Abstract
During productive herpes simplex virus type 1 (HSV-1) infection, a subset of viral delayed-early (DE) and late (L) genes require the immediate-early (IE) protein ICP27 for their expression. However, the cis-acting regulatory sequences in DE and L genes that mediate their specific induction by ICP27 are unknown. One viral L gene that is highly dependent on ICP27 is that encoding glycoprotein C (gC). We previously demonstrated that this gene is posttranscriptionally transactivated by ICP27 in a plasmid cotransfection assay. Based on our past results, we hypothesized that the gC gene possesses a cis-acting inhibitory sequence and that ICP27 overcomes the effects of this sequence to enable efficient gC expression. To test this model, we systematically deleted sequences from the body of the gC gene and tested the resulting constructs for expression. In so doing, we identified a 258-bp “silencing element” (SE) in the 5′ portion of the gC coding region. When present, the SE inhibits gC mRNA accumulation from a transiently transfected gC gene, unless ICP27 is present. Moreover, the SE can be transferred to another HSV-1 gene, where it inhibits mRNA accumulation in the absence of ICP27 and confers high-level expression in the presence of ICP27. Thus, for the first time, an ICP27-responsive sequence has been identified in a physiologically relevant ICP27 target gene. To see if the SE functions during viral infection, we engineered HSV-1 recombinants that lack the SE, either in a wild-type (WT) or ICP27-null genetic background. In an ICP27-null background, deletion of the SE led to ICP27-independent expression of the gC gene, demonstrating that the SE functions during viral infection. Surprisingly, the ICP27-independent gC expression seen with the mutant occurred even in the absence of viral DNA synthesis, indicating that the SE helps to regulate the tight DNA replication-dependent expression of gC.
Herpes simplex virus type 1 (HSV-1) is a widespread human herpesvirus that is responsible for several clinically important diseases, including recurrent orolabial lesions, corneal blindness, and encephalitis (46). As HSV-1 is quite readily studied in the laboratory, this virus also serves as a useful model to investigate the fundamental mechanisms used by herpesviruses to replicate in their host cells. One active area of HSV-1 research is studying how the virus regulates the expression of its ∼80 genes during productive infection (reviewed in reference 37). During such infections, viral gene transcription is mediated in the cell nucleus by the host RNA polymerase II (Pol II). Expression of the viral genes occurs in a temporally regulated cascade in which the five viral immediate-early (IE) genes are expressed first, followed by the delayed-early (DE) and finally the late (L) genes. Transcription of IE genes occurs soon after infection and does not require the synthesis of any new viral proteins. In contrast, expression of the ∼75 DE and L genes is strongly dependent on the synthesis of viral IE proteins, four of which (ICP0, ICP4, ICP22, and ICP27) are known regulators of gene expression. ICP4 is particularly important for expression of DE and L genes; in its absence, DE/L transcription does not proceed efficiently, and viral mRNA levels are drastically reduced (5, 8, 52). ICP27 also stimulates DE/L gene expression, but unlike ICP4, it is differentially required among the DE/L genes (23, 33, 38). Some, such as the L gene encoding gC (33), are highly dependent on ICP27, while others are moderately or weakly dependent (47, 49). At least one DE gene, that encoding the single-stranded DNA-binding protein ICP8, is expressed independently of ICP27 (35, 49).
The mechanism or mechanisms by which ICP27 stimulates expression of DE/L genes is not clear. A leading model is that ICP27 promotes DE/L gene expression by stimulating the export of viral mRNAs from the nucleus to the cytoplasm (19, 40, 41, 44). Viral DE/L genes may require such a factor, since they are predominantly intron-less and thus do not have access to the major pathway for cellular mRNA export, in which export is linked to pre-mRNA splicing (20). ICP27 has several properties that are consistent with a possible role as an mRNA export factor: it binds RNA (15, 28, 40), shuttles between the nucleus and cytoplasm (27, 40), and interacts with the Aly and NXF1 components of the cellular mRNA export system (2, 3, 19). Moreover, it promotes mRNA export in microinjected Xenopus oocytes (19) and HSV-1-infected mammalian cells (3, 40).
