Abstract
The Epstein-Barr virus (EBV)-encoded LMP1 oncogene has a role in transformation, proliferation, and metastasis of several EBV-associated tumors. Furthermore, LMP1 is critically involved in transformation and growth of EBV-immortalized B cells in vitro. The oncogenic properties of LMP1 are attributed to its ability to upregulate anti-apoptotic proteins and growth signals. The transcriptional regulation of LMP1 is dependent on the context of cellular and viral proteins present in the cell. Here, we investigated the effect of several signaling pathways on the regulation of LMP1 expression. Inhibition of p38 signaling, using p38-specific inhibitors SB203580 and SB202190, downregulated LMP1 in estrogen-induced EREB2.5 cells. Similarly, p38 inhibition decreased trichostatin A-induced LMP1 expression in P3HR1 cells. Exogenous expression of p38 in lymphoblastoid cell lines (LCLs) led to an increase in LMP1 promoter activity in reporter assays, and this activation was mediated by the previously identified CRE site in the promoter. Inhibition of p38 by SB203580 and p38-specific small interfering RNA (siRNA) also led to a modest decrease in endogenous LMP1 expression in LCLs. Chromatin immunoprecipitation indicated decreased binding of CREB-ATF1 to the CRE site in the LMP1 promoter after inhibition of the p38 pathway in EREB2.5 cells. Taken together, our results suggest that an increase in p38 activation upregulates LMP1 expression. Since p38 is activated in response to stimuli such as stress or possibly primary infection, a transient upregulation of LMP1 in response to p38 may allow the cells to escape apoptosis. Since the p38 pathway itself is activated by LMP1, our results also suggest the presence of an autoregulatory loop in LMP1 upregulation.
Epstein-Barr virus (EBV) is a human B-lymphocryptovirus that infects approximately 90% of the world's adult population and is the causative agent of infectious mononucleosis (IM). EBV is associated with several human malignancies such as Burkitt's lymphoma (BL), Hodgkin's lymphoma (HL), nasopharyngeal carcinoma (NPC), nasal T/NK lymphoma (NL), peripheral T-cell lymphoma, gastric carcinoma, and lymphoproliferative diseases in immunocompromised patients (57). EBV establishes latent infection in human B cells and transforms them in culture. The EBV-encoded LMP1 oncogene is critically involved in the EBV immortalization of B cells and their persistence in vitro, and it has the ability to transform rodent fibroblast and human cell lines in culture (4). LMP1 expression is thought to contribute to the genesis and growth of EBV-associated tumors. The oncogenic ability of LMP1 is attributed to its upregulation of anti-apoptotic proteins and growth signaling pathways (55).
LMP1 transcription is regulated by both viral and cellular factors. EBV uses different programs of viral gene expression in order to drive naïve B cells into resting memory cells. These patterns of viral gene expression are referred to as latency types I, II, and III. EBV-associated tumors also exhibit similar patterns of EBV gene expression (58). LMP1 is expressed in both latency types II and III (4). The expression of LMP1 in latency III infected B cells is dependent on the viral EBNA2 protein (32). Since EBNA2 is unable to bind DNA itself, additional factors are required to target EBNA2 to DNA. These include the Jκ recombination signal binding protein (RBP-Jκ) (24, 25, 41, 61, 65), the Ets-related PU.1 factor (30, 52), a POU domain protein (52), and AP-2 site binding factor(s) (29). In latency II cells, LMP1 expression occurs in the absence of EBNA2 and has been the subject of several investigations. While a number of EBNA2-independent LMP1 activators have been identified, none of them seem to be critical for LMP1 expression in latency II. Recent data suggest that LMP1 expression in latency II cells is mediated by cell signaling pathways that are activated by LMP1 itself (21, 40).
The complexity of LMP1 expression is further emphasized in a study by Lam et al. (37). This study showed that LMP1 expression levels of individual cells in a clone of EBV lymphoblasts can range over 100-fold. It was also shown that the variation observed with LMP1 was at the level of transcript and independent of EBNA2 expression levels. Other studies suggest cyclical fluctuations in LMP1 expression, in an individual cell, over time (3, 39, 40). Thus, the transcriptional regulation of LMP1 appears to be regulated by multiple factors in each latency type. The question we then asked ourselves is whether variation observed in LMP1 transcription may be stimulated by signaling pathways that are triggered in response to the extracellular environment.
In our previous studies we have extensively investigated the LMP1 promoter sequence, also referred to as the LMP1 regulatory sequence (LRS) or ED-L1. The cyclic AMP (cAMP) response element (CRE) in the promoter has been shown to be one of the critical sites for LMP1 transctivation. Both EBNA2-dependent and -independent transactivation of the LMP1 promoter require an intact CRE in the LMP1 regulatory sequence. Mutations in this site lead to a drastic decrease in promoter activity in reporter assays (17, 53), and sequence variations in the site correlate with varied LMP1 expression levels in tumors and tumor cell lines (28). CRE binds the heterodimeric transcription factor CREB-ATF1 (53). CREB and ATF1 are members of the CRE family of transcription factors that activate transcription when phosphorylated at specific serine/threonine residues (26). Several upstream serine/threonine kinases are responsible for the phosphorylation of these factors (27, 56, 59). This led us to investigate the possible role of signaling pathways that regulate CRE-binding factors in LMP1 regulation. The effect of different serine/threonine kinase inhibitors on LMP1 transcription was investigated here, and the overall result suggests that the p38 signaling pathway regulates the LMP1 promoter activity through the binding of CREB-ATF1 to the CRE site. We hypothesize that the activation of LMP1 by the p38 pathway may promote cell survival at the early stages of infection and in response to extracellular stimuli. Further, our study suggests the presence of an additional positive autoregulatory loop in LMP1 expression.
MATERIALS AND METHODS
Cell culture and treatment.
EREB2.5 is a transformed lymphoblastoid cell line expressing a conditional mutant of EBNA2 (ER-EBNA2), and its activity is regulated by estrogen (34). P3HR1 is an EBV-positive EBNA2-deficient BL cell line. DG75 (BL) is an EBV-negative B-cell lymphoma. WW1-LCL (23) is an EBV-positive lymphoblastoid cell line (LCL). The cells were maintained as suspension cultures in RPMI 1640 medium supplemented with 10% fetal calf serum, 100 U/ml penicillin, and 100 μg/ml streptomycin (Sigma). In addition, the EREB2.5 cell line was supplemented with 1 μM β-estradiol (β-estradiol-water soluble; Sigma). To inactivate EBNA2 in EREB2.5 cells, β-estradiol was withdrawn from the medium for 48 or 72 h. To reactivate EBNA2, 1 μM β-estradiol was added to the culture medium again. Trichostatin A (TSA) (Wako) was added at 100 ng/ml to P3HR1 cells. The serine/threonine kinase inhibitors were added to the medium 1 h prior to stimulation with β-estradiol or TSA at the following final concentrations: 50 nM staurosporine, 5 μM bisindolylmaleimide, 10 μM H89, 1 μM KN93, 20 μM SB203580, 20 μM PD98059, and 10 μM SB202190. The inhibitors were purchased from Calbiochem and dissolved in dimethyl sulfoxide (DMSO) for the experiments shown in Fig. 1. SB203580-hydrochloride (Calbiochem) dissolved in water was used for the experiments in the following sections. 5,6-Dichloro-1-ß-d-ribofuranosyl benzimidazole (DRB) (Sigma) was dissolved in DMSO and used at 100 μM.
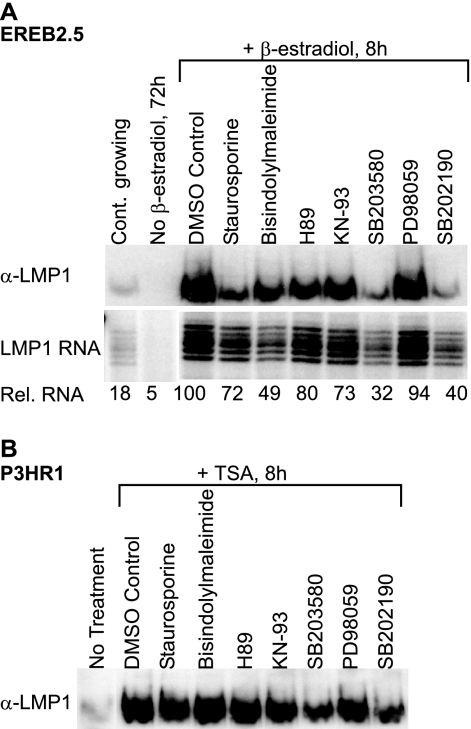
FIG. 1.
