Abstract
Ebola virus (EBOV) cellular attachment and entry is initiated by the envelope glycoprotein (GP) on the virion surface. Entry of this virus is pH dependent and associated with the cleavage of GP by proteases, including cathepsin L (CatL) and/or CatB, in the endosome or cell membrane. Here, we characterize the product of CatL cleavage of Zaire EBOV GP (ZEBOV-GP) and evaluate its relevance to entry. A stabilized recombinant form of the EBOV GP trimer was generated using a trimerization domain linked to a cleavable histidine tag. This trimer was purified to homogeneity and cleaved with CatL. Characterization of the trimeric product by N-terminal sequencing and mass spectrometry revealed three cleavage fragments, with masses of 23, 19, and 4 kDa. Structure-assisted modeling of the cathepsin L-cleaved ZEBOV-GP revealed that cleavage removes a glycosylated glycan cap and mucin-like domain (MUC domain) and exposes the conserved core residues implicated in receptor binding. The CatL-cleaved ZEBOV-GP intermediate bound with high affinity to a neutralizing antibody, KZ52, and also elicited neutralizing antibodies, supporting the notion that the processed intermediate is required for viral entry. Together, these data suggest that CatL cleavage of EBOV GP exposes its receptor-binding domain, thereby facilitating access to a putative cellular receptor in steps that lead to membrane fusion.
Ebola virus (EBOV) is a member of the Filoviridae family and causes severe hemorrhagic fever in humans and nonhuman primates, with case fatality rates of up to 90%. Virus entry and attachment is mediated by a single envelope glycoprotein (GP) as a class I fusion protein, which is proteolytically processed during maturation into two subunits, GP1 and GP2. The GP1 N terminus contains a putative receptor-binding domain (RBD) (2, 9, 11, 12), and the GP2 C terminus contains a fusion peptide, two heptad-repeat regions, and a transmembrane domain. GP1 and GP2 are linked by a disulfide bond (Cys53-Cys609) and form trimers of heterodimers on the surface of virions. EBOV GP is also extensively glycosylated, especially within a region of GP1 termed the mucin-like domain (MUC domain), which contains multiple N- and O-linked glycans. We and others have previously shown the MUC domain of GP1 to be cytotoxic and to induce cell rounding (17, 21), and deletion of this region increases pseudovirus infectivity compared to that of full-length GP (11). The MUC domain, however, is also known to enhance cell binding through the human macrophage C-type lectin specific for galactose and N-acetylglucosamine (hMGL) (18), suggesting that glycans in this domain may be involved in the initial cellular attachment. Several other studies have identified factors that enhance cell binding and/or infectivity, including folate receptor α (4), β integrins (19), C-type lectins DC-SIGN and L-SIGN (1), and Tyro3 family members (16). However, the critical cellular receptor(s) thought to interact directly with the GP1 RBD have yet to be identified.
Following virus uptake into host cells, which is presumed to occur via receptor-mediated endocytosis (13), the virion is transported to acidified endosomes where GP is exposed to a low pH and enzymatic processing. EBOV entry is pH dependent (19); however, unlike influenza virus, for which a low pH alone induces the conformational changes that lead to membrane fusion (20), recent studies indicate that proteolysis by endosomal cathepsin L (CatL) and CatB (active only at pH 5 to 6) is a dependent step for EBOV entry (5, 14). Although the intermediate EBOV GP generated by CatL cleavage is known to have increased binding and infectivity to target cells (7), little else is known about the cleavage product, specifically where the proteolytic sites are within GP and whether the cleaved product is immunogenic. Recently, Dube and colleagues have proposed a model for CatL cleavage based on thermolysin cleavage (6). However, thermolysin is nonphysiological in this setting and is a member of the metalloenzyme-protease family, whereas CatL is a member of the cysteine-protease family and essential for EBOV entry. In this study, we have characterized the physiological CatL cleavage of the Zaire EBOV GP (ZEBOV-GP) trimer and explored the effect of cleavage on the immunological properties of the GP trimer. To generate this intermediate, we expressed and purified a recombinant form of the Ebola GP trimer ectodomain that had been stabilized with a trimerization motif derived from T4 fibritin (foldon) and purified to homogeneity. The recombinant protein was cleaved with CatL, and the stable cleavage intermediate was characterized biochemically and immunologically. We identified several sites of CatL cleavage within the ZEBOV-GP ectodomain which are different than those observed with thermolysin. The cleaved intermediate product retained binding to the EBOV-neutralizing antibody KZ52 and elicited EBOV-neutralizing antibodies in vaccinated mice. Our data, in conjunction with the recently determined structure of the ZEBOV-GP ectodomain (10), shed light on the critical role of CatL processing in GP structure and function.
MATERIALS AND METHODS
Cell lines and plasmids.
Recombinant baculovirus vectors containing ZEBOV-GP (1976 Mayinga; GenBank accession no. AAC54887) or Sudan EBOV GP (SEBOV-GP, 1976 Boniface; GenBank accession no. AAB37096) were generated by cloning the appropriate EBOV GP into a baculovirus backbone vector (BD Pharmingen, San Diego, CA). A foldon trimerization sequence from bacteriophage T4 fibritin was cloned in place of the transmembrane region at the C terminus, followed by a thrombin cleavage site and His tag. The purified protein contained the following additional C-terminal residues: GSGYIPEAPRDGQAYVRKDGEWVLLSTFLGGSLVPRGSPHHHHHH, with the foldon trimerization domain shown in italics, the thrombin cleavage site underlined, and the His tag in boldface. Hi5 insect cells were maintained in Express Five serum-free media (Invitrogen, Carlsbad, CA). Pseudotyped EBOV GP plasmids included Zaire GP (GenBank accession no. AAC54887), Sudan GP (GenBank accession no. AAB37096), Ivory Coast GP (GenBank accession no. ACI28632), Reston GP (GenBank accession no. AAC54889), and from the newest EBOV strain, Bundibugyo GP (BEBOV-GP; GenBank accession no. ACI28624), which were synthesized using human-preferred codons as described previously (8) by GeneArt (Regensburg, Germany).
Baculovirus production.