However, despite ICP27's clear linkage to mRNA export, not all of its stimulatory effects on DE/L gene expression can be explained by effects on mRNA export. First, the mRNA export model does not explain why viral DE/L genes vary so widely in their ICP27-dependence, since nearly all of these genes lack introns. Second, there is genetic evidence that suggests that the mRNA export function of ICP27 has only a limited effect on HSV-1 gene expression. Specifically, deletion of the sequences encoding ICP27's leucine-rich nuclear export sequence (NES) or RGG box RNA-binding domain in recombinant viruses has been shown to abrogate ICP27's mRNA export function (3, 17). However, these mutations have only a modest effect on expression of many ICP27-dependent genes, as revealed by metabolic labeling of viral proteins in mutant-infected cells (21). Third, the mRNA export model predicts that, in the absence of ICP27, dependent transcripts should accumulate in the nucleus. Although this has been observed for a few transcripts (40), several ICP27-dependent mRNAs, such as those encoding gC, UL8 and UL24, are drastically reduced in their overall levels in the absence of ICP27 (30, 33, 49). This suggests that, for some dependent genes, ICP27's primary effect may be to promote mRNA accumulation. In this regard, one study has suggested that ICP27 can enhance the transcription rate of some ICP27-responsive genes (16). There is also evidence that ICP27 helps to recruit Pol II to viral replication compartments (4, 22), which are believed to be sites of viral transcription. Additionally, ICP27 has been shown to stimulate the utilization of certain HSV-1 poly(A) sites, thus promoting mRNA 3′-end formation and the accumulation of viral transcripts (24, 25). However, the relative contribution of these various mechanisms to ICP27-dependent viral gene expression is presently unclear.
To investigate how ICP27 enhances viral mRNA accumulation, we have been studying the highly ICP27-dependent gC gene (also known as UL44) as a model. This gene is a “true-L” gene, meaning that its expression is highly dependent on viral DNA synthesis (7, 11). In an earlier study, we found that ICP27 can strongly transactivate the gC gene in a plasmid cotransfection assay (31). Moreover, transactivation in this system was found to be posttranscriptional and independent of the gC gene's promoter or poly(A) site. As part of our past study, we constructed plasmid pgCΔpro, in which the human cytomegalovirus (HCMV) IE promoter drives transcription of the gC gene. Unexpectedly, despite the presence of the powerful CMV promoter, the gC gene was expressed very poorly as mRNA in transfected cells. However, in the presence of ICP27, it was efficiently expressed. Based on these data, we hypothesized that the gC gene harbors a cis-acting posttranscriptional inhibitory sequence that prevents its expression as stable mRNA. We further hypothesized that ICP27 overcomes the effect of this sequence to enable mRNA accumulation. Here, we report that we have identified such an element in the 5′ portion of the gC coding region. Furthermore, this sequence can be transferred to another gene, where it functions to confer both silencing in the absence of ICP27 and high-level expression in the presence of ICP27. Thus, for the first time, an ICP27-responsive sequence has been identified in a physiologically relevant ICP27 target gene. Importantly, we show that this sequence also functions during HSV-1 infection to regulate the ICP27-dependent expression of the gC gene.
MATERIALS AND METHODS
Cells, viruses, and infections.
Vero (African green monkey kidney cells) were obtained from the American Type Culture Collection (ATCC). V27 cells are derivatives of Vero cells that have been stably transfected with the HSV-1 ICP27 (33) gene. Vero cells were grown as monolayers in Dulbecco modified Eagle medium (DMEM) containing 5% heat-inactivated fetal bovine serum (FBS), 50 U/ml penicillin, and 50 μg/ml streptomycin. The medium for V27 cells was the same, except that it also contained 300 μg/ml G418. All tissue culture reagents were purchased from Life Technologies/Invitrogen (Carlsbad, CA), except FBS, which was purchased from Atlas Biologicals (Fort Collins, CO).
