Abstract
The Env protein from gibbon ape leukemia virus (GaLV) has been shown to be incompatible with human immunodeficiency virus type 1 (HIV-1) in the production of infectious pseudotyped particles. This incompatibility has been mapped to the C-terminal cytoplasmic tail of GaLV Env. Surprisingly, we found that the HIV-1 accessory protein Vpu modulates this incompatibility. The infectivity of HIV-1 pseudotyped with murine leukemia virus (MLV) Env was not affected by Vpu. However, the infectivity of HIV-1 pseudotyped with an MLV Env with the cytoplasmic tail from GaLV Env (MLV/GaLV Env) was restricted 50- to 100-fold by Vpu. A Vpu mutant containing a scrambled membrane-spanning domain, VpuRD, was still able to restrict MLV/GaLV Env, but mutation of the serine residues at positions 52 and 56 completely alleviated the restriction. Loss of infectivity appeared to be caused by reduced MLV/GaLV Env incorporation into viral particles. The mechanism of this downmodulation appears to be distinct from Vpu-mediated CD4 downmodulation because Vpu-expressing cells that failed to produce infectious HIV-1 particles nonetheless continued to display robust surface MLV/GaLV Env expression. In addition, if MLV and HIV-1 were simultaneously introduced into the same cells, only the HIV-1 particle infectivity was restricted by Vpu. Collectively, these data suggest that Vpu modulates the cellular distribution of MLV/GaLV Env, preventing its recruitment to HIV-1 budding sites.
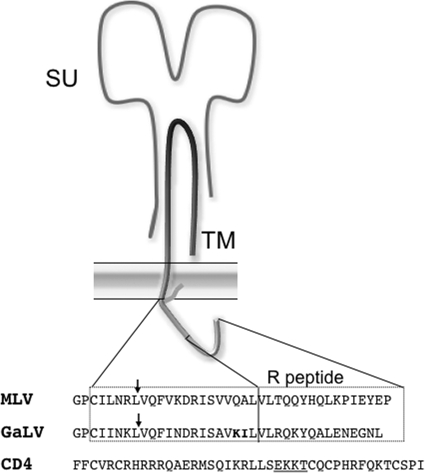
The gammaretrovirus gibbon ape leukemia virus (GaLV) has been widely used for gene therapy because of its wide host cell tropism and nonpathogenicity (1, 6, 10, 12, 13, 20). The host cell receptor for GaLV Env has been cloned and identified as a sodium-dependent phosphate transporter protein (25, 26). Like other retroviruses, GaLV encodes a single transmembrane surface glycoprotein (GaLV Env), which is cleaved into surface (SU) and transmembrane (TM) subunits (Fig. 1). The TM domain of GaLV Env contains a short 30-amino-acid C-terminal cytoplasmic tail. Although GaLV Env functions well when coupled (pseudotyped) with murine leukemia virus (MLV)-based retroviral vectors, it has been shown to be completely incompatible with HIV-1 (4, 35). When GaLV Env is expressed with HIV-1, essentially no infectious HIV-1 particles are produced (4, 35). The mechanism for this infectivity downmodulation is unknown, but the component of GaLV Env responsible for the restriction has been mapped to the cytoplasmic tail. Replacing the cytoplasmic tail of GaLV Env with the equivalent sequence from MLV Env ameliorates the restriction. Likewise, replacing the cytoplasmic tail of MLV Env with that from GaLV Env confers the restriction (4).
FIG. 1.
Schematic of MLV Env protein. Sequences are the C-terminal cytoplasmic tails of MLV Env, GaLV Env, and human CD4. GaLV sequences in boldface are residues that have been shown to modulate the HIV-1 incompatibility (4). Underlined sequences in CD4 are amino acids required for Vpu-mediated downmodulation (2, 15). Arrows denote the location of MLV/GaLV tail substitution. SU, surface domain; TM, transmembrane domain.
Vpu is an 81-amino-acid HIV-1 accessory protein produced from the same mRNA as the HIV-1 Env gene. The N terminus of Vpu contains a membrane-spanning domain, followed by a 50-amino-acid cytoplasmic domain. Vpu is unique to HIV-1 and a few closely related SIV strains. The best-characterized roles for Vpu in the HIV-1 life cycle are modulation of host proteins CD4 and tetherin (also known as BST-2, CD317, and HM1.24) (24, 38, 39). Vpu promotes the degradation of CD4 in the endoplasmic reticulum through a proteasome-dependent mechanism (29). The cytoplasmic tail of Vpu physically interacts with the cytoplasmic tail of CD4 and recruits the human β-transducing repeat-containing protein (β-TrCP) and E3 ubiquitin ligase components to polyubiquitinate and ultimately trigger the degradation of CD4 (18). Two serine residues at positions 52 and 56 of Vpu are phosphorylated by casein kinase-2 and are required for CD4 degradation (31, 32). The membrane-spanning domain of Vpu is not specifically required for CD4 degradation. A mutant protein containing a scrambled membrane-spanning sequence, VpuRD, is still able to trigger the degradation of CD4 (32). The region of CD4 that is targeted by Vpu is approximately 17 to 13 amino acids from the C terminus in the cytoplasmic tail (Fig. 1) (2, 15).
In addition to degrading CD4, Vpu has also long been known to result in enhanced viral release (EVR) in certain cell lines (14, 36). Recently, the type I interferon-induced host protein tetherin was identified as being responsible for this Vpu-modulated restriction (24, 38). In the absence of Vpu, tetherin causes particles to remain tethered (hence the name) to the host cell postfission. Although Vpu counteracts the function of tetherin, the exact mechanism has not been fully elucidated. However, the mechanism for tetherin antagonism appears to be distinct from that for modulating CD4. Mutation of the serines 52 and 56 of Vpu abolish CD4 degradation, but only reduce EVR activity (5, 17, 21, 32). Some EVR activity remains even when much of the Vpu cytoplasmic tail is deleted (30). In addition, many mutations in the membrane-spanning domain, such as VpuRD, do not affect CD4 degradation and yet completely abolish EVR activity (27, 30, 37). The critical residues in tetherin for recognition by Vpu appear to be in the membrane-spanning domain and not the cytoplasmic tail (9, 19, 28). Although β-TrCP is required for complete EVR activity, there is no consensus whether the degradation of tetherin is proteasome or lysosome mediated (5, 7, 21) or whether degradation is required at all. In some cases there can be some EVR activity in the absence of tetherin degradation (17, 22).