GPs were produced by cotransfection of baculovirus transfer vector with BaculoGold-linearized baculovirus DNA (BD Pharmingen) into Spodoptera frugiperda (Sf9) cells (Invitrogen) using a BaculoGold transfection buffer set (BD Pharmingen) and subsequently were amplified in the same cells according to the manufacturer's instructions.
Generation of CatL-cleaved EBOV GP.
ZEBOV-GP with the transmembrane segment deleted was generated using recombinant baculovirus infection of Hi5 insect cells for 48 h at 27°C with gentle shaking. The secreted protein was concentrated using a Pall Centramate system and purified by nickel affinity chromatography (GE Healthcare, Piscataway, NJ), including an ATP (5 mM) and MgCl (3 mM) (Sigma) column wash to remove a contaminating protein interaction. The His tag was removed by thrombin digestion (3 U/mg protein) (Novagen, Madison, WI) and further purified on a Superose6 column (GE Healthcare). Peak EBOV-GP fractions were pooled and cleaved with recombinant Cathepsin L (40 μg/mg protein) (CalBiochem, San Diego, CA) for 90 min at 37°C in 100 mM sodium acetate and 1 mM EDTA at pH 5.5 in a reaction volume of 10 ml, concentrated, and purified on a Superdex200 column (GE Healthcare).
SDS-PAGE, Western blot, and Native Blue analysis.
Purified protein was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and visualized with InstantBlue stain (Novexin, Cambridge, United Kingdom). Protein was analyzed in parallel by Western blot analysis using Ebola GP-specific rabbit immune sera (1:2,000) and a goat anti-rabbit secondary antibody conjugated to horseradish peroxidase (HRP; 1:3,000). All antibodies were diluted in 2.5% skim milk and 0.5% Tween in Tris-buffered saline (TBS). Purified protein was analyzed by Native Blue gel electrophoresis using 4 to 12% Nupage Ready gels (Invitrogen). Molecular mass standards (0.4 μg/μl) included thyroglobulin (669 kDa), ferritin (440 kDa), and aldolase (158 kDa) (Amersham Biosciences, Piscataway, NJ).
PNGaseF treatment.
Purified CatL-cleaved ZEBOV-GP (10 μg) was denatured with 1× glycoprotein denaturing buffer (New England BioLabs [NEB], Ipswich, MA) at 100°C for 10 min and treated with PNGaseF (150 U/μg protein; NEB) in 1× G7 reaction buffer (NEB), supplemented with 1% NP-40, (NEB) and incubated at 37°C for 1 h.
Matrix-assisted laser desorption ionization mass spectrometry (MALDI-MS) analysis.
Protein samples were analyzed using the modified thin-layer method described by Cadene and Chait (3). The matrix used was a saturated solution of α-cyano-4-hydroxycinnamic acid in a 3:1:2 (vol/vol/vol) mixture of formic acid-water-isopropanol. The protein solution was mixed 1:5 with matrix solution and spotted on the sample plate prepared with a thin layer of α-cyano-4-hydroxycinnamic acid and allowed to dry. Standard solutions of 6 mM apomyoglobin and 15 mM bovine serum albumin were mixed with matrix and spotted in the same way for use as calibrants. Spectra were acquired on a Voyager-DE Pro (Applied Biosystems, Foster City, CA) operating in linear mode. Instrument settings were as follows: accelerating voltage, 25,000 V; grid voltage, 93%; extraction delay time, 1,050 ns; laser intensity, 2,200 to 2,700.
N-terminal sequencing.
Proteins were adsorbed onto polyvinylidene difluoride (PVDF) membrane using a ProSorb sample preparation cartridge (Applied Biosystems) and washed with 0.1% trifluoroacetic acid (TFA). The PVDF disk was removed from the cartridge and sequenced on a Procise 494 protein sequencer (Applied Biosystems) using the standard protocol from the manufacturer.
Surface plasmon resonance.
Kinetic parameters of KZ52 and CatL-cleaved ZEBOV-GP were determined with a Biacore 3000 surface plasmon resonance spectrometer (GE Healthcare). KZ52 was immobilized on CM5 sensor chips using standard amine coupling to surface densities of 474, 246, 241, 238, 154, and 151 response units. CatL-cleaved ZEBOV-GP and SEBOV-GP were injected sequentially over the coupled sensor chips with increasing concentrations of 31.25, 125, 250, and 500 nM at a flow rate of 30 μl/min for 300 s at 25°C. The dissociation rates of the complexes were monitored for 600 s at a flow rate of 30 μl/min. The buffer used for preparation of the ZEBOV-GP samples and buffer blanks contained 10 mM HEPES (pH 7.4), 150 mM NaCl, 3 mM EDTA, 0.005% surfactant P-20, and 0.1% carboxymethyldextran. The same buffer was used as running buffer during the binding studies. A 1:1 Langmuir binding model with drifting baseline (BiaEvaluation 4.1) was used to fit data globally and to extract kinetic parameters of interaction with ZEBOV-GP.
Vaccination.
Female BALB/c mice (6 to 8 weeks old; Jackson Laboratories, Bar Harbor, ME) were immunized intramuscularly with 20 μg of purified CatL-cleaved ZEBOV-GP or uncleaved ZEBOV-GP in 100 μl of phosphate-buffered saline (PBS; pH 7.4) at weeks 0, 4, and 8. The collected immune sera were analyzed for neutralizing activity. All animal experiments were conducted in full compliance with all relevant federal guidelines and NIH policies.
Production of GP-pseudotyped lentiviral vectors and measurement of neutralizing activity of immune sera.
Ebola GP-pseudotyped lentiviral vectors expressing a luciferase reporter gene were produced as described previously (8, 22). Briefly, 293T cells were cotransfected with 7 μg of pCMVΔR8.2, 7 μg of pHR′CMV-Luc, and 125 ng cytomegalovirus enhancer with HTLV-1 R region (CMV/R) ZEBOV-GP, CMV/R SEBOV-GP, CMV/R Ivory Coast EBOV GP (ICEBOV-GP), CMV/R Reston EBOV GP (REBOV-GP), CMV/R BEBOV-GP, or vesicular stomatitis virus GP (VSV-GP). Cells were transfected overnight and then washed the following day, and fresh medium was added. Supernatants were harvested 48 h later, filtered through a 0.45-μm syringe filter, aliquoted, and stored at −80°C. For neutralization assays, immune sera were mixed with 100 μl of pseudovirus at various dilutions and added to 786-O cells (ATCC, Manassas, VA) in 96-well dishes (1.5 × 103 cells/well). Plates were washed, and fresh medium was added 14 to 16 h later. Following infection for 48 h, cells were lysed in mammalian cell lysis buffer (Promega, Madison, WI). A standard quantity of cell lysate was used in a luciferase assay with luciferase assay reagent (Promega) according to the manufacturer's protocol. The 50% inhibitory concentrations (IC50s) were calculated using GraphPad Prism4 (GraphPad, San Diego, CA). Preimmune sera mixed with pseudovirus were used to calculate the neutralizing activity.