Inhibition of serine/threonine kinases inhibits LMP1 induction in EREB2.5 and P3HR1 cells to various levels. (A) Estrogen-starved EREB2.5 cells were treated with specific inhibitors of the serine/threonine kinase pathways 1 h prior to induction with β-estradiol. The cells were harvested after 8 h and assayed by RNase protection assay and immunoblot analysis. Immunoblot analysis was carried out with an LMP1-specific antibody. Equal protein loading was confirmed by Ponceau S staining of the membrane prior to immunoblotting. The RNA samples (40 μg) were hybridized with an antisense LMP1 riboprobe. The protected fragments were separated by 8% denaturing PAGE. The gel was exposed to a PhosphorImager screen and visualized with a Typhoon 9200 scanner. The relative RNA values were calculated by the Image Quant software with respect to the DMSO (solvent) control, which was assigned a value of 100. All RNA samples shown originate from the same scan and have been subjected to the same digital processing. The results are representative of at least six independent experiments. (B) P3HR1 cells were treated with specific inhibitors of the serine/threonine kinase pathways 1 h prior to induction with TSA. After 8 h, the cells were harvested and assayed for LMP1 expression by immunoblotting as described for panel A. The results are representative of at least six independent experiments.
Plasmids, transfections, and reporter assays.
The p38α expression vector was a kind gift from Jiahuai Han (Scripps Research Institute, La Jolla, CA). The p38β expression vector was generously provided by Ingrid Fleming (Johann Wolfgang Goethe Universität, Germany) with permission from Gang Pei (Shanghai Institutes for Biological Sciences, China). The LRS(−634)Luc, LRS(−634)(CREmut)Luc (28), and pgProbeLRS(−106)ΔLuc (15) plasmids have been described previously. The LMP1 regulatory sequence (LRS) is defined as positions 169019 to 169692 of the B95−8 EBV DNA (GenBank accession no. AJ507799), which corresponds to positions −634 to +40 relative to the LMP1 transcription initiation site (+1).
Transfections were carried out by electroporation as described previously (53). Briefly, 5 × 106 cells in 250 μl of RPMI 160 medium were cotransfected with 10 μg of reporter plasmids and 8 μg of p38α or p38β expression vectors or the corresponding molar amount of the empty pcDNA3 vector. The cells were harvested after 24 h for the luciferase assay. Reporter gene activity was measured with the luciferase assay system (Promega) and a Glomax luminometer (Turner Biosystems) according to the manufacturers' instructions.
Small interfering RNA (siRNA) transfections were carried out in a similar manner with a few modifications. WW1-LCL was maintained in antibiotic-free medium for 24 h before transfection. Cells (5 × 106) in 300 μl of antibiotic-free medium were cotransfected with 10 μg of carrier DNA (GFP expression vector) and 0.1 nmol of SignalSilence Pool p38 MAPK siRNA or SignalSilence Control siRNA (Cell Signaling Technology). The cells were incubated for either 24 or 48 h before harvest. LDS loading sample buffer (Invitrogen) supplemented with 50 mM dithiothreitol (DTT) was directly added to the cell pellets for immunoblot analysis.
Quantitative RT-PCR.
After cell treatments, total RNA was prepared from 1 × 105 to 2 × 105 cells, using the Qiagen RNeasy Plus mini kit as instructed by the manufacturer. Quantitative reverse transcription-PCR (RT-PCR) of LMP1 and EBNA2 was carried out as described by Bell et al. (2) with a few modifications. The SuperScript III RTS First-Strand cDNA synthesis kit (Invitrogen) was used according to the manufacturer's protocol. An EBV-specific cDNA primer mix (2 μM) (2) was added to each reaction mixture. For quantitative PCR of LMP1 and EBNA2, the fluorescent power SYBRGreen kit was used according to the manufacturer's protocol (Applied Biosystems) with a primer pair amplifying LMP1 and EBNA2 of B95-8 origin (2) in combination with a melting curve analysis to ensure the specificity of the PCR primers. The amount of each transcript was normalized against the GAPDH gene transcript and quantified with the commercially available predeveloped assay reagent from Applied Biosystems, and the PCRs were performed in an ABI HT7900 instrument (Applied Biosystems).
Chromatin immunoprecipitation (ChIP) assay.
The EREB2.5 cells were estrogen starved for 48 h, and SB203580-hydrochloride was added (80 μM) 1 h prior to the addition of 1 μM β-estradiol. ChIP extract was prepared 4 h post-estrogen induction from cells with and without SB203580 treatment as well as estrogen-starved and continuously growing EREB2.5 cells. ChIP was carried out according to the protocol provided by Upstate Biotechnology, Inc., with minor modifications. Briefly, the ChIP extract was sonicated to between 200- and 350-bp DNA fragments on a Diagenode Bioruptor according to the manufacturer's protocol (Diagenode). CREB-1 (C-21) and normal rabbit IgG antibodies (Santa Cruz) were incubated with the extract for 30 min in an ultrasonic water bath (Branson), and the DNA was prepared as described by Nelson et al. (46). The level of immunoprecipitated DNA was determined by quantitative PCR using primer pairs that amplified the region encompassing the LRS CRE site of EREB2.5 origin (primers: GGCAGAGTAGTGTGAGAGGCTTATG and GTAACGCGTGTTTCTGGGGA) and the fluorescent power SYBRGreen kit (Applied Biosystems). PCR was carried out in an ABI HT7900 instrument (Applied Biosystems) in combination with a melting curve analysis to ensure the specificity of the PCR primers.
Immunoblot analysis.
Treated cells (5 × 106) were directly resuspended in LDS loading buffer (Invitrogen) supplemented with 50 mM DTT and sonicated for 10 min on a Bioruptor sonicator (Diagenode). The samples were subjected to SDS-PAGE in 10% Bis-Tris gels (Invitrogen) according to standard protocols. The membranes were stained with 0.1% Ponceau S (Sigma) in 5% acetic acid to confirm equal loading and transfer of proteins. The immunoblots were probed with antibodies against phosphorylated and total p38 (Cell Signaling), LMP-1 (CS1-4) and EBNA2 (PE2) (Dako A/S), GAPDH and α-tubulin (Santa Cruz), and HRP-conjugated secondary antibodies (Cell Signaling). The immunoreactive protein bands were visualized by chemiluminescence reagents (Pierce) and detected using a ChemiDoc instrument (Bio-Rad) as instructed by the manufacturers.
RNase protection assay.
After cell treatments and harvests, total RNA was prepared with TRI REAGENT LS reagent (Sigma) as instructed by the manufacturer and treated with RQ1 DNase (Promega). RNase protection assays were carried out as described previously (64) with minor modifications. Briefly, 32P-labeled antisense LMP1 riboprobe was transcribed in vitro from pgProbeLRS(−106)ΔLuc for 1 h at 37°C. The template was digested with DNase for 15 min at 37°C. The riboprobe was purified using a CHROMA SPIN-100 column (Clontech), and 1 × 106 cpm was added to 40 μg of each RNA sample. The samples were hybridized at 50°C overnight followed by RNase digestion for 1 h at 25°C. The samples were analyzed by electrophoresis in a 10% denaturing polyacrylamide gel at 1,500 V for 1.5 h. The RNA bands were visualized by exposure to a PhosphorImager screen and scanned with a Thyphone 9200 (Amersham). The relative RNA values were calculated by the Image Quant software with respect to the DMSO (solvent) control, which was assigned a value of 100.
RNA stability assay.
The EREB2.5 cells were estrogen starved, and 1 μM β-estradiol was added after 48 h. DRB (100 μM) and SB203580-hydrochloride (80 μM) were added 3 h postinduction, and RNA and immunoblot samples were collected at several time points as indicated (see Fig. 6). The samples were analyzed by quantitative RT-PCR as described above.
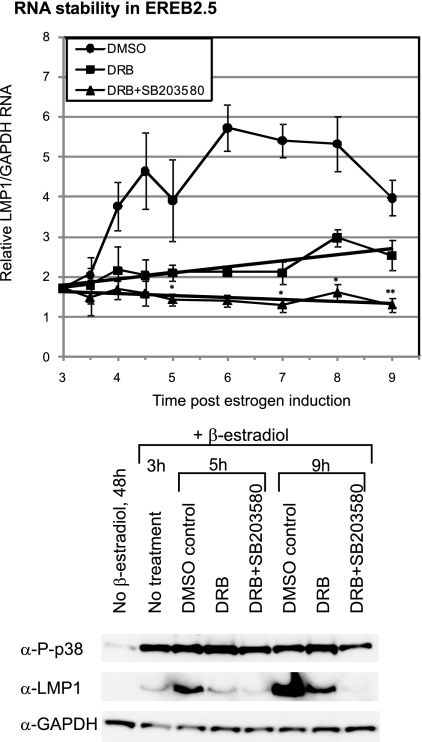
FIG. 6.
Inhibition of p38 activity does not markedly affect LMP1 RNA levels in the absence of RNA synthesis. Estrogen-starved cells were induced with estrogen for 3 h. The cells were then treated with DRB alone (100 μM dissolved in DMSO), DRB and the p38 inhibitor SB203580.HCl (80 μM dissolved in dH2O), or just DMSO as a control. Samples were collected at the times indicated in the figure and subjected to quantitative RT-PCR and immunoblot analysis. The LMP1 RNA level was normalized against GAPDH RNA. The results shown are the means of four independent experiments, and the T bars indicate the standard errors of the means. The asterisks indicate statistical significance in differences relati7ve to the corresponding DRB samples, obtained by a two-tailed paired t test. Immunoblot analysis was used to detect phosphorylated p38 (P-p38), LMP1, and GAPDH protein levels in the cells. Equal protein loading was confirmed by monitoring the GAPDH levels.
