Abstract
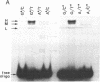
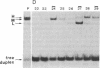
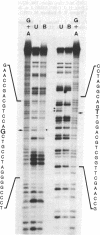
G.T mispairs, the sole mismatch type that can arise in "resting" mammalian DNA (through spontaneous hydrolytic deamination of 5-methylcytosine), are corrected in vivo with high efficiency and mostly to a G.C. We identified a protein factor, present in HeLa cell extracts, that binds selectively to DNA substrates containing this mismatch. The partially purified protein was shown by gel-filtration chromatography and UV cross-linking experiments to have an apparent molecular mass of 200 kDa. Its binding to G.T mispairs was not influenced by sequences flanking the mismatch, but methylation of guanines either within the mismatch itself or in its immediate vicinity abolished the formation of the protein-DNA complex. The protein appears to lack both endo- and exonuclease activities and requires neither magnesium nor zinc nor ATP for binding. We discuss the possible role of this protein in a repair pathway, which helps mammalian cells counter the mutagenic effect of the hydrolytic deamination of 5-methylcytosine.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bird A. P. CpG-rich islands and the function of DNA methylation. Nature. 1986 May 15;321(6067):209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- Brown T. C., Jiricny J. A specific mismatch repair event protects mammalian cells from loss of 5-methylcytosine. Cell. 1987 Sep 11;50(6):945–950. doi: 10.1016/0092-8674(87)90521-6. [DOI] [PubMed] [Google Scholar]
- Brown T. C., Jiricny J. Different base/base mispairs are corrected with different efficiencies and specificities in monkey kidney cells. Cell. 1988 Aug 26;54(5):705–711. doi: 10.1016/s0092-8674(88)80015-1. [DOI] [PubMed] [Google Scholar]
- Chodosh L. A., Carthew R. W., Sharp P. A. A single polypeptide possesses the binding and transcription activities of the adenovirus major late transcription factor. Mol Cell Biol. 1986 Dec;6(12):4723–4733. doi: 10.1128/mcb.6.12.4723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerfler W., Langner K. D., Knebel D., Weyer U., Dobrzanski P., Knust-Kron B. Site-specific promoter methylations and gene inactivation. Prog Clin Biol Res. 1985;198:133–155. [PubMed] [Google Scholar]
- Fried M. G., Crothers D. M. CAP and RNA polymerase interactions with the lac promoter: binding stoichiometry and long range effects. Nucleic Acids Res. 1983 Jan 11;11(1):141–158. doi: 10.1093/nar/11.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner-Garden M., Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987 Jul 20;196(2):261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- Jiricny J., Wood S. G., Martin D., Ubasawa A. Oligonucleotide duplexes containing inosine, 7-deazainosine, tubercidin, nebularine and 7-deazanebularine as substrates for restriction endonucleases HindII, SalI and TaqI. Nucleic Acids Res. 1986 Aug 26;14(16):6579–6590. doi: 10.1093/nar/14.16.6579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M., Wagner R., Radman M. Mismatch repair of deaminated 5-methyl-cytosine. J Mol Biol. 1987 Mar 5;194(1):155–159. doi: 10.1016/0022-2836(87)90724-8. [DOI] [PubMed] [Google Scholar]
- Lieb M. Recombination in the lambda repressor gene: evidence that very short patch (VSP) mismatch correction restores a specific sequence. Mol Gen Genet. 1985;199(3):465–470. doi: 10.1007/BF00330759. [DOI] [PubMed] [Google Scholar]
- Lindahl T. DNA repair enzymes. Annu Rev Biochem. 1982;51:61–87. doi: 10.1146/annurev.bi.51.070182.000425. [DOI] [PubMed] [Google Scholar]
- Lu A. L., Welsh K., Clark S., Su S. S., Modrich P. Repair of DNA base-pair mismatches in extracts of Escherichia coli. Cold Spring Harb Symp Quant Biol. 1984;49:589–596. doi: 10.1101/sqb.1984.049.01.066. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollack Y., Kasir J., Shemer R., Metzger S., Szyf M. Methylation pattern of mouse mitochondrial DNA. Nucleic Acids Res. 1984 Jun 25;12(12):4811–4824. doi: 10.1093/nar/12.12.4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggs A. D., Jones P. A. 5-methylcytosine, gene regulation, and cancer. Adv Cancer Res. 1983;40:1–30. doi: 10.1016/s0065-230x(08)60678-8. [DOI] [PubMed] [Google Scholar]
- Su S. S., Lahue R. S., Au K. G., Modrich P. Mispair specificity of methyl-directed DNA mismatch correction in vitro. J Biol Chem. 1988 May 15;263(14):6829–6835. [PubMed] [Google Scholar]
- Su S. S., Modrich P. Escherichia coli mutS-encoded protein binds to mismatched DNA base pairs. Proc Natl Acad Sci U S A. 1986 Jul;83(14):5057–5061. doi: 10.1073/pnas.83.14.5057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R. Y., Kuo K. C., Gehrke C. W., Huang L. H., Ehrlich M. Heat- and alkali-induced deamination of 5-methylcytosine and cytosine residues in DNA. Biochim Biophys Acta. 1982 Jun 30;697(3):371–377. doi: 10.1016/0167-4781(82)90101-4. [DOI] [PubMed] [Google Scholar]
- Zell R., Fritz H. J. DNA mismatch-repair in Escherichia coli counteracting the hydrolytic deamination of 5-methyl-cytosine residues. EMBO J. 1987 Jun;6(6):1809–1815. doi: 10.1002/j.1460-2075.1987.tb02435.x. [DOI] [PMC free article] [PubMed] [Google Scholar]