Abstract
Coinfection with human T-cell lymphotropic virus type 2 (HTLV-2) and human immunodeficiency virus type 1 (HIV-1) has been reported to have either a slowed disease course or to have no effect on progression to AIDS. In this study, we generated a coinfection animal model and investigated whether HTLV-2 could persistently infect macaques, induce a T-cell response, and impact simian immunodeficiency virus SIVmac251-induced disease. We found that inoculation of irradiated HTLV-2-infected T cells into Indian rhesus macaques elicited humoral and T-cell responses to HTLV-2 antigens at both systemic and mucosal sites. Low levels of HTLV-2 provirus DNA were detected in the blood, lymphoid tissues, and gastrointestinal tracts of infected animals. Exposure of HTLV-2-infected or naïve macaques to SIVmac251 demonstrated comparable levels of SIVmac251 viral replication, similar rates of mucosal and peripheral CD4+ T-cell loss, and increased T-cell proliferation. Additionally, neither the magnitude nor the functional capacity of the SIV-specific T-cell-mediated immune response was different in HTLV-2/SIVmac251 coinfected animals versus SIVmac251 singly infected controls. Thus, HTLV-2 targets mucosal sites, persists, and importantly does not exacerbate SIVmac251 infection. These data provide the impetus for the development of an attenuated HTLV-2-based vectored vaccine for HIV-1; this approach could elicit persistent mucosal immunity that may prevent HIV-1/SIVmac251 infection.
Human T-cell lymphotropic virus type 2 (HTLV-2) was discovered in 1982 and recognized as the second human retrovirus found (29). HTLV-2 is closely related to the first human retrovirus discovered, HTLV-1 (49, 50), a pathogenic virus that causes adult T-cell leukemia/lymphoma (ATLL) and an inflammatory neurologic disorder called HTLV-1-associated myelopathy or tropical spastic paraparesis (HAM/TSP) (22, 45).
HTLV-2 is prevalent in Amerindian populations of North and South America and in Africa (57). The prevalence of HTLV-2 is generally low; however, in the past 20 years, an epidemic of HTLV-2 infection has occurred among intravenous drug users (8, 24, 54, 57). HTLV-2 establishes a lifelong infection and replicates at low levels in most infected individuals. While anecdotal cases of TSP/HAM-like neurological manifestations (1, 44) and hematopoietic diseases, such as large granular lymphoma (LGL), in HTLV-2-infected individuals have been reported (3, 37-39, 46), the extent to which HTLV-2 can induce disease in humans remains unclear. Indeed, even in the condition of immune deficiency, such as infection with human immunodeficiency virus type 1 (HIV-1), HTLV-2 coinfection has not been reported to be associated with cancer or neurological diseases. However, more studies are necessary to fully understand the role of HTLV-2 in human disease. While HTLV-1 infection has been connected with an accelerated course of disease in HIV-1 coinfected patients (2, 34), HTLV-2 has been reported to either have no effect (26) or suggested to exert a potential protective role during HIV-1 infection (12, 23). This protective role is thought to be due to a maintenance of CD4+ T cells, lowering immune activation, and delayed progression to AIDS (4, 5). In addition, modulation of cytokine and chemokine networks by HTLV-2 has been suggested to contribute to the control of HIV-1 infection (12, 36, 47). Since studies on the immunological interactions between HIV-1 and HTLV-2 have been performed in patients coinfected with HIV-1 and HTLV-2 in the chronic phase of HIV-1 disease, little is known about the effects of HTLV-2 infection during acute HIV-1 replication, mucosal CD4+ T-cell depletion, or HIV-1-specific immune responses. Furthermore, the potential protective effect of an HTLV-2 vector that would target both CD4+ and CD8+ T cells and induce a low-grade persistent infection makes HTLV-2 an interesting potential vaccine platform for an HIV-1 vaccine.
Current HIV-1 vaccine strategies have focused on viral vectors delivering HIV-1 antigens. These vectors stimulate strong, systemic antigen-specific responses but are unable to protect from infection, since they generate only limited mucosal responses and do not persist. The only vaccine approach that has conferred protection in the simian immunodeficiency virus SIVmac251 macaque model is a live attenuated virus (17), suggesting that persistent expression of viral antigens in mucosal and lymphoid tissues may be necessary. An HTLV-2 vector expressing HIV-1 antigens at mucosal sites that stimulates and maintains T-cell responses in the gut may confer protection from infection by quickly eliminating cells infected by the founder virus at the portal of entry. This study establishes that the Indian rhesus macaque model for HTLV-2 infection is a suitable model to test this hypothesis, as it demonstrates that HTLV-2 targets systemic, lymphoid, as well as mucosal tissues of rhesus macaques. HTLV-2 infection induces humoral as well as cell-mediated immune responses, and importantly, T-cell responses can be found at both systemic and mucosal sites. In this study, we demonstrate that the viral and T-cell dynamics of macaques dually infected with HTLV-2 and SIVmac251 are similar to those of macaques singly infected with SIVmac251.
MATERIALS AND METHODS
Experimental HTLV-2 and SIVmac251 infection.
The 11 Indian rhesus macaques (RMs) used in this study were housed and cared for under the guidelines of the Association for the Assessment and Accreditation of Laboratory Animal Care International. RMs were housed at the Advanced Biosciences Laboratory in Rockville, MD. Seven RMs were inoculated with 108 HTLV-2-infected lethally irradiated cells of human (Mo-T) or rhesus (M304) origin. Mo-T cells are an HTLV-2-producing cell line that was derived from a male patient with hairy cell leukemia (29). To create an HTLV-2-expressing rhesus macaque cell line, we cocultured cells from macaque M304 with gamma-irradiated Mo-T cells at a 1:1 ratio. Cells were maintained in RPMI 1640 complete medium supplemented with 10% heat-inactivated fetal calf serum (FCS), 1% penicillin-streptomycin, and 20 U/ml recombinant human interleukin 2 (IL-2). HTLV-2 infection was documented by measuring extracellular p19 Gag production by an antigen capture assay and by intracellular p19 Gag staining. The cell line derived from this culture was designated RhM304. Animals M304 and L900 were inoculated with HTLV-2-infected cells from animal M304, i.e., an autologous or allogeneic infection, respectively, while animal M214 received a heterologous infection with Mo-T cells. Both heterologous and autologous infections produced a persistent HTLV-2 infection; therefore, the remaining HTLV-2 infections were performed using irradiated Mo-T cells. Four additional animals (M893, M897, M905, and M906) were infected with HTLV-2; 10 months after HTLV-2 infection, these four animals and four naïve macaques were infected intrarectally with SIVmac251 (105 50% tissue culture infective dose [TCID50]). All SIV-infected macaques were haplotyped, two macaques were MamuA01+ (animals M905 and P160), one was an HTLV-2 preinfected animal, and the other a naïve control. All of the animals were B08 negative, and one of the HTLV-2 preinfected animals was positive for B17 (animal M893).
Blood and tissue collection.
Mononuclear cells were isolated from blood, rectal, jejunal pinch biopsy, lymph node biopsy, and bone marrow aspirate specimens. Mononuclear cells were separated from whole blood and bone marrow aspirate specimens by density gradient centrifugation (Ficoll). Lymph nodes were homogenized and passed through a 100-μm cell strainer, and mononuclear cells were purified by density gradient centrifugation (Ficoll). Rectal and jejunal pinch biopsy specimens were treated with 1 mM ultrapure dithiothreitol (Invitrogen, Carlsbad, CA) for 20 min followed by incubation in 0.1 M EDTA solution in calcium-magnesium-free HBSS with penicillin-streptomycin for 60 min to remove the epithelial layer. Lamina propria lymphocytes were separated, following the removal of the intraepithelial lymphocytes, by incubating with collagenase D (400 U/ml; Boehringer Mannheim, Mannheim, Germany) and DNase (1 μg/ml; Invitrogen, Carlsbad, CA) for 2 h at 37°C in Iscove's modified Dulbecco's medium supplemented with 10% fetal bovine serum (FBS) and penicillin-streptomycin. The dissociated mononuclear cells were then placed over 42% Percoll (General Electric Healthcare, Piscataway, NJ) and centrifuged at 2,000 rpm for 30 min at 4°C. Immunophenotypic studies were performed by polychromatic flow cytometry (as detailed below) on mononuclear cells derived from all tissues. All cells were stained on the day of sampling, except for cells from the jejunum and rectum, which were allowed to rest overnight and then stained.
Immunophenotyping and intracellular cytokine assay.
Four- and seven-color flow cytometric analysis was performed on mononuclear cells from blood and tissues. Surface staining was performed for 30 min at room temperature with antibodies to CD3 (clone SP34-2), CD4 (L200), chemokine (C-C motif) receptor 5 (CCR5) (clone 3A9), and CD95 (clone DX2), all obtained from BD Biosciences (San Diego, CA). Anti-CD28 (clone CD28.2) was obtained from eBiosciences (San Diego, CA), and anti-CD8 (clone 3B5) was obtained from Invitrogen (Carlsbad, CA). Following surface staining, the cells were permeabilized and stained with anti-Ki67 (clone B56 from BD Biosciences). To monitor SIV- and HTLV-2-specific immune responses, lymphocytes were resuspended at 106 cells per ml in RPMI 1640 complete medium with 10% heat-inactivated FCS in combination with anti-CD28 (Biosource International, Camarillo, CA), anti-CD49d (Becton Dickinson, San Jose, CA), and monensin (BD Biosciences). The cells were incubated with medium only or with peptide pools of 15-mers overlapping by 11 amino acids at a concentration of 1 μg/ml derived from the entire HTLV-2 Tax, HTLV-2 Gag, SIVmac251 Gag, and SIVmac251 Env protein sequence. Stimulation with either phorbol myristate acid (PMA) and ionomycin A23187 (Sigma-Aldrich, St. Louis, MO) or staphylococcal endotoxin B (SEB) (Toxin Technologies, Sarasota, FL) was used as a positive control. Cells were stimulated for 5 h, washed, and stained for CD3, CD4, and CD8 for 30 min at room temperature. Following surface staining, lymphocytes were permeabilized with FACS Perm/Wash solution (BD Biosciences), and stained intracellularly for gamma interferon (IFN-γ) (clone B27), tumor necrosis factor alpha (TNF-α) (clone MAB11), macrophage inflammatory protein 1α (MIP1α) (clone 11A3), and IL-2 (clone MQ1-17H12) (all from BD Biosciences), in addition to IL-17 (clone eBio64DEC17) from eBioscience. All cells were fixed with 1% paraformaldehyde, and at least 100,000 events were acquired on either a FACSCalibur or LSRII. Data analysis was performed with Flowjo (Treestar, CA).
