and costained for 19S Rpt6 (red) and 20S α6 (green), with Hoescht stain in blue. (B) HCMV-infected HFFs (MOI of 5) were fixed at 48 h p.i. and costained with antibodies to 19S Rpn12 (green) and UL57 (red), 19S Rpn10 (green) and UL57 (red), 19S Rpt6 (red) and UL57 (green), or 20S α6 (green) and 11S REGγ (red).
The relocalization of the proteasome subunits occurs after the onset of viral DNA synthesis.
Since this relocalization of the proteasome subunits is first detected shortly after the onset of viral DNA replication, we wanted to determine whether the relocalization is dependent on viral DNA synthesis. HCMV DNA replication begins ∼16 h p.i., requiring both viral and cellular proteins (for a review, see reference 41). Six core viral replication proteins (i.e., UL44, UL57, UL54, UL70, UL102, and UL105) along with UL84, UL112-113, and UL114 are necessary for efficient viral DNA replication, as measured in transient-expression assays. The UL112-113 phosphoproteins localize adjacent to promyelocytic leukemia protein-associated oncogenic domains (PODs) at sites of input viral genome accumulation and IE transcription of viral genes, where it then recruits the core replication proteins in forming the prereplication centers (1, 44). These foci eventually coalesce into large replication centers that continue to grow as viral replication proceeds.
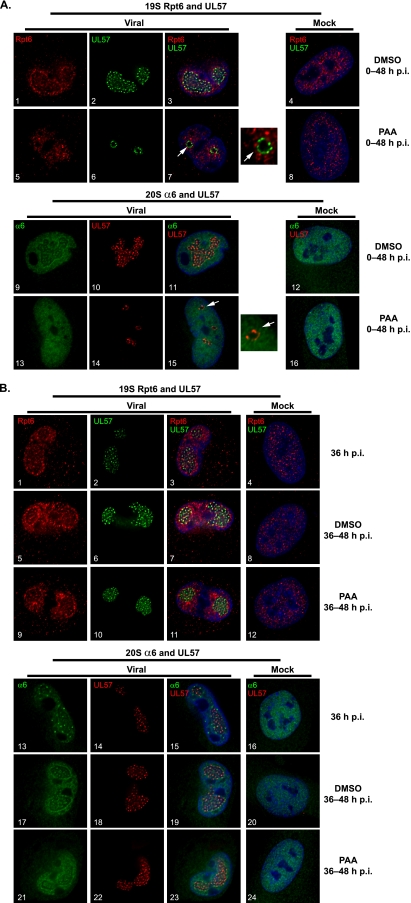
Mock- or HCMV-infected HFFs were treated with the viral DNA polymerase inhibitor PAA at 0 to 48 h p.i. or 36 to 48 h p.i. and then fixed at 48 h p.i. for IFA with antibodies to 19S subunits Rpt1, Rpt6, Rpn10, and 20S α6. As representative examples, 19S Rpt6 and 20S α6 are shown costained with UL57 in Fig. 7. The presence of the drug had no significant effect on the localization of 19S Rpt6 or 20S α6 in the mock-infected cells with treatment at either 0 to 48 h p.i. (Fig. 7A, panels 8 and 16) or 36 to 48 h p.i. (Fig. 7B, panels 12 and 24). Addition of PAA at the onset of infection severely limited viral replication, as only prereplication center foci were observed by UL57 staining (Fig. 7A, panels 6 and 14), compared to the large replication centers in the untreated samples (Fig. 7A, panels 2 and 10). Interestingly, both 19S Rpt6 and 20S α6 remain diffusely localized throughout the infected nuclei and were excluded from the prereplication center foci in the presence of the inhibitor (Fig. 7A, white arrows in panels 7 and 15 and adjacent magnified sections). This is consistent with the relocalization of the proteasome subunits occurring after the onset of viral DNA synthesis.
FIG. 7.
Relocation of proteasome subunits to the replication center periphery occurs after the onset of viral DNA replication. (A) Mock- or HCMV-infected HFFs (MOI of 2) were treated with DMSO or PAA at 0 to 48 h p.i. and fixed at 48 h p.i. for IFA with antibodies to 19S Rpt6, 20S α6, and UL57. (B) DMSO or PAA was added at 36 to 48 h p.i. to inhibit viral DNA synthesis during this 12-h period. Cells were fixed at 36 h p.i. prior to treatment and at 48 h p.i. and processed for IFA with antibodies to 19S Rpt6, 20S α6, and UL57. Cells were imaged by deconvolution microscopy with 0.2-μm sections at a magnification of 1,000× under oil immersion conditions. Midsectional planes of representative cells are shown merged with Hoescht stain (blue). Magnified images of the prereplication foci (as indicated by the white arrow) are shown adjacent to subpanels 7 and 15 in panel A.