RESULTS
LMP1 expression is downregulated in response to the inhibition of the p38 signaling pathway.
We have previously shown that the heterodimeric transcription factor CREB-ATF1 activates the LMP1 promoter via a CRE site (53). Here, we investigated the effect of inhibiting several signaling pathways involved in the activation of CRE-binding transcription factors on LMP1 expression in B cells. To this end, the EREB2.5 cell line was used. The EREB2.5 cell line is a useful tool in the study of LMP1, as it is conditional for the activation of EBNA2. Withdrawal of estrogen results in inactivation of EBNA2 followed by downregulation of the LMP1 promoter and cell cycle arrest (34). LMP1 expression and cell proliferation are induced by the addition of β-estradiol to the medium. Several known inhibitors of serine/threonine kinases were added to the culture medium of estrogen-starved EREB2.5 cells 1 h before stimulation with β-estradiol, and the cells were collected 8 h after stimulation. LMP1 protein and RNA levels were determined using immunoblotting and an RNase protection assay (Fig. 1A). The LMP1 protein and RNA levels in the control (DMSO) were much higher than those observed in continuously growing EREB2.5 cells. Notably, a large transient increase in LMP1 levels after estrogen induction of estrogen-starved cells has been reported by others and is independent of DMSO (32, 42) (Fig. 2A). The RNA and protein levels of LMP1 after the different treatments generally correlated with each other. Treatment with staurosporine, which is a broad range inhibitor of serine/threonine kinases (49), led to a decrease in LMP1 protein level and a lower level of inhibition of LMP1 RNA, relative to the DMSO control. This indicates that at least one serine/threonine kinase pathway may have a role in LMP1 regulation. Bisindolylmaleimide (an inhibitor of PKC) (60), H89 (an inhibitor of PKA) (6), and KN-93 (an inhibitor of CaM Kinase) (43) treatments led to some downregulation of LMP1 expression relative to the DMSO control, indicating that these kinase pathways are partially contributing to LMP1 regulation. On the other hand, PD098059 (an inhibitor of MEK1) (1) did not appear to have an effect on LMP1 induction. Treatments with SB203580 and SB202190, which are inhibitors of the p38 MAPK pathway (9, 19), gave rise to the most pronounced downregulation of LMP1 induction as indicated by its RNA and protein levels. The results are compatible with the notion that the p38 signaling pathway may play a more significant role in LMP1 regulation than other pathways investigated here.
FIG. 2.
Inhibition of p38 activation inhibits LMP1 induction in EREB2.5 and P3HR1 cells. (A) EREB2.5 cells were estrogen starved for 48 h. The cells were then treated with the p38 inhibitor (SB203580.HCl dissolved in distilled water [dH2O]) or just dH2O as a control 1 h prior to activation with β-estradiol (1 μM). Samples were collected at 2, 4, and 8 h after activation and subjected to quantitative RT-PCR and immunoblot analysis. Immunoblot analysis was used to detect phosphorylated p38 (P-p38), LMP1, total p38, EBNA2, and GAPDH protein levels in the cells. Equal protein loading was confirmed by monitoring the GAPDH levels. The results are representative of at least six independent experiments. The LMP1 RNA level in the treated cells was analyzed using quantitative RT-PCR. The LMP1 RNA level was normalized against that of GAPDH RNA, and the change in LMP1 RNA was measured relative to that of the continuously growing EREB2.5 cells (set to 1). The results are the averages of three independent experiments, and the T bars indicate the standard errors of the means. (B) P3HR1 cells were treated with the p38 inhibitor (SB203580.HCl dissolved in dH2O) or dH2O as a control 1 h prior to treatment with TSA (100 ng/ml). Samples were collected at 2, 4, and 8 h after activation and subjected to quantitative RT-PCR and immunoblot analysis as described for panel A. (C) To determine the effect of β-estradiol and TSA on the phosphorylation of p38 in the absence of EBV, DG75 cells were treated with 1 μM β-estradiol or 100 ng/ml TSA and samples were collected at 2, 4, and 8 h. The samples were subjected to immunoblotting using phosphorylated p38 (P-p38)-, total p38-, and GAPDH-specific antibodies. The results are representative of two independent experiments. (D) To illustrate the relative levels of LMP1 in the samples used in this study, protein samples from different cell lines and treatments were subjected to electrophoresis on the same gel followed by immunoblotting using LMP1-, P-p38-, and GAPDH-specific antibodies.
To confirm these results, a different experimental system was utilized. TSA treatment of the EBNA2-deficient P3HR1 cell line induces LMP1 transcription (54). The effect of treatment with the inhibitors used above on TSA-induced LMP1 expression in this cell line was also investigated. Under these conditions, a downregulation of LMP1 protein induction relative to the DMSO control was detectable only after inhibition of the p38 pathway (Fig. 1B). Taken together, the results indicate a role for the p38 signaling pathway in positively regulating LMP1 expression. Furthermore, this upregulation can occur even in the absence of EBNA2, as demonstrated in P3HR1 cells.
Increased phosphorylation of p38 upregulates LMP1 expression.
The inhibitors SB203580 and SB202190 block the biological activity of p38 kinase by binding to the inactive form of p38 and reducing its rate of activation (i.e., phosphorylation) (9, 19). The terms activation and phosphorylation are used interchangeably in this study when referring to the p38 status. To further investigate the role of the p38 pathway in the regulation of LMP1 expression, the phosphorylation status of the p38 protein (P-p38) was monitored by immunoblot analysis in estrogen-induced EREB2.5 cells, as well as in TSA-treated P3HR1 cells over 8 h. In parallel, SB203580 was added 1 h before induction to the estrogen-treated EREB2.5 cells, as well as to the TSA-treated P3HR1, to determine the contribution of p38 signaling to LMP1 transcriptional activity over time. Notably, we found that DMSO treatment alone could induce some p38 phosphorylation (data not shown). While the contribution of DMSO to p38 phosphorylation in estrogen-induced cells and TSA-induced P3HR1 was relatively marginal, we decided to avoid complication of data due to this problem by using the water-soluble SB203580-hydrochloride to inhibit p38 activation. LMP1 expression levels, as well as EBNA2 and total p38, were also monitored by immunoblotting. Immunoblot analysis of GAPDH was used to confirm equal protein loading. After 48 h of estrogen withdrawal, LMP1 was not detectable, indicating successful inactivation of transcription. After the addition of estrogen, the level of LMP1 expression increased progressively at 2, 4, and 8 h, to a level that was significantly higher than that found in continuously growing EREB2.5 cells. Phosphorylated p38 was present in both continuously growing and estrogen-starved EREB2.5 cells, but the level increased markedly after estrogen induction. The highest level of p38 phosphorylation was detected 4 h after estrogen induction. Treatment with SB203580 led to the inhibition, although not the complete inhibition, of p38 phosphorylation relative to that in the untreated samples (H2O controls). This is probably due to the very high level of phosphorylated p38 present after estrogen induction. The inhibition of p38 phosphorylation led to a decreased LMP1 induction relative to that in the control untreated samples. Thus, the large increase in LMP1 levels after estrogen induction mostly depends on the high levels of phosphorylated p38. The total level of p38 protein remained relatively constant, indicating that the p38 phosphorylation increase was not a result of an increase in the total protein level. The p38 inhibition did not appear to affect EBNA2 expression. The LMP1 RNA levels were also determined in all samples using RT-PCR, which is expected to be more quantitative than an RNase protection assay (Fig. 2A), and showed the same pattern in response to p38 inhibition as that observed for the LMP1 protein, with a 2- and 3-fold LMP1 decrease at 4 and 8 h, respectively. Our data suggest that p38 is involved in LMP1 transcription regulation.
A similar investigation was carried out with TSA-induced P3HR1 cells (Fig. 2B). An increase in the level of active p38 was observed over the 8-h period after TSA induction, and this correlated with an increase in LMP1 expression. Again, inhibition of p38 phosphorylation led to an inhibition of LMP1 protein and RNA levels. Therefore, our results indicate that an increase in p38 signaling upregulates LMP1 expression.
The experiments above indicated that after each treatment, the level of p38 phosphorylation is increased relative to that in the untreated cells. This raised the question of the cause of p38 activation in each case. To investigate whether estrogen and TSA activate p38 in the absence of other EBV factors, the EBV-negative cell line DG75 was also treated with these agents (Fig. 2C). Immunoblot analysis of DG75 cells treated with estrogen showed that estrogen does not lead to increased p38 phosphorylation in DG75. Hence, the induction of p38 phosphorylation in EREB2.5 cells after estrogen stimulation is most likely a result of EBNA2 activation of the cells. TSA treatment in the DG75 context resulted in increased p38 phosphorylation, indicating that TSA is most likely responsible for p38 activation in P3HR1 cells.
We aimed to determine if p38 activation in EREB2.5 occurs as a direct response to EBNA2 and independently of protein synthesis. To do so, EREB2.5 cells were estrogen starved and then induced with estrogen as described above but in the presence of the protein synthesis inhibitor cycloheximide. The level of active p38 did increase after induction in the presence of cycloheximide; however, cycloheximide treatment in the absence of estrogen induction also caused p38 activation, making it impossible to elucidate the source of p38 activation (data not shown). Nevertheless, two lines of evidence suggest that the p38 activation may be independent of protein synthesis. First, p38 is activated above the levels of continuously growing cells already at 2 h after induction (Fig. 2A). Second, an increase in p38 activation precedes a high level of LMP1 expression, which is directly transactivated by EBNA2 itself in the absence of secondary protein synthesis (32).