The wild-type (WT) HSV-1 strain used in these studies was KOS1.1 (14). The ICP27-null mutant d27-1 has been described (33), and the ΔCA, d27ΔCA, and rΔCA mutants are described below. Infections were carried out at a multiplicity of infection (MOI) of 10 PFU per cell, in phosphate-buffered saline (PBS) containing 0.1% glucose and 0.1% heat-inactivated newborn calf serum. Viral adsorption was for 1 h at 37°C, at which time the viral inoculum was replaced with 199 medium containing 2% heat-inactivated newborn calf serum, 50 U/ml penicillin, and 50 μg/ml streptomycin. Infections were incubated at 37°C. In some experiments, phosphonoacetic acid (PAA) was added to the overlay media at a concentration of 400 μg/ml.
To construct viral mutants, a homologous recombination procedure was used (33). The d27ΔCA mutant was constructed in V27 cells by cotransfecting infectious d27-1 DNA with a HindIII-PstI restriction fragment from pgCA1A2 (described below). To identify a viral isolate having the desired ΔCA deletion, DNA from viral plaques was screened by PCR using primers bracketing the deletion. A positive isolate was plaque purified and designated the d27ΔCA mutant. To engineer the ΔCA mutant, infectious DNA from the d27ΔCA mutant was cotransfected into V27 cells with a PstI restriction fragment from plasmid pPs27pd1 (36), which contains the WT ICP27 gene. ICP27-positive recombinants were selected by growth on Vero cells, and a positive isolate was designated the ΔCA mutant. To engineer the rΔCA mutant, infectious DNA from the ΔCA mutant was cotransfected into Vero cells with a HindIII-PstI restriction fragment from plasmid pgC (31), bearing the WT gC gene. Plaques were screened by PCR as described above to identify an isolate having the WT gC gene. This virus was designated the rΔCA mutant.
Plasmids.
For the cotransfection assay, pgCΔpro (31) or its derivatives were cotransfected alone or with ICP27 expression plasmid pC27 (27), except when the plasmid pM27 (34) was used to express ICP27 (see Fig. 4C). Derivatives of pgCΔpro were made in the following ways. Plasmid pgCProPA has the endogenous gC poly(A) site replaced by that from the simian virus 40 (SV40) late poly(A) region, as well as a deletion of much of the gC gene's 5′ untranslated region (UTR). To engineer this plasmid, the gC promoter was removed from pgCPA (31) by digestion with KpnI and NheI. A KpnI-NheI fragment from pgCΔPro containing the CMV IE promoter was then ligated into the promoterless pgCPA fragment, resulting in pgCProPA. To make pPro5UTR, pgCΔpro was digested with NheI, and the DNA ends were made blunt by a DNA fill-in reaction with the Klenow fragment of Escherichia coli DNA polymerase. XhoI linkers were ligated on, and the DNA was cut with XhoI, which also cuts pgCΔpro at the junction between the CMV IE promoter and gC gene. The resulting large fragment was gel purified and self-ligated, resulting in pPro5UTR. To engineer pProgC5d+, pgCΔpro was digested with NheI, and the DNA ends were made blunt, as described above. SphI linkers were ligated on, and the DNA was cut with SphI. The large DNA fragment was then gel purified and self-ligated to create pProgC5d+. To engineer pProUL45ko, plasmid pUL45ko was first constructed by digesting plasmid pgCAge (31) with SphI and AgeI, blunting the resulting DNA ends with T4 DNA polymerase, and self-ligating the large fragment. To make pProUL45ko, pgCΔpro and pUL45ko were digested with KpnI and NheI, and the small and large fragments of each, respectively, were purified and ligated together. The resulting plasmid was named pProUL45ko. To make pgCΔproNE, pgCΔpro was digested with NheI and EcoNI. Following digestion, the DNA ends were made blunt using the Klenow fragment. The large DNA fragment was purified and self-ligated to yield pgCΔproNE. To make pgCΔproES, pgCΔpro was digested with EcoNI and SfiI. Following digestion, the DNA ends were blunted using T4 DNA polymerase. The large fragment was gel purified and