To address whether ongoing viral DNA replication is necessary to maintain the proteasome subunits at the rim of the replication center, PAA was added at 36 h p.i., which is after the initiation of viral DNA synthesis, and cells were analyzed by IFA at 48 h p.i. At 36 h p.i., some enhancement of both 19S Rpt6 and 20S α6 could be visualized at the rim of the replication center, with some punctate foci observed for 20S α6 (Fig. 7B, panels 1 and 13). The accumulation of the proteasome subunits at the periphery of the replication center continued with the addition of DMSO from 36 to 48 h p.i. (Fig. 7B, panels 5 to 7 and 17 to 19). No significant differences were seen in the localization of 19S Rpt6 in the presence of PAA from 36 to 48 h p.i. (Fig. 7B, panels 9 to 11), whereas the localization of 20S α6 was slightly more diffuse within and around the replication centers with PAA (Fig. 7B, panels 21 to 23). The drug treatment had no effect on the localization of the proteasomes in mock-infected cells (Fig. 7B, panels 12 and 24). Taken together, these data indicate that ongoing viral DNA replication is not required for the maintenance of the proteasome subunits at this site. Similar results were also obtained using GCV (data not shown).
Active proteasomes accumulate around the viral DNA replication center in regions of RNA synthesis.
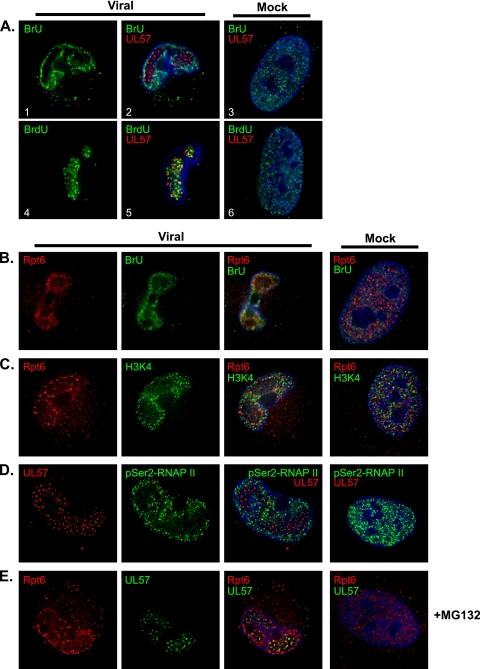
The same shift in localization to the rim of the replication center has previously been reported for the phosphorylated forms of RNAP II in HCMV-infected cells (57), further suggesting that the proteasomes may be involved in transcriptional processes during the infection. To assess whether RNA synthesis is occurring at the periphery of the replication center, BrU pulse-labeling experiments were performed. HFFs infected with HCMV or mock infected were pulse-labeled with 1 mM BrU at 36 h p.i. for 20, 30, or 60 min. Cells were then fixed and processed by IFA. In the uninfected cells, BrU localized diffusely throughout the nucleus, with signal intensities increasing with pulse length; a representative cell with a 30-min BrU pulse is shown costained with UL57 (Fig. 8A, panel 3). In the HCMV-infected cells, the incorporated BrU located primarily around the viral replication center, as marked by UL57 (Fig. 8A; panels 1 and 2 show the 30-min BrU labeling). Whether BrU incorporated into cellular or viral transcripts could not be discerned by this assay. As a complement, mock- or virus-infected cells were pulse-labeled with BrdU (60 min) and costained for UL57 (Fig. 8A, panels 4 to 6) to localize the area of DNA synthesis. Given the virus-mediated shutoff of cellular DNA synthesis, the BrdU would primarily be incorporated into replicating viral DNA. While the area of BrdU incorporation is diffusely spread through the nucleus in the uninfected cells (Fig. 8A, panel 6), the BrdU in the infected cells localized specifically within the viral replication center as marked by UL57 (Fig. 8A, panels 4 and 5). Taken together, these data indicate that ongoing RNA and DNA syntheses occur in spatially distinct areas in the infected cell, with RNA synthesis mainly occurring outside the viral replication center and viral DNA synthesis occurring inside the replication center.
FIG. 8.
The perireplication center region is transcriptionally active. Mock- or virus-infected HFFs (MOI of 2) were treated as indicated and processed by IFA with Hoescht stain (in blue). Midsectional planes of representative cells imaged by deconvolution microscopy with 0.2-μm sections at a magnification of 1,000× under oil immersion conditions are shown. (A) Cells were pulsed with BrU (30 min) or BrdU (60 min) at 36 h p.i. and then fixed for IFA. Incorporated BrU or BrdU is shown in green and UL57 is shown in red. (B) Cells were pulsed with BrU at 36 h p.i. for 30 min and stained for 19S Rpt6 (red) and incorporated BrU (green). (C and D) Cells were fixed at 48 h p.i. and stained for 19S Rpt6 (red) and H3K4 (green) (C) or UL57 (red) and pSer2-RNAP II by the H5 antibody (green) (D). (E) Cells were treated with MG132 from 36 to 48 h p.i. and then fixed. Cells stained for 19S Rpt6 (red) and UL57 (green) are shown.
Mock- or virus-infected cells were further examined by IFA to determine whether the relocalization of the proteasome subunits during the infection correspond to areas of active transcription. In Fig. 8B, cells pulsed with BrU at 36 h p.i. for 30 min were assayed for BrU and 19S Rpt6 localization. Comparable staining patterns were observed, with both BrU and Rpt6 localizing diffusely throughout the nucleus in the uninfected cell while localizing at the periphery of the replication center in the infected cell (Fig. 8B). Other markers associated with active transcription were also analyzed, including histone H3 trimethylated at residue K4 (H3K4) and pSer2-RNAP II (shown at 48 h p.i. in Fig. 8C and D, respectively). Both H3K4 and pSer2-RNAP II also shifted from a diffusely nuclear localization in the uninfected cells to the perireplication center region in the infected cells, as indicated by costaining with 19S Rpt6 and UL57 in Fig. 8C and D, respectively. These results provide further support that proteasomes localize to active transcription areas during the infection.