To be able to compare phosphorylated p38 (P-p38) and LMP1 levels in the cell lines and cell treatments used above, immunoblot analysis was carried out on one blot (Fig. 2D). Two EBV-positive latency III cell lines, namely, WW1-LCL and B95-8, were also included as a reference. The LMP1 and phosphorylated p38 levels in EREB2.5 cells induced with estrogen for 24 h decreased in comparison to the levels in cells 8 h after induction, indicating upregulation after estrogen induction to be transient. The decrease of LMP1 levels back to those seen in continuously growing cells has also been reported previously (32, 42). Various levels of phosphorylated p38 were present in all cell lines. The phosphorylated p38 levels were higher in EBV-positive latency III cell lines WW1-LCL and B95-8 relative to those in continuously growing EREB and untreated P3HR1 cells. The levels of LMP1 in WW1-LCL and B95-8, however, did not correlate with their relative levels of phosphorylated p38. These data illustrate the notion that a higher level of phosphorylated p38 leads to LMP1 upregulation but is not a direct determinant of LMP1 level in different cell lines.
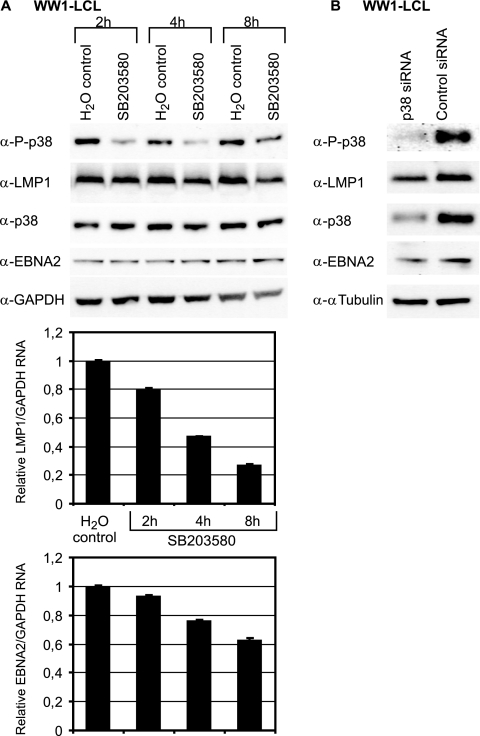
Inhibition of the p38 pathway downregulates LMP1 in LCLs.
The regulation of LMP1 by phosphorylated p38 in both EREB2.5 and P3HR1 occurs under conditions of high levels of phosphorylated p38 relative to those in continuously growing EBV-positive B cells. To address whether p38 signaling has a role in LMP1 regulation in EBV-positive B cells, we investigated the effect of p38 signaling inhibition in LCLs. To this end, WW1-LCL was also treated with SB203580 over a period of 8 h and LMP1 and EBNA2 expression was analyzed by immunoblotting and RT-PCR (Fig. 3A). Sequencing of the LMP1 promoter in the WW1-LCL virus originating from the WW1-BL cell line showed the presence of the same CRE variant found in the B95-8 virus (data not shown). Immunoblot analysis of WW1-LCL showed that phosphorylated p38 was present in the continuously dividing cells and was downregulated by SB203580 treatment. A small decrease in LMP1 expression was observed in response to p38 signaling inhibition. RT-PCR analysis of LMP1 mRNA indicated that the p38 inhibitor led to a more distinct downregulation of LMP1 RNA relative to protein. However, since immunoblotting is not a very quantitative method, no conclusions can be drawn about the actual level of decrease in LMP1 protein levels. Interestingly, a smaller decrease in EBNA2 RNA was observed after 8 h of treatment with the inhibitor while EBNA2 protein levels were stable over time. This is expected, as EBNA2 has a 24-h half-life (22). Thus, EBNA2 is not involved in the downregulation of LMP1 in this experiment.
FIG. 3.
Inhibition of p38 activity downregulates LMP1 levels in an LCL. (A) WW1-LCL was treated with the p38 inhibitor (SB203580.HCl dissolved in dH2O) or dH2O as a control, and samples were collected at 2, 4, and 8 h. Immunoblot analysis was used to detect phosphorylated p38 (P-p38), LMP1, total p38, EBNA2, and GAPDH protein levels in the cells. Equal protein loading was confirmed by monitoring the GAPDH levels. The results are representative of five independent experiments. LMP1 and EBNA2 RNA levels in the treated cells were analyzed using quantitative RT-PCR. The RNA levels were normalized against the GAPDH RNA, and the change in RNA was measured relative to that of the untreated WW1-LCL cells (set to 1). The results are the averages of three independent experiments, and the T bars indicate the standard errors of the means. (B) WW1-LCL was transfected with 0.1 nmol of p38 siRNA or control siRNA. The transfected cells were harvested after 24 h. Western blot analysis was carried out with antibodies against LMP1, phosphorylated p38 (P-p38), total p38, and α-tubulin as a loading control. The results are representative of three independent experiments.
It has been reported that the cells in an LCL population express a wide range of LMP1 protein levels. To investigate whether LMP1 downregulation in response to p38 inhibition was specific to a subpopulation of WW1-LCL, the cells were treated with and without SB203580 for 6 h, labeled with an anti-LMP1 primary and a fluorescent secondary antibody, and analyzed by flow cytometry. The mean expression of LMP1 in SB203580-treated cells was decreased relative to that in untreated WW1-LCL; this was a general effect on the cell population and was not associated with the high-LMP1-expressing subpopulation (data not shown). Overall, the contribution of p38 signaling to LMP1 expression in WW1-LCL was not as pronounced as that seen in induced EREB2.5 cells. This is probably due to relatively lower levels of active p38 and LMP1 in LCLs with respect to that in estrogen-induced-EREB2.5 cells (Fig. 2D). Similar results were observed in an LCL immortalized with the B95.8 virus (data not shown).
In our experimental system, the SB203580 treatment generally ceased to inhibit p38 phosphorylation after 8 h. To determine whether inhibition of p38 over a longer time period would induce a more pronounced decrease of LMP1 expression, we aimed to silence p38 expression, thereby decreasing the level of phosphorylated p38. p38-targeted siRNA was transiently transfected in WW1-LCL, and the cells were analyzed by immunoblotting and RT-PCR for LMP1 expression (Fig. 3B). The level of p38 knockdown was verified by the same methods. Already at 24 h after transfection, total p38 protein and phosphorylated p38 levels decreased considerably. A minor downregulation of the level of LMP1 protein was observed (Fig. 3B), but a change in the LMP1 RNA level was not detected (data not shown). After 48 h and 72 h only a very small or no change in the LMP1 protein and RNA levels could be detected (data not shown). Together, the data indicate that inhibition of the p38 pathway modestly downregulates LMP1 expression, but the effect is short-lived.
The LMP1 promoter is activated in response to exogenous p38 expression.
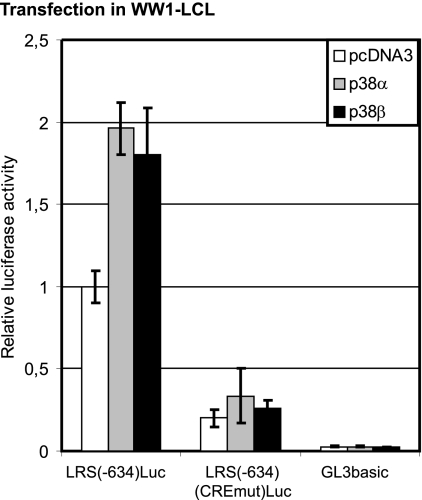
To address whether LMP1 regulation by p38 activation is at the transcription level, transient transfection of LMP1 reporters was utilized. While the most obvious way to activate p38 would be to expose the cells to environmental stress, the complexity and intertwinement of the numerous stress-activated pathways in the cells made this an undesirable means of p38 activation. We therefore used exogenous overexpression of the p38 protein in order to increase the total levels of phosphorylated p38 protein. Both p38α and p38β isoforms were cotransfected with LMP1 promoter reporters into the EBV-positive cell line WW1-LCL. These isoforms of p38 are targeted by the p38 inhibitors used in this study (10). To assess the involvement of the CRE element in the LMP1 promoter as a mediator of LMP1 regulation by p38 signaling, an LMP1 reporter with a mutation in this site was used. The transfected cells were assayed for luciferase activity 24 h posttransfection (Fig. 4). The wild-type LMP1 promoter [LRS(−634)Luc] doubled in activity in response to the overexpression of both p38 isoforms. The activity of the mutated LMP1 promoter construct [LRS(−634)(CREmut)Luc], however, did not increase significantly in response to p38 overexpression. Our results suggest that the p38 pathway does in fact upregulate LMP1 transcription through factors binding to the CRE site of the LMP1 promoter. While a doubling of promoter activity may seem a modest increase, it should be considered that it is against the background of an already active promoter. Since the LMP1 reporter is already activated by the endogenous EBNA2, a doubling of the activity is a considerable increase. Overexpression of p38 in the EBV-negative cell line DG75 failed to activate the LMP1 reporters (data not shown). Overall, our results indicate that the p38 pathway upregulates a transcriptionally active LMP1 promoter but cannot recruit all the required coactivators by itself to activate the promoter.