Infection of primary dendritic cells.
Plasmacytoid dendritic cells (pDCs) were isolated from the peripheral blood mononuclear cells (PBMCs) of uninfected rhesus macaques. Non-pDC lineages were first removed using a commercially available kit (Miltenyi Biotec, Auburn, CA) followed by positive selection of BDCA-4 (NP-1) cells. A portion of the cells were fixed immediately, permeabilized, and stained for the presence of Tax-2. The remaining pDCs were infected with HTLV-2 harvested from the supernatant of Mo-T cells by incubating the virus with pDCs for 3 h, washing, and replating in six-well plates at 5 ×105/well. The pDCs were confirmed to be CD123+ by flow cytometry using an anti-CD123 antibody (clone 7G3; BD Bioscience). These pDCs were cultured in RPMI 1640 medium supplemented with 10% human AB serum and IL-3 (10 ng/ml), and the cells were harvested, permeabilized, and stained for Tax-2 either 2 or 6 days later.
Serological analysis and intracellular staining for HTLV-2 antigens.
The presence of antibodies to viral antigens was determined by Western blotting the sera collected from infected animals. Blot strips were obtained from ZeptoMetrix Corporation (Buffalo, NY). The blots contain multiple viral proteins, including HTLV Gag (p19), a unique HTLV-1 envelope recombinant protein rgp46-I, and GD21, a common, yet specific HTLV-1 and HTLV-2 gp41 epitope of the envelope protein. Each strip also includes an internal sample control to minimize the risk of false-negative results due to operational errors. The assay was carried out according to the manufacturer's instructions.
Intracellular staining for viral antigens was performed using a mouse anti-p19 Gag antibody (Zeptometrix, Buffalo, NY) or a rabbit anti-Tax2 antibody (40). Briefly, cell pellets were fixed in 100 μl IC fixation buffer (eBioscience). Intracellular staining was performed in 100 μl of permeabilization buffer using a 1:250 dilution of the anti-p19 Gag antibody or 1:100 dilution of the anti-Tax2 antibody and incubated for 30 min in the dark at room temperature. The cells were washed twice and stained with Alexa Fluor 488-labeled anti-mouse IgG antibody or Alexa Fluor 488-labeled anti-rabbit antibody (Invitrogen, Carlsbad CA) for 30 min in the dark at room temperature. Next, the cells were washed and resuspended in 1% paraformaldehyde (Electron Microscopy Sciences, Hatfield, PA) in phosphate-buffered saline (PBS). Four-parameter flow cytometric analysis was performed using a FACSCalibur with CELLQuest software.
Viral load.
SIV RNA in plasma samples and SIV DNA in the blood and tissue samples of macaques was quantified by nucleic acid sequence-based amplification (NASBA) as previously described (51). Quantification of HTLV-2 DNA in blood and tissue samples from animals L900, M214, and M304 was performed by extracting genomic DNA with the DNeasy tissue kit (Qiagen, Valencia, CA) according to the manufacturer's protocol, except for the DNA elution step. DNA was eluted with 10 mM Tris (pH 8.0). Five hundred nanograms of genomic DNA was subjected to real-time PCR. The TaqMan probe and PCR primers were designed within the gag gene of HTLV-2 (53). The TaqMan probe and primer sequences are as follows: probe, 5′-FAM-AGGCGTGGACACCCAAGGACAAAAC-TAMRASp-3′ (FAM is 6-carboxyfluorescein); forward primer, 5′-GGGAGATGCTCCGGACATG-3′; reverse primer, 5′-CGTGGTTGGACCACAAGGA-3′. Reaction conditions were as follows. The 25-μl PCR mixture for HTLV-2 and macaque albumin DNA consisted of 500 ng of DNA extracted from tissues or PBMCs, 200 nM primers, 100 nM probe, and 2× TaqMan Universal PCR Mastermix (Applied Biosystems, Carlsbad, CA). Amplification was performed using an ABI Prism 7500 sequence detector system (Applied Biosystems). The normalized value of the HTLV-2 proviral DNA load was calculated as HTLV-2 DNA copy number/macaque albumin gene copy number × 2 × 106 and expressed as the number of HTLV-2 proviral DNA copies per 106 cells (15).
The HTLV-2 proviral DNA load on animals M893, M897, M905, and M906 was determined in quadruplicate for each sample using real-time quantitative PCR with SYBR green intercalation as described elsewhere (33). This assay had excellent amplification efficiency, as determined by diluted DNA standards included on each plate, had a lower limit of detection of a single copy per reaction mixture, and had good interassay reliability with an R2 of 0.94 for HTLV-2-infected subjects. Serial samples from the same animal were included in the same testing batch to minimize interassay effects on serial viral loads. Cell input was determined by HLA-DQA quantification in the same aliquot (33).
A nested quantitative PCR was also used to amplify a 128-nucleotide sequence in the HTLV-2 Tax gene. DNA was extracted from blood and lymph node samples. DNA extraction was performed using the DNeasy blood and tissue kit (Qiagen, Valencia, CA) according to the manufacturer's instructions. The PCR mixture included 250 ng of DNA, the Platinum High Fidelity Supermix (Invitrogen, Carlsbad, CA), and primers specific for either β-actin (forward, 5′-CGGTTGGCCTTGGGGTTCAGGGGG-3′; reverse, 5′-ATCGTGGGGCGCCCCAGGCACCA-3′) or the HTLV-2 tax gene (forward,5′-TGGATACCCCGTCTACGTGT-3′; reverse,5′-GAGCTGACAACGCGTCCATC-3′; nested forward, 5′-GTGTTTGGCGATTGTGTACA-3′; nested reverse, 5′-CCATCGATGGGGTCCCA-3′). The primers were designed using Primer3 software.
Immunohistochemistry and quantitative image analysis.
All slides were stained using the Dako autostainer (Dako Inc., Carpinteria, CA). Slides were visualized with epifluorescence illumination using a Zeiss Axioplan 2 microscope (Carl Zeiss) and appropriate filters. Digital images were captured and analyzed by using a Zeiss Axiocam System and Openlab software (Inprovision) as previously described (14, 19). The primary antibodies used included monoclonal anti-CD4 mouse serum (clone IF6; Vector, Burlingame, CA), monoclonal anti-CD8 mouse serum (clone IA5; Leica Microsystems, Bannockburn, IL), and polyclonal anti-Ki67 rabbit serum (Lab Vision, Fremont, CA). For all primary antibodies, slides were subjected to an antigen retrieval step consisting of incubation in AR10 (Biogenex Inc., San Ramon, CA) for 2 min at 123°C in the digital decloaking chamber (Biocare Medical Inc., Concord, CA) followed by cooling to 90°C before rinsing in running water and a final buffer rinse. Primary antibodies were replaced by normal (healthy) rabbit IgG (Zymed Inc., South San Francisco, CA) or mouse IgG (Dako Inc., Carpinteria, CA) and included with each staining series as a negative control. Binding of CD4 and Ki-67 was detected simultaneously using Alexa Fluor 488-labeled polyclonal goat anti-rabbit IgG (Molecular Probes, Eugene, OR) and Alexa Fluor 568-labeled polyclonal goat anti-mouse IgG (Molecular Probes, Eugene, OR) for 1 h. For CD8 and Ki67 staining, Envision mouse and rabbit polymer (Dako Inc., Carpinteria, CA) were used. Binding of CD8 and Ki-67 was detected by diaminobenzidine (DAB) (CD8) and vector SG (Ki67) (Dako Inc., Carpinteria, CA). All the control experiments gave appropriate results with minimal nonspecific staining (data not shown). Slides were visualized with epifluorescence illumination using a Zeiss Axioplan 2 microscope (Carl Zeiss Inc., Thornwood, NY) and appropriate filters. Digital images were captured and analyzed by using a Zeiss Axiocam System and Openlab software (Inprovision Inc., Waltham MA). Only clearly positive cells were considered positive. The number of positive cells is presented as the number of cells per square millimeter.
ELISA.
An HTLV-1/2 enzyme-linked immunosorbent assay (ELISA) kit from Advanced Biosciences Laboratories (Kensington, MD) was used to quantify HTLV-2 p24 in serum. Briefly, plates were coated with affinity-purified HTLV-2 p24. Standards and samples were added to the plate, any antibodies in the sera bound to the HTLV-2 antigen, the plates were washed, and the quantity of HTLV-2 p24 is detected by an enzymatic reaction. A value is considered positive if its optical density is at least twice the value of a known negative macaque serum sample read at 450 nm. A MIP1α ELISA kit from R&D Systems (Minneapolis, MN) was used to quantify MIP1α; this assay uses a quantitative sandwich assay technique, where a monoclonal antibody specific to MIP1α is precoated on the plate. Standards and plasma samples from the macaques were added to the plates. If MIP1α is present, the immobilized antibody binds, the plate is then washed, and a polyclonal antibody is used to sandwich the MIP1α. The quantity of MIP1α is detected using an enzymatic reaction. The minimum MIPα detectable by the kit is 10 pg/ml.