Given the inhibition of viral transcript expression upon proteasome inhibition, we were interested in determining whether inhibition of proteasome activity also affected the localization of the subunits. To address this possibility, proteasome localization was analyzed in cells treated with MG132 from 36 to 48 h p.i. and then fixed. The localization of proteasome subunits 19S Rpt1 and Rpt6, 20S α6, and 11S REGγ were assessed, and Rpt6 is shown costained with UL57 as a representative example in Fig. 8E. Rpt6 localization was not significantly altered with MG132, as the shift to the perireplication center region was still observed (Fig. 8E). Together, these results indicate that at later times in the infection, ongoing transcriptional activity occurs outside the viral replication center at regions where proteasome subunits also locate during later times of the infection.
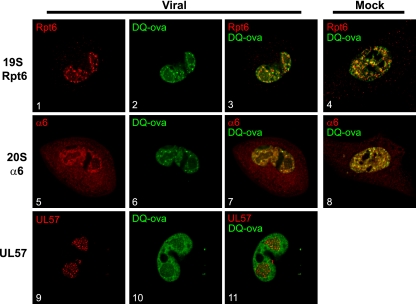
To determine if the proteasome is active at these perireplication center sites, the nuclei of mock- or virus-infected HFFs were microinjected with DQ-ova at 40 h p.i. DQ-ova is conjugated to BODIPY FL-dye, which is self-quenching and exhibits a low background signal but displays green fluorescence upon proteolytic degradation, allowing the localization of proteolytic activity to be tracked. Cells were fixed 30 min postinjection and processed for IFA with antibodies to 19S Rpt6, 20S α66, and UL57 (Fig. 9). Coinjections with the proteasome inhibitor Sal A (150 nM) sufficiently blocked DQ-ova fluorescence, confirming that the observed fluorescence was proteasome dependent (data not shown). In the mock-infected cells, DQ-ova localized throughout the nucleus, correlating with the localization of both 19S Rpt6 and 20S α6 (Fig. 9, panels 4 and 8, respectively). In the virus-infected cells, the localization of DQ-ova also correlated with that of 19S Rpt6 and 20S α6 (Fig. 9, panels 3 and 7, respectively). DQ-ova fluorescence was also increased in areas outside of the viral replication center, although faint staining within the replication center was still observed (Fig. 9, panels 10 and 11). Thus, this perireplication center region supports both transcriptional and proteolytic activity during later times of the infection.
FIG. 9.
Enhanced proteolytic activity occurs in the perireplication center region as HCMV infection progresses. HCMV- or mock-infected HFFs were microinjected with DQ-ova (green) at 40 h p.i. Cells were fixed 30 min postinjection and stained for 19S Rpt6, 20S α6, or UL57 (shown in red). Midsectional planes of representative cells imaged by deconvolution microscopy with 0.2-μm sections at a magnification of 600× under oil immersion conditions are shown.
The 19S non-ATPase base subunit Rpn2 relocalizes to the viral DNA replication center.
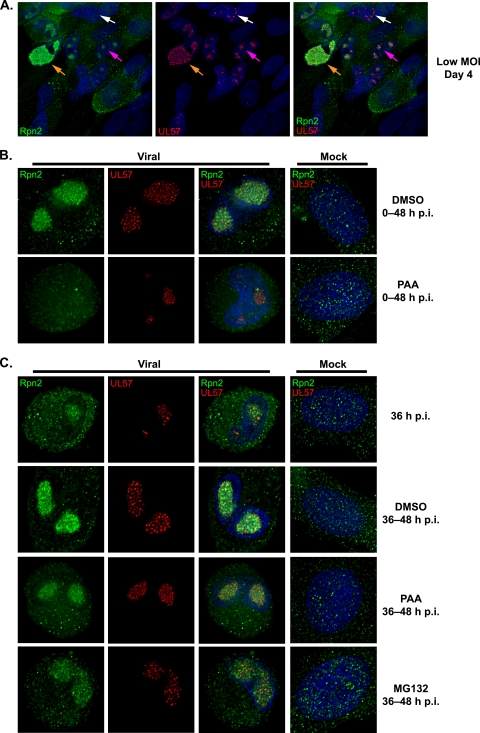
While several 19S, 20S, and 11S proteasome subunits relocalized around the replication center during the infection, a marked difference in localization pattern was observed for the 19S non-ATPase base subunit Rpn2. In mock-infected cells, Rpn2 appeared diffusely distributed throughout the cytosol and nucleus (Fig. 10B and C). However, in cells infected with HCMV at an MOI of 2, Rpn2 was observed within the viral replication center beginning at ∼30 h p.i. and continued to accumulate there as the infection progressed (Fig. 10 and data not shown). To better assess whether Rpn2 relocalization coincides with viral replication center formation, low-MOI infections, which allowed for the closer observation of replication centers at various stages of development, were analyzed for Rpn2, UL44, and UL57 localization by IFA. Cells fixed on day 4 and stained for Rpn2 and UL57 are shown in Fig. 10A as a representative example. As indicated by the white arrow in Fig. 10A, UL57 is present at the prereplication foci but not Rpn2. Rpn2 was not present at the replication centers until they were slightly more developed (Fig. 10A, pink arrow) and continued to accumulate there as the replication centers grew and coalesced (Fig. 10A, orange arrow). Thus, Rpn2 is not brought in with the core replication machinery and likely does not play a role in the initial formation of the replication center.