We demonstrate here that Vpu is responsible for the incompatibility between HIV-1 and GaLV Env. Glycoproteins containing the cytoplasmic tail from GaLV Env are prevented from being incorporated into HIV-1 particles by Vpu, effectively reducing infectious particle production by 50- to 100-fold. The serines at positions 52 and 56 are required for this restriction, but the membrane-spanning domain is not. Although the mechanism for this restriction appears similar to CD4 degradation, there are apparent differences. Vpu does not prevent surface expression, and it does not prevent its incorporation into MLV particles. Therefore, the mechanism of restriction appears to involve a system that does not rely directly on global protein degradation.
MATERIALS AND METHODS
Plasmid constructs.
The ecotropic Friend MLV Env and yellow fluorescent protein (YFP)-tagged MLV Env constructs were kindly provided by Walther Mothes (Yale University) (34). The codon-optimized late domain deficient HIV-1 Gag construct used in scanning electron microscopy (SEM) images has been described previously (11). The MLV/GaLV Env expression vectors were constructed by using oligonucleotide generated linkers to replace the ClaI to EcoRI (located 100 nucleotides upstream and just downstream of the stop codon, respectively) fragment of the parent MLV Env expression constructs. The NL4-3 derived HIV-CMV-GFP was kindly provided by Vineet KewalRamani at NCI-Frederick (40). The HIV-1 packaging construct cytomegalovirus (CMV) ΔR8.2 containing all HIV-1 accessory genes was obtained from Addgene (plasmid 12263) (40). The HIV-1 construct CMV ΔR8.91 lacking Vpr, Vif, Vpu, and Nef was previously described (40). The HIV-1 packaging construct lacking Vif and Vpr was engineered by replacing the SalI-XbaI fragment of CMV ΔR8.91 with the equivalent sequence from CMV ΔR8.2. The construct lacking Vpu was engineered by replacing the SalI-BamHI fragment of CMV ΔR8.2 with the equivalent sequence from CMV ΔR8.91. The construct lacking Nef was engineered by replacing the BamHI-XbaI fragment of CMV ΔR8.2 with the equivalent sequence from CMV ΔR8.91. The construct lacking Vpu and Nef was engineered by replacing the SalI-XbaI fragment of CMV ΔR8.2 with the equivalent sequence from CMV ΔR8.91. The construct lacking Vif, Vpr, and Nef was engineered by replacing the SalI-BamHI fragment of CMV ΔR8.91 with the equivalent sequence from CMV ΔR8.2. The construct lacking Vif, Vpr, and Vpu was engineered by replacing the BamHI-XbaI fragment of CMV ΔR8.91 with the equivalent sequence from CMV ΔR8.2. The reporter construct pSIN18.cPPT.hEF1a.EGFP.WPRE was designed by Michal Gropp and Benjamin Reubinoff (8). Vpu was added to the HIV-CMV-GFP provirus by replacing the BamHI-SalI fragment of with the equivalent sequence from CMV ΔR8.2 to create HIV-CMV-GFP+Vpu. The HIV-CMV-GFP+VpuRD and CMV-GFP+Vpu52/56 were engineered by PCR mutagenesis using HIV-CMV-GFP+Vpu as a parent. The mutations introduced have been previously described (31). The green fluorescent protein (GFP)-tagged MLV Env and MLV/GaLV Env were generated by replacing the YFP sequence with an eGFP sequence from pEGFP-N1 (Clontech). The MLV packaging construct expressing GagPol was kindly provided by Walther Mothes. The MLV reporter construct expressing TdTomato was constructed by introducing the TdTomato sequence from pRSET-B TdTomato (33), kindly provided by Roger Tsien, into the MLV reporter vector pQCXIP (Clontech).
Cells.
The 293FT cell line was obtained from Invitrogen. The cell line expressing the ecotropic MLV Env receptor, 293T mCAT-1, was kindly provided by Walther Mothes. Both cell lines were maintained in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum, 2 mM l-glutamine, 1 mM sodium pyruvate, 10 mM nonessential amino acids, and 0.5 mg of G418/ml.
SEM.
The method for imaging the distribution of MLV Env on the cell surface has been described previously (11). Briefly, cells were plated onto coverslips coated with patterned gold and transfected with a late domain defective HIV-1 Gag expression vector and a YFP-tagged Env expression vector using FuGene 6 (Roche). Transfected cells were fixed with 4% paraformaldehyde at ca. 20 h posttransfection, and the locations of individual transfected cells on the finder grids were recorded. Cells were labeled with primary mouse anti-GFP (Sigma) and 12-nm gold-conjugated anti-mouse secondary antibody (Jackson ImmunoResearch). Cells were then fixed with 2.5% glutaraldehyde, dehydrated in ethanol, critical point dried, coated with carbon, and imaged with a Hitach S-4700 FE-SEM.
Infectivity assays.
Infectivity assays using the CMV ΔR8.2 and its derivatives were performed by transfecting a 35-mm dish of 293FT cells with 400 ng of packaging construct, 400 ng of pSIN18.cPPT.hEF1a.EGFP.WPRE, and 200 ng of MLV Env or MLV/GaLV Env using FuGene 6. The medium was replaced 24 h posttransfection to remove residual transfection reagent. Supernatant was collected 48 h posttransfection and filtered through 0.45-μm-pore-size filters, and 1 ml of medium was added to fresh 293T mCAT-1 cells in the presence of 10 μg of Polybrene/ml. Cells were collected 48 h later, fixed with 4% paraformaldehyde, and analyzed to determine the percent infected by flow cytometry at the University of Missouri Cell and Immunobiology Core facility. Infectivity assays using HIV-CMV-GFP and its derivatives were performed as described above, except that the transfections were carried out with 500 ng of the proviral DNA and 500 ng of the Env expression construct. Infectivity assays that combined both HIV-1 and MLV proviruses were performed with 800 ng of the HIV-CMV-GFP or HIV-CMV-GFP+Vpu, 40 ng of the MLV GagPol expression construct, 40 ng of pQCXIP-TdTomato, and 120 ng of MLV Env or MLV/GaLV Env.
Western blots.
Transfections for westerns were performed as described for infectivity assays. Viral supernatants were filtered with a 0.45-μm-pore-size filter and pelleted through a 20% sucrose solution. Viral pellets were resuspended in 1× SDS-PAGE loading buffer, and the equivalent of 0.5 ml of supernatant was analyzed by 10% discontinuous SDS-PAGE. Cells were washed, pelleted, and resuspended in SDS-1% PAGE loading buffer, and ca. 4% of the cell suspension was analyzed in parallel with the supernatant. Proteins were transferred by using the iBlot dry blotting system (Invitrogen). Membranes were blocked with 5% nonfat dry milk and probed with goat anti-MLV Env (kindly provided by Alan Rein, NCI-Frederick) diluted 1:10,000 or anti-HIV CA hybridoma medium diluted 1:1,000 (obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: HIV-1 p24 hybridoma [183-H12-5C]) from Bruce Chesebro (3). Blots were then probed with horseradish peroxidase-conjugated anti-goat antibody diluted 1:20,000 or anti-mouse antibody diluted 1:10,000 (both from Sigma).
Surface labeling.
Transfections for protein surface labeling were performed as for the infectivity assays except that GFP-tagged Env expression vectors, which are more appropriate for flow cytometry, were used. The medium was changed 24 h posttransfection to remove the residual transfection reagent. Media and cells were collected 48 h posttransfection. Infectivity from the supernatant was assayed as described for infectivity assays. Cells were resuspended with phosphate-buffered saline (PBS) containing 1 mM EDTA, chilled to 4°C, blocked with 5% goat serum, and labeled with Alexa Fluor 647-conjugated anti-GFP antibody (Invitrogen) diluted 1:1,000 in PBS containing 1% goat serum for 1 h at 4C. After a washing step, the cells were fixed with 4% paraformaldehyde and analyzed by flow cytometry.
Statistical analysis.
Student's t test, analysis of variance (ANOVA), and Tukey-Kramer HSD analyses were performed using the statistical software JMP (SAS Institute, Inc., Cary, NC).
RESULTS
The GaLV Env tail does not exclude glycoproteins from HIV-1 assembly sites.
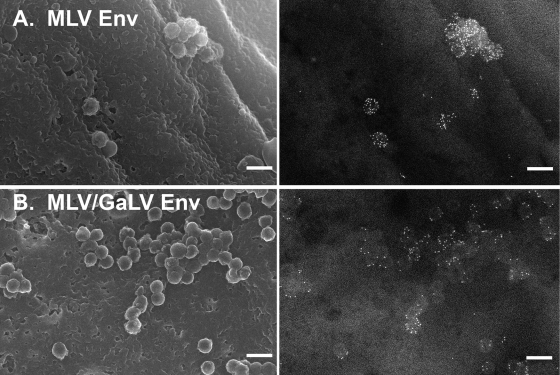
Our lab developed a novel SEM technique for visualizing recruitment or exclusion of surface proteins to HIV-1 assembly and/or budding sites. With this technique, we have demonstrated that certain foreign viral glycoproteins, including MLV Env, are efficiently recruited to HIV-1 assembly sites (Fig. 2A) (11). Because MLV Env glycoproteins containing the cytoplasmic tail from GaLV Env have been shown to be incompatible with HIV-1 in infectivity assays, we predicted that an Env protein with a GaLV tail would be excluded from HIV-1 assembly sites. To test this hypothesis, we replaced the last 30 amino acids of a YFP-tagged ecotropic MLV Env (34) with the equivalent sequence from GaLV Env (henceforth referred to as MLV/GaLV Env) and observed its distribution on the cell surface relative to HIV-1 assembly sites. Surprisingly, MLV/GaLV Env was not excluded from HIV-1 assembly sites and in some instances appeared recruited to them (Fig. 2B).
FIG. 2.
Distribution of MLV Env relative to HIV-1 assembly sites. 293T mCAT-1 cells were cotransfected with a plasmid expressing late domain-defective HIV-1 Gag and a plasmid expressing YFP-tagged MLV Env (A) or YFP-tagged MLV/GaLV Env (B). Env was labeled with 12-nm gold and imaged by SEM. Left, secondary electron images of HIV-1 assembly sites. Right, backscatter electron images of gold-labeled Env. Scale bars, 200 nm.
The GaLV Env tail is not inherently incompatible with HIV-1.
In previous studies equivalent MLV/GaLV Env chimeras were found to be >1,000-fold less infectious than the parent MLV Env when coupled with HIV-1 (4, 35). To determine whether our MLV/GaLV Env chimera could form infectious particles with HIV-1, we performed a single-round infectivity assay combining MLV/GaLV Env with a minimal HIV-1 provirus lacking all nonessential accessory proteins, HIV-CMV-GFP (23). This provirus contains a CMV-driven GFP, which serves as a reporter, in place of the Nef gene. This provirus was cotransfected with either wild-type ecotropic MLV Env or the MLV/GaLV Env chimera into 293FT cells. Although the YFP-tagged Env is capable of forming infectious particles, a non-YFP tagged Env was used for this infectivity experiment. Infectious particles were collected 2 days posttransfection, filtered, and used to transduce 293T mCAT-1 cells (a stable cell line expressing the mCAT-1 receptor). Infectivity was determined by counting the number of fluorescent cells by flow cytometry. The infectivity of MLV/GaLV Env with HIV-1 was reduced compared to MLV Env but was considerably higher than had been previously observed (4, 35).
HIV-1 incompatibility with GaLV is modulated by Vpu.
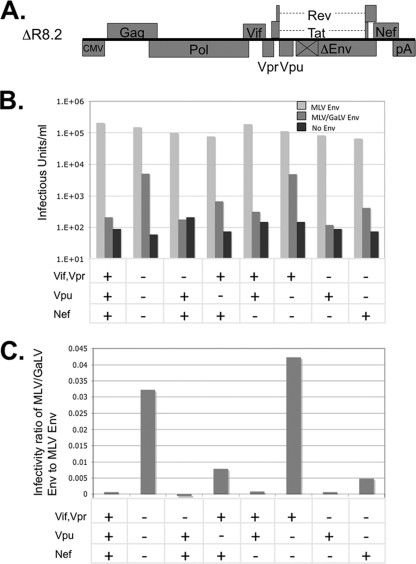
Because these findings with MLV/GaLV Env conflict with the results of previous studies, we tested our MLV/GaLV chimera with the HIV-1 construct that was found to be incompatible with MLV/GaLV Env previously, ΔR8.2 (Fig. 3A) (35, 40). Two obvious differences exist between our minimal proviral construct and ΔR8.2: (i) ΔR8.2 contains a CMV promoter in place of the 5′ LTR and packaging sequence; and (ii) ΔR8.2 contains the accessory Vif, Vpr, Vpu, and Nef genes. Because ΔR8.2 does not produce a packageable genomic RNA, it was cotransfected with a reporter plasmid that contained the viral packaging sequence and a GFP reporter gene, pSIN18.cPPT.hEF1a.EGFP.WPRE (8). To determine whether the accessory genes influenced the incompatibility between HIV-1 and MLV/GaLV Env, we also tested a derivative of ΔR8.2 that lacks the four accessory genes, ΔR8.91 (40). When coupled with wild-type MLV Env, both constructs produced high virus titers. However, when coupled with MLV/GaLV Env, only the construct lacking accessory genes was able to produce a significant virus titer (Fig. 3B and C). This observation indicated that one or more of the accessory genes in ΔR8.2 prevent HIV-1 from forming infectious particles with MLV/GaLV Env. To determine which of the accessory genes contributed to the restriction, we made a series of constructs lacking one or more accessory genes and tested each of these derivatives for infectivity (Fig. 3B and C). All of the ΔR8.2 derivatives were able to efficiently produce infectious particles when combined with MLV Env. However, the infectivity of all four constructs that contained Vpu dropped to background levels when combined with MLV/GaLV Env. The four constructs that contained Nef also displayed a reduction in infectivity when coupled with MLV/GaLV Env, although the restriction was not as striking as with Vpu. Vif and Vpr did not appear to contribute to the MLV/GaLV Env restriction. Because Vpu appeared to be the major contributor to the incompatibility between HIV-1 and MLV/GaLV Env, this protein was selected for further study.
FIG. 3.
HIV-1 accessory genes modulate MLV/GaLV Env restriction. (A) Schematic of HIV-1 packaging constructs ΔR8.2. (B) ΔR8.2 or its derivatives containing the accessory genes shown were cotransfected with reporter vector pSIN18.cPPT.hEF1a.EGFP.WPRE and either MLV Env or MLV/GaLV Env. Viral supernatant was transferred from transfected 293FT cells to 293T mCAT-1 cells, and infectivity was determined by flow cytometry. (C) Output of the same experiment with the infectivity of each construct expressed as the ratio of infections with MLV/GaLV Env to infections with MLV Env.
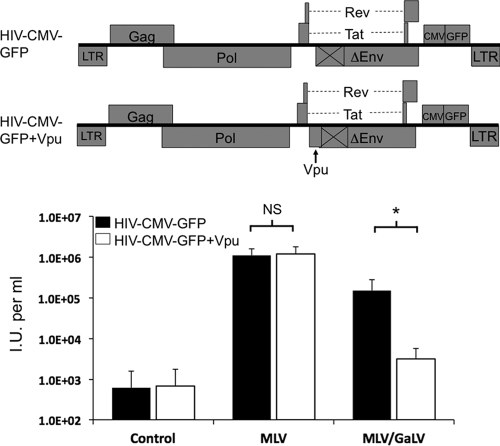
To confirm that Vpu mediated the restriction, we reintroduced Vpu back into the HIV-CMV-GFP provirus background (Fig. 4). The proviruses, containing or lacking Vpu, were cotransfected with MLV Env or MLV/GaLV Env into 293FT cells. The supernatant was collected after 48 h, filtered, and transferred to 293T mCAT-1 cells, and the infectivity was measured by flow cytometry (Fig. 4). Because 293FT cells do not constitutively express tetherin, Vpu should not enhance viral release in these cells through the antagonism of tetherin. The addition of Vpu slightly increased the infectivity of HIV-1 with MLV Env, but the difference was not significant (two-tailed Student t test, n = 7, P = 0.76). The addition of Vpu significantly diminished the infectivity of HIV-1 with MLV/GaLV Env (two-tailed Student t test, n = 7, P = 0.02), with an average drop in infectivity of >60-fold.
FIG. 4.
Vpu modulates HIV-1 infectivity with MLV/GaLV Env. Infectivity of MLV Env, MLV/GaLV Env, or an empty DNA vector (control) pseudotyped with HIV-1 was determined in the absence (▪) or presence (□) of Vpu. Infectivity was measured as infectious units (I.U.) per ml of viral supernatant transferred from transfected 293FT cells to 293T mCAT-1 cells 48 h posttransfection. The averages and standard deviations of infectious units per milliliter of seven independent experiments with the same combinations are shown. Significant differences (*, P < 0.05) and nonsignificant differences (NS) were determined by two-tailed Student's t test for each envelope type in the presence or absence of Vpu.
Vpu prevents MLV/GaLV Env from being incorporated into HIV-1 particles.
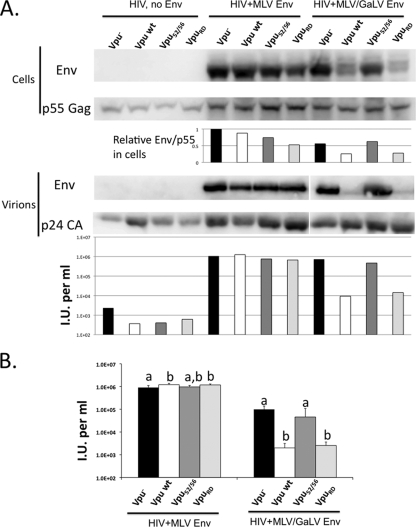
To analyze the nature of the Vpu-mediated restriction of MLV/GaLV Env, we introduced two Vpu mutants into HIV-CMV-GFP (Fig. 5). The first, Vpu52/56, contains two alanine substitutions at positions 52 and 56 of the cytoplasmic domain. The second mutant, VpuRD, contains a scrambled Vpu membrane-spanning domain. Each provirus was transfected along with MLV Env or MLV/GaLV Env, and the expression, incorporation, and infectivity was determined for each. HIV-1 protein production and particle release were not affected by the expression of Vpu or MLV/GaLV Env in this context (Fig. 5A). In the presence of wild-type Vpu, a moderate reduction in the amount of MLV/GaLV Env in expressing cells was observed (Fig. 5A, cells, compare lanes 9 and 10). This reduction varied between experiments where on two occasions there was no reduction, but on four occasions there was a two- to fourfold reduction (as shown in Fig. 5A). In contrast, the Vpu-mediated reduction of MLV/GaLV Env in virions was consistent and drastic. In six of six experiments, Vpu caused an acute reduction in the amount of MLV/GaLV Env in virions (Fig. 5A, virions, compare lanes 9 and 10), and in half of these experiments the MLV/GaLV Env in virions was undetectable. This lack of MLV/GaLV Env in virions correlated with the loss in infectivity (Fig. 5A, bottom). The HIV-CMV-GFP provirus containing VpuRD incorporated MLV Env but not MLV/GaLV Env into virions, whereas the provirus containing Vpu52/56 incorporated both MLV and MLV/GaLV Env. The infectivity experiment was repeated six times, and the average infectivity and standard deviations are reported (Fig. 5B). As was suggested by the immunoblot analysis, Vpu type had a significant effect on the ability of MLV/GaLV Env to form infectious particles with HIV-1 (ANOVA, F3,20 = 19.88, P < 0.0001). Wild-type Vpu or VpuRD significantly reduced the infectivity of MLV/GaLV Env pseudotyped particles compared to no Vpu or Vpu52/56 (Fig. 5B). There was also a variation in infectivity among Vpu constructs pseudotyped with MLV Env, but the difference was very slight (ANOVA, F3,20 = 5.04, P = 0.0092). Because Vpu52/56 fails to recruit the E3 ubiquitin ligase complex during CD4 downmodulation (18), these data are consistent with Vpu targeting MLV/GaLV Env for degradation in a manner synonymous with CD4 degradation. However, the reduction in MLV/GaLV Env expression levels appeared modest compared to the loss in incorporation and infectivity.
FIG. 5.
Vpu prevents MLV/GaLV Env from being incorporated into HIV-1 particles. (A) 293FT cells were transfected with HIV-CMV-GFP containing no Vpu (Vpu−), Vpu wt, VpuRD, or Vpu52/56, and MLV Env or MLV/GaLV Env. For the upper panel, Western blot analysis performed on the transfected cells and pelleted viral supernatants. The lower panel shows the infectivity output from the same experiment. Infectious units per ml were normalized to p24 levels. (B) The averages and standard deviations of infectious units per milliliter of six independent experiments with the same combinations are shown. Significantly different means for each Env treatment are indicated by unique letters (Tukey-Kramer HSD, P < 0.05).
Vpu does not prevent the surface expression of MLV/GaLV Env.
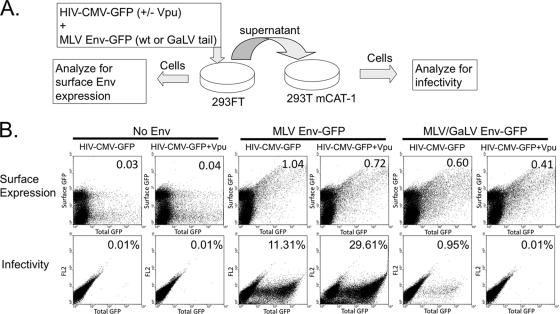
Although we did not observe a severe decrease in MLV/GaLV Env expression levels, it is plausible that Vpu prevents MLV/GaLV Env from reaching the plasma membrane by triggering its degradation at a posttranslational stage or by interfering with its trafficking. To test whether Vpu prevents MLV/GaLV Env from reaching the plasma membrane, we transfected HIV-CMV-GFP (with or without Vpu), along with MLV Env or MLV/GaLV Env tagged with GFP. The transfected cells were stained live with an Alexa Fluor 647-conjugated antibody against GFP at 4°C and analyzed by flow cytometry to assay for Env expression on the cell surface (Fig. 6). Scatter plots denote total Env surface staining (y axis) to total transfection level (x axis). Although HIV-CMV-GFP and Env-GFP both contribute to the overall mean cellular GFP fluorescent intensity (x axis), the signal from Env-GFP was negligible compared to the signal from HIV-CMV-GFP. To ensure only surface GFP was stained, we also analyzed cells transfected with HIV-CMV-GFP alone (first two columns). A portion of cells exhibited nonspecific staining, but the staining occurred equally in untransfected cells (not shown) and did not correlate with increased cytoplasmic GFP expression. Both MLV and MLV/GaLV Env displayed robust surface expression (y axis), which directly correlated with overall cellular expression levels (x axis). The presence of Vpu did not considerably alter the surface expression of MLV or MLV/GaLV Env. The supernatant from the same transfection was transferred onto fresh 293T mCAT-1 cells to assay for infectivity. The supernatant from the cells expressing Vpu and MLV/GaLV Env contained ∼95-fold fewer infectious particles than cells lacking Vpu. Although in some instances we have witnessed a minor (two- to fourfold) reduction in relative MLV/GaLV Env surface staining in the presence of Vpu, we chose to show this experiment because it demonstrates that the restriction in infectivity is not dependent on significant MLV/GaLV Env surface downmodulation.
FIG. 6.
Vpu does not prevent MLV/GaLV Env surface expression. (A) Schematic of surface labeling experiment. 293FT cells were transfected HIV-CMV-GFP (+/− Vpu) in the presence or absence of MLV Env-GFP or MLV/GaLV Env-GFP. At 48 h posttransfection, cells were collected and stained live for surface GFP expression with Alexa Fluor 647-conjugated anti-GFP antibody, and the supernatant was transferred to 293T mCAT-1 cells and assayed for infectivity. (B) Surface expression and infectivity of Env proteins. The average surface GFP intensity/total GFP intensity of transfected cells is reported in the upper right-hand corner of surface expression scatter plots. Infectivity scatter plots show infections on the x axis; the y axis (FL2) is not relevant for this experiment, but is used to maintain visual consistency. Infectivity is shown in each plot as the percentage of the 293T mCAT-1 cells infected with HIV-CMV-GFP.
HIV-1 Vpu does not prevent MLV/GaLV Env from being incorporated into MLV particles.
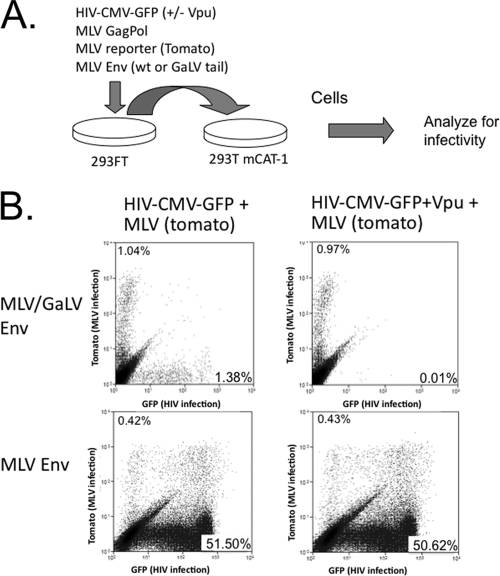
It was shown previously that the cotransfection of ΔR8.2 (containing the HIV-1 accessory genes) and an MLV viral construct, along with GaLV Env, resulted in negligible infectious HIV-1 production but normal levels of infectious MLV particle production (4). These data suggested that the incompatibility between HIV-1 and MLV/GaLV Env was not the result of global downregulation of the GaLV Env protein. To confirm this finding, we configured a fluorescence-based dual infectivity assay using HIV-CMV-GFP, which produces fluorescent green infected cells, and an MLV packaging construct and genome containing TdTomato, which produces fluorescent red infected cells. The plasmids for producing these single-round infectious particles were cotransfected in 293FT cells with MLV or MLV/GaLV Env and in the presence or absence of Vpu (Fig. 7). Because the MLV structural components are more efficient at producing infectious particles with MLV/GaLV Env than HIV-1, the ratio of HIV-1 components to MLV components was adjusted so that roughly equivalent amounts of HIV-1 and MLV infections were produced in the absence of Vpu (Fig. 7B, top left panel). When the parallel transfection was performed with a Vpu-containing HIV-1 provirus, the output of infectious HIV-1 particles was dramatically reduced but the output of infectious MLV particles was essentially unchanged (Fig. 7, top right). As a control, the same ratio of packaging components was transfected into cells, along with MLV Env. HIV-1 was much more efficient at forming infectious particles with MLV Env than with MLV/GaLV Env, resulting in a much higher ratio of HIV-1 to MLV infections. However, the inclusion of Vpu did not alter the output of infectious HIV-1 or MLV particles with MLV Env.
FIG. 7.
Vpu does not prevent the production of infectious MLV particles. (A) Schematic of dual infection assay. 293FT cells were transfected with HIV-1 and MLV assembly components, along with MLV or MLV/GaLV Env. At 48 h posttransfection the supernatant was transferred to 293T mCAT-1 cells. The ratio of HIV-1 to MLV was adjusted so that similar HIV-1 and MLV infectious particles were produced with MLV/GaLV Env and so that same ratio was used in each of the four transfections. (B) Flow cytometry output of 293T mCAT-1 infections. MLV infections display red fluorescence (y axis), and HIV-1 infections display green fluorescence (x axis). Infectivity is shown in each plot as percentage of the 293T mCAT-1 cells infected, excluding double-positive cells.
DISCUSSION
There have been many examples of virus-glycoprotein pairs that do not efficiently produce infectious pseudotyped virus together. For example, HIV-1 does not pseudotype efficiently with Rous sarcoma virus (RSV) Env, influenza virus hemagglutinin, or lymphocytic choriomeningitis glycoprotein (4, 16). However, most of these virus-glycoprotein pairs do yield some infectious pseudotyped particles, albeit at an inefficient level. The incompatibility between HIV-1 and GaLV Env (specifically Env proteins containing the GaLV cytoplasmic domain) is unique because it is so absolute, yielding essentially no infectious particles. It is now clear that this extreme incompatibility is the result of at least two factors. First, HIV-1 shows a general incompatibility with the GaLV Env cytoplasmic tail that is not related to Vpu (Fig. 4). This incompatibility has not been addressed here but is likely the result of poor fusogenicity. Second, MLV/GaLV Env incorporation into HIV-1 particles is prevented by the HIV-1 accessory protein Vpu (and possibly Nef), resulting in a further 50- to 100-fold decrease in infectivity.
The mechanism for how Vpu modulates MLV/GaLV Env is only partially elucidated here. In some ways, the mechanism seems similar to the downmodulation of CD4 by Vpu. As with CD4, the sequence in Env responsible for the Vpu modulation exists in the C-terminal cytoplasmic tail. Also like CD4, the conserved 52/56 serine residues in the cytoplasmic tail of Vpu are required for the modulation, but the precise membrane-spanning sequence is dispensable. However, whereas CD4 is prevented from accumulating on the cell surface and is degraded by Vpu, neither appears to occur to a significant extent with MLV/GaLV Env. The reduction in MLV/GaLV Env expression level is more modest than previously reported (4), but it should be noted that the present studies were performed with a different Env background, the cells were not treated with cycloheximide to prevent new protein synthesis, and Nef, which likely targets MLV/GaLV Env independently, was not included in the majority of our experiments.
Although the ultimate downmodulation of infectious particle production by Vpu appears to be due to lack of Env incorporation into viral particles, it is not clear how this is accomplished. It is particularly intriguing that Vpu does not prevent MLV/GaLV Env from producing infectious particles with MLV packaging proteins. There are two explanations for this observation. Either Vpu alters MLV/GaLV Env trafficking in a way that affects the two types of retroviruses differently, or Vpu alters MLV/GaLV Env in a way that would affect both viruses, but MLV structural components are able to overcome this restriction. For example, it is plausible that Vpu binds the cytoplasmic tail of MLV/GaLV Env and sequesters it away from assembly sites, but MLV Gag binds with greater affinity to the cytoplasmic tail to subvert this sequestration.
The observation that HIV-1 Vpu causes an incompatibility with an unrelated viral glycoprotein is surprising. This incompatibility seems to be specific and robust, but it is not obvious how or why HIV-1 would acquire the ability to target a specific foreign viral glycoprotein. HIV-1 and GaLV do not infect the same host, so it is not likely that downmodulation of GaLV Env provides a selective advantage for HIV-1. The targeting of GaLV Env could be the result of a coincidental similarity between the GaLV Env cytoplasmic tail and a natural target of Vpu. GaLV Env has certain similarities with CD4; both are type 1 surface glycoproteins that are targeted by Vpu through elements in their cytoplasmic tails. However, the cytoplasmic tails of CD4 and GaLV Env display no obvious sequence similarity (Fig. 1), although it is plausible that they contain a conserved structural element not recognized in the primary sequence. One could postulate that GaLV Env mimics some host cell protein that is a natural target of Vpu, but there is at present no experimental evidence to support this speculation.
Acknowledgments
We thank Walther Mothes, Vineet KewalRemani, Alan Rein, and the AIDS Research and Reference Reagent Program for reagents. SEM studies were performed at the University of Missouri Electron Microscopy Core facility.
This research was supported by U.S. Public Health Service grant AI73098 and the Arnold and Mabel Beckman Foundation Young Investigator Program.
Footnotes
Published ahead of print on 30 December 2009.
REFERENCES
- 1.Beard, B. C., P. Mezquita, J. C. Morris, and H. P. Kiem. 2006. Efficient transduction and engraftment of G-CSF-mobilized peripheral blood CD34+ cells in nonhuman primates using GALV-pseudotyped gammaretroviral vectors. Mol. Ther. 14:212-217. [DOI] [PubMed] [Google Scholar]
- 2.Chen, M. Y., F. Maldarelli, M. K. Karczewski, R. L. Willey, and K. Strebel. 1993. Human immunodeficiency virus type 1 Vpu protein induces degradation of CD4 in vitro: the cytoplasmic domain of CD4 contributes to Vpu sensitivity. J. Virol. 67:3877-3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chesebro, B., K. Wehrly, J. Nishio, and S. Perryman. 1992. Macrophage-tropic human immunodeficiency virus isolates from different patients exhibit unusual V3 envelope sequence homogeneity in comparison with T-cell-tropic isolates: definition of critical amino acids involved in cell tropism. J. Virol. 66:6547-6554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Christodoulopoulos, I., and P. M. Cannon. 2001. Sequences in the cytoplasmic tail of the gibbon ape leukemia virus envelope protein that prevent its incorporation into lentivirus vectors. J. Virol. 75:4129-4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Douglas, J. L., K. Viswanathan, M. N. McCarroll, J. K. Gustin, K. Fruh, and A. V. Moses. 2009. Vpu directs the degradation of the human immunodeficiency virus restriction factor BST-2/Tetherin via a βTrCP-dependent mechanism. J. Virol. 83:7931-7947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gaspar, H. B., K. L. Parsley, S. Howe, D. King, K. C. Gilmour, J. Sinclair, G. Brouns, M. Schmidt, C. Von Kalle, T. Barington, M. A. Jakobsen, H. O. Christensen, A. Al Ghonaium, H. N. White, J. L. Smith, R. J. Levinsky, R. R. Ali, C. Kinnon, and A. J. Thrasher. 2004. Gene therapy of X-linked severe combined immunodeficiency by use of a pseudotyped gammaretroviral vector. Lancet 364:2181-2187. [DOI] [PubMed] [Google Scholar]
- 7.Goffinet, C., I. Allespach, S. Homann, H. M. Tervo, A. Habermann, D. Rupp, L. Oberbremer, C. Kern, N. Tibroni, S. Welsch, J. Krijnse-Locker, G. Banting, H. G. Krausslich, O. T. Fackler, and O. T. Keppler. 2009. HIV-1 antagonism of CD317 is species specific and involves Vpu-mediated proteasomal degradation of the restriction factor. Cell Host Microbe 5:285-297. [DOI] [PubMed] [Google Scholar]
- 8.Gropp, M., P. Itsykson, O. Singer, T. Ben-Hur, E. Reinhartz, E. Galun, and B. E. Reubinoff. 2003. Stable genetic modification of human embryonic stem cells by lentiviral vectors. Mol. Ther. 7:281-287. [DOI] [PubMed] [Google Scholar]
- 9.Gupta, R. K., S. Hue, T. Schaller, E. Verschoor, D. Pillay, and G. J. Towers. 2009. Mutation of a single residue renders human tetherin resistant to HIV-1 Vpu-mediated depletion. PLoS Pathog. 5:e1000443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horn, P. A., M. S. Topp, J. C. Morris, S. R. Riddell, and H. P. Kiem. 2002. Highly efficient gene transfer into baboon marrow repopulating cells using GALV-pseudotype oncoretroviral vectors produced by human packaging cells. Blood 100:3960-3967. [DOI] [PubMed] [Google Scholar]
- 11.Jorgenson, R. L., V. M. Vogt, and M. C. Johnson. 2009. Foreign glycoproteins can be actively recruited to virus assembly sites during pseudotyping. J. Virol. 83:4060-4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kiem, H. P., P. A. McSweeney, B. Bruno, M. Goerner, G. Buron, J. Morris, R. Storb, and A. D. Miller. 1999. Improved gene transfer into canine hematopoietic repopulating cells using CD34-enriched marrow cells in combination with a gibbon ape leukemia virus-pseudotype retroviral vector. Gene Ther. 6:966-972. [DOI] [PubMed] [Google Scholar]
- 13.Kiem, H. P., J. E. Rasko, J. Morris, L. Peterson, P. Kurre, and R. G. Andrews. 2002. Ex vivo selection for oncoretrovirally transduced green fluorescent protein-expressing CD34-enriched cells increases short-term engraftment of transduced cells in baboons. Hum. Gene Ther. 13:891-899. [DOI] [PubMed] [Google Scholar]
- 14.Klimkait, T., K. Strebel, M. D. Hoggan, M. A. Martin, and J. M. Orenstein. 1990. The human immunodeficiency virus type 1-specific protein vpu is required for efficient virus maturation and release. J. Virol. 64:621-629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lenburg, M. E., and N. R. Landau. 1993. Vpu-induced degradation of CD4: requirement for specific amino acid residues in the cytoplasmic domain of CD4. J. Virol. 67:7238-7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lewis, B. C., N. Chinnasamy, R. A. Morgan, and H. E. Varmus. 2001. Development of an avian leukosis-sarcoma virus subgroup A pseudotyped lentiviral vector. J. Virol. 75:9339-9344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mangeat, B., G. Gers-Huber, M. Lehmann, M. Zufferey, J. Luban, and V. Piguet. 2009. HIV-1 Vpu neutralizes the antiviral factor Tetherin/BST-2 by binding it and directing its beta-TrCP2-dependent degradation. PLoS Pathog. 5:e1000574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Margottin, F., S. P. Bour, H. Durand, L. Selig, S. Benichou, V. Richard, D. Thomas, K. Strebel, and R. Benarous. 1998. A novel human WD protein, h-beta TrCp, that interacts with HIV-1 Vpu connects CD4 to the ER degradation pathway through an F-box motif. Mol. Cell 1:565-574. [DOI] [PubMed] [Google Scholar]
- 19.McNatt, M. W., T. Zang, T. Hatziioannou, M. Bartlett, I. B. Fofana, W. E. Johnson, S. J. Neil, and P. D. Bieniasz. 2009. Species-specific activity of HIV-1 Vpu and positive selection of tetherin transmembrane domain variants. PLoS Pathog. 5:e1000300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller, A. D., J. V. Garcia, N. von Suhr, C. M. Lynch, C. Wilson, and M. V. Eiden. 1991. Construction and properties of retrovirus packaging cells based on gibbon ape leukemia virus. J. Virol. 65:2220-2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mitchell, R. S., C. Katsura, M. A. Skasko, K. Fitzpatrick, D. Lau, A. Ruiz, E. B. Stephens, F. Margottin-Goguet, R. Benarous, and J. C. Guatelli. 2009. Vpu antagonizes BST-2-mediated restriction of HIV-1 release via beta-TrCP and endo-lysosomal trafficking. PLoS Pathog. 5:e1000450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miyagi, E., A. J. Andrew, S. Kao, and K. Strebel. 2009. Vpu enhances HIV-1 virus release in the absence of Bst-2 cell surface down-modulation and intracellular depletion. Proc. Natl. Acad. Sci. U. S. A. 106:2868-2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mulky, A., T. V. Cohen, S. V. Kozlov, B. Korbei, R. Foisner, C. L. Stewart, and V. N. KewalRamani. 2008. The LEM domain proteins emerin and LAP2alpha are dispensable for human immunodeficiency virus type 1 and murine leukemia virus infections. J. Virol. 82:5860-5868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neil, S. J., T. Zang, and P. D. Bieniasz. 2008. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature 451:425-430. [DOI] [PubMed] [Google Scholar]
- 25.O'Hara, B., S. V. Johann, H. P. Klinger, D. G. Blair, H. Rubinson, K. J. Dunn, P. Sass, S. M. Vitek, and T. Robins. 1990. Characterization of a human gene conferring sensitivity to infection by gibbon ape leukemia virus. Cell Growth Differ. 1:119-127. [PubMed] [Google Scholar]
- 26.Olah, Z., C. Lehel, W. B. Anderson, M. V. Eiden, and C. A. Wilson. 1994. The cellular receptor for gibbon ape leukemia virus is a novel high-affinity sodium-dependent phosphate transporter. J. Biol. Chem. 269:25426-25431. [PubMed] [Google Scholar]
- 27.Paul, M., S. Mazumder, N. Raja, and M. A. Jabbar. 1998. Mutational analysis of the human immunodeficiency virus type 1 Vpu transmembrane domain that promotes the enhanced release of virus-like particles from the plasma membrane of mammalian cells. J. Virol. 72:1270-1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rong, L., J. Zhang, J. Lu, Q. Pan, R. P. Lorgeoux, C. Aloysius, F. Guo, S. L. Liu, M. A. Wainberg, and C. Liang. 2009. The transmembrane domain of BST-2 determines its sensitivity to down-modulation by human immunodeficiency virus type 1 Vpu. J. Virol. 83:7536-7546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schubert, U., L. C. Anton, I. Bacik, J. H. Cox, S. Bour, J. R. Bennink, M. Orlowski, K. Strebel, and J. W. Yewdell. 1998. CD4 glycoprotein degradation induced by human immunodeficiency virus type 1 Vpu protein requires the function of proteasomes and the ubiquitin-conjugating pathway. J. Virol. 72:2280-2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schubert, U., S. Bour, A. V. Ferrer-Montiel, M. Montal, F. Maldarell, and K. Strebel. 1996. The two biological activities of human immunodeficiency virus type 1 Vpu protein involve two separable structural domains. J. Virol. 70:809-819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schubert, U., P. Henklein, B. Boldyreff, E. Wingender, K. Strebel, and T. Porstmann. 1994. The human immunodeficiency virus type 1 encoded Vpu protein is phosphorylated by casein kinase-2 (CK-2) at positions Ser52 and Ser56 within a predicted alpha-helix-turn-alpha-helix-motif. J. Mol. Biol. 236:16-25. [DOI] [PubMed] [Google Scholar]
- 32.Schubert, U., and K. Strebel. 1994. Differential activities of the human immunodeficiency virus type 1-encoded Vpu protein are regulated by phosphorylation and occur in different cellular compartments. J. Virol. 68:2260-2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shaner, N. C., R. E. Campbell, P. A. Steinbach, B. N. Giepmans, A. E. Palmer, and R. Y. Tsien. 2004. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22:1567-1572. [DOI] [PubMed] [Google Scholar]
- 34.Sherer, N. M., M. J. Lehmann, L. F. Jimenez-Soto, A. Ingmundson, S. M. Horner, G. Cicchetti, P. G. Allen, M. Pypaert, J. M. Cunningham, and W. Mothes. 2003. Visualization of retroviral replication in living cells reveals budding into multivesicular bodies. Traffic 4:785-801. [DOI] [PubMed] [Google Scholar]
- 35.Stitz, J., C. J. Buchholz, M. Engelstadter, W. Uckert, U. Bloemer, I. Schmitt, and K. Cichutek. 2000. Lentiviral vectors pseudotyped with envelope glycoproteins derived from gibbon ape leukemia virus and murine leukemia virus 10A1. Virology 273:16-20. [DOI] [PubMed] [Google Scholar]
- 36.Terwilliger, E. F., E. A. Cohen, Y. C. Lu, J. G. Sodroski, and W. A. Haseltine. 1989. Functional role of human immunodeficiency virus type 1 Vpu. Proc. Natl. Acad. Sci. U. S. A. 86:5163-5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tiganos, E., J. Friborg, B. Allain, N. G. Daniel, X. J. Yao, and E. A. Cohen. 1998. Structural and functional analysis of the membrane-spanning domain of the human immunodeficiency virus type 1 Vpu protein. Virology 251:96-107. [DOI] [PubMed] [Google Scholar]
- 38.Van Damme, N., D. Goff, C. Katsura, R. L. Jorgenson, R. Mitchell, M. C. Johnson, E. B. Stephens, and J. Guatelli. 2008. The interferon-induced protein BST-2 restricts HIV-1 release and is downregulated from the cell surface by the viral Vpu protein. Cell Host Microbe 3:245-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Willey, R. L., F. Maldarelli, M. A. Martin, and K. Strebel. 1992. Human immunodeficiency virus type 1 Vpu protein regulates the formation of intracellular gp160-CD4 complexes. J. Virol. 66:226-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zufferey, R., D. Nagy, R. J. Mandel, L. Naldini, and D. Trono. 1997. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat. Biotechnol. 15:871-875. [DOI] [PubMed] [Google Scholar]