Statistical analyses.
Comparisons between groups were performed using the Wilcoxon rank sum test for continuous factors or a repeated-measure analysis of variance with a Dunn's multiple comparison posttest. Graphical analysis was performed using GraphPad Prism, and error bars on graphs represent standard errors of the means.
RESULTS
HTLV-2 establishes a persistent infection in rhesus macaques and replicates in lymphoid and mucosal tissue.
To determine the optimum method for establishing an HTLV-2 infection in rhesus macaques, we inoculated three monkeys with 108 lethally irradiated, HTLV-2-producing cells of either human (Mo-T) or rhesus (RhM304) origin. We performed a heterologous infection in one animal (M214), which was given the HTLV-2 cell line Mo-T (Table 1). This cell line was originally generated from cells obtained from a male patient with a T-cell variant of hairy cell leukemia (29). It contains a replication-competent genome of HTLV-2 and two defective HTLV-2 genomes. To perform an autologous infection, we cultured PBMCs from one macaque (M304) with lethally irradiated Mo-T cells and generated a HTLV-2-expressing macaque cell line, named RhM304. HTLV-2 infection and gene expression in RhM304 and Mo-T cells were confirmed by flow cytometric staining for Gag and by PCR (Fig. 1A and B). RhM304 cells were confirmed to be cells of nonhuman primate origin with a diploid set of 22 chromosomes. RhM304 cells were lethally irradiated and used to infect animal M304 and a third macaque, L900. Two of the animals, M214 and M304, became persistently infected and seroconverted; antibodies to HTLV-2 p24 were detected by ELISA (Fig. 1C). Eighteen months after inoculation, the animals were sacrificed, and blood, bone marrow, spleen, lymph nodes, intestinal, vaginal, and brain tissue samples were collected along with spinal fluid. HTLV-2 proviral DNA was amplified from the spleen, ileum, and jejunum (Fig. 1D), whereas proviral DNA was not detected in the brain or in the spinal cord of any of the animals (data not shown). HTLV-2 was also demonstrated to infect primary rhesus macaque dendritic cells. Plasmacytoid dendritic cells were isolated from blood from a naïve rhesus macaque and infected with cell-free HTLV-2 in vitro. Figure 1E shows Tax staining in macaque CD123+ dendritic cells after 6 days in culture. Altogether, these findings demonstrate that HTLV-2 infects rhesus macaques and replicates in lymphoid and mucosal tissues.
TABLE 1.
Inoculation of macaquesa
| Animal code | HTLV-2 cell line | Type of inoculation |
|---|---|---|
| L900 | RhM304 | Allogeneic |
| M304 | RhM304 | Autologous |
| M214 | Mo-T | Heterologous |
| M893 | Mo-T | Heterologous |
| M897 | Mo-T | Heterologous |
| M905 | Mo-T | Heterologous |
| M906 | Mo-T | Heterologous |
Each macaque was inoculated intravenously with 108 RhM304 or Mo-T cells in 20 ml of PBS.
FIG. 1.
HTLV-2 establishes an infection in rhesus macaques, replicating in lymphoid and mucosal tissues. (A) Intracellular HTLV-2 Gag p19 expression in the Mo-T and RhM304 cell lines measured by flow cytometry. The isotype control (ctrl) is shown in blue, and p19 Gag expression is shown in black. (B) Number of HTLV-2 genome copies per 106 cells in Mo-T and RhM304 cells. (C) Serum antibodies to p24 Gag measured by ELISA 1 to 3 months after HTLV-2 infection in animals L900, M214, and M304. OD, optical density. (D) HTLV-2 proviral DNA load in the spleen, ileum, and jejunum from animals M214 and M304. (E) Tax protein expression in plasmacytoid dendritic cells infected in vitro with cell-free HTLV-2 (black) or uninfected plasmacytoid dendritic cells (blue).
HTLV-2 induces humoral and cell-mediated immune responses in rhesus macaques.
As both autologous and heterologous HTLV-2-expressing cells were infectious in rhesus macaques, we inoculated 4 additional animals (M893, M897, M905, and M906) with the irradiated Mo-T cell line (Table 1) and monitored T-cell dynamics and humoral and cellular immune responses to HTLV-2 in these animals. The presence of antibodies specific for viral antigens was evaluated by Western blotting in all the HTLV-2-infected macaques. All four animals mounted antibody responses against HTLV-2 antigens within 30 days of infection, which persisted for over 6 months (Fig. 2A). HTLV-2 proviral DNA was measured for the first 18 weeks after HTLV-2 infection (Table 2). HTLV-2 tax DNA was amplified by nested PCR from T cells obtained from the lymph nodes of all four animals at 3 months postinfection (Fig. 2B), but not in the DNA of uninfected macaques used as negative controls. Interestingly, both CD8+ and CD8− cells contained HTLV-2 tax, which suggests that HTLV-2 can replicate in both T-cell types in macaques (12). Importantly, HTLV-2 infection did not induce a loss of CD4+ or CD8+ T cells in vivo. A repeated-measure analysis of variance with square root-transformed counts over time demonstrated that the number of CD8+ T cells increased significantly over time (P = 0.007), while the number of CD4+ T cells displayed a trend toward significance (P = 0.049) (Fig. 2C). HTLV-2-specific T-cell responses in blood and the gastrointestinal tract were measured by intracellular cytokine staining for IFN-γ, TNF-α, and IL-2 in response to peptide pools of 2 major antigenic proteins of HTLV-2 Gag and Tax. Representative flow cytometric plots of IFN-γ and/or TNF-α staining in CD8+ T cells isolated from the blood and gut before and after stimulation with HTLV-2 Gag or Tax are shown in Fig. 2D and E, respectively. Flow cytometric analysis of the average frequency of CD8+ T cells producing cytokines demonstrated low levels of antigen-specific responses in the blood and gut (Fig. 2F and G). HTLV-2-specific responses peaked 3 to 4 months postinfection, and very low level responses were detectable up to 10 months postinfection (data not shown).
FIG. 2.
HTLV-2 infection stimulates humoral and cell-mediated responses. (A) Serum antibodies specific to several HTLV-2 proteins measured by Western blotting in animals M893, M897, M905, and M906. The months postinfection (p.i.) are shown below the blot. (B) Nested semiquantitative PCR to amplify either HTLV-2 tax or β-actin. DNA was extracted from the lymph nodes of macaques 3 months after HTLV-2 infection, and the mononuclear cells were CD8 enriched (+) or CD8 depleted (−). (C) CD4+ and CD8+ T-cell counts in blood from HTLV-2-infected macaques. Repeated-measure analysis of variance with square root-transformed counts over time demonstrated that CD8+ T cells significantly increase over time with a P value of 0.0017, while the CD4+ cells show a trend toward significance with a P value of 0.049. (D and E) Representative flow cytometric pseudocolor plots showing the frequency of IFN-γ and/or TNF-α production by CD8+ T cells in blood (D) or the gastrointestinal (GI) tract (E) after stimulation with HTLV-2 Tax and Gag overlapping peptides or in unstimulated cells. (F and G) Mean HTLV-2-specific CD4+ and CD8+ T-cell responses, after background subtraction, 10 weeks after HTLV-2 infection in blood (F) and the gastrointestinal tract (G). The frequency of Gag- or Tax-specific cells was measured by intracellular cytokine staining for IFN-γ/TNF-α or IL-2.
TABLE 2.
HTLV-2 proviral load in the infected macaques
| Animal code | Time after HTLV-2 infection (no. of wks) | HTLV-2 proviral DNA load/106 PBMCs |
|---|---|---|
| M893 | 3 | 15 |
| 6 | 7 | |
| 12 | 22 | |
| 18 | 8 | |
| M897 | 3 | 15 |
| 6 | 38 | |
| 12 | 12 | |
| 18 | 7 | |
| M905 | 3 | 28 |
| 6 | 7 | |
| 12 | 54 | |
| 18 | NDa | |
| M906 | 3 | 16 |
| 12 | 18 | |
| 18 | ND |
ND, not detected.
Comparable viral and cellular dynamics in blood from dually HTLV-2/SIVmac251 infected or SIVmac251 singly infected macaques.