FIG. 4.
Exogenous expression of p38 activates the LMP1 promoter. The p38α expression vector, the p38β expression vector, or the control vector (pcDNA3) was transfected into WW1-LCL, together with the LRS(−634)Luc, the LRS(−634)(CREmut)Luc, or the control vector (GL3basic) reporter plasmid as shown. The relative luciferase activity is given as fold activation with respect to the LRS(−634)Luc activity in the absence of p38 expression (set to 1). The results shown are the mean results of five independent transfections, and the T bars indicate the standard errors of the means.
The upregulation of LMP1 transcription by p38 is mediated by CREB-ATF1.
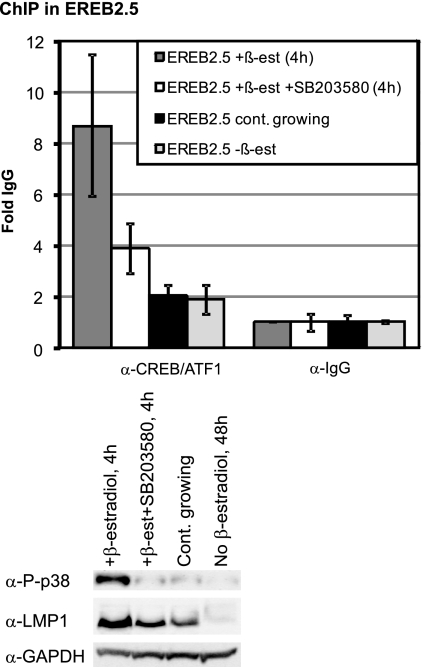
Since the mutation in the CRE of the LMP1 reporter rendered it irresponsive to p38-mediated activation, the role of CRE binding factors in p38 regulation of the LMP1 promoter was investigated in vivo. Phosphorylation of CREB and ATF1 by upstream kinases such as p38 is thought to stimulate their binding to target promoters (44). To determine the effect of p38 on CREB-ATF1 binding to the LMP1 promoter, a ChIP assay was carried out on estrogen-induced EREB2.5 cells treated with or without the p38 inhibitor SB203580 4 h postinduction with an antibody that detects both CREB and ATF1 (Fig. 5). Notably, the CRE site in the EREB2.5 virus variant which originates from the P3HR1 virus binds the CREB-ATF1 dimer, although not as efficiently as the wild-type B95-8 virus. Since in previous experiments the very high level of phosphorylated p38 in estrogen-induced EREB2.5 at 4 h could not be fully inhibited by SB203580, we used a higher concentration of the inhibitor in this experiment. Continuously growing and estrogen-starved EREB2.5 cells were also included as a control. The chromatin pull-down with an antibody that recognizes both CREB and ATF1 (CREB/ATF1) was normalized against pull-down with a normal rabbit IgG (background). The ChIP indicated an 8-fold CREB-ATF1 binding to the LMP1 promoter 4 h after estrogen induction, and the level of this binding was reduced to half in SB203580-treated EREB2.5 cells. Both continuously growing and estrogen-starved EREB2.5 cells had a lower level of CREB binding to the LMP1 promoter. The low enrichment (2-fold, relative to that of the IgG pull-down) of the LMP1 promoter in continuously growing cells reflects the lower efficiency of CREB-ATF1 binding to the CRE variant in this cell line. Surprisingly, the estrogen-starved EREB2.5 cells had the same level of LMP1 enrichment after CREB/ATF1 pull-down as the continuously growing cells. This may be explained by the ability of CREB-ATF1 to bind promoters even in their inactive form (44). The level of LMP1 and phosphorylated p38 was monitored by immunoblot analysis. The expression levels of LMP1 correlated with the levels of CREB binding to the LMP1 promoter and phosphorylation status of p38. Our attempts at ChIP with several antibodies against phosphorylated CREB failed. A likely explanation may be that the active (phosphorylated) site of CREB at the LMP1 promoter is covered as a result of interaction with the transcriptional protein complex(es). The ChIP data suggest that transcriptional regulation of LMP1 by p38 is mediated by the regulation of CREB-ATF1 binding to the LMP1 promoter.
FIG. 5.
Inhibition of p38 activity downregulates CREB/ATF1 binding to the LMP1 promoter. EREB2.5 cells were estrogen starved for 48 h. The cells were then treated with the p38 inhibitor (SB203580.HCl dissolved in dH2O) (80 μM) or just dH2O as a control 1 h prior to activation with β-estradiol (1 μM). Chromatin immunoprecipitation (ChIP) of CREB/ATF1 at the LMP1 promoter (primers flanking the CRE in the promoter) was then performed on the treated cells 4 h post-estrogen induction, as well as estrogen-starved and continuously growing EREB2.5 cells. Results of real-time PCR analysis of ChIP assays with an antibody specific to CREB and ATF1 or control rabbit IgG are shown. The results are expressed as fold IgG where the IgG was set to 1. Samples were analyzed in triplicate, and the results are the average results of four independent experiments; the T bars indicate the standard errors of the means. Immunoblot analysis was used to detect phosphorylated p38 (P-p38), LMP1, and GAPDH protein levels in the treated cells. Equal protein loading was confirmed by monitoring the GAPDH levels.
Inhibition of the p38 pathway does not play a major role in LMP1 RNA turnover.
The p38 pathway has been implicated in the regulation of mRNA stability of several genes in a specific manner (20). To investigate if this mechanism is also relevant to LMP1 regulation by p38, the stability of LMP1 RNA after the inhibition of the p38 activity was investigated. To this end, estrogen-starved EREB2.5 cells were induced by β-estradiol for 3 h and then treated with 5,6-dichloro-1-ß-d-ribofuranosylbenzimidazole (DRB) (38), an inhibitor of mRNA synthesis. This was carried out in the presence or absence of the p38 inhibitor, SB203580 (80 μM), and samples were collected at different time points for quantitative RT-PCR and immunoblot analysis as indicated (Fig. 6). By doing so, we aimed to investigate whether the p38 inhibitor had an impact on LMP1 RNA stability in the absence of transcription. In the control cells (DMSO), LMP1 RNA and protein levels increased over time, as observed in previous experiments. In cells treated with DRB alone, the LMP1 RNA levels did increase slightly, indicating that despite the high concentration of DRB used (100 μM) there was some residual transcription. Consistently, a small increase in LMP1 protein level was observed at 9 h, relative to 5 h in the DRB-treated sample. In DRB-plus-SB203580-treated samples, p38 phosphorylation (P-p38) was lower than in the DRB-treated samples both at 5 h and more visibly so at 9 h. However, p38 phosphorylation was not fully inhibited. This is probably due to the counteracting induction of p38 phosphorylation by DRB, indicated by the increased level of phosphorylated p38 in DRB-treated samples relative to that in the corresponding DMSO controls. In the DRB-plus-SB203580-treated samples, the LMP1 RNA level decreased slightly relative to that with DRB treatment alone. This decrease was shown to be statistically significant in a two-tailed paired t test. However, it was difficult to conclude if this decrease was due to a decrease in LMP1 RNA stability or the further inhibition of residual transcriptional activity by SB203580 in these cells. Even in the SB203580-treated cells, the LMP1 RNA appeared to be fairly stable over time. The relative high stability of LMP1 RNA with respect to other EBV latent proteins has been reported previously (50). Thus, p38 phosphorylation seemed to contribute very little, if anything, to LMP1 RNA stability. Taken together, our data indicate that the p38 upregulation of LMP1 is mainly through the regulation of its transcriptional activity.
DISCUSSION
EBNA2 is indisputably the physiological activator of LMP1 transcription in latency III EBV-positive cells (63). This transactivation is dependent on cellular transcription factors and is mediated through several EBNA2-responsive elements at the LMP1 promoter. In the latency II pattern of gene expression, a few transcription factors, including STAT, IRF7, ATF4, ATF2, and NF-κB (5, 21, 31, 40, 47), have been identified as candidates for LMP1 transactivation. It is becoming apparent that LMP1 transactivation in latency II may require the cooperation of several transcription factors, none of which seem to be absolutely critical in promoter activation. The emerging pattern seems to be that these transcription factors are all LMP1 induced themselves, pointing to LMP1 as its own transactivator. The ability of LMP1 to activate its own transcription has been shown by Goormachtigh et al. (21). Therefore, LMP1 transcriptional regulation is regulated by multiple cellular and viral factors. One of the remaining questions is whether LMP1 expression is also regulated in response to signaling pathways that are triggered by environmental stimuli.
In order to narrow our investigation of signaling pathways affecting LMP1 expression, we took advantage of the extensive studies on the LMP1 promoter sequence. The TATA-proximal cAMP response element (CRE) in the LMP1 promoter appears to be one of the more critical sites in LMP1 transactivation according to numerous studies (28, 53). Accordingly, cAMP increases endogenous LMP1 expression (17). The CRE mediates binding of transcriptional factors that are activated by phosphorylation in response to upstream kinases (14). Therefore, the effect of different serine/threonine kinase inhibitors on LMP1 transcription was investigated in our study. Here, we show that the p38 signaling pathway is involved in LMP1 transcription upregulation.
The p38 kinase is a member of the mitogen-activated protein kinases (MAPK) (59). The MAPK group of kinases comprises key mediators of transcriptional responses to extracellular signals that include growth factors, hormones, cytokines, and environmental stress (62). In B cells specifically, p38 MAPK activation participates in different pathways leading to either proapoptotic or proliferative outcomes (8). There are four members of the p38 MAPK family (p38α, p38β, p38γ, and p38δ) with different substrate specificities (10). The phosphorylation of p38α and p38β is specifically inhibited by SB203580 and SB202190 (10), indicating that these isoforms are involved in LMP1 regulation in our studies.
In reporter studies, the overexpression of p38α and p38β upregulated the wild-type LMP1 promoter activity but not the CRE-mutated LMP1 promoter. Both p38α and p38β can induce CREB and ATF1 phosphorylation by an indirect phosphorylation cascade through their substrates MAPKAP-2 (56) and MSK1 (13). The ATF1-CREB heterodimer has been identified as the main activating transcription factor that binds CRE in LMP1 (28, 53). The ChIP assay of the LMP1 promoter confirmed that the CRE binding factors CREB-ATF1 are involved in mediating p38 regulation of LMP1 expression. Notably, both ATF4 (CREB2) and JNK signaling through ATF2-c-Jun have also been proposed to be involved in LMP1 autoactivation via this CRE site (21, 40). In our previous studies of CRE-binding factors, ATF2-c-Jun binding to this site could not be confirmed (28), and ATF4 has consistently been absent in our binding analyses of this site in both EBV-positive and -negative nuclear extracts (28, 53). It is possible that the in vitro conditions of our electrophoretic mobility shift assay (EMSA) experiments do not allow for the detection of the binding of these factors to the promoter. It is also possible that these transcription factors activate the promoter through other sites in the LMP1 promoter. Our results also suggest that the p38 pathway does not play a considerable role in LMP1 RNA stability; however, additional mechanisms of LMP1 promoter regulation by p38 cannot be excluded by the present study.
LMP1 upregulation by p38 has several biological implications. One of the main oncogenic characteristics of LMP1 is its ability to inhibit apoptosis by upregulating anti-apoptotic proteins and promoting growth signals (55). Considering that p38 signaling can be activated by cytokines and environmental stress, which can lead to apoptosis, upregulation of LMP1 by the cytokine- or stress-activated p38 may allow EBV-positive cells to evade apoptosis. This would be a survival mechanism for LMP1-expressing tumors that need to escape apoptosis induced by their environment. In this way, LMP1 converts a cellular defense mechanism to the advantage of the virus.
The regulation of LMP1 by p38 may have an additional function. The events following EBNA2 activation of estrogen-starved EREB2.5 cells are reported to mimic those of EBV infection of B cells (35). It is possible that transient induction of p38 activity during early EBV infection is required for a transient LMP1 upregulation, thereby ensuring survival and evading apoptosis following infection. This mechanism would be a useful tool in successful infection, regardless of whether p38 signaling is triggered by viral factors or extracellular stimuli.
Interestingly, the p38 signaling pathway is a downstream target of LMP1 itself (12, 16) and is responsible for several characteristics exerted by LMP1. Therefore, this study presents yet another autoregulatory loop in LMP1 expression. In the LCLs investigated in our study, p38 inhibition by SB203580 and siRNA led to a low downregulation of LMP1, confirming the presence of an autoregulatory loop. The fact that p38 inhibition modestly inhibits LMP1 expression, and that this downregulation is transient, supports the notion that LMP1 transcriptional activity is dependent on several transcriptional factors, and if necessary the lack of one can eventually be compensated for by other factors. Overall, the contribution of the p38 to the autoregulation of LMP1 does not seem to be critical for LMP1 expression.
This study did not elucidate the cause of the high level of transient p38 activity in EREB2.5 after EBNA2 activation. It was, however, shown that it was not as a result of estrogen treatment. Some evidence suggests that EBNA2 itself may be responsible for the p38 activation. First, p38 activation occurred shortly after EBNA2 activation and preceded the high LMP1 expression and is probably independent of protein synthesis. Second, a recent study indicates that the transient high level of LMP1 expression after EBNA2 activation is restricted to type 1 EBNA2, which is the type present in this investigation (35). EBV strains are classified as type 1 or 2 according to the variation in their sequence. The most prominent and biologically relevant variation between these two types is the sequence of EBNA2 (36, 42). The transient high levels of LMP1 expression are not observed following induction with the type 2 EBNA2 in an experimental system similar to that used here (42). Our finding that the high level of LMP1 is induced by the high levels of p38 activity in our experiments, together with the previous report that transient high levels of LMP1 are not observed with type 2 EBNA2, leads us to speculate that the type 1 EBNA2 is responsible for p38 activation. Exactly how EBNA2 would mediate this activity is outside the scope of the current investigation. If true, this hypothesis would resolve a current paradox. Namely, the type 1 EBV isolate is much more efficient at immortalizing B cells in vitro, a characteristic that is mainly due to the variation in the EBNA2 sequence (7). The timely induction of a high level of LMP1 by type 1 EBNA2 is proposed to be important in the survival of B cells driven into the cell cycle by EBNA2 after infection and partly responsible for the more efficient immortalizing ability of type 1 EBV. On the other hand, the two variants of EBNA2 protein are quite similar in the domain that has been defined as the transactivating region (RBP-Jκ interacting region) (11, 36), and LCLs established by the two EBV strains express comparable levels of LMP1 (7). Our hypothesis suggests that the transforming efficiency differences between the two EBNA2 variants may partly be the result of transient activation of the p38 pathway by type 1 EBNA2 leading to a transient increase in LMP1 level and increased survival efficiency. Since the cells infected by EBV in the host are exposed to a large array of environmental stimuli that could trigger the p38 signaling pathway transiently, the type 2 virus need not be less efficient in establishing infection in the body, explaining the equal abundance of both EBV strains in Africa and other regions of the world.
Notably, a high level of LMP1 is detrimental to the fate of EBV-positive cells. A high level of LMP1 expression has been shown to induce cytostasis (18, 33, 51) and inhibits the activity of viral and cellular promoters in the absence of cytostasis (45). Recently, it has also been shown that high LMP1 levels facilitate epitope presentation and T-cell recognition of infected cells (3), which is unfavorable for virus survival. To avoid the cytotoxic effects of LMP1, mechanisms of LMP1 downregulation have been developed in EBV-positive cells. LMP1 transcription is downregulated by IRF5 in latency II cells (48), and recent findings indicate that the LMP1 protein downregulates itself by inducing autophagy (39). Conceivably, high levels of LMP1 expression are advantageous to the survival of EBV-positive cells at times and are then regulated to lower levels, thus avoiding cytostasis and immune detection. The transient activation of LMP1 by p38 signaling reported by the present study may possibly be a reflection of situations where high LMP1 levels are required transiently for cell survival and infection efficiency.
In summary, we have reported a stress- or cytokine-activated regulatory pathway involved in upregulation of LMP1 transcription and a positive regulatory loop. Our data support a mechanism whereby the viral oncogene is regulated in response to the cellular environment, allowing it to escape apoptosis and attain higher infection efficiency. This finding also points out the need to study LMP1 regulatory mechanisms with considerations for the cell's physiological environment, in order to obtain a more complete picture of its regulation in infection and tumorigenesis.
Acknowledgments
The EREB2.5 cell line was a kind gift from Bettina Kempkes. The p38α and p38β expression vectors were generously provided by Jiahuai Han and Ingrid Fleming with permission from Gang Pei. We thank Susann Li and Frida Oddhammar for their contributions to the preliminary stages of this work.
This study was supported by grants from the Swedish Medical Research Council (project 5667), the Swedish Cancer Society, Swedish Society for Medical Research, Assar Gabrielssons Foundation, and the Sahlgrenska University Hospital.
Footnotes
Published ahead of print on 6 January 2010.
REFERENCES
- 1.Alessi, D. R., A. Cuenda, P. Cohen, D. T. Dudley, and A. R. Saltiel. 1995. PD 098059 is a specific inhibitor of the activation of mitogen-activated protein kinase kinase in vitro and in vivo. J. Biol. Chem. 270:27489-27494. [DOI] [PubMed] [Google Scholar]
- 2.Bell, A. I., K. Groves, G. L. Kelly, D. Croom-Carter, E. Hui, A. T. Chan, and A. B. Rickinson. 2006. Analysis of Epstein-Barr virus latent gene expression in endemic Burkitt's lymphoma and nasopharyngeal carcinoma tumour cells by using quantitative real-time PCR assays. J. Gen. Virol. 87:2885-2890. [DOI] [PubMed] [Google Scholar]
- 3.Brooks, J. M., S. P. Lee, A. M. Leese, W. A. Thomas, M. Rowe, and A. B. Rickinson. 2009. Cyclical expression of EBV latent membrane protein 1 in EBV-transformed B cells underpins heterogeneity of epitope presentation and CD8+ T cell recognition. J. Immunol. 182:1919-1928. [DOI] [PubMed] [Google Scholar]
- 4.Cahir-McFarland, E., and E. Kieff. 2005. Epstein-Barr Virus latent infection membrane protein 1, p. 553-570. In E. S. Robertson (ed.), Epstein-Barr virus. Caister Academic Press, Norwich, England.
- 5.Chen, H., J. M. Lee, Y. Zong, M. Borowitz, M. H. Ng, R. F. Ambinder, and S. D. Hayward. 2001. Linkage between STAT regulation and Epstein-Barr virus gene expression in tumors. J. Virol. 75:2929-2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chijiwa, T., A. Mishima, M. Hagiwara, M. Sano, K. Hayashi, T. Inoue, K. Naito, T. Toshioka, and H. Hidaka. 1990. Inhibition of forskolin-induced neurite outgrowth and protein phosphorylation by a newly synthesized selective inhibitor of cyclic AMP-dependent protein kinase, N-[2-(p-bromocinnamylamino)ethyl]-5-isoquinolinesulfonamide (H-89), of PC12D pheochromocytoma cells. J. Biol. Chem. 265:5267-5272. [PubMed] [Google Scholar]
- 7.Cohen, J. I., F. Wang, J. Mannick, and E. Kieff. 1989. Epstein-Barr virus nuclear protein 2 is a key determinant of lymphocyte transformation. Proc. Natl. Acad. Sci. U. S. A. 86:9558-9562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Craxton, A., G. Shu, J. D. Graves, J. Saklatvala, E. G. Krebs, and E. A. Clark. 1998. p38 MAPK is required for CD40-induced gene expression and proliferation in B lymphocytes. J. Immunol. 161:3225-3236. [PubMed] [Google Scholar]
- 9.Cuenda, A., J. Rouse, Y. N. Doza, R. Meier, P. Cohen, T. F. Gallagher, P. R. Young, and J. C. Lee. 1995. SB 203580 is a specific inhibitor of a MAP kinase homologue which is stimulated by cellular stresses and interleukin-1. FEBS Lett. 364:229-233. [DOI] [PubMed] [Google Scholar]
- 10.Cuenda, A., and S. Rousseau. 2007. p38 MAP-kinases pathway regulation, function and role in human diseases. Biochim. Biophys. Acta 1773:1358-1375. [DOI] [PubMed] [Google Scholar]
- 11.Dambaugh, T., K. Hennessy, L. Chamnankit, and E. Kieff. 1984. U2 region of Epstein-Barr virus DNA may encode Epstein-Barr nuclear antigen 2. Proc. Natl. Acad. Sci. U. S. A. 81:7632-7636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dawson, C. W., G. Tramountanis, A. G. Eliopoulos, and L. S. Young. 2003. Epstein-Barr virus latent membrane protein 1 (LMP1) activates the phosphatidylinositol 3-kinase/Akt pathway to promote cell survival and induce actin filament remodeling. J. Biol. Chem. 278:3694-3704. [DOI] [PubMed] [Google Scholar]
- 13.Deak, M., A. D. Clifton, L. M. Lucocq, and D. R. Alessi. 1998. Mitogen- and stress-activated protein kinase-1 (MSK1) is directly activated by MAPK and SAPK2/p38, and may mediate activation of CREB. EMBO J. 17:4426-4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Cesare, D., and P. Sassone-Corsi. 2000. Transcriptional regulation by cyclic AMP-responsive factors. Prog. Nucleic Acid Res. Mol. Biol. 64:343-369. [DOI] [PubMed] [Google Scholar]
- 15.Dufva, M., J. Flodin, A. Nerstedt, U. Ruetschi, and L. Rymo. 2002. Epstein-Barr virus nuclear antigen 5 inhibits pre-mRNA cleavage and polyadenylation. Nucleic Acids Res. 30:2131-2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eliopoulos, A. G., N. J. Gallagher, S. M. Blake, C. W. Dawson, and L. S. Young. 1999. Activation of the p38 mitogen-activated protein kinase pathway by Epstein-Barr virus-encoded latent membrane protein 1 coregulates interleukin-6 and interleukin-8 production. J. Biol. Chem. 274:16085-16096. [DOI] [PubMed] [Google Scholar]
- 17.Fahraeus, R., L. Palmqvist, A. Nerdstedt, S. Farzad, L. Rymo, and S. Lain. 1994. Response to cAMP levels of the Epstein-Barr virus EBNA2-inducible LMP1 oncogene and EBNA2 inhibition of a PP1-like activity. EMBO J. 13:6041-6051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Floettmann, J. E., K. Ward, A. B. Rickinson, and M. Rowe. 1996. Cytostatic effect of Epstein-Barr virus latent membrane protein-1 analyzed using tetracycline-regulated expression in B cell lines. Virology 223:29-40. [DOI] [PubMed] [Google Scholar]
- 19.Frantz, B., T. Klatt, M. Pang, J. Parsons, A. Rolando, H. Williams, M. J. Tocci, S. J. O'Keefe, and E. A. O'Neill. 1998. The activation state of p38 mitogen-activated protein kinase determines the efficiency of ATP competition for pyridinylimidazole inhibitor binding. Biochemistry 37:13846-13853. [DOI] [PubMed] [Google Scholar]
- 20.Frevel, M. A., T. Bakheet, A. M. Silva, J. G. Hissong, K. S. Khabar, and B. R. Williams. 2003. p38 mitogen-activated protein kinase-dependent and -independent signaling of mRNA stability of AU-rich element-containing transcripts. Mol. Cell. Biol. 23:425-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goormachtigh, G., T. S. Ouk, A. Mougel, D. Tranchand-Bunel, E. Masy, C. Le Clorennec, J. Feuillard, G. W. Bornkamm, C. Auriault, E. Manet, V. Fafeur, E. Adriaenssens, and J. Coll. 2006. Autoactivation of the Epstein-Barr virus oncogenic protein LMP1 during type II latency through opposite roles of the NF-kappaB and JNK signaling pathways. J. Virol. 80:7382-7393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grasser, F. A., P. Haiss, S. Gottel, and N. Mueller-Lantzsch. 1991. Biochemical characterization of Epstein-Barr virus nuclear antigen 2A. J. Virol. 65:3779-3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gregory, C. D., R. J. Murray, C. F. Edwards, and A. B. Rickinson. 1988. Downregulation of cell adhesion molecules LFA-3 and ICAM-1 in Epstein-Barr virus-positive Burkitt's lymphoma underlies tumor cell escape from virus-specific T cell surveillance. J. Exp. Med. 167:1811-1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grossman, S. R., E. Johannsen, X. Tong, R. Yalamanchili, and E. Kieff. 1994. The Epstein-Barr virus nuclear antigen 2 transactivator is directed to response elements by the J kappa recombination signal binding protein. Proc. Natl. Acad. Sci. U. S. A. 91:7568-7572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Henkel, T., P. D. Ling, S. D. Hayward, and M. G. Peterson. 1994. Mediation of Epstein-Barr virus EBNA2 transactivation by recombination signal-binding protein J kappa. Science 265:92-95. [DOI] [PubMed] [Google Scholar]
- 26.Herdegen, T., and J. D. Leah. 1998. Inducible and constitutive transcription factors in the mammalian nervous system: control of gene expression by Jun, Fos and Krox, and CREB/ATF proteins. Brain Res. Brain Res. Rev. 28:370-490. [DOI] [PubMed] [Google Scholar]
- 27.Iordanov, M., K. Bender, T. Ade, W. Schmid, C. Sachsenmaier, K. Engel, M. Gaestel, H. J. Rahmsdorf, and P. Herrlich. 1997. CREB is activated by UVC through a p38/HOG-1-dependent protein kinase. EMBO J. 16:1009-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jansson, A., P. Johansson, S. Li, and L. Rymo. 2007. Activity of the LMP1 gene promoter in Epstein-Barr virus-transformed cell lines is modulated by sequence variations in the promoter-proximal CRE site. J. Gen. Virol. 88:1887-1894. [DOI] [PubMed] [Google Scholar]
- 29.Jansson, A., P. Johansson, W. Yang, L. Palmqvist, A. Sjoblom-Hallen, and L. Rymo. 2007. Role of a consensus AP-2 regulatory sequence within the Epstein-Barr virus LMP1 promoter in EBNA2 mediated transactivation. Virus Genes 35:203-214. [DOI] [PubMed] [Google Scholar]
- 30.Johannsen, E., E. Koh, G. Mosialos, X. Tong, E. Kieff, and S. R. Grossman. 1995. Epstein-Barr virus nuclear protein 2 transactivation of the latent membrane protein 1 promoter is mediated by J kappa and PU. 1. J. Virol. 69:253-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johansson, P., A. Jansson, U. Ruetschi, and L. Rymo. 2009. Nuclear factor-kappaB binds to the Epstein-Barr virus LMP1 promoter and upregulates its expression. J. Virol. 83:1393-1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaiser, C., G. Laux, D. Eick, N. Jochner, G. W. Bornkamm, and B. Kempkes. 1999. The proto-oncogene c-myc is a direct target gene of Epstein-Barr virus nuclear antigen 2. J. Virol. 73:4481-4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaykas, A., and B. Sugden. 2000. The amino-terminus and membrane-spanning domains of LMP-1 inhibit cell proliferation. Oncogene 19:1400-1410. [DOI] [PubMed] [Google Scholar]
- 34.Kempkes, B., M. Pawlita, U. Zimber-Strobl, G. Eissner, G. Laux, and G. W. Bornkamm. 1995. Epstein-Barr virus nuclear antigen 2-estrogen receptor fusion proteins transactivate viral and cellular genes and interact with RBP-J kappa in a conditional fashion. Virology 214:675-679. [DOI] [PubMed] [Google Scholar]
- 35.Kempkes, B., D. Spitkovsky, P. Jansen-Durr, J. W. Ellwart, E. Kremmer, H. J. Delecluse, C. Rottenberger, G. W. Bornkamm, and W. Hammerschmidt. 1995. B-cell proliferation and induction of early G1-regulating proteins by Epstein-Barr virus mutants conditional for EBNA2. EMBO J. 14:88-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kieff, E., and A. B. Rickinson. 2001. Epstein-Barr virus and its replication, p. 2511-2574. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott-Raven Publishers, Philadelphia, PA.
- 37.Lam, N., M. L. Sandberg, and B. Sugden. 2004. High physiological levels of LMP1 result in phosphorylation of eIF2 alpha in Epstein-Barr virus-infected cells. J. Virol. 78:1657-1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Laub, O., E. B. Jakobovits, and Y. Aloni. 1980. 5,6-Dichloro-1-beta-ribofuranosylbenzimidazole enhances premature termination of late transcription of simian virus 40 DNA. Proc. Natl. Acad. Sci. U. S. A. 77:3297-3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee, D. Y., and B. Sugden. 2008. The latent membrane protein 1 oncogene modifies B-cell physiology by regulating autophagy. Oncogene 27:2833-2842. [DOI] [PubMed] [Google Scholar]
- 40.Lee, D. Y., and B. Sugden. 2008. The LMP1 oncogene of EBV activates PERK and the unfolded protein response to drive its own synthesis. Blood 111:2280-2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ling, P. D., D. R. Rawlins, and S. D. Hayward. 1993. The Epstein-Barr virus immortalizing protein EBNA-2 is targeted to DNA by a cellular enhancer-binding protein. Proc. Natl. Acad. Sci. U. S. A. 90:9237-9241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lucchesi, W., G. Brady, O. Dittrich-Breiholz, M. Kracht, R. Russ, and P. J. Farrell. 2008. Differential gene regulation by Epstein-Barr virus type 1 and type 2 EBNA2. J. Virol. 82:7456-7466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mamiya, N., J. R. Goldenring, Y. Tsunoda, I. M. Modlin, K. Yasui, N. Usuda, T. Ishikawa, A. Natsume, and H. Hidaka. 1993. Inhibition of acid secretion in gastric parietal cells by the Ca2+/calmodulin-dependent protein kinase II inhibitor KN-93. Biochem. Biophys. Res. Commun. 195:608-615. [DOI] [PubMed] [Google Scholar]
- 44.Mayr, B., and M. Montminy. 2001. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat. Rev. Mol. Cell Biol. 2:599-609. [DOI] [PubMed] [Google Scholar]
- 45.Narbonnet, S., and B. Mariame. 2006. The Epstein-Barr virus oncoprotein LMP1 inhibits the activity of viral or cellular promoters without inducing cytostasis. Virology 350:381-393. [DOI] [PubMed] [Google Scholar]
- 46.Nelson, J. D., O. Denisenko, and K. Bomsztyk. 2006. Protocol for the fast chromatin immunoprecipitation (ChIP) method. Nat. Protoc. 1:179-185. [DOI] [PubMed] [Google Scholar]
- 47.Ning, S., A. M. Hahn, L. E. Huye, and J. S. Pagano. 2003. Interferon regulatory factor 7 regulates expression of Epstein-Barr virus latent membrane protein 1: a regulatory circuit. J. Virol. 77:9359-9368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ning, S., L. E. Huye, and J. S. Pagano. 2005. Interferon regulatory factor 5 represses expression of the Epstein-Barr virus oncoprotein LMP1: braking of the IRF7/LMP1 regulatory circuit. J. Virol. 79:11671-11676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ruegg, U. T., and G. M. Burgess. 1989. Staurosporine, K-252 and UCN-01: potent but nonspecific inhibitors of protein kinases. Trends Pharmacol. Sci. 10:218-220. [DOI] [PubMed] [Google Scholar]
- 50.Sample, J., and E. Kieff. 1990. Transcription of the Epstein-Barr virus genome during latency in growth-transformed lymphocytes. J. Virol. 64:1667-1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sandberg, M. L., A. Kaykas, and B. Sugden. 2000. Latent membrane protein 1 of Epstein-Barr virus inhibits as well as stimulates gene expression. J. Virol. 74:9755-9761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sjoblom, A., A. Jansson, W. Yang, S. Lain, T. Nilsson, and L. Rymo. 1995. PU box-binding transcription factors and a POU domain protein cooperate in the Epstein-Barr virus (EBV) nuclear antigen 2-induced transactivation of the EBV latent membrane protein 1 promoter. J. Gen. Virol. 76(Part 11):2679-2692. [DOI] [PubMed] [Google Scholar]
- 53.Sjoblom, A., W. Yang, L. Palmqvist, A. Jansson, and L. Rymo. 1998. An ATF/CRE element mediates both EBNA2-dependent and EBNA2-independent activation of the Epstein-Barr virus LMP1 gene promoter. J. Virol. 72:1365-1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sjoblom-Hallen, A., W. Yang, A. Jansson, and L. Rymo. 1999. Silencing of the Epstein-Barr virus latent membrane protein 1 gene by the Max-Mad1-mSin3A modulator of chromatin structure. J. Virol. 73:2983-2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Soni, V., E. Cahir-McFarland, and E. Kieff. 2007. LMP1 TRAfficking activates growth and survival pathways. Adv. Exp. Med. Biol. 597:173-187. [DOI] [PubMed] [Google Scholar]
- 56.Tan, Y., J. Rouse, A. Zhang, S. Cariati, P. Cohen, and M. J. Comb. 1996. FGF and stress regulate CREB and ATF-1 via a pathway involving p38 MAP kinase and MAPKAP kinase-2. EMBO J. 15:4629-4642. [PMC free article] [PubMed] [Google Scholar]
- 57.Tao, Q., L. S. Young, C. B. Woodman, and P. G. Murray. 2006. Epstein-Barr virus (EBV) and its associated human cancers-genetics, epigenetics, pathobiology and novel therapeutics. Front. Biosci. 11:2672-2713. [DOI] [PubMed] [Google Scholar]
- 58.Thorley-Lawson, D. A. 2005. EBV the prototypical human tumor virus-just how bad is it? J. Allergy Clin. Immunol. 116:251-261. [DOI] [PubMed] [Google Scholar]
- 59.Tibbles, L. A., and J. R. Woodgett. 1999. The stress-activated protein kinase pathways. Cell. Mol. Life Sci. 55:1230-1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Toullec, D., P. Pianetti, H. Coste, P. Bellevergue, T. Grand-Perret, M. Ajakane, V. Baudet, P. Boissin, E. Boursier, F. Loriolle, et al. 1991. The bisindolylmaleimide GF 109203X is a potent and selective inhibitor of protein kinase C. J. Biol. Chem. 266:15771-15781. [PubMed] [Google Scholar]
- 61.Waltzer, L., F. Logeat, C. Brou, A. Israel, A. Sergeant, and E. Manet. 1994. The human J kappa recombination signal sequence binding protein (RBP-J kappa) targets the Epstein-Barr virus EBNA2 protein to its DNA responsive elements. EMBO J. 13:5633-5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Whitmarsh, A. J. 2007. Regulation of gene transcription by mitogen-activated protein kinase signaling pathways. Biochim. Biophys. Acta 1773:1285-1298. [DOI] [PubMed] [Google Scholar]
- 63.Zetterberg, H., and L. Rymo. 2005. EBNA2 transcription regulation in EBV latency, p. 439-462. In E. S. Robertson (ed.), Epstein-Barr virus. Caister Academic Press, Norwich, England.
- 64.Zetterberg, H., M. Stenglein, A. Jansson, A. Ricksten, and L. Rymo. 1999. Relative levels of EBNA1 gene transcripts from the C/W, F and Q promoters in Epstein-Barr virus-transformed lymphoid cells in latent and lytic stages of infection. J. Gen. Virol. 80(Part 2):457-466. [DOI] [PubMed] [Google Scholar]
- 65.Zimber-Strobl, U., L. J. Strobl, C. Meitinger, R. Hinrichs, T. Sakai, T. Furukawa, T. Honjo, and G. W. Bornkamm. 1994. Epstein-Barr virus nuclear antigen 2 exerts its transactivating function through interaction with recombination signal binding protein RBP-J kappa, the homologue of Drosophila Suppressor of Hairless. EMBO J. 13:4973-4982. [DOI] [PMC free article] [PubMed] [Google Scholar]