self-ligated, resulting in pgCΔproES. To engineer pgCΔproSS, pgCΔpro was digested with SfiI and SphI, followed by a T4 DNA polymerase-mediated DNA blunting reaction. The large fragment was gel purified and self-ligated to give pgCΔproSS. To make plasmids pgCΔproNA1, pgCΔproA1A2, and pgCΔproA2E, oligonucleotide-directed mutagenesis was used to engineer two pgCΔpro derivatives with substitution mutations creating AfeI sites. The mutagenesis was carried out using the QuikChange site-directed mutagenesis kit (Stratagene). Mutant pgCΔproA1 has alterations that change codons 44/45 of gC from AGC/GAG to AGC/GCT. Mutant pgCΔproA2 has alterations that change codons 130/131 from CGC/GAC to AGC/GCT. To construct pgCΔproNA1, pgCΔproA1 was digested with NheI and AfeI, the ends were blunted with T4 DNA polymerase, and the large fragment was self-ligated. To construct pgCΔproA2E, a comparable strategy was used on pgCproA2, utilizing AfeI and EcoNI as the restriction enzymes. To engineer pgCΔproA1A2, pgCΔproA1 and pgCΔproA2 were digested with AfeI and SphI, and the large fragment from pgCΔproA1 was ligated to the small fragment from pgCΔproA2 to yield pgCΔproA1A2. Plasmid pgCA1A2 was constructed to engineer the d27ΔCA virus mutant. This plasmid has the A1A2 deletion introduced into the background of pgC (31).
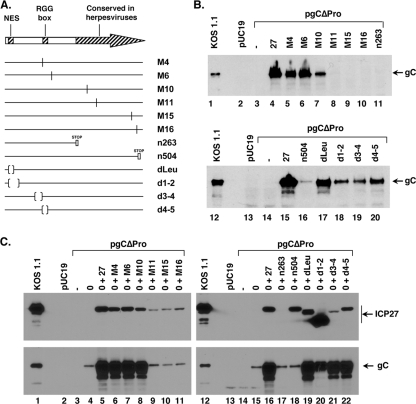
FIG. 4.
Identification of sequences in ICP27 needed for gC transactivation. (A) Schematic diagram of ICP27 mutant plasmids. (Top) The arrow represents the WT ICP27 protein. The locations of the nuclear export sequence (NES), RGG-box RNA-binding domain, and C-terminal region conserved in mammalian herpesviral ICP27 homologs are shown. (Bottom) The lines denote mutant forms of ICP27. Vertical lines indicate codon substitution mutations, bars denote stop codon mutations, and parentheses represent in-frame deletions. (B) pgCΔpro cotransfection assay. The pgCΔpro plasmid was transfected alone, or with WT or ICP27 mutant plasmids. At 2 days posttransfection, total proteins were collected and analyzed for gC expression by immunoblotting. (C) pgCΔpro cotransfection assay in the presence of ICP0. The pgCΔpro plasmid was transfected alone, with an ICP0 expression plasmid, or with an ICP0 expression plasmid plus WT or mutant ICP27 plasmids. At 2 days posttransfection, total proteins were collected and analyzed for gC and ICP27 expression by immunoblotting.
For analysis of the HSV-1 UL42 gene, we engineered pBUL42Δpro, which contains the UL42 transcription unit under the control of the CMV IE promoter. This plasmid was constructed by the following steps. First, a 2,890-bp BstEII restriction fragment from the HSV-1 genome containing the UL42 gene was blunted using the Klenow fragment. It was then cloned into the SmaI site of pUC19 to give pBUL42. To delete the endogenous UL42 promoter, pBUL42 was digested with RsrII, which cleaves at a site within the UL42 gene's 5′ UTR. The DNA ends were then blunted using the Klenow fragment, BamHI linkers were ligated on, and the DNA was cut with BamHI. In addition to digesting residual linker DNA, BamHI also cuts in the vector sequence just upstream of the 5′ end of the UL42 insert. The large DNA fragment of this digestion was self-ligated to give a promoterless UL42 gene. This plasmid was designated pBUL42dpro. A plasmid having the CMV IE promoter driving UL42 transcription was next made. To do this, the CMV IE promoter was obtained from pRL-CMV (Promega) as a 753-bp restriction BglII-HindIII restriction fragment and cloned into the BamHI site of pBUL42dpro using BglII linkers. This plasmid was designated pBUL42Δpro. For Northern blot detection of UL42 mRNA, we used the insert of plasmid pUL42. This plasmid was generated by PCR using HSV-1 DNA as a template and oligonucleotides GATCGAATTCTTCCCCTGGCGGTGTGGCCC and GATCGAATTCCGAACGTGGTGGGCGTGGCA as primers. The PCR product was digested with EcoRI, and the ∼600-bp UL42 sequence was cloned into the EcoRI site of pUC19.