The effects of preexisting HTLV-2 infection on the acute phases of HIV-1 infection in humans are unknown. We therefore studied the effect of HTLV-2 infection on SIVmac251 replication and the SIVmac251-specific immune response in macaques coinfected with HTLV-2 and SIVmac251 to address these questions. As summarized in the study design (Fig. 3A), 10 months after HTLV-2 infection, we challenged macaques with SIVmac251 intrarectally. The persistence of HTLV-2 infection was confirmed by nested PCR in blood, before and after SIVmac251 infection in the macaques preinfected with HTLV-2 (Fig. 3B). HTLV-2 tax DNA was amplified from all four persistently infected macaques, while the uninfected control animal P182 (not infected with HTLV-2) was negative (Fig. 3B). Serum antibodies to HTLV-2 proteins were also present before and after SIV infection (Fig. 3C). The viral burden and T-cell dynamics in HTLV-2-infected macaques, superinfected with SIVmac251, were characterized and compared to SIVmac251 singly infected controls. SIV RNA in the plasma was quantified by reverse transcription-PCR (RT-PCR) (Fig. 4A). Peak viremia occurred at 14 days postinfection, with an average of 1.1 × 108 copies/ml for the HTLV-2/SIVmac251 group and 1.2 × 108 copies/ml in the controls. Set point viremia occurred by 60 days postinfection with an average of 8.4 × 106 copies/ml in the HTLV-2/SIVmac251 group and 7.2 × 106 copies/ml in the controls. Neither peak viral load nor set point viremia was significantly different between the two groups. These data support the clinical findings in HTLV-2/HIV coinfected patients, where HTLV-2 infection was not found to influence HIV viral loads (56). SIVmac251 infection caused a sharp decline in the number of CD4+ T cells in blood in the animals of both groups; this decline stabilized by 30 days postinfection (Fig. 4B). Absolute CD4+ T-cell counts were not significantly different between the groups. Memory CD4+ T cells, particularly those that express CCR5, are the primary targets of HIV-1 and SIVmac251 infection. These target T cells are rapidly lost in the acute phase of the infection. HTLV-2-infected CD8+ T cells secrete MIP1α, a natural ligand for CCR5. The production of MIP1α by HTLV-2-infected CD8+ T cells isolated from coinfected individuals has been shown to inhibit HIV-1 replication (12, 13). Thus, MIP1α secretion by HTLV-2-infected cells and its binding to CCR5 on circulating or mucosal CD4+ T cells may theoretically protect this cell type from the typical rapid depletion observed during SIVmac251 and HIV-1 infections. To examine whether this is the case, we determined the frequency of CD4+ CCR5+ T cells in blood. To control for animal-to-animal variation, the frequency of CD4+ CCR5+ T cells is displayed as a percentage of baseline value over the course of the infection. A severe depletion of CD4+ CCR5+ cells was observed in both groups by 21 days postinfection (Fig. 4C), and no difference in the rate of decline of CD4+ CCR5+ cells was observed between the coinfected animals and controls. Activation and proliferation drive T-cell differentiation from naïve to memory and effector cells. The phenotype of T cells in HTLV-2/SIVmac251 coinfected animals was compared to the phenotype of T cells in SIVmac251 singly infected controls. Expression of CD28 along with CD95 was used to determine the differentiation status of CD4+ and CD8+ T cells. T cells that express only CD28 were considered naïve, cells dually positive for CD28 and CD95 were characterized as central memory cells, and cells positive for CD95 only were categorized as effector/effector memory cells (48). A sharp decline in the fraction of CD4+ central memory T cells was observed in both groups (similar to CD4+ CCR5+ T cells), which then stabilized between day 30 and day 60 (Fig. 4D). An increase in the absolute number of CD8+ T cells was observed following SIVmac251 infection in both groups (Fig. 4E); this increase was mainly due to an expansion of CD8+ effector cells (graphed as a fraction of the baseline level in Fig. 4F). The CD8+ T-cell expansion coincides with the postpeak decline in viremia, suggesting a contribution of CD8+ T cells to the partial control of acute viremia, as observed during HIV infection (9, 30). The acute increase in the percentage of CD8+ effectors peaked at 21 days postinfection and then declined transiently, only to continue to increase in the chronic phase; this is likely due to the continued generalized immune activation observed during chronic HIV-1 and SIVmac251 infections. Altogether, there were no significant differences between the two groups of CD8+ T cells (Fig. 4E and F).
FIG. 3.
Persistent HTLV-2 infection despite SIV coinfection. (A) Study design. Four macaques were infected with HTLV-2. Ten months postinfection, these 4 macaques along with 4 naïve animals were challenged with SIVmac251. Blood, lymphoid, and mucosal tissues were sampled before infection and for up to 90 days after SIVmac251 infection. (B) Semiquantitative nested PCR for HTLV-2 tax DNA or β-actin amplified from the peripheral blood of HTLV-2-infected animals (M893, M897, M905, and M906) and uninfected control (P182) before and after SIVmac251 infection. The number of weeks after HTLV-2 infection is shown below the blots. (C) Serum antibodies to HTLV-2 measured by Western blotting before and after SIVmac251 infection.
FIG. 4.
Similar viral and cellular dynamics in HTLV-2/SIVmac251 coinfected and SIVmac251 monoinfected macaques. (A) SIVmac251 viral load in HTLV-2 coinfected macaques (shown in black) and controls (shown in red) measured by the number of SIVmac251 RNA copies/ml of plasma. (B) Average CD4+ T-cell count in blood following SIVmac251 infection in HTLV-2/SIVmac251-infected macaques (black) and SIVmac251-infected macaques (red). (C) Percentage of baseline CD4+ CCR5+ T-cell count following SIVmac251 infection in HTLV-2/SIVmac251-infected macaques (shown in black) and SIVmac251-infected animals (shown in red). (D) Percentage of baseline CD4+ memory T-cell count in blood following SIVmac251 infection. Memory cells are defined as cells that express both CD28 and CD95. (E) Average CD8+ T-cell count following SIVmac251 infection. (F) Percentage of baseline CD8+ effector/effector memory T-cell count in blood after SIVmac251 infection. CD8+ effector/effector memory T cells are CD8+ CD28− CD95+.
SIVmac251 replicates and induces CD4+ T-cell loss in the lymphoid organs and gastrointestinal tracts of HTLV-2 coinfected macaques.
The tropism of HTLV-2 for T cells and its potential ability to activate T cells in lymphoid and mucosal sites raised the possibility that HTLV-2 infection may increase SIVmac251 replication by creating more viral targets. We compared the number of viral DNA copies and CD4+ T-cell depletion in lymphoid and mucosal sites in HTLV-2/SIVmac251 coinfected macaques and SIVmac251 singly infected controls. Bone marrow aspirate specimens, along with lymph node, jejunum, and rectum biopsy specimens were collected before and 10 and 30 days after SIVmac251 infection. SIVmac251 DNA was amplified by real-time PCR on DNA extracted from lymph nodes and the gastrointestinal tract at both 10 and 30 days postinfection (Fig. 5A). The level of SIVmac251 viral DNA in the tissues, including the gastrointestinal tract, was not significantly different between the two groups. Similar to the measurements in blood, we monitored the frequency of CD4+ T cells in the lymphoid organs (bone marrow and lymph nodes; Fig. 5B) and the gastrointestinal tract (jejunum and rectum; Fig. 5C). Both macaques coinfected with HTLV-2 and SIVmac251 and macaques singly infected with SIVmac251 showed a decline in the frequency of CD4+ T cells in both lymphoid organs at 30 days postinfection, but there was no significant difference between the groups. A significant depletion of lamina propria CD4+ T cells was observed in the jejunum and rectum (Fig. 5C). However, the extent of the depletion was severe in both groups; these observations were confirmed by immunohistochemical staining which enumerated the absolute number of CD4-expressing cells in the rectum (Fig. 5D). Interestingly, immunohistochemical staining demonstrated a significantly greater number of CD4+ cells in the rectum before SIV infection in animals preinfected with HTLV-2 (P < 0.05). Overall, we observed no difference in the number of cells in the rectum expressing CD8+ (Fig. 5E) or the frequency of CD8+ T cells (data not shown) in the singly or dually infected animals before or after SIV infection. Thus, at least in rhesus macaques, coinfection with HTLV-2 did not attenuate SIVmac251 replication or the loss of CD4+ T cells at mucosal sites (Fig. 5).
FIG. 5.
HTLV-2 preinfection did not significantly affect either the viral load or the rate of CD4+ T-cell loss after SIVmac251 infection. (A) Viral load in the lymph nodes, jejunum, and rectum in HTLV-2/SIVmac251 coinfected macaques (black) and SIVmac251 monoinfected macaques (white) quantified as the number of SIV DNA copies/106 cells 10 and 30 days after SIVmac251 infection. (B) The frequency of CD3+ CD4+ T cells in the bone marrow and lymph nodes of HTLV-2/SIVmac251 coinfected and SIVmac251 singly infected animals before infection (baseline) and 10 and 30 days after SIVmac251 infection. (C) The frequency of CD3+ CD4+ T cells in the jejunum and rectum in HTLV-2/SIVmac251 coinfected and SIVmac251 singly infected animals before infection (baseline) and 10 and 30 days after SIVmac251 infection. (D) Number of CD4+ expressing cells, enumerated by immunohistochemical staining of paraformaldehyde-fixed tissue sections from the rectum. A repeated-measure analysis of variance demonstrated that the difference between the baseline (before infection) values for HTLV-2/SIVmac251 coinfected macaques and SIVmac251 singly infected macaques is significantly different (P < 0.05). (E) Number of CD8+ expressing cells, enumerated by immunohistochemical staining of paraformaldehyde-fixed rectum tissue sections.
Similar levels of spontaneous MIP1α production in HTLV-2/SIVmac251 coinfected and SIV singly infected macaques.
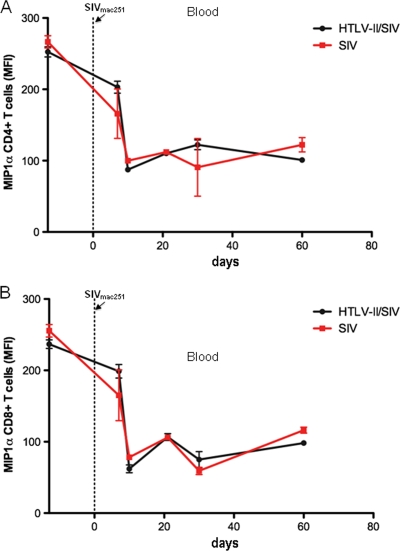
Early in HIV-1 infection, viruses that utilize the chemokine receptor CCR5 for entry into T cells and macrophages dominate (52). HIV-1 and SIVmac251 entry can be inhibited by the binding of the natural ligands for CCR5; these CC chemokines include MIP1α, MIP1β, and RANTES (16, 35). HTLV-2-infected T cells have been shown to spontaneously secrete MIP1α; the induction of MIP1α is likely the result of the viral protein Tax transactivating the chemokine promoter (36). To characterize the ability of CD4+ and CD8+ T cells to spontaneously produce MIP1α, we cultured PBMCs for 5 h in the presence of monensin and performed intracellular cytokine staining for MIP1α. Both the MIP1α expression per cell (mean fluorescence intensity [MFI]) and the frequency of MIP1α production (data not shown) decreased after SIVmac251 infection (Fig. 6A and B). However, no difference was observed in the levels of spontaneous MIP1α produced by CD4+ or CD8+ T cells from HTLV-2/SIVmac251 coinfected and SIVmac251 singly infected controls at any time point before or after SIVmac251 infection (Fig. 6A and B). In addition, no correlation was observed between the levels of spontaneous MIP1α produced and viral load in either coinfected macaques or controls. MIP1α was not detected in the plasma samples from all macaques by ELISA.