RESULTS
Expression, purification, and characterization of CatL-cleaved ZEBOV-GP.
CatL cleavage of Ebola virus GP is a critical and essential step in virus entry; however, important properties of the intermediate EBOV GP cleavage product are currently undefined. To characterize the intermediate ZEBOV-GP resulting from CatL cleavage, we first purified the full-length, mucin-containing ZEBOV-GP ectodomain expressed in baculovirus using His-affinity purification. Insect protein glycosylation pathways are not necessarily equivalent to mammalian pathways; any lack of complex glycosylation is not expected to interfere with proteolytic enzyme cleavage, although the greater accessibility to CatL could lead to false-positive cleavage not observed in vivo. This method of protein expression nonetheless provides a well-defined biochemical system to analyze proposed mechanisms of proteolysis. The recombinant trimer GP was stabilized with a foldon trimerizing domain at the C terminus followed by a thrombin cleavage site and a His tag (Fig. 1A). After affinity purification, we found it necessary to remove the His tag by thrombin cleavage to prevent nonspecific aggregation. The sequence of the C-terminal tag for generating stable trimer GP was therefore important, and we found that the most stable protein was generated using the sequence of a foldon trimerizing domain followed by a thrombin cleavage site and His tag (FTH) (Fig. 1A). Other recombinant trimer GPs were generated using a C-terminal tag of thrombin cleavage site followed by a foldon trimerizing domain and a His tag (TFH) or the native GP sequence; however, these proteins were unstable and therefore not pursued. Following thrombin digestion to remove the His tag (while retaining the foldon trimerization domain), we further purified the protein using size exclusion chromatography. The protein eluted as a single peak, consistent with an apparent molecular mass of approximately 600 kDa (Fig. 1B and C). The apparent molecular weight was confirmed by dynamic light scattering analysis using DYNAMICS V6 software (Wyatt Technology Corporation, Santa Barbara, CA), which determines the apparent molecular weight derived from the measured hydrodynamic radius (in nm) and empirical relationships between radius and molar mass (Fig. 1D). The estimated molecular weight was determined using the globular molecular family setting. Peak gel filtration fractions analyzed by SDS-PAGE (Fig. 1C, lanes 1 to 3) and Western blotting (Fig. 1C, lanes 4 to 6) show the reduced GP1 and GP2 at approximately 120 and 25 kDa, respectively. The calculated molecular mass of the protein component of the thrombin-cleaved trimer is 214.5 kDa, and the ectodomain of ZEBOV-GP is predicted to have up to 17 N-linked glycosylation sites (http://www.cbs.dtu.dk/services/NetNGlyc/) and potentially 15 O-linked glycosylation sites (http://www.cbs.dtu.dk/services/NetOGlyc/), which together would suggest an average molecular mass of 2 kDa for each of the potential N-linked sites and 0.5 kDa for the O-linked sites. The high apparent molecular weight of the trimer likely results from this large number of glycans, which in addition to adding mass are highly hydrated and extend in a nonspherical way from the protein surface, thereby resulting in an increased Stokes radius. This elongated shape of the trimer, based on our sedimentation velocity analysis (data not shown), also contributes to the high apparent molecular weight. Thus, our results are consistent with purification of an intact trimer. To further determine the protein's oligomeric state, we performed Native Blue gel electrophoresis analysis and observed a GP1:GP2 trimer band at approximately 600 kDa (Fig. 1C, lanes 7 to 9), with no dimer or monomer bands detected. Together, these results confirm that stable, trimeric ZEBOV-GP was generated.
FIG. 1.
Characterization of recombinant full-length ZEBOV-GP. (A) Schematic of EBOV GP plasmid construct containing the GP1 region (orange; residues 31 to 500), with the RBD (dark-dotted orange; residues 54 to 201) and the MUC domain (white-dotted orange; residues 309 to 500), the GP2 region (green; residues 501 to 647), signal sequence (red; residues 1 to 30) and foldon domain (blue). The furin cleavage site is shown directly prior to GP2 at residue 500, and the thrombin cleavage site is shown directly prior to the His tag. N-linked glycan sites are shown as “Y,” and disulfide bonds are shown in red as “S”. (B) His affinity-purified EBOV GP was thrombin digested and resolved by gel filtration size-exclusion chromatography. The approximate peak size was 600 kDa. Molecular mass standards shown are as follows: thyroglobulin, 669 kDa; ferritin, 440 kDa; aldolase, 158 kDa; ovalbumin, 43 kDa; and ribonuclease A, 13.7 kDa. (C) Gel filtration peak fractions were separated by SDS-PAGE and Coomassie stained (left) or transferred to nitrocellulose membrane and Western blotted with an Ebola-specific antibody detecting both GP1 and GP2 (middle) and show GP1 and GP2 at approximately 120 and 25 kDa, respectively. Peak fractions were also analyzed by Native Blue gel electrophoresis to confirm that the protein was trimeric (right) and showed an approximately 600-kDa GP1:GP2 trimer protein. (D) Dynamic light scattering analysis of full-length recombinant ZEBOV-GP. Molecular weight (MW; in thousands) and polydispersity percent (Pd%) of peak fractions following thrombin digestion and size exclusion chromatography on a Superose6 column.
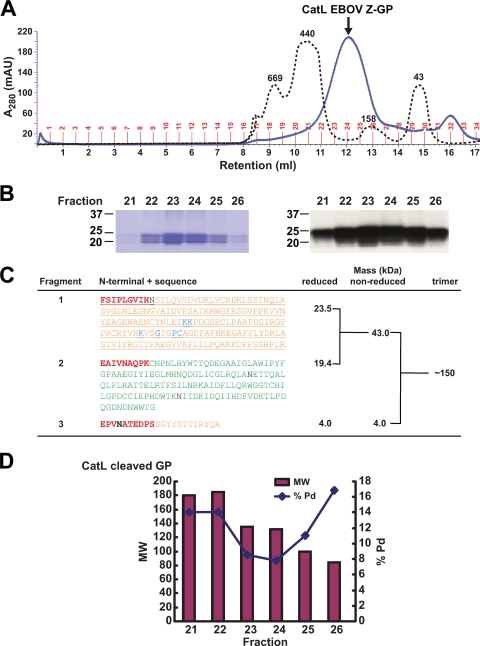
To characterize the intermediate CatL-cleaved ZEBOV-GP product, we purified CatL-digested protein by size exclusion chromatography (Superdex 200 10/30 model) and observed a sharp single peak centered at 12.7 ml, an elution volume consistent with a molecular mass of approximately 150 kDa (Fig. 2A). The peak gel filtration fractions were analyzed by SDS-PAGE (Fig. 2B, left panel) and Western blotting (Fig. 2B, right panel) and showed two major bands at ∼19 and 23 kDa and a less-prominent band at ∼21 kDa. The peak fractions were pooled and analyzed by N-terminal sequencing and MS to map the CatL cleavage sites and estimate the C termini. N-terminal sequencing results revealed three distinct termini in equal ratios, starting at residue 31 after the signal sequence, residue 501 directly following the furin cleavage site, and at residue 201 at the C terminus of the putative RBD (Fig. 2C). MS analyses showed that the disulfide bond between GP1:GP2 was maintained following CatL cleavage, with a nonreduced mass of 43 kDa and reduced masses of 23.5 and 19.4 kDa, respectively (Fig. 2C). A minor peak of 21.6 kDa was also reported by MS, and all three masses correspond to the fragments in Fig. 2B. MS analysis of the third fragment, in both nonreduced and reduced samples, revealed a mass of approximately 4 kDa and indicates that the fragment remained noncovalently associated with the rest of the GP1:GP2 complex (Fig. 2C). Analysis by gel filtration and dynamic light scattering showed that the trimer mass was approximately 150 kDa, which fits well with our observed monomeric mass of approximately 47 kDa, being comprised of the 43-kDa GP1:GP2 disulfide-linked fragments and the 4-kDa non-covalently associated fragment (Fig. 2C). This is consistent with the removal of most of the glycosylation on GP and loss of the MUC domain by CatL, leading to a more compact molecule with a smaller Stokes radius. The protein polydispersity percent is an indicator of homogeneity with respect to oligomeric state. The polydispersity of CatL-cleaved ZEBOV-GP was lower in peak fractions compared to uncleaved ZEBOV-GP (Fig. 1D and 2D). These combined results confirm that we successfully generated the CatL-cleaved EBOV GP and characterized three CatL cleavage products, with two fragments in GP1 and one fragment of intact GP2.
FIG. 2.
Characterization of CatL-cleaved EBOV GP. (A) Thrombin-digested EBOV GP was cleaved with CatL and resolved on a Superdex200 column. Molecular mass standards shown are the same as for Fig. 1B. The approximate size was 150 kDa by dynamic light scatter analysis. (B) The reduced protein size is shown by SDS-PAGE Coomassie staining (left) and Western blotting using an Ebola virus-specific antibody (right). (C) The three N terminals resulting from CatL cleavage of ZEBOV-GP are shown in boldface and red type, with the GP1 sequences in orange and GP2 in green. N-linked glycan sites (black) and residues important for virus entry (blue) are also shown. The estimated C termini were calculated based on mass analyses of reduced and nonreduced CatL cleavage reactions and dynamic light scatter (shown as trimer) to confirm the protein trimeric state and mass. (D) Dynamic light scattering analysis of recombinant CatL-processed ZEBOV-GP. Molecular weight (MW; in thousands) and polydispersity percent (Pd%) of peak fractions following cleavage and size exclusion chromatography on a Superdex200 column.
Modeling the CatL-cleaved ZEBOV-GP structure.
From the masses reported by our MS analysis and the N-terminal sequencing results, we estimated the approximate length of each fragment generated from CatL cleavage. These fragments were then mapped onto the recently reported crystal structure of the prefusion ZEBOV-GP trimer complexed to the human neutralizing antibody KZ52 (10). An MS analysis of nonreduced, PNGaseF-treated CatL-cleaved ZEBOV-GP resulted in a mass of 35.1 kDa. The weight difference between the 43-kDa glycosylated and the 35.1-kDa deglycosylated GP1:GP2 fragment suggested an N-linked glycan contribution of 7.9 kDa. If one assumes a single CatL cleavage site between Arg200 and Glu201, the calculated weight of the first GP1 fragment is 18.6 kDa. Subtracting this from the 35.1-kDa deglycosylated GP1:GP2 total results in a GP2 fragment calculated mass of 16.5 kDa. A GP2 fragment of this size beginning at the furin cleavage site (Glu502) would end at approximately residue Gly647, directly adjacent to the Gly-Ser-Gly linker leading into the C-terminal foldon (Fig. 3A). The GP1 fragment is predicted to have one N-linked glycosylation site, and the GP2 fragment is predicted to have two N-linked glycosylation sites. The small 4-kDa fragment resulting from CatL cleavage starts at Glu201, and within five residues there is a potential N-linked glycosylation site, at Asn204. However, since the estimated N-linked glycan contribution is approximately 2 kDa, it is possible that the majority of this fragment is composed of carbohydrate, with N-terminal sequencing confirming that this peptide extends at least to Ser210 and may possibly extend up to Ala222.
FIG. 3.
Mapping and modeling of the ZEBOV-GP CatL cleavage site. (A) Schematic of the EBOV GP sequence aligned with secondary structure features. Alpha-helices and beta-strands are numbered and shown in gray (or outlined in a black dotted line for tentative assignment), and the three N termini at resides 31, 201, and 501 are shown in boldface and red. The N-terminal CatL cleavage site in GP1 is shown at residue 201 (red arrow) and the C-terminal site approximately at residue 222 (dotted red arrow), while the C-terminal of GP2 extends to residue 647 (dotted red arrow). The disordered loop between beta-strands 13 and 14 is shown by a black dotted line. The furin cleavage site is depicted by a black arrow at residue 502. GP1 (orange), GP2 (green), N-glycans (black), and residues important for entry (blue) are labeled as in Fig. 2. The region deleted upon CatL cleavage, including the entire MUC domain, is highlighted in blue. (B) The modeled CatL-cleaved monomer structure is shown based on the recently reported structure and Protein Data Bank file number 3CSY (10). Beta-strands and alpha-helices of the head region are numbered, with GP1 and GP2 residues shown in orange and green, respectively. The first GP1 CatL cleavage site is shown in the disordered loop between β13 and β14, with the latter beta-strand being covalently associated to beta-strand 9. This model assumes there is no conformational change following processing by CatL. (C) The EBOV GP trimer structure (upper panel) (10) is shown by ribbons with a transparent surface representation and red lines depicting the CatL cleavage site in the β13-14 loop. As reported in reference 10, GP1 (shades of orange) forms a chalice, while GP2 (shades of green) girdles the base of GP1. The modeled CatL-cleaved EBOV trimer structure (lower panel) (based on reference 10 and using the same color scheme as the upper panel) shows the removal of the glycan cap while maintaining the GP2 and the intact fusion peptide, and it also assumes that no conformational change results from the cleavage process. All graphic representations were produced with PyMOL.
Our analysis shows that CatL cleaves at a minimum of two different sites, resulting in an approximately 150-kDa trimer, only one-fourth of the original apparent 600-kDa size. When mapped onto the crystal structure of ZEBOV-GP, the first cleavage site at Glu201 is in a flexible loop between GP1 β-strands 13 and 14 (disordered in the ZEBOV-GP structure). The second cleavage site is between Ser210 and Ala222, either before or after β-strand 14. The observation that the 4-kDa fragment remains associated with the trimer suggests that it has secondary structure; therefore, the cleavage probably occurs after β-strand 14. In contrast, thermolysin cleavage is observed to completely remove residues 191 to 501, including β-strand 14 (6). Our two reported CatL cleavage sites coupled with the natural furin cleavage result in removal of the membrane distal “glycan cap” region of the trimer as well as the entire MUC domain. A third potential cleavage site mapped to the region of Gly647 toward the end of GP2 directly before the foldon domain (or the transmembrane region in wild-type protein); however, the site is likely protected because of its expected proximity to the viral membrane. A comparison between the modeled CatL-cleaved trimer ZEBOV-GP structure and the uncleaved prefusion trimer ZEBOV-GP structure is displayed in Fig. 3C.
CatL-cleaved ZEBOV-GP elicits a neutralizing antibody response.
There are very few Ebola virus-neutralizing antibodies and presently none that protect efficiently against virus challenge in nonhuman primates. Given that a critical region for virus entry is exposed upon CatL cleavage, we hypothesized that the CatL-cleaved ZEBOV-GP intermediate product may elicit a neutralizing antibody response. To test this hypothesis, we immunized two groups of mice with CatL-cleaved ZEBOV-GP and compared the antibody response to that elicited by uncleaved ZEBOV-GP. Both ZEBOV-GPs (CatL cleaved and uncleaved) elicited a neutralizing antibody response, and the CatL-cleaved ZEBOV-GP immunogen elicited a titer that was 3-fold higher than that of the uncleaved ZEBOV-GP immunogen, with respective IC50 titers of 2,837 and 948 (Fig. 4A). Though the difference is modest, the data nonetheless confirm that the cleaved core is a target of neutralization and can elicit neutralizing antibody responses. To determine the specificity of the antisera, we examined its activity against other EBOV species. This finding was of interest because the surface residues in contact with the six critical virus entry residues are highly conserved among EBOV species (Fig. 5). Using Ebola GP-pseudotyped virus for each species, we observed the highest cross-neutralization with ICEBOV-GP (Fig. 4B). Limited neutralization of other Ebola species (Sudan, Bundibugyo, and Reston) was observed while the control VSV-G pseudotyped virus showed no neutralization (Fig. 4B). These results demonstrate that vaccination with the CatL-cleaved ZEBOV-GP intermediate elicits a neutralizing antibody response but that the breadth of immunity is limited, being highest in the most closely related virus.
FIG. 4.
Ability of CatL-cleaved trimeric EBOV-GP ZEBOV-GP to elicit neutralizing antibodies and bind KZ52. (A) The neutralization by antisera from groups of five mice immunized and boosted at weeks 0, 4, and 8 with 20 μg of either CatL-cleaved or uncleaved ZEBOV-GP in 50 μl PBS and an equal volume of Ribi adjuvant was analyzed by incubating the mouse sera (collected 10 days after each vaccination) with an ZEBOV-GP pseudotyped lentiviral reporter vector expressing luciferase. The neutralization activity was calculated based on the luciferase activity relative to values reported for preimmune mouse sera. CatL-cleaved ZEBOV-GP induced the highest titer of neutralizing antibody response, while the uncleaved ZEBOV-GP was 3-fold less potent. (B) Mouse sera from CatL-cleaved ZEBOV-GP mice were analyzed for neutralizing activity against all five EBOV-GP strains individually pseudotyped or a VSV control with lentiviral reporter vectors expressing luciferase. Matched Zaire GP and Ivory Coast GP pseudotyped viruses showed the highest neutralization, while Sudan GP, Bundibugyo GP, and Reston GP pseudotyped viruses showed limited neutralization. The VSV control showed no neutralization. (C) Surface plasmon resonance analysis of CatL-cleaved ZEBOV-GP (upper panel) and SEBOV-GP (lower panel) showed that KZ52 binds both CatL-cleaved EBOV GP strains with a binding affinity of 1.5 nM and 63 nM, respectively.
FIG. 5.
Filovirus GPs receptor-binding domain sequence alignment. EBOV-GP species Bundibugyo (BEBOV), Ivory Coast (ICEBOV), Zaire (ZEBOV), Reston (REBOV), and Sudan (SEBOV) and Marburg virus strains Angola (MARB-A) and Popp (MARB-P) were sequence aligned in the receptor-binding domain. Residues in red are conserved among all filoviruses, and the six conserved core residues are marked with an asterisk. Residues that are located in close surface proximity to the six conserved residues are boxed in red.
ZEBOV-GP CatL binds to KZ52 human neutralizing antibody.
The structure of the prefusion form of ZEBOV-GP complexed to the human neutralizing antibody KZ52 has been reported (10). To determine whether the CatL-cleaved ZEBOV-GP binds to KZ52, we analyzed the binding properties of the CatL-cleaved intermediate protein using surface plasmon resonance. Any conformational change that may potentially be initiated by CatL cleavage did not disrupt binding to KZ52, and we observed strong binding affinities of 1.5 nM for CatL-cleaved ZEBOV-GP and 63 nM for CatL-cleaved SEBOV-GP, when bound to KZ52 (Fig. 4C). The KZ52 epitope on ZEBOV-GP is complex and discontinuous, comprising residues from both GP1 and GP2. Therefore, the observed high affinity of KZ52 to the CatL-cleaved ZEBOV-GP suggests that no large conformational changes have been triggered by this cleavage. Moreover, the foldon domain at the C terminus of our construct is removed by CatL, preventing it from hindering any potential conformational change in GP1:GP2. Thus, proteolysis alone is likely insufficient to directly trigger the conformational changes that precipitate membrane fusion, suggesting that a secondary factor may be required.
CatL cleavage exposes the conserved EBOV GP core and critical entry residues.
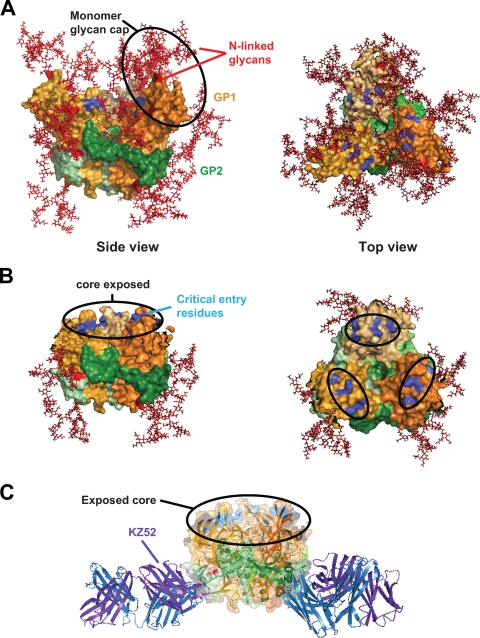
To further characterize the CatL-cleaved intermediate EBOV GP product, we examined how cleavage would affect residues that are critical for virus entry. Six residues identified as being essential for virus entry (10) were mapped onto the modeled CatL-cleaved ZEBOV-GP structure and were shown to be completely exposed within the conserved core (Fig. 6B), which was previously occluded by five N-linked glycan sites in GP1 (Fig. 6A) and further concealed by extensive glycosylation of the MUC domain (eight N-linked glycans and 11 predicted O-linked glycan sites). Furthermore, modeling of the CatL-cleaved ZEBOV-GP structure complexed to KZ52 shows that the conserved core remains exposed (Fig. 6C), with no interference from KZ52 predominantly binding to GP2 at the structure base. These modeling results indicate that critical residues within the conserved RBD are exposed as a direct result of CatL cleavage.
FIG. 6.
Receptor-binding residues modeled on CatL-cleaved EBOV GP trimer structure. (A) Surface representation of the ZEBOV-GP trimer structure (as reported in reference 10) depicting N-glycan sites in the head region (red) (side view; left panel) and residues important for virus entry (blue) (top view; right panel). GP1 is shown in shades of orange and GP2 in shades of green. (B) The surface-modeled CatL-cleaved EBOV-GP trimer structure (based on reference 10) reveals the complete removal of all N-linked glycans (red) from the head region surface (side view; left panel) and exposes the conserved core of the RBD and critical residues for virus entry (blue) (top view; right panel). The approximately 20-Å by 15-Å footprint reported by Lee et al., which contains residues important for virus entry, is indicated in the black circled regions of the top view (right panel). (C) Using a ribbon diagram and transparent surface rendering, the model of CatL-cleaved ZEBOV-GP complexed to the heavy (blue) and light (purple) chains of KZ52 depicts unobstructed binding of KZ52 to the processed EBOV GP and supports the surface plasmon resonance binding data. The color scheme of EBOV GP is the same as in Fig. 5. All graphic representations were produced with PyMOL.
DISCUSSION
Proteolysis of the Ebola surface glycoprotein by cathepsins in the host cell endosome is a critical step for viral entry (5, 14). In this study, we characterized the products generated by CatL cleavage of ZEBOV-GP and examined their relevance to entry and membrane fusion. Using N-terminal sequencing and mass spectrometry analyses of CatL-cleaved ZEBOV-GP, we identified three CatL cleavage sites, confirmed the furin cleavage site, and observed three cleavage products. The first CatL cleavage occurs at Glu201 between β-strands 13 and 14, the second between Ser210 and Ala222, most likely after β-strand 14, and a third potential site may occur close to Gly647 near the C-terminal end of GP2 but is likely protected by the close proximity of the viral membrane. The known furin cleavage was also observed at Glu502. The first two cleavages result in the removal of the membrane distal “glycan cap” and the entire MUC domain. This confirms a hypothesis by Lee et al., who predicted a CatL cleavage site within the disordered β13-14 loop (residues 190 to 213) based on their crystal structure of the GP ectodomain (10). However, in contrast to thermolysin cleavage of EBOV-ZEBOV-GP, it appears that the outer strand of the head, β-strand 14, remains as part of the trimer after CatL cleavage. Two cleavage products were observed by MS under nonreducing conditions (43 kDa and 4 kDa), and three main products were observed under reducing conditions (23.5, 19.4, and 4 kDa). These products result from disulfide-bonded GP1 and GP2 and a non-covalently associated 4-kDa fragment. Most of the extensive glycosylation of GP is removed by CatL cleavage, with MS analysis of PNGaseF-treated, CatL-cleaved GP showing that only 7.9 kDa of glycosylation per GP1:GP2 monomer remains after cleavage.