FIG. 10.
Proteasome subunit 19S Rpn2 relocalizes to the viral replication center after the onset of viral DNA replication. Cells were fixed at the times indicated and stained for 19S Rpn2 (green), UL57 (red), and Hoescht (blue). Midsectional planes of representative cells imaged by deconvolution microscopy with 0.2-μm sections are shown. (A) HFFs infected with HCMV (MOI of 0.01) were fixed daily. Representative cells at different stages of infection from day 4 are shown. Examples of prereplication center foci (white arrows), midsize replication centers (pink arrows), and a large coalesced replication center (orange arrows) are indicated. Cells were imaged at a magnification of 400× under oil immersion conditions. (B) Mock- or virus-infected HFFs (MOI of 2) were treated with DMSO or PAA at the onset of infection and fixed at 48 h p.i. Cells were imaged at a magnification of 1,000× under oil immersion conditions. (C) Mock- or virus-infected HFFs (MOI of 2) were treated with DMSO, PAA, or MG132 from 36 to 48 h p.i. Cells were fixed prior to treatment at 36 h p.i. and after treatment at 48 h p.i. Images were taken at a magnification of 1,000× under oil immersion conditions.
Rpn2 localization was also assessed in the presence of PAA or GCV to determine whether the relocalization is dependent on viral DNA synthesis. With the addition of PAA at 0 to 48 h p.i., replication center foci containing UL57 were observed, but Rpn2 remained diffusely nuclear and cytoplasmic and did not accumulate in the foci with UL57 (Fig. 10B). These data suggest that the Rpn2 relocalization also occurs after the onset of viral DNA synthesis. Similar results were also seen with GCV (data not shown). Addition of PAA at 36 to 48 h p.i. (after the onset of viral DNA synthesis) appeared to prevent further accumulation of Rpn2 in the viral replication center, as levels remained comparable to those seen at 36 h p.i. (Fig. 10C). Further accumulation of Rpn2 into the replication center was similarly inhibited with the addition of the proteasome inhibitor MG132 at 36 to 48 h p.i. (Fig. 10C). These data indicate that the relocalization of Rpn2 to the replication center is dependent on the initiation of viral DNA synthesis. Further accumulation of the subunit, but not maintenance once relocalized, also requires ongoing viral DNA synthesis and proteasome proteolytic activity.
DISCUSSION
In this report, we extend the studies from our lab and others on the interactions between HCMV and the ubiquitin-proteasome degradation pathway (4, 8, 15, 22, 23, 26-28, 31, 39, 46, 50, 51, 58, 59, 61, 62). Previous studies have shown HCMV infection to be negatively affected by inhibition of the proteasome (31, 46, 50), and in the experiments presented here, we have elucidated the temporal and molecular basis of this inhibition. We show that proteasome proteolytic activity is important for viral transcription and that as the infection progresses, the proteasome subunits accumulate in regions of RNA synthesis at the periphery of the viral DNA replication center in the nucleus.
We found that proteasome inhibition during the initial 12 h p.i. had little effect on IE gene expression, although IE2-86 expression decreased thereafter either after release from the drug or when the inhibitor was added at later times during the infection. Previous studies reported a decrease in both IE1-72 and IE2-86 expression when the proteasome inhibitor was added at the onset of infection and maintained throughout the infection time course (31, 46, 50). This discrepancy, especially with regard to IE1-72 expression, may be attributed to a difference in the MOI (31) and/or cell lines (50) used in these assays. The recent study by Sadanari et al. (50) showed that the effect of proteasome inhibitors on IE gene expression can be cell type specific, with inhibition in glioma and astrocytoma cell lines and activation in a neuroblastoma cell line. Although proteasome activity did not appear necessary to facilitate efficient viral entry or initial IE expression in the context of the experiments presented in this study, the requirement may also vary depending on MOI and cell type. The effects on IE2-86 expression upon proteasome inhibition have been more consistently observed, with decreased expression seen in multiple cell lines infected with HCMV at different MOIs, although little to no effect is seen at higher MOIs (31, 46, 50). The observed modest decrease in IE2-86 expression with the proteasome inhibitor present 0 to 12 h p.i. may in part account for the associated suppression of viral early and late protein expression, as there may not be sufficient transactivation of the viral early and late genes or cellular genes. Interestingly, early and late protein expression did not fully recover even after a 60-h release from MG132. This suggests that the switch to early gene expression and/or viral DNA synthesis may be critically dependent on the proteasome-mediated events that occur during the first 12 h p.i. For instance, cellular factors that are normally degraded to facilitate efficient viral replication (e.g., Daxx, unphosphorylated Rb) would not be degraded. Proteasome inhibition may also cause further cell cycle deregulation that is not conducive for viral replication.