FIG. 6.
No difference in spontaneous MIPα expression by T cells in blood from HTLV-2/SIVmac251 coinfected and SIVmac251 monoinfected animals. (A) Mean fluorescence intensity (MFI) of MIP1α in CD3+ CD4+ T cells in blood from HTLV-2/SIVmac251 coinfected animals (black) and SIVmac251 singly infected animals (red) before or after SIVmac251 infection. (B) Mean fluorescence intensity of MIP1α in CD3+ CD8+ T cells in blood from HTLV-2/SIVmac251 coinfected animals and SIVmac251 singly infected animals SIVmac251 before or after SIVmac251 infection.
HTLV-2 preinfection does not affect T-cell proliferation in SIVmac251-infected macaques.
Increased T-cell proliferation is a hallmark of HTLV infections (18). Thus, we monitored the level of proliferation by staining for the nuclear antigen Ki67 in CD4+ and CD8+ T cells from blood and tissue samples following SIVmac251 infection. An increase in the percentage of CD4+ and CD8+ T cells expressing Ki67 was observed in blood following SIVmac251 infection; this peaked between 21 and 30 days postinfection (Fig. 7A and B). The frequency of proliferating T cells declined between 30 to 60 days but continued to be elevated in the chronic phase. The magnitude of the increased acute proliferation was greater in CD8+ T cells, and a significantly higher level of Ki67 expression remained at 90 days postinfection compared to the pre-SIVmac251 infection levels (P = 0.0078). However, there were no significant differences in the magnitude of proliferating CD4+ or CD8+ T cells in blood from HTLV-2 preinfected and control macaques (Fig. 7A and B). The frequency of CD4 and CD8+ Ki67+ cells increased in the bone marrow with kinetics similar to the kinetics observed in blood, and the lymph nodes also demonstrated an increase in activating and proliferating CD4+ and CD8+ T cells (Fig. 7C and D). Interestingly, the enumeration of CD4+ cells expressing Ki67 in rectal tissues demonstrated significantly more proliferating cells in HTLV-2 preinfected animals compared with uninfected controls (i.e., before SIV infection; Fig. 7E). This increased turnover may account for the increased number of CD4+ T cells measured by immunohistochemical staining in the rectum before SIV infection (Fig. 5D). SIVmac251 infection caused an increase in mucosal CD8+ T-cell proliferation; however, the extent of this increase was similar in both HTLV-2 preinfected animals and controls measured by immunohistochemical staining for CD8+ and Ki67 in the rectum (Fig. 7F). Overall, SIVmac251 infection was associated with a significant increase in the level of proliferating T cells in blood and tissues. HTLV-2 preinfection did not affect the magnitude or duration of this proliferation in most tissues.
FIG. 7.
Proliferation of T cells in blood and tissues from HTLV-2/SIVmac251 coinfected animals and SIVmac251 monoinfected animals. (A and B) Frequency of CD3+ CD4+ T cells (A) and CD3+ CD8+ T cells (B) expressing Ki67 in blood from HTLV-2/SIVmac251 coinfected animals (black) and SIVmac251 singly infected animals (red) after SIVmac251 infection. A Wilcoxon signed-rank test demonstrated that the level of CD8+ Ki67 expression continued to be significantly greater in the chronic phase compared to preinfection levels (B). (C and D) Frequency of CD3+ CD4+ T cells (C) and CD3+ CD8+ T cells (D) expressing Ki67 in the lymphoid tissues (bone marrow and lymph nodes) of HTLV-2/SIVmac251 coinfected animals and SIVmac251 singly infected animals before infection (baseline) and 10 and 30 days after SIVmac251 infection. (E) Number of cells expressing both CD4 and Ki67 in the rectum in HTLV-2/SIVmac251 coinfected macaques and SIVmac251 singly infected macaques before infection (baseline) and 10 and 30 days after SIVmac251 infection. Repeated-measure analysis of variance demonstrated that the difference between HTLV-2/SIVmac251 coinfected macaques and SIVmac251 singly infected macaques at baseline is significantly different (P = 0.0007). (F) Number of cells expressing both CD8 and Ki67 in the rectum in HTLV-2/SIVmac251 coinfected macaques and SIVmac251 singly infected macaques before infection (baseline) and 10 and 30 days after SIVmac251 infection.
Limited polyfunctional SIVmac251 responses in infected animals despite HTLV-2 preinfection.
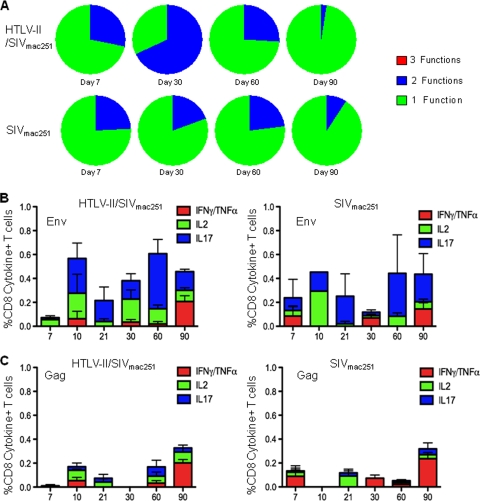
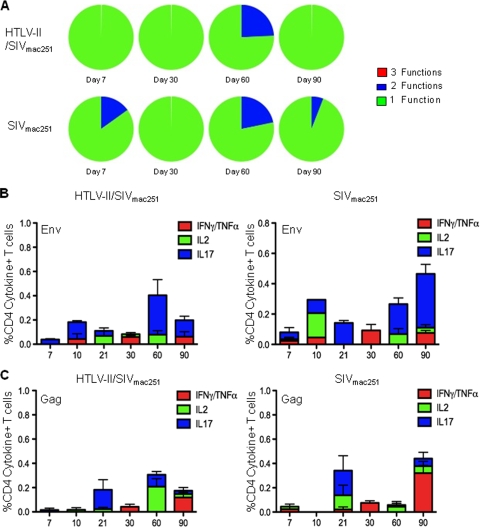
HIV-1 infection induces measurable virus-specific T-cell responses; however, these responses are not of sufficient quality and/or quantity to eliminate virus replication (7). The goal of any vaccination strategy for HIV-1 is to induce immune responses that can eliminate or at least reduce HIV-1 replication so that both transmission and disease progression are prevented. Live attenuated viruses with low levels of virus replication may prime and continually boost immune responses, creating a protective immune response and successfully immunizing the individual. In order to evaluate HTLV-2 as a potential platform for an HIV-1 vaccine, we sought to determine whether HTLV-2 preinfection impacted either the quantity or quality of the SIVmac251-specific response. Previous studies have indicated that HTLV-2 infection and specifically, its transactivating Tax protein, can induce IFN-γ production (11, 12). The quality of an HIV-1- or SIVmac251-specific immune response has been determined by monitoring the ability of CD4+ and CD8+ T cells to respond to viral antigens by secreting multiple cytokines and chemokines. Using polychromatic flow cytometry, we assessed the ability of CD4+ and CD8+ T cells to simultaneously produce the cytokines IFN-γ and/or TNF-α, IL-2, and IL-17 and evaluated the magnitude of the SIVmac251-specific response. Overlapping peptide pools to SIVmac251 Gag and SIVmac251 Env were used to stimulate mononuclear cells from blood. The proportion of CD8+ and CD4+ T cells responding to SIVmac251 Env stimulation by concurrently producing multiple cytokines (3 or 2 functions) was not different in HTLV-2/SIVmac251 coinfected animals compared to SIVmac251 singly infected controls (Fig. 8A and 9A). Similarly, no difference in polyfunctional responses was observed in CD8+ T cells stimulated with SIVmac251 Gag (data not shown). Figures 8B and C show the magnitude of the mean SIVmac251-specific response following SIVmac251 infection. Overall, the immune response to SIVmac251 Env was of greater magnitude than the response to Gag; however, no clear differences in the magnitude of the response to either antigen was observed between HTLV-2/SIVmac251 coinfected animals and controls. Figure 9B and C show the magnitude of the mean SIVmac251-specific response in CD4+ T cells. Again, there were no differences between the groups. In all, these data demonstrated that preinfection with HTLV-2 neither enhances nor limits the SIVmac251-specific immune responses.
FIG. 8.
HTLV-2 preinfection does not affect the quality or quantity of the SIVmac251-specific CD8+ T-cell response. Flow cytometric analysis of CD3+ CD8+ T cells expressing IFN-γ and/or TNF-α, IL-2, and IL-17 in blood was performed after stimulation with overlapping pools of SIV Gag and SIV Env peptides or in unstimulated cells. (A) Polyfunctional cytokine response to overlapping SIVmac251 Env peptides produced by CD3+ CD8+ T cells in blood from HTLV-2/SIVmac251 coinfected macaques and SIVmac251 singly infected macaques 7, 30, 60, and 90 days after SIVmac251 infection. The cytokines measured after stimulation were as follows: IL-2, IL-17, and IFN-γ stained along with TNF-α. Within the pie charts, the green slice represents the proportion of cells producing 1 cytokine considered to have 1 function (i.e., either IL-2 or IL-17 or IFN-γ/TNF-α), the blue slice represents the proportion of cells concurrently producing 2 cytokines considered to have 2 functions, and the red slice represents the proportion of cells producing 3 cytokines or 3 functions (i.e., cells concurrently producing IFN-γ and/or TNF-α, IL-2, and IL-17). (B) Average total cytokine (IFN-γ and/or TNF-α, IL-2, IL-17) production, after background subtraction, in CD3+ CD8+ T cells from blood in response to stimulation with SIV Env overlapping peptide pool after SIVmac251 infection. Cytokine production in HTLV-2/SIVmac251 coinfected macaques and SIVmac251 singly infected macaques is shown. (C) Average total cytokine (IFN-γ and/or TNF-α, IL-2, IL-17) production, after background subtraction, in CD3+ CD8+ T cells from blood in response to stimulation with SIV Gag overlapping peptide pool after SIVmac251 infection. Cytokine production in HTLV-2/SIVmac251 coinfected macaques and SIVmac251 singly infected macaques is shown.