A recent study by Dube and coworkers characterized ZEBOV-GP cleavage and priming by thermolysin, a nonphysiological metalloprotease (6). They observed a single cleavage site between residues 190 and 194, generating a 19-kDa GP1 fragment beginning at residue 33, with the GP2 fragment extending up to the transmembrane domain. However, we observe the more physiological CatL cleavage at two different locations. It is not surprising that we observed a different cleavage pattern, since CatL and thermolysin are members of completely different protease families (cysteine and metalloprotease, respectively) and function at different pHs (5.5 and 7.5, respectively). Whether further processing of the CatL-cleaved GP by CatB generates a cleavage site at residue 190 remains to be determined in future studies. Dube and coworkers also predicted a 20-kDa fragment, likely to include residues 33 to 200. The difference between the reported 20-kDa fragment and our 23.5-kDa fragment is likely due to the contribution of an N-linked glycosylation site contained within the GP1 N-terminal fragment. Nevertheless, modeling or predictions of the processed intermediate structure by us and others agree that the MUC domain, the glycan cap, and β-strand 15 of the head domain are removed upon CatL processing.
CatL cleavage of the ZEBOV-GP trimer is required for viral entry (5, 14). One way in which CatL likely facilitates entry is that removal of the glycan cap domain and the MUC domain and extensive glycosylation provide much greater access to the receptor-binding domain. This is evident from studies showing higher levels of cell binding of Ebola virus upon CatL treatment (7) and is also supported by our modeling of the CatL-cleaved GP. As CatL cleavage optimally occurs in the acidic milieu of the endosome, it is also possible that a direct interaction between CatL-cleaved EBOV-GP and a cellular receptor may take place in the endosome; however, future studies are required to determine the cellular receptor(s) and site of binding. The EBOV GP1 residues critical to virus entry have been proposed by multiple groups (2, 11, 12), with the recently reported ZEBOV-GP trimer structure identifying a cluster of six residues (K114, K115, K140, G143, P146, and C147) located in an approximately 20- by 15-Å footprint on the inner surface of the GP1 trimer (10). These six residues, when mapped onto our CatL-processed GP model, become completely exposed on the surface of the trimer intermediate. Furthermore, these six residues are conserved among all EBOV species, and two of the six residues are conserved in Marburg virus strains (Fig. 5). Recently, Dube and coworkers employed mutational studies to confirm the importance of residues K114, K115, and K140 and also identified K95 as another critical residue (6). However, receptor binding was not completely obliterated by the combined mutations of these four lysine residues, indicating that other residues of GP1 may also be involved in EBOV entry (6).
Our study is the first to report the immunological properties of the CatL-cleaved ZEBOV-GP intermediate. Importantly, the cleaved ZEBOV-GP elicited 3-fold more potent neutralization activity in the sera of vaccinated mice, slightly greater than that of uncleaved EBOV GP (Fig. 4). CatL cleavage, through exposure of the RBD and residues critical for virus entry, likely facilitates access of neutralizing antibodies to the GP receptor-binding site. We employed uncleaved EBOV-GP pseudovirus in evaluating the neutralizing activity of mouse sera following immunization, principally to mimic the initial interaction between EBOV GP and the immune system. The neutralizing sera elicited by immunization with CatL-cleaved EBOV GP may well have been more potent had matched CatL-treated pseudovirus been used to analyze the neutralizing activity. Furthermore, the polyclonal neutralizing immune serum response was generated against the entire CatL-cleaved EBOV-GP surface; thus, a monoclonal antibody directed specifically against the conserved core residues may provide a broader neutralizing response in Ebola species than an antibody directed to regions with higher sequence variability.
Our results also indicate that binding to the neutralizing antibody KZ52 was not disrupted by CatL cleavage. This strongly argues that another cathepsin, CatB, and/or secondary factors are likely required to trigger fusion peptide exposure and GP-mediated membrane fusion (Fig. 7). This proposal is supported by two differing models of GP-mediated fusion: that of Chandran et al., who propose that CatL and B processing alone is sufficient to trigger membrane fusion, and that of Schornberg et al., who propose that CatL and CatB as well as additional secondary factors are required to trigger the membrane fusion event. However, all reported models and our study agree that CatL processing alone is insufficient to trigger and initiate the required conformational change that leads to membrane fusion. A recent study examining cathepsins in the endosome suggested that α5β1 integrins play a role in regulating the activation of cathepsins and may thus be important for EBOV virus entry (15). Interestingly, the regulation was shown to occur at the level of cathepsin processing, after binding and prior to membrane fusion (15).
FIG. 7.
Schematic model of ZEBOV-GP entry and fusion events. Attachment and binding of the virion (purple) to the host cell membrane is the first step required for EBOV-GP virus entry and replication, followed by endocytosis of the virion into vesicles. The critical step of virus entry is CatL/CatB cleavage at acidic pH 5 to 6, which acts to prime the GP for subsequent membrane fusion. Virus on the cell surface may also encounter factors that are present in CatL-containing endosomes. A recent study implicated α5β1 integrins in the regulation of double chain (DC) CatB and L expression. Our results indicate that CatL processing alone is insufficient to trigger the conformational change and suggest that CatB and/or additional cellular factors are required to initiate the conformational change leading to the final membrane fusion step and release of the nucleocapsid into the cytoplasm.
This study provides the first reported specific CatL-cleavage sites in ZEBOV-GP and demonstrates that exposure of the conserved core of GP likely facilitates receptor binding and elicits a potent neutralizing antibody response. Our results confirm that these conserved determinants are required for viral entry and suggest that CatB and/or other secondary factors are involved in triggering the conformational changes that lead to EBOV GP-mediated membrane fusion.
Acknowledgments
We thank Ati Tislerics for manuscript preparation, Brenda Hartman and Michael Cichanowski for the preparation of figures, and members of the Virological Laboratory, Vaccine Research Center, for helpful advice and discussions. We acknowledge Cammi Bittner, Ralph Hopkins, and James Hartley of the Advanced Technology Program, SAIC-Frederick, Inc., National Cancer Institute at Frederick, MD, for recombinant baculovirus protein production. We thank Mary Ann Gawinowicz for performing the mass spectrometry and N-terminal sequencing analyses. We also thank Young Do Kwon for help performing the surface plasmon resonance measurements.
This research was supported by the Intramural Research Program, Vaccine Research Center, NIAID, NIH.
Footnotes
Published ahead of print on 6 January 2010.
REFERENCES
- 1.Alvarez, C. P., F. Lasala, J. Carrillo, O. Muñiz, A. L. Corbí, and R. Delgado. 2002. C-type lectins DC-SIGN and L-SIGN mediate cellular entry by Ebola virus in cis and in trans. J. Virol. 76:6841-6844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brindley, M. A., L. Hughes, A. Ruiz, P. B. McCray, Jr., A. Sanchez, D. A. Sanders, and W. Maury. 2007. Ebola virus glycoprotein 1: identification of residues important for binding and postbinding events. J. Virol. 81:7702-7709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cadene, M., and B. T. Chait. 2000. A robust, detergent-friendly method for mass spectrometric analysis of integral membrane proteins. Anal. Chem. 72:5655-5658. [DOI] [PubMed] [Google Scholar]
- 4.Chan, S. Y., C. J. Empig, F. J. Welte, R. F. Speck, A. Schmaljohn, J. F. Kreisberg, and M. A. Goldsmith. 2001. Folate receptor-α is a cofactor for cellular entry by Marburg and Ebola viruses. Cell 106:117-126. [DOI] [PubMed] [Google Scholar]
- 5.Chandran, K., N. J. Sullivan, U. Felbor, S. P. Whelan, and J. M. Cunningham. 2005. Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection. Science 308:1643-1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dube, D., M. B. Brecher, S. E. Delos, S. C. Rose, E. W. Park, K. L. Schornberg, J. H. Kuhn, and J. M. White. 2009. The primed ebolavirus glycoprotein (19-kilodalton GP1,2): sequence and residues critical for host cell binding. J. Virol. 83:2883-2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaletsky, R. L., G. Simmons, and P. Bates. 2007. Proteolysis of the Ebola virus glycoproteins enhances virus binding and infectivity. J. Virol. 81:13378-13384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kong, W.-P., C. Hood, Z.-Y. Yang, C. J. Wei, L. Xu, A. Garcia-Sastre, T. M. Tumpey, and G. J. Nabel. 2006. Protective immunity to lethal challenge of the 1918 pandemic influenza virus by vaccination. Proc. Natl. Acad. Sci. U. S. A. 103:15987-15991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuhn, J. H., S. R. Radoshitzky, A. C. Guth, K. L. Warfield, W. Li, M. J. Vincent, J. S. Towner, S. T. Nichol, S. Bavari, H. Choe, M. J. Aman, and M. Farzan. 2006. Conserved receptor-binding domains of Lake Victoria marburgvirus and Zaire ebolavirus bind a common receptor. J. Biol. Chem. 281:15951-15958. [DOI] [PubMed] [Google Scholar]
- 10.Lee, J. E., M. L. Fusco, A. J. Hessell, W. B. Oswald, D. R. Burton, and E. O. Saphire. 2008. Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor. Nature 454:177-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Manicassamy, B., J. Wang, H. Jiang, and L. Rong. 2005. Comprehensive analysis of ebola virus GP1 in viral entry. J. Virol. 79:4793-4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mpanju, O. M., J. S. Towner, J. E. Dover, S. T. Nichol, and C. A. Wilson. 2006. Identification of two amino acid residues on Ebola virus glycoprotein 1 critical for cell entry. Virus Res. 121:205-214. [DOI] [PubMed] [Google Scholar]
- 13.Sanchez, A. 2007. Analysis of filovirus entry into Vero e6 cells, using inhibitors of endocytosis, endosomal acidification, structural integrity, and cathepsin (B and L) activity. J. Infect. Dis. 196(Suppl. 2):S251-S258. [DOI] [PubMed] [Google Scholar]
- 14.Schornberg, K., S. Matsuyama, K. Kabsch, S. Delos, A. Bouton, and J. White. 2006. Role of endosomal cathepsins in entry mediated by the Ebola virus glycoprotein. J. Virol. 80:4174-4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schornberg, K. L., C. J. Shoemaker, D. Dube, M. Y. Abshire, S. E. Delos, A. H. Bouton, and J. M. White. 2009. Alpha5beta1-integrin controls ebolavirus entry by regulating endosomal cathepsins. Proc. Natl. Acad. Sci. U. S. A. 106:8003-8008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shimojima, M., A. Takada, H. Ebihara, G. Neumann, K. Fujioka, T. Irimura, S. Jones, H. Feldmann, and Y. Kawaoka. 2006. Tyro3 family-mediated cell entry of Ebola and Marburg viruses. J. Virol. 80:10109-10116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Simmons, G., R. J. Wool-Lewis, F. Baribaud, R. C. Netter, and P. Bates. 2002. Ebola virus glycoproteins induce global surface protein down-modulation and loss of cell adherence. J. Virol. 76:2518-2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takada, A., K. Fujioka, M. Tsuiji, A. Morikawa, N. Higashi, H. Ebihara, D. Kobasa, H. Feldmann, T. Irimura, and Y. Kawaoka. 2004. Human macrophage C-type lectin specific for galactose and N-acetylgalactosamine promotes filovirus entry. J. Virol. 78:2943-2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takada, A., C. Robison, H. Goto, A. Sanchez, K. G. Murti, M. A. Whitt, and Y. Kawaoka. 1997. A system for functional analysis of Ebola virus glycoprotein. Proc. Natl. Acad. Sci. U. S. A. 94:14764-14769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.White, J. M., and I. A. Wilson. 1987. Anti-peptide antibodies detect steps in a protein conformational change: low-pH activation of the influenza virus hemagglutinin. J. Cell Biol. 105:2887-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang, Z.-Y., H. J. Duckers, N. J. Sullivan, A. Sanchez, E. G. Nabel, and G. J. Nabel. 2000. Identification of the Ebola virus glycoprotein as the main viral determinant of vascular cell cytotoxicity and injury. Nat. Med. 6:886-889. [DOI] [PubMed] [Google Scholar]
- 22.Yang, Z.-Y., B. K. Chakrabarti, L. Xu, B. Welcher, W.-P. Kong, K. Leung, A. Panet, J. R. Mascola, and G. J. Nabel. 2004. Selective modification of variable loops alters tropism and enhances immunogenicity of human immunodeficiency virus type 1 envelope. J. Virol. 78:4029-4036. [DOI] [PMC free article] [PubMed] [Google Scholar]