The effect on viral early and late gene expression became less pronounced as the proteasome inhibitor was added later during the infection time course, such that addition at late times no longer affected early gene expression, while late gene expression was impacted to varying degrees. The proteasome inhibitors appeared to prevent further accumulation of both viral RNA and protein. If the inhibitor was added after expression had peaked, then gene expression was no longer affected. Although DNA synthesis decreased during intervals that the proteasome inhibitor was present, the effect was relatively modest (2- to 3-fold inhibition in accumulation over a 24-h period). Whether this inhibition is due to a direct effect on DNA synthesis or decreased expression of the viral early genes, which encode the majority of the viral DNA replication proteins, remains to be determined.
While several studies have localized IE transcription to the viral transcriptosomes, which are subnuclear foci that contain the input viral genome as well as several cellular transcription factors, cyclin-dependent kinases, and RNAP II that form adjacent to POD/ND10 structures (24, 25, 30, 57), the site of viral late gene transcription has not been established, although it has been hypothesized to occur around the replication center, given the localization of phosphorylated RNAP II there at later times of infection (57). We have further identified H3K4 and newly labeled BrU transcripts to localize outside the viral replication center, further indicating the perireplication center region to be transcriptionally active. Although we could not discriminate whether the BrU incorporated into viral or cellular transcripts in the BrU pulse-labeling assays, we provide further evidence that the majority of ongoing transcriptional activity is occurring outside the viral replication center at later times of the infection.
Interestingly, all of the proteasome subunits tested by IFA, except for Rpn2, showed enhanced localization around the replication center as the infection progressed into early-late times, when the majority of viral transcription occurs. In accord with this temporal relocalization, there was a requirement for initiation of viral DNA synthesis. Taken together with the increases in both the activity and abundance of the proteasome and the effect of proteasome inhibitors, these results further suggest that proteasome activity is required to facilitate efficient viral transcription. This would be consistent with the large body of evidence that the proteasome and its proteolytic activity play a major role in cellular transcription (9, 20, 41) as well as the studies linking proteasome activity with viral transcription. At this time, we cannot exclude a nonproteolytic role for specific subunits, as the 19S subunits also possess chaperone activities and can aid in remodeling protein complexes during transcription, modifying histones, and recruiting cotransactivator complexes (9, 13, 20, 32, 34, 40). The HIV-1 promoter is a notable example of transcription regulation mediated by both proteolytic and nonproteolytic mechanisms (33, 42, 43, 55). Moreover, a study with adenovirus reported the selective and independent recruitment of the APIS (i.e., 19S ATPase base subunits) and 20S complexes to the adenovirus E1A transactivation domain CR3 (47) and their roles in transcription initiation and elongation. Those authors further showed that treatment with proteasome inhibitors inhibited E1A transcription (47). A requirement for proteasome proteolytic activity in the removal of stalled RNAP II complexes has been implicated for efficient HSV-1 early and late gene transcription (10, 35). Although the necessity for the specific removal of RNAP II does not appear to be the case during HCMV infection, given that both Ser2- and Ser5-phosphorylated forms of RNAP II increase during the infection (reference 57 and this study), proteasome proteolytic activity may be required for the removal of other proteins involved in transcription.
The relocalization of the proteasome subunits to the HCMV replication center periphery is reminiscent of the enrichment of proteasomes in the VICE (virus-induced chaperone-enriched) domains that develop adjacent to the replication center in HSV-1-infected cells. Along with proteasomes, the domains contain various cellular chaperone proteins (i.e., Hsp70, Hsp40, Hsp90, and Hsc70) and ubiquitin-conjugated substrates (6, 7, 10, 35-37). It is proposed that the VICE domains regulate and sequester misfolded or modified proteins to prevent innate antiviral responses (i.e., apoptosis or unfolded protein response) from being activated (6) as well as facilitate the removal of stalled RNAP II complexes from HSV-1 templates when RNA synthesis is occurring at high levels (10).
The only proteasome subunit that appeared to accumulate within the HCMV DNA replication center was Rpn2. Interestingly, recent studies regarding 19S base assembly have shown that Rpn2/S1 is the only proteasome subunit that is not incorporated into a subcomplex prior to formation of the whole base complex, and thus it may exist as a free pool (29). It has been found that many cellular proteins, including p53, RPA, PCNA Nbs1, Rad50, Atrip, Chk1, cdk7, and MAT-1, accumulate in the viral replication center (12, 16, 38). Whether the accumulation of Rpn2 in the replication center is associated with a role in viral DNA synthesis or simply reflects its existence as a free pool is an important question to address.
While HCMV has developed multiple mechanisms to subvert and inhibit cellular functions to create an environment more conducive for viral replication, it is dependent on host factors and machinery to facilitate a productive infection. Given the many cellular systems the ubiquitin-proteasome pathways regulate, it is a convenient target for the virus to modify and manipulate. In this report, we have shown yet another facet in which HCMV utilizes the UPS in enhancing viral replication. The importance of the UPS at all stages of the HCMV infection supports further studies into this pathway as a potential antiviral target.
Acknowledgments
We are grateful to Bradley Moore for the sample of Sal A, to William Britt for the UL97, UL85, and UL86 antibodies, and to David Rose for his help with the microinjection assays. We appreciate the use of the microscopy resources at the UCSD School of Medicine Light Microscopy Facility. We also thank the members of the Spector lab for their support and comments on the manuscript.
This work was supported by NIH grants CA073490 and CA034729.
Footnotes
Published ahead of print on 30 December 2009.
REFERENCES
- 1.Ahn, J.-H., W.-J. Jang, and G. S. Hayward. 1999. The human cytomegalovirus IE2 and UL112-113 proteins accumulate in viral DNA replication compartments that initiate from the periphery of promyelocytic leukemia protein-associated nuclear bodies (PODs or ND10). J. Virol. 73:10458-10471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bajorek, M., and M. H. Glickman. 2004. Keepers at the final gates: regulatory complexes and gating of the proteasome channel. Cell. Mol. Life Sci. 61:1579-1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banks, L., D. Pim, and M. Thomas. 2003. Viruses and the 26S proteasome: hacking into destruction. Trends Biochem. Sci. 28:452-459. [DOI] [PubMed] [Google Scholar]
- 4.Biswas, N., V. Sanchez, and D. H. Spector. 2003. Human cytomegalovirus infection leads to accumulation of geminin and inhibition of the licensing of cellular DNA replication. J. Virol. 77:2369-2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blanchette, P., and P. E. Branton. 2009. Manipulation of the ubiquitin-proteasome pathway by small DNA tumor viruses. Virology 384:317-323. [DOI] [PubMed] [Google Scholar]
- 6.Burch, A. D., and S. K. Weller. 2005. Herpes simplex virus type 1 DNA polymerase requires the mammalian chaperone hsp90 for proper localization to the nucleus. J. Virol. 79:10740-10749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burch, A. D., and S. K. Weller. 2004. Nuclear sequestration of cellular chaperone and proteasomal machinery during herpes simplex virus type 1 infection. J. Virol. 78:7175-7185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cantrell, S. R., and W. A. Bresnahan. 2005. Interaction between the human cytomegalovirus UL82 gene product (pp71) and hDaxx regulates immediate-early gene expression and viral replication. J. Virol. 79:7792-7802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collins, G. A., and W. P. Tansey. 2006. The proteasome: a utility tool for transcription? Curr. Opin. Genet. Dev. 16:197-202. [DOI] [PubMed] [Google Scholar]
- 10.Dai-Ju, J. Q., L. Li, L. A. Johnson, and R. M. Sandri-Goldin. 2006. ICP27 interacts with the C-terminal domain of RNA polymerase II and facilitates its recruitment to herpes simplex virus 1 transcription sites, where it undergoes proteasomal degradation during infection. J. Virol. 80:3567-3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Demartino, G. N., and T. G. Gillette. 2007. Proteasomes: machines for all reasons. Cell 129:659-662. [DOI] [PubMed] [Google Scholar]
- 12.Dittmer, D., and E. S. Mocarski. 1997. Human cytomegalovirus infection inhibits G1/S transition. J. Virol. 71:1629-1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ezhkova, E., and W. P. Tansey. 2004. Proteasomal ATPases link ubiquitylation of histone H2B to methylation of histone H3. Mol. Cell 13:435-442. [DOI] [PubMed] [Google Scholar]
- 14.Finley, D. 2009. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu. Rev. Biochem. 78:477-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fortunato, E. A., V. Sanchez, J. Y. Yen, and D. H. Spector. 2002. Infection of cells with human cytomegalovirus during S phase results in a blockade to immediate-early gene expression that can be overcome by inhibition of the proteasome. J. Virol. 76:5369-5379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fortunato, E. A., and D. H. Spector. 1998. p53 and RPA are sequestered in viral replication centers in the nuclei of cells infected with human cytomegalovirus. J. Virol. 72:2033-2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fraser, K. A., and S. A. Rice. 2005. Herpes simplex virus type 1 infection leads to loss of serine-2 phosphorylation on the carboxyl-terminal domain of RNA polymerase II. J. Virol. 79:11323-11334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao, G., and H. Luo. 2006. The ubiquitin-proteasome pathway in viral infections. Can. J. Physiol. Pharmacol. 84:5-14. [DOI] [PubMed] [Google Scholar]
- 19.Glickman, M. H., and A. Ciechanover. 2002. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol. Rev. 82:373-428. [DOI] [PubMed] [Google Scholar]
- 20.Hegde, A. N., and S. C. Upadhya. 2006. Proteasome and transcription: a destroyer goes into construction. Bioessays 28:235-239. [DOI] [PubMed] [Google Scholar]
- 21.Hirsch, C., and H. Ploegh. 2000. Intracellular targeting of the proteasome. Trends Cell Biol. 10:268-272. [DOI] [PubMed] [Google Scholar]
- 22.Hofmann, H., H. Sindre, and T. Stamminger. 2002. Functional interaction between the pp71 protein of human cytomegalovirus and the PML-interacting protein human Daxx. J. Virol. 76:5769-5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hwang, J., and R. F. Kalejta. 2007. Proteasome-dependent, ubiquitin-independent degradation of Daxx by the viral pp71 protein in human cytomegalovirus-infected cells. Virology 367:334-338. [DOI] [PubMed] [Google Scholar]
- 24.Ishov, A. M., and G. G. Maul. 1996. The periphery of nuclear domain 10 (ND10) as site of DNA virus deposition. J. Cell Biol. 134:815-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ishov, A. M., R. M. Stenberg, and G. G. Maul. 1997. Human cytomegalovirus immediate early interaction with host nuclear structures: definition of an immediate transcript environment. J. Cell Biol. 138:5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ishov, A. M., O. V. Vladimirova, and G. G. Maul. 2002. Daxx-mediated accumulation of human cytomegalovirus tegument protein pp71 at ND10 facilitates initiation of viral infection at these nuclear domains. J. Virol. 76:7705-7712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kalejta, R. F., J. T. Bechtel, and T. Shenk. 2003. Human cytomegalovirus pp71 stimulates cell cycle progression by inducing the proteasome-dependent degradation of the retinoblastoma family of tumor suppressors. Mol. Cell. Biol. 23:1885-1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kalejta, R. F., and T. Shenk. 2003. Proteasome-dependent, ubiquitin-independent degradation of the Rb family of tumor suppressors by the human cytomegalovirus pp71 protein. Proc. Natl. Acad. Sci. U. S. A. 100:3263-3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaneko, T., J. Hamazaki, S.-I. Iemura, K. Sasaki, K. Furuyama, T. Natsume, K. Tanaka, and S. Murata. 2009. Assembly pathway of the mammalian proteasome base subcomplex is mediated by multiple specific chaperones. Cell 137:914-925. [DOI] [PubMed] [Google Scholar]
- 30.Kapasi, A. J., and D. H. Spector. 2008. Inhibition of the cyclin-dependent kinases at the beginning of the human cytomegalovirus infection specifically alters the levels and localization of the RNA polymerase II carboxyl-terminal domain kinases cdk9 and cdk7 at the viral transcriptosome. J. Virol. 82:394-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaspari, M., N. Tavalai, T. Stamminger, A. Zimmermann, R. Schilf, and E. Bogner. 2008. Proteasome inhibitor MG132 blocks viral DNA replication and assembly of human cytomegalovirus. FEBS Lett. 582:666-672. [DOI] [PubMed] [Google Scholar]
- 32.Kinyamu, H. K., W. N. Jefferson, and T. K. Archer. 2008. Intersection of nuclear receptors and the proteasome on the epigenetic landscape. Environ. Mol. Mutagen. 49:83-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lassot, I., D. Latreille, E. Rousset, M. Sourisseau, L. K. Linares, C. Chable-Bessia, O. Coux, M. Benkirane, and R. E. Kiernan. 2007. The proteasome regulates HIV-1 transcription by both proteolytic and nonproteolytic mechanisms. Mol. Cell 25:369-383. [DOI] [PubMed] [Google Scholar]
- 34.Lee, D., E. Ezhkova, B. Li, S. G. Pattenden, W. P. Tansey, and J. L. Workman. 2005. The proteasome regulatory particle alters the SAGA coactivator to enhance its interactions with transcriptional activators. Cell 123:423-436. [DOI] [PubMed] [Google Scholar]
- 35.Li, L., L. A. Johnson, J. Q. Dai-Ju, and R. M. Sandri-Goldin. 2008. Hsc70 focus formation at the periphery of HSV-1 transcription sites requires ICP27. PLoS One 3:e1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Livingston, C. M., N. A. Deluca, D. E. Wilkinson, and S. K. Weller. 2008. Oligomerization of ICP4 and rearrangement of heat shock proteins may be important for herpes simplex virus type 1 prereplicative site formation. J. Virol. 82:6324-6336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Livingston, C. M., M. F. Ifrim, A. E. Cowan, and S. K. Weller. 2009. Virus-induced chaperone-enriched (VICE) domains function as nuclear protein quality control centers during HSV-1 infection. PLoS Pathog. 5:e1000619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luo, M. H., K. Rosenke, K. Czornak, and E. A. Fortunato. 2007. Human cytomegalovirus disrupts both ataxia telangiectasia mutated protein (ATM)- and ATM-Rad3-related kinase-mediated DNA damage responses during lytic infection. J. Virol. 81:1934-1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marshall, K. R., K. V. Rowley, A. Rinaldi, I. P. Nicholson, A. M. Ishov, G. G. Maul, and C. M. Preston. 2002. Activity and intracellular localization of the human cytomegalovirus protein pp71. J. Gen. Virol. 83:1601-1612. [DOI] [PubMed] [Google Scholar]
- 40.Mittenberg, A., T. Moiseeva, and N. Barlev. 2008. Role of proteasomes in transcription and their regulation by covalent modifications. Front. Biosci. 13:7184. [DOI] [PubMed] [Google Scholar]
- 41.Mocarski, E. S., T. Shenk, and R. F. Pass. 2007. Cytomegaloviruses, p. 2701-2772. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 5th ed., vol. 2. Wolters Kluwer Health/Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 42.Nelbock, P., P. J. Dillon, A. Perkins, and C. A. Rosen. 1990. A cDNA for a protein that interacts with the human immunodeficiency virus tat transactivator. Science 248:1650-1653. [DOI] [PubMed] [Google Scholar]
- 43.Ohana, B., P. A. Moore, S. M. Ruben, C. D. Southgate, M. R. Green, and C. A. Rosen. 1993. The type 1 human immunodeficiency virus Tat binding protein is a transcriptional activator belonging to an additional family of evolutionary conserved genes. Proc. Natl. Acad. Sci. U. S. A. 90:138-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Penfold, M. E., and E. S. Mocarski. 1997. Formation of cytomegalovirus DNA replication compartments defined by localization of viral proteins and DNA synthesis. Virology 239:46-61. [DOI] [PubMed] [Google Scholar]
- 45.Pickart, C. M., and R. E. Cohen. 2004. Proteasomes and their kin: proteases in the machine age. Nat. Rev. Mol. Cell Biol. 5:177-187. [DOI] [PubMed] [Google Scholar]
- 46.Prösch, S., C. Priemer, C. Höflich, C. Liebenthaf, N. Babel, D. H. Krüger, and H.-D. Volk. 2003. Proteasome inhibitors: a novel tool to suppress human cytomegalovirus replication and virus-induced immune modulation. Antivir. Ther. (London) 8:555-567. [PubMed] [Google Scholar]
- 47.Rasti, M., R. J. A. Grand, A. F. Yousef, M. Shuen, J. S. Mymryk, P. H. Gallimore, and A. S. Turnell. 2006. Roles for APIS and the 20S proteasome in adenovirus E1A-dependent transcription. EMBO J. 25:2710-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rechsteiner, M., and C. P. Hill. 2005. Mobilizing the proteolytic machine: cell biological roles of proteasome activators and inhibitors. Trends Cell Biol. 15:27-33. [DOI] [PubMed] [Google Scholar]
- 49.Reed, S. H., and T. G. Gillette. 2007. Nucleotide excision repair and the ubiquitin proteasome pathway: do all roads lead to Rome? DNA Repair (Amsterdam) 6:149-156. [DOI] [PubMed] [Google Scholar]
- 50.Sadanari, H., J. Tanaka, Z. Li, R. Yamada, K. Matsubara, and T. Murayama. 2009. Proteasome inhibitor differentially regulates expression of the major immediate early genes of human cytomegalovirus in human central nervous system-derived cell lines. Virus Res. 142:68-77. [DOI] [PubMed] [Google Scholar]
- 51.Saffert, R. T., and R. F. Kalejta. 2006. Inactivating a cellular intrinsic immune defense mediated by Daxx is the mechanism through which the human cytomegalovirus pp71 protein stimulates viral immediate-early gene expression. J. Virol. 80:3863-3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Salvant, B. S., E. A. Fortunato, and D. H. Spector. 1998. Cell cycle dysregulation by human cytomegalovirus: influence of the cell cycle phase at the time of infection and effects on cyclin transcription. J. Virol. 72:3729-3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sanchez, V., A. K. McElroy, and D. H. Spector. 2003. Mechanisms governing maintenance of cdk1/cyclin B1 kinase activity in cells infected with human cytomegalovirus. J. Virol. 77:13214-13224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shackelford, J., and J. S. Pagano. 2005. Targeting of host-cell ubiquitin pathways by viruses. Essays Biochem. 41:139-156. [DOI] [PubMed] [Google Scholar]
- 55.Shibuya, H., K. Irie, J. Ninomiya-Tsuji, M. Goebl, T. Taniguchi, and K. Matsumoto. 1992. A new human gene encoding a positive modulator of HIV Tat-mediated transactivation. Nature 357:700-702. [DOI] [PubMed] [Google Scholar]
- 56.Tamashiro, J. C., L. J. Hock, and D. H. Spector. 1982. Construction of a cloned library of the EcoRI fragments from the human cytomegalovirus genome (strain AD169). J. Virol. 42:547-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tamrakar, S., A. J. Kapasi, and D. H. Spector. 2005. Human cytomegalovirus infection induces specific hyperphosphorylation of the carboxyl-terminal domain of the large subunit of RNA polymerase II that is associated with changes in the abundance, activity, and localization of cdk9 and cdk7. J. Virol. 79:15477-15493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tortorella, D., B. Gewurz, D. Schust, M. Furman, and H. Ploegh. 2000. Down-regulation of MHC class I antigen presentation by HCMV; lessons for tumor immunology. Immunol. Invest. 29:97-100. [DOI] [PubMed] [Google Scholar]
- 59.Tran, K., J. A. Mahr, J. Choi, J. G. Teodoro, M. R. Green, and D. H. Spector. 2008. Accumulation of substrates of the anaphase-promoting complex (APC) during human cytomegalovirus infection is associated with the phosphorylation of Cdh1 and the dissociation and relocalization of the APC subunits. J. Virol. 82:529-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.White, E. A., C. L. Clark, V. Sanchez, and D. H. Spector. 2004. Small internal deletions in the human cytomegalovirus IE2 gene result in nonviable recombinant viruses with differential defects in viral gene expression. J. Virol. 78:1817-1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wiebusch, L., M. Bach, R. Uecker, and C. Hagemeier. 2005. Human cytomegalovirus inactivates the G0/G1-APC/C ubiquitin ligase by Cdh1 dissociation. Cell Cycle 4:1435-1439. [DOI] [PubMed] [Google Scholar]
- 62.Wiebusch, L., R. Uecker, and C. Hagemeier. 2003. Human cytomegalovirus prevents replication licensing by inhibiting MCM loading onto chromatin. EMBO Rep. 4:42-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wing, B. A., and E. S. Huang. 1995. Analysis and mapping of a family of 3′-coterminal transcripts containing coding sequences for human cytomegalovirus open reading frames UL93 through UL99. J. Virol. 69:1521-1531. [DOI] [PMC free article] [PubMed] [Google Scholar]