FIG. 9.
SIVmac251-specific CD4+ T-cell responses are similar in HTLV-2/SIVmac251 coinfected animals and SIVmac251 monoinfected animals. (A) Polyfunctional cytokine response to overlapping SIVmac251 Env peptides produced by CD3+ CD4+ T cells in blood from HTLV-2/SIVmac251 coinfected macaques and SIVmac251 singly infected macaques 7, 30, 60, and 90 days after SIVmac251 infection. The cytokines measured after stimulation were as follows: IL-2, IL-17, and IFN-γ stained along with TNF-α. Within the pie charts, the green slice represents the proportion of cells producing 1 cytokine considered to have 1 function (i.e., either IL-2 or IL-17 or IFN-γ/TNF-α), the blue slice represents the proportion of cells concurrently producing 2 cytokines considered to have 2 functions, and the red slice represents the proportion of cells producing 3 cytokines or 3 functions (i.e., cells concurrently producing IFN-γ and/or TNF-α and IL-2 and IL-17). (C) Average total cytokine (IFN-γ and/or TNF-α, IL-2, IL-17) production, after background subtraction, in CD3+ CD4+ T cells from blood in response to stimulation with SIV Env overlapping peptide pool after SIVmac251 infection. Cytokine production in HTLV-2/SIVmac251 coinfected macaques and SIVmac251 singly infected macaques is shown.(D) Average total cytokine (IFN-γ and/or TNF-α, IL-2, IL-17) production, after background subtraction, in CD3+ CD4+ T cells from blood in response to stimulation with SIV Gag overlapping peptide pool after SIVmac251 infection. Cytokine production in HTLV-2/SIVmac251 coinfected macaques and SIVmac251 singly infected macaques is shown.
DISCUSSION
HTLV-2 infection is prevalent among intravenous drug users (IDU) coinfected with HIV-1 (56). The effects of HTLV-2 infection on progression to AIDS are controversial, as multiple studies have suggested a protective role of HTLV-2 in patients coinfected with HTLV-2 and HIV-1, while others have reported no beneficial effect. In the former studies, maintenance of CD4+ T cells and lower levels of virus replication and immune activation have been reported in dually infected HTLV-2/HIV-1 individuals versus patients infected with HIV-1 only (4, 12, 23). Additionally, several IDU long-term nonprogressors with stable CD4+ counts, in the absence of antiretroviral therapy, have been reported to be infected with HTLV-2 (56, 58). A possible mechanism underlying the milder disease course has been attributed to the increased expression of β chemokines in HTLV-2-infected patients (12, 36, 47), particularly MIP1α, a chemokine that inhibits viral entry by binding to CCR5. Despite the fact that it has been many years since the discovery of HTLV-2/HIV-1 coinfection, little is known about the effects of HTLV-2 on the HIV-specific response and on viral and T-cell dynamics in blood and tissues. Clinical studies of dually infected patients have provided useful information, but lengths of infection, routes of infection, pathogenicity of virus isolates, and drug use, all of which can affect the immune system, are not easily controlled for. Indeed, cross-sectional studies demonstrating differences in immunological parameters, such as immune activation and cytokine secretion in dually infected patients versus controls (4), may be more reflective of the levels of HIV-1 replication than an effect of HTLV-2 infection. Thus, an animal model where the route, dose, and length of each infection are controlled can provide useful information. Here, we have established models of HTLV-2 infection and HTLV-2/SIVmac251 coinfection in rhesus macaques. HTLV-2 infection of macaques was persistent, as demonstrated by the durability of serum antibodies to HTLV-2 antigens and the detection of proviral DNA in blood and tissues from the infected macaques. The data demonstrated that HTLV-2 persists at low levels in the lymphoid organs and importantly in the mucosal tissues of macaques. The low HTLV-2 viral loads observed in macaques are similar to HTLV-2-infected humans where the provirus DNA levels have been reported to be undetectable or a few genome copies per 106 PBMCs (6, 31, 43, 56). Repeated exposures to HTLV-2 may induce a more robust infection in the blood of rhesus macaques similar to HTLV-1- and STLV-1-infected animals (21, 55). Unlike other coinfection studies with SIV and either HTLV-1 or STLV-1 performed in nonhuman primates, we observed no indication of pathology associated with HTLV-2 infection (21, 55).
Recent observations that primary dendritic cells are permissive for HTLV-1 and that infected dendritic cells can efficiently transfer virus to CD4+ T cells in vitro (28), together with reports that HTLV-1-infected dendritic cells have been isolated from infected individuals (27, 28), suggest that dendritic cells may play a role in HTLV-1 transmission and/or persistence. In the present work, we show that HTLV-2 can also infect rhesus macaque dendritic cells in vitro. HTLV-2 infection was not associated with significant changes in CD4+ and CD8+ T-cell counts. Coinfection with HTLV-2 did not affect the level of SIVmac251 RNA or DNA in plasma or tissues. HTLV-2 infection has been demonstrated to affect the activation of signal transducers and activators of transcription (STAT) genes in HTLV-2/HIV-1 coinfected patients (10), and the IFN-γ gene promoter can be activated by the HTLV-2 trans-activator Tax protein (11). However, unlike HTLV-1 (41), HTLV-2 does not induce constitutive STAT-5 activation (42). We questioned whether HTLV-2 preinfection would affect the SIVmac251-specific immune response. However, we observed no difference in the magnitude or functional capacity of the antigen-specific immune response in dually or singly infected macaques. Accordingly, the rate of decline of systemic and mucosal CD4+ T cells and specifically CD4+ CCR5+ T cells, the targets for SIVmac251 infection, were indistinguishable in singly and dually infected macaques. These results indicate that at least in this experimental model, HTLV-2 preinfection did not affect the ability of SIVmac251 to infect CD4+ T cells, replicate to high titers, and induce a profound loss of CD4+ T cells. HTLV-2 also infects CD8+ T cells, which secrete β chemokines, such as MIP1α, a natural ligand for CCR5 that inhibits HIV-1 and SIVmac251 infection. SIVmac251 infection caused a decline in both the percentage and mean florescence intensity of MIP1α; however, in T cells, there was no difference between the levels of MIP1α spontaneously produced in HTLV-2/SIVmac251-infected animals and in controls. HTLV-1 is known to stimulate the proliferation of T cells, and during HTLV-1 infection, the clonal expansion of T cells can lead to neoplastic transformation (32). In the case of HTLV-2, long-term infection has been associated with increased T-cell number (3), but no clear association with hematological diseases is proven. Thus, we evaluated the frequency of proliferating CD4+ and CD8+ T cells in the blood and tissues of macaques preinfected with HTLV-2. SIVmac251 infection caused a generalized increase in activation and T-cell proliferation. The magnitude of this increase peaked in the acute phase but remained elevated into the chronic phase. HTLV-2 preinfection did not appear to exacerbate this proliferation; except for a modest increase in CD4 proliferation in the rectums of animals preinfected with HTLV-2. However, the increased number of proliferating CD4+ T cells at this site did not impact either the level of local or systemic virus replication or the rate of CD4+ T-cell loss after SIV infection.
The finding that HTLV-2 infects and persists in lymphoid and mucosal tissue suggests that an attenuated HTLV-2 may be able to target antigens to the site of SIVmac251/HIV-1 replication. The induction of effective and persistent T-cell responses at the portal of entry of SIVmac251/HIV-1 could limit infection by the founder virus and prevent virus dissemination to distal sites. A similar concept has been explored using rhesus cytomegalovirus (RhCMV) in rhesus macaques (20, 25). Live SIVmac251-based attenuated vaccines have demonstrated their efficacy at preventing infection in macaques, but their safety for use in humans is a concern (17). An HTLV-2-based vaccine for HIV-1 may provide an opportunity to confirm whether persistent antigen expression is a requirement for an effective vaccine for HIV-1. HTLV-2 vectored vaccines, even if attenuated, would pose regulatory issues. However, should their efficacy be proven, strategies to increase their safety, could also be developed.
Acknowledgments
We thank Teresa Habina for editorial assistance; P. D. Markham, S. Orndorff, D. Weiss, and J. Treece of Advanced BioScience Laboratories, Inc., Kensington, MD, and Katherine McKinnon for flow cytometric support.
Footnotes
Published ahead of print on 13 January 2010.
REFERENCES
- 1.Araujo, A., and W. W. Hall. 2004. Human T-lymphotropic virus type II and neurological disease. Ann. Neurol. 56:10-19. [DOI] [PubMed] [Google Scholar]
- 2.Bartholomew, C., W. Blattner, and F. Cleghorn. 1987. Progression to AIDS in homosexual men co-infected with HIV and HTLV-I in Trinidad. Lancet 2:1469. [DOI] [PubMed] [Google Scholar]
- 3.Bartman, M. T., Z. Kaidarova, D. Hirschkorn, R. A. Sacher, J. Fridey, G. Garratty, J. Gibble, J. W. Smith, B. Newman, A. E. Yeo, and E. L. Murphy. 2008. Long-term increases in lymphocytes and platelets in human T-lymphotropic virus type II infection. Blood 112:3995-4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bassani, S., M. Lopez, C. Toro, V. Jimenez, J. M. Sempere, V. Soriano, and J. M. Benito. 2007. Influence of human T cell lymphotropic virus type 2 coinfection on virological and immunological parameters in HIV type 1-infected patients. Clin. Infect. Dis. 44:105-110. [DOI] [PubMed] [Google Scholar]
- 5.Beilke, M. A., K. P. Theall, M. O'Brien, J. L. Clayton, S. M. Benjamin, E. L. Winsor, and P. J. Kissinger. 2004. Clinical outcomes and disease progression among patients coinfected with HIV and human T lymphotropic virus types 1 and 2. Clin. Infect. Dis. 39:256-263. [DOI] [PubMed] [Google Scholar]
- 6.Beilke, M. A., V. L. Traina-Dorge, M. Sirois, A. Bhuiyan, E. L. Murphy, J. M. Walls, R. Fagan, E. L. Winsor, and P. J. Kissinger. 2007. Relationship between human T lymphotropic virus (HTLV) type 1/2 viral burden and clinical and treatment parameters among patients with HIV type 1 and HTLV-1/2 coinfection. Clin. Infect. Dis. 44:1229-1234. [DOI] [PubMed] [Google Scholar]
- 7.Betts, M. R., M. C. Nason, S. M. West, S. C. De Rosa, S. A. Migueles, J. Abraham, M. M. Lederman, J. M. Benito, P. A. Goepfert, M. Connors, M. Roederer, and R. A. Koup. 2006. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood 107:4781-4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Biglione, M., O. Vidan, R. Mahieux, M. de Colombo, M. de los Angeles de Basualdo, M. Bonnet, G. Pankow, M. A. De Efron, A. Zorrilla, F. Tekaia, E. Murphy, G. de The, and A. Gessain. 1999. Seroepidemiological and molecular studies of human T cell lymphotropic virus type II, subtype b, in isolated groups of Mataco and Toba Indians of northern Argentina. AIDS Res. Hum. Retroviruses 15:407-417. [DOI] [PubMed] [Google Scholar]
- 9.Borrow, P., H. Lewicki, B. H. Hahn, G. M. Shaw, and M. B. A. Oldstone. 1994. Virus-specific CD8+ cytotoxic T-lymphocyte activity associated with control of viremia in primary human immunodeficiency virus type 1 infection. J. Virol. 68:6103-6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bovolenta, C., E. Pilotti, M. Mauri, B. Panzeri, M. Sassi, P. Dall'Aglio, U. Bertazzoni, G. Poli, and C. Casoli. 2002. Retroviral interference on STAT activation in individuals coinfected with human T cell leukemia virus type 2 and HIV-1. J. Immunol. 169:4443-4449. [DOI] [PubMed] [Google Scholar]
- 11.Brown, D. A., F. B. Nelson, E. L. Reinherz, and D. J. Diamond. 1991. The human interferon-gamma gene contains an inducible promoter that can be transactivated by tax I and II. Eur. J. Immunol. 21:1879-1885. [DOI] [PubMed] [Google Scholar]
- 12.Casoli, C., E. Pilotti, and U. Bertazzoni. 2007. Molecular and cellular interactions of HIV-1/HTLV coinfection and impact on AIDS progression. AIDS Rev. 9:140-149. [PubMed] [Google Scholar]
- 13.Casoli, C., E. Vicenzi, A. Cimarelli, G. Magnani, P. Ciancianaini, E. Cattaneo, P. Dall'Aglio, G. Poli, and U. Bertazzoni. 2000. HTLV-II down-regulates HIV-1 replication in IL-2-stimulated primary PBMC of coinfected individuals through expression of MIP-1alpha. Blood 95:2760-2769. [PubMed] [Google Scholar]
- 14.Cecchinato, V., E. Tryniszewska, Z. M. Ma, M. Vaccari, A. Boasso, W. P. Tsai, C. Petrovas, D. Fuchs, J. M. Heraud, D. Venzon, G. M. Shearer, R. A. Koup, I. Lowy, C. J. Miller, and G. Franchini. 2008. Immune activation driven by CTLA-4 blockade augments viral replication at mucosal sites in simian immunodeficiency virus infection. J. Immunol. 180:5439-5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chung, H. K., T. Unangst, J. Treece, D. Weiss, and P. Markham. 2008. Development of real-time PCR assays for quantitation of simian betaretrovirus serotype-1, -2, -3, and -5 viral DNA in Asian monkeys. J. Virol. Methods 152:91-97. [DOI] [PubMed] [Google Scholar]
- 16.Cocchi, F., A. L. DeVico, A. Garzino-Demo, S. K. Arya, R. C. Gallo, and P. Lusso. 1995. Identification of RANTES, MIP-1 alpha, and MIP-1 beta as the major HIV-suppressive factors produced by CD8+ T cells. Science 270:1811-1815. [DOI] [PubMed] [Google Scholar]
- 17.Desrosiers, R. C. 2004. Prospects for an AIDS vaccine. Nat. Med. 10:221-223. [DOI] [PubMed] [Google Scholar]
- 18.Dezzutti, C. S., D. R. Sasso, D. L. Rudolph, and R. B. Lal. 1998. Down-regulation of interleukin-10 expression and production is associated with spontaneous proliferation by lymphocytes from human T lymphotropic virus type II-infected persons. J. Infect. Dis. 177:1489-1496. [DOI] [PubMed] [Google Scholar]
- 19.Favre, D., S. Lederer, B. Kanwar, Z. M. Ma, S. Proll, Z. Kasakow, J. Mold, L. Swainson, J. D. Barbour, C. R. Baskin, R. Palermo, I. Pandrea, C. J. Miller, M. G. Katze, and J. M. McCune. 2009. Critical loss of the balance between Th17 and T regulatory cell populations in pathogenic SIV infection. PLoS Pathog. 5:e1000295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Franchini, G. 2009. Choosing the right memory T cell for HIV. Nat. Med. 15:244-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fultz, P. N., T. McGinn, I. C. Davis, J. W. Romano, and Y. Li. 1999. Coinfection of macaques with simian immunodeficiency virus and simian T cell leukemia virus type I: effects on virus burdens and disease progression. J. Infect. Dis. 179:600-611. [DOI] [PubMed] [Google Scholar]
- 22.Gessain, A., F. Barin, J.-C. Vernant, O. Gout, L. Maurs, A. Calendar, and G. de The. 1985. Antibodies to human T-lymphotropic virus type I in patients with tropical spastic paraparesis. Lancet ii:407-410. [DOI] [PubMed] [Google Scholar]
- 23.Giacomo, M., E. G. Franco, C. Claudio, C. Carlo, D. A. Anna, D. Anna, and F. Franco. 1995. Human T-cell leukemia virus type II infection among high risk groups and its influence on HIV-1 disease progression. Eur. J. Epidemiol. 11:527-533. [DOI] [PubMed] [Google Scholar]
- 24.Hall, W. W., R. Ishak, S. W. Zhu, P. Novoa, N. Eiraku, H. Takahashi, M. C. Ferreira, V. Azevedo, M. O. Ishak, O. C. Ferreira, C. Monken, and T. Kurata. 1996. Human T lymphotropic virus type II (HTLV-II): epidemiology, molecular properties, and clinical features of infection. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 13(Suppl 1):S204-S214. [DOI] [PubMed] [Google Scholar]
- 25.Hansen, S. G., C. Vieville, N. Whizin, L. Coyne-Johnson, D. C. Siess, D. D. Drummond, A. W. Legasse, M. K. Axthelm, K. Oswald, C. M. Trubey, M. Piatak, Jr., J. D. Lifson, J. A. Nelson, M. A. Jarvis, and L. J. Picker. 2009. Effector memory T cell responses are associated with protection of rhesus monkeys from mucosal simian immunodeficiency virus challenge. Nat. Med. 15:293-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hershow, R. C., N. Galai, K. Fukuda, J. Graber, D. Vlahov, G. Rezza, R. S. Klein, J. Des, C. Vitek, R. Khabbaz, S. Freels, R. Zuckerman, P. Pezzotti, and J. E. Kaplan. 1996. An international collaborative study of the effects of coinfection with human T-lymphotropic virus type II on human immunodeficiency virus type 1 disease progression in injection drug users. J. Infect. Dis. 174:309-317. [DOI] [PubMed] [Google Scholar]
- 27.Hishizawa, M., K. Imada, T. Kitawaki, M. Ueda, N. Kadowaki, and T. Uchiyama. 2004. Depletion and impaired interferon-alpha-producing capacity of blood plasmacytoid dendritic cells in human T-cell leukaemia virus type I-infected individuals. Br. J. Haematol. 125:568-575. [DOI] [PubMed] [Google Scholar]
- 28.Jones, K. S., C. Petrow-Sadowski, Y. K. Huang, D. C. Bertolette, and F. W. Ruscetti. 2008. Cell-free HTLV-1 infects dendritic cells leading to transmission and transformation of CD4(+) T cells. Nat. Med. 14:429-436. [DOI] [PubMed] [Google Scholar]
- 29.Kalyanaraman, V. S., M. G. Sarngadharan, M. Robert-Guroff, I. Miyoshi, D. Golde, and R. C. Gallo. 1982. A new subtype of human T-cell leukemia virus (HTLV-II) associated with a T-cell variant of hairy cell leukemia. Science 218:571-573. [DOI] [PubMed] [Google Scholar]
- 30.Koup, R. A., J. T. Safrit, Y. Cao, C. A. Andrew, G. McLeod, W. Borkowsky, C. Farthing, and D. D. Ho. 1994. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J. Virol. 68:4650-4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kwaan, N., T. H. Lee, D. M. Chafets, C. Nass, B. Newman, J. Smith, G. Garratty, and E. L. Murphy. 2006. Long-term variations in human T lymphotropic virus (HTLV)-I and HTLV-II proviral loads and association with clinical data. J. Infect. Dis. 194:1557-1564. [DOI] [PubMed] [Google Scholar]
- 32.Lairmore, M. D., and G. Franchini. 2007. Human T-cell leukemia/lymphoma virus types 1 and 2, p. 2071-2106. In D. M. Knipe and P. M. Howley (ed.), Fields virology. Lippincott Williams & Wilkins, Philadelphia, PA.
- 33.Lee, T. H., D. M. Chafets, M. P. Busch, and E. L. Murphy. 2004. Quantitation of HTLV-I and II proviral load using real-time quantitative PCR with SYBR Green chemistry. J. Clin. Virol. 31:275-282. [DOI] [PubMed] [Google Scholar]
- 34.Lefrere, J. J., A. M. Courouce, M. Mariotti, E. Wattel, O. Prou, F. Bouchardeau, and P. Lambin. 1990. Rapid progression to AIDS in dual HIV-1/HTLV-I infection. Lancet 336:509. [DOI] [PubMed] [Google Scholar]
- 35.Lehner, T., Y. Wang, M. Cranage, L. Tao, E. Mitchell, C. Bravery, C. Doyle, K. Pratt, G. Hall, M. Dennis, L. Villinger, and L. Bergmeier. 2000. Up-regulation of beta-chemokines and down-modulation of CCR5 co-receptors inhibit simian immunodeficiency virus transmission in non-human primates. Immunology 99:569-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lewis, M. J., V. W. Gautier, X. P. Wang, M. H. Kaplan, and W. W. Hall. 2000. Spontaneous production of C-C chemokines by individuals infected with human T lymphotropic virus type II (HTLV-II) alone and HTLV-II/HIV-1 coinfected individuals. J. Immunol. 165:4127-4132. [DOI] [PubMed] [Google Scholar]
- 37.Loughran, T. P., Jr., T. Coyle, M. P. Sherman, G. Starkebaum, G. D. Ehrlich, F. W. Ruscetti, and B. J. Poiesz. 1992. Detection of human T-cell leukemia/lymphoma virus, type II, in a patient with large granular lymphocyte leukemia. Blood 80:1116-1119. [PubMed] [Google Scholar]
- 38.Loughran, T. P., Jr., M. P. Sherman, F. W. Ruscetti, S. Frey, T. Coyle, R. A. Montagna, B. Jones, G. Starkebaum, and B. J. Poiesz. 1994. Prototypical HTLV-I/II infection is rare in LGL leukemia. Leuk. Res. 18:423-429. [DOI] [PubMed] [Google Scholar]
- 39.Martin, M. P., R. J. Biggar, G. Hamlin-Green, S. Staal, and D. Mann. 1993. Large granular lymphocytosis in a patient infected with HTLV-II. AIDS Res. Hum. Retroviruses 9:715-719. [DOI] [PubMed] [Google Scholar]
- 40.Meertens, L., S. Chevalier, R. Weil, A. Gessain, and R. Mahieux. 2004. A 10-amino acid domain within human T-cell leukemia virus type 1 and type 2 tax protein sequences is responsible for their divergent subcellular distribution. J. Biol. Chem. 279:43307-43320. [DOI] [PubMed] [Google Scholar]
- 41.Migone, T. S., J. X. Lin, A. Cereseto, J. C. Mulloy, J. J. O'Shea, G. Franchini, and W. J. Leonard. 1995. Constitutively activated JAK-STAT pathway in T-cells transformed with HTLV-I. Science 269:79-81. [DOI] [PubMed] [Google Scholar]
- 42.Mulloy, J. C., T.-S. Migone, T. M. Ross, N. Ton, P. L. Green, W. J. Leonard, and G. Franchini. 1998. Human and simian T-cell leukemia viruses type 2 (HTLV-2 and STLV-2pan-p) transform T cells independently of Jak/STAT activation. J. Virol. 72:4408-4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Murphy, E. L., T. H. Lee, D. Chafets, C. C. Nass, B. Wang, K. Loughlin, and D. Smith. 2004. Higher human T lymphotropic virus (HTLV) provirus load is associated with HTLV-I versus HTLV-II, with HTLV-II subtype A versus B, and with male sex and a history of blood transfusion. J. Infect. Dis. 190:504-510. [DOI] [PubMed] [Google Scholar]
- 44.Orland, J. R., J. Engstrom, J. Fridey, R. A. Sacher, J. W. Smith, C. Nass, G. Garratty, B. Newman, D. Smith, B. Wang, K. Loughlin, and E. L. Murphy. 2003. Prevalence and clinical features of HTLV neurologic disease in the HTLV Outcomes Study. Neurology 61:1588-1594. [DOI] [PubMed] [Google Scholar]
- 45.Osame, M., M. Matsumoto, K. Usuku, S. Izumo, N. Ijichi, H. Amitani, M. Tara, and A. Igata. 1987. Chronic progressive myelopathy associated with elevated antibodies to human T-lymphotropic virus type I and adult T-cell leukemialike cells. Ann. Neurol. 21:117-122. [DOI] [PubMed] [Google Scholar]
- 46.Pawson, R., T. F. Schulz, E. Matutes, and D. Catovsky. 1997. The human T-cell lymphotropic viruses types I/II are not involved in T prolymphocytic leukemia and large granular lymphocytic leukemia. Leukemia 11:1305-1311. [DOI] [PubMed] [Google Scholar]
- 47.Pilotti, E., L. Elviri, E. Vicenzi, U. Bertazzoni, M. C. Re, S. Allibardi, G. Poli, and C. Casoli. 2007. Postgenomic up-regulation of CCL3L1 expression in HTLV-2-infected persons curtails HIV-1 replication. Blood 109:1850-1856. [DOI] [PubMed] [Google Scholar]
- 48.Pitcher, C. J., S. I. Hagen, J. M. Walker, R. Lum, B. L. Mitchell, V. C. Maino, M. K. Axthelm, and L. J. Picker. 2002. Development and homeostasis of T cell memory in rhesus macaque. J. Immunol. 168:29-43. [DOI] [PubMed] [Google Scholar]
- 49.Poiesz, B. J., F. W. Ruscetti, A. F. Gazdar, P. A. Bunn, J. D. Minna, and R. C. Gallo. 1980. Detection and isolation of type C retrovirus particles from fresh and cultured lymphocytes of a patient with cutaneous T-cell lymphoma. Proc. Natl. Acad. Sci. U. S. A. 77:7415-7419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Poiesz, B. J., F. W. Ruscetti, M. S. Reitz, V. S. Kalyanaraman, and R. C. Gallo. 1981. Isolation of a new type C retrovirus (HTLV) in primary uncultured cells of a patient with Sezary T-cell leukaemia. Nature 294:268-271. [DOI] [PubMed] [Google Scholar]
- 51.Romano, J. W., K. G. Williams, R. N. Shurtliff, C. Ginocchio, and M. Kaplan. 1997. NASBA technology: isothermal RNA amplification in qualitative and quantitative diagnostics. Immunol. Invest. 26:15-28. [DOI] [PubMed] [Google Scholar]
- 52.Scarlatti, G., E. Tresoldi, A. Bjorndal, R. Fredriksson, C. Colognesi, H. K. Deng, M. S. Malnati, A. Plebani, A. G. Siccardi, D. R. Littman, E. M. Fenyo, and P. Lusso. 1997. In vivo evolution of HIV-1 co-receptor usage and sensitivity to chemokine-mediated suppression. Nat. Med. 3:1259-1265. [DOI] [PubMed] [Google Scholar]
- 53.Shimotohno, K., Y. Takahashi, N. Shimizu, T. Gojobori, D. W. Golde, I. S. Chen, M. Miwa, and T. Sugimura. 1985. Complete nucleotide sequence of an infectious clone of human T-cell leukemia virus type II: an open reading frame for the protease gene. Proc. Natl. Acad. Sci. U. S. A. 82:3101-3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Switzer, W. M., F. L. Black, D. Pieniazek, R. J. Biggar, R. B. Lal, and W. Heneine. 1996. Endemicity and phylogeny of the human T cell lymphotropic virus type II subtype A from the Kayapo Indians of Brazil: evidence for limited regional dissemination. AIDS Res. Hum. Retroviruses 12:635-640. [DOI] [PubMed] [Google Scholar]
- 55.Traina-Dorge, V. L., L. N. Martin, R. Lorino, E. L. Winsor, and M. A. Beilke. 2007. Human T cell leukemia virus type 1 up-regulation after simian immunodeficiency virus-1 coinfection in the nonhuman primate. J. Infect. Dis. 195:562-571. [DOI] [PubMed] [Google Scholar]
- 56.Turci, M., E. Pilotti, P. Ronzi, G. Magnani, A. Boschini, S. G. Parisi, D. Zipeto, A. Lisa, C. Casoli, and U. Bertazzoni. 2006. Coinfection with HIV-1 and human T-cell lymphotropic virus type II in intravenous drug users is associated with delayed progression to AIDS. J. Acquir. Immune Defic. Syndr. 41:100-106. [DOI] [PubMed] [Google Scholar]
- 57.Vrielink, H., and H. W. Reesink. 2004. HTLV-I/II prevalence in different geographic locations. Transfus. Med. Rev. 18:46-57. [DOI] [PubMed] [Google Scholar]
- 58.Willy, R. J., C. M. Salas, G. E. Macalino, and J. D. Rich. 1999. Long-term non-progression of HIV-1 in a patient coinfected with HTLV-II. Diagn. Microbiol. Infect. Dis. 35:269-270. [DOI] [PubMed] [Google Scholar]