To introduce the SE into the UL42 gene, we utilized a PCR approach. The SE sequence was amplified from pgCΔpro using the primers GATCAGGCCTGAGGCCCCCACATCGGGGTC and GATCAGGCCTGCGGCGGTCGCACCACACGG, which have StuI sites near their 5′ ends. Following the reaction, the PCR product was cleaved with StuI and cloned into the AfeI site of pBUL42Δpro. A plasmid having the desired insert in the sense orientation was designated pUL42-SE; one having it in the reverse orientation was designated pUL42-rSE.
For analysis of the ICP27 sequences involved in stimulating gC expression, we used plasmids encoding mutant forms of ICP27 to transactivate the gC genes on pgCΔpro or pgC (31). The mutant ICP27 plasmids have point mutations (pM4, pM6, pM10, pM11, pM15, and pM16), nonsense mutations (pBH-263R and pBH-504R), and in-frame deletions (pBH27ΔLeu, pBSd1-2, pMd3-4, and pMd4-5) (21, 26, 34-36). The plasmid pSHZ (29) was used to express ICP0.
Transfections and RNA analysis.
Transfections for RNA analysis were carried out on 10-cm2 dishes of Vero cells by using the calcium phosphate technique (9, 32) or Lipofectamine 2000 (Life Technologies/Invitrogen, Carlsbad, CA), using the manufacturer's recommended protocol. When the calcium phosphate technique was used, 12 μg of pgCΔpro or derivative were used, as well as 12 μg of pC27, when applicable. For transfections lacking either or both of the constituents described above, pUC19 was substituted in order to maintain the total amount of DNA in each transfection at 48 μg. When Lipofectamine 2000 was used, the total amount of DNA transfected per dish was 24 μg. This included 12 μg of pgCΔpro or derivative, and when applicable, 12 μg of pC27. The transfection for protein analysis in Fig. 4B was carried out using the calcium phosphate technique as previously described (31). The transfection experiment in Fig. 4C was done using Lipofectamine 2000. Total RNA was prepared using Trizol reagent (Life Technologies/Invitrogen, Carlsbad, CA) and the protocol supplied by the manufacturer. All RNA preparations were treated with RNase-free DNase (Roche Diagnostics Corporation, Indianapolis, IN) to remove contaminating DNA. Northern blotting was done using either formaldehyde or glyoxal as RNA denaturing agents during gel electrophoresis, as previously described (42). To detect gC mRNA, two hybridization probes were used. In Fig. 1B and C, a HindIII-PstI fragment from pgC, corresponding to the entire 3.0-kb HSV-1 insert of pgC, was the probe. In Fig. 1D, 5, and 6, an 813-bp EcoRV-EcoNI fragment from pgC was the probe. For detection of UL42 mRNA, the 599-bp EcoRI fragment from pUL42 was used. The hybridization probe for US11 mRNA was a 483-bp PCR fragment derived from the US11 coding region. This product was generated from HSV-1 DNA using the primers ATGAGCCAGACCCAACCCCCG and TACAGACCCGCGAGCCGTACG. Radioactive labeling of the probes with 32P was done using a random primer labeling kit (Life Technologies/Invitrogen, Carlsbad, CA). To see whether the gC transcript was spliced, we carried out reverse transcription (RT)-PCR analysis as described previously (42).
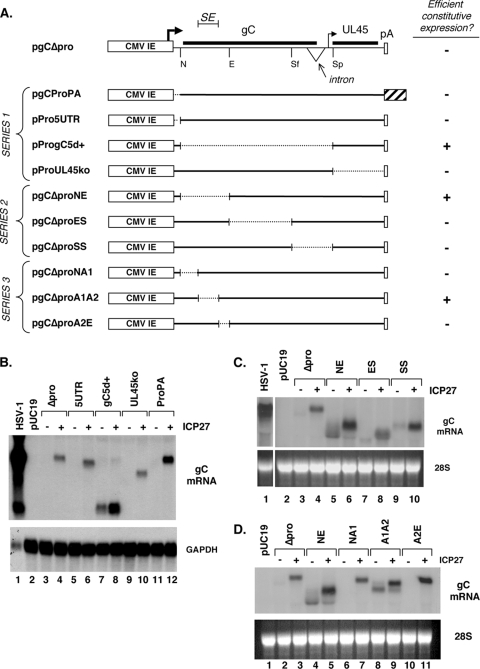
FIG. 1.
Mapping of a silencing element in the gC gene. (A) Analysis of gC mRNA expression from pgCΔpro and its derivatives. (Top) Schematic of the gC transcription unit on plasmid pgCΔpro, including the position of the CMV IE promoter and transcription start site, gC (UL44) coding region, intron, UL45 mRNA transcription start, UL45 coding region, and endogenous poly(A) sequence. Also shown are restriction sites used to construct various deletion derivatives. N, NheI; E, EcoNI; Sf, SfiI; Sp, SphI. (Bottom) Schematics of derivative plasmids are shown, with broken lines indicating deletions. The large striped bar denotes an SV40 sequence containing the late-region poly(A) site. Pluses and minuses indicate whether the transfected genes efficiently express gC mRNA in transfected Vero cells in the absence of ICP27. (B to D). Northern blotting results. The parental construct pgCΔpro or mutant derivatives (series 1 to 3, in panels B to D, respectively) were transfected into Vero cells in the absence or presence of pC27, an ICP27 expression construct. Total RNA was harvested at 2 days posttransfection and analyzed for gC mRNA expression by Northern blotting.
Immunoblotting.
Immunoblotting was carried out as previously described (31). Mouse monoclonal antibodies for gC (H1104), ICP27 (H1113 and H1119), ICP0 (H1112), ICP4 (H1114), and ICP8 (H1115) were obtained from Rumbaugh-Goodwin Institute for Cancer Research (Plantation, FL). For analysis of gC, ICP0, ICP4, and ICP8, the appropriate antibody was used at a dilution of 1:300, 1:1,000, 1:5,000, and 1:300, respectively. For the ICP27 analysis in Fig. 4B and C, a mixture of H1113 and H1119 was used at dilutions of 1:1,000 and 1:5,400, respectively. For the ICP27 analysis in Fig. 6 and 7, H1119 was used at a dilution of 1:5,400. Levels of gCsec, a variant form of gC that has an alternate C-terminal sequence, were determined using a 1:1,000 dilution of rabbit polyclonal antiserum (42). US11 rabbit antisera was a kind gift of J. J. Diaz and was used at a dilution of 1:1,000. A mouse monoclonal antibody specific for β-actin was purchased from Abcam (Cambridge, MA) and used at a dilution of 1:10,000.
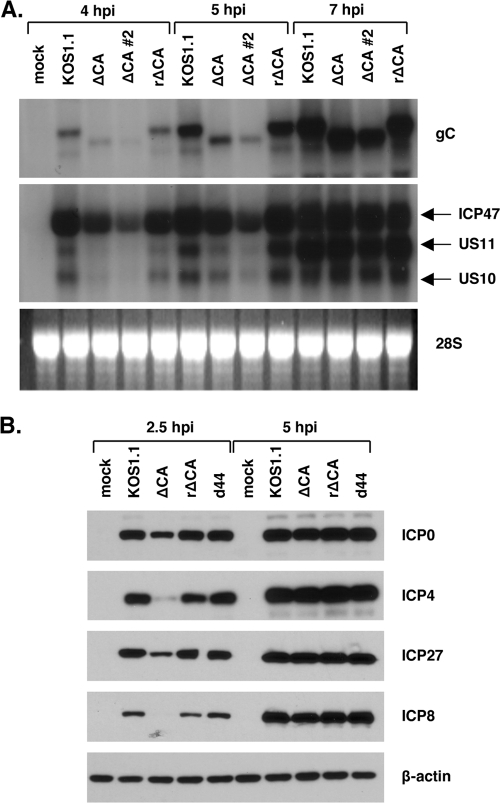
FIG. 6.
The SE deletion leads to a general delay in viral gene expression. (A) Expression of L genes in ΔCA mutant-infected cells. Vero cells were mock infected or infected with the viruses shown, and total cell RNA was isolated at 4, 5, and 7 hpi. The RNA was analyzed by Northern blotting using probes for true-L transcripts gC and US11. Note that the US11 probe also hybridizes to the IE ICP47 (US12) transcript and the L US10 transcript. (B) Expression of IE and DE proteins in ΔCA mutant-infected cells. Vero cells were mock infected or infected with the viruses shown. Total cell proteins were harvested at 2.5 and 5 hpi. Expression of IE (ICP0, ICP4, and ICP27) or DE (ICP8) proteins was analyzed by immunoblotting.
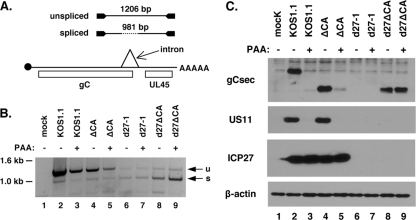
FIG. 7.
ICP27- and DNA replication-independent expression of gC in d27ΔCA mutant-infected cells. (A). RT-PCR assay to detect gC mRNA splicing. (Bottom) A diagram of gC mRNA is shown, with cap, open reading frames, intron, and poly(A) tail indicated. (Top) The RT-PCR products arising from unspliced and spliced gC mRNA (1,206 and 981 bp, respectively) are represented. (B). RT-PCR assay. RNA samples isolated at 5 hpi were analyzed by the RT-PCR assay. A negative image of the ethidium bromide-stained gel is shown. u, unspliced gC mRNA; s, spliced gC mRNA. (C) Expression of gCsec-related proteins. Vero cells were infected at an MOI of 10 with the viruses indicated, in the absence or presence of 400 μg/ml PAA. Total proteins were harvested at 16 hpi and analyzed by immunoblotting for expression of the gCsec-related proteins using an antibody that binds to the unique C terminus of this gC variant. US11, ICP27, and β-actin were also analyzed by immunoblotting.
RESULTS
Identification of a silencing element (SE) in the gC gene.
We previously found that the gC gene on plasmid pgCΔpro (Fig. 1A) is not expressed efficiently as mRNA when this plasmid is transfected into Vero cells (31). However, gC mRNA is abundantly expressed if ICP27 is coexpressed. The poor constitutive expression of the gC gene in this construct was surprising, given that its transcription is controlled by the powerful CMV IE promoter. Based on these past results, we speculated that the gC transcription unit contains an inhibitory sequence and that ICP27 overcomes the effects of this sequence to allow gC mRNA accumulation. This hypothesis predicts that the gC gene would be efficiently expressed in the absence of ICP27 if the putative inhibitory sequence is deleted. To test this prediction, we constructed a series of pgCΔpro derivatives in which segments of the gene were systematically deleted (Fig. 1A, series 1). Since the very 3′ end of the gene could not be deleted without removing the endogenous poly(A) site, we tested this region by replacing it with a sequence containing the SV40 late-region poly(A) site. To carry out the analysis, pgCΔpro or the various derivatives were transfected into Vero cells in the absence or presence of pC27, an ICP27 expression plasmid. Total cell RNA was isolated 2 days later, and gC mRNA levels were analyzed by Northern blotting (Fig. 1B). As expected, gC mRNA expression from pgCΔpro was inefficient (Fig. 1B, lane 3) but could be transactivated by ICP27 (Fig. 1B, lane 4). Notably, expression of all constructs was enhanced by ICP27, despite the systematic deletion/substitution of the entire transcription unit. This suggests that the gC gene harbors two or more distinct sequences that confer its responsiveness to ICP27. However, one of the pgCΔpro derivatives, pProgC5d+, did have a significant change in its expression properties, in that it showed relatively efficient basal expression of gC mRNA in the absence of ICP27 (lane 7). This suggests that the region deleted in pProgC5d+ contains an inhibitory sequence.
To investigate the putative inhibitory sequence further, we made a second series of deletions, focusing on the region deleted in pProgC5d+ (Fig. 1A, series 2). Analysis of these plasmids in the cotransfection assay indicated that of the three new mutants, only pgCΔproNE exhibited a high level of constitutive gC mRNA expression (Fig. 1C, lane 5). It was also apparent from this experiment that when ICP27 was coexpressed, there was an increase in the electrophoretic mobility of the gC transcript (Fig. 1C, lane 6). This is consistent with ICP27's ability to promote the retention of the 225-nucleotide intron in the gC transcript (42). Note that this effect was not seen in the experiment involving pProgC5d+ (Fig. 1B), since the deletion in this construct removes the intron.
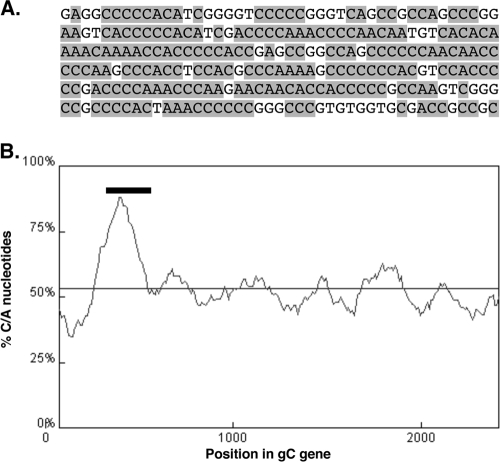
The inhibitory element was mapped still further using a third set of deletions (Fig. 1A, series 3). Of the three new mutants, only pgCΔproA1A2 was able to efficiently express gC mRNA in the absence of ICP27 (Fig. 1D, lane 8). Again, when ICP27 was coexpressed, there was an increase in the size of the gC transcript due to retention of the gC intron. Together, these mapping experiments demonstrate that the gC gene contains an inhibitory element that maps to the 258 bp deleted in pgCΔproA1A2. For the remainder of this report, we will refer to this sequence as the gC gene's “silencing element” (SE). The SE is contained entirely within the coding region of the gC gene and corresponds to codons 45 to 130. The DNA sequence of the SE is shown in Fig. 2A. Analysis of the SE by the RNA structural prediction program Mfold (54) did not reveal any particularly stable RNA structures that could form in SE-encoded RNA. However, a visual inspection of the sequence suggested that the SE is somewhat unusual in its nucleotide composition, being extremely rich in C and A nucleotides (77%). This was verified by a computer analysis that shows that the SE maps to a region of the gene that is quite high in C+A content compared to the rest of the gC transcription unit (Fig. 2B). We also noted that the SE contains numerous homopolymeric runs of up to seven C residues.
FIG. 2.
The SE is rich in C and A nucleotides. (A) DNA sequence of the SE. C and A residues are shaded. (B) Percent (C+A) nucleotide content of the gC transcription unit as determined by a DNA sequence analysis tool (http://molbiol-tools.ca/Jie_Zheng/; window size, 150; step size, 10). The thin horizontal line indicates the average (C+A) content for the entire transcript unit (53.4%). The bar indicates the position to which the SE was mapped.
The SE can function in a heterologous context.
The results described above show that the SE inhibits mRNA expression from the gC gene in transfected cells. We were curious to know whether the SE could function in the context of a different gene. To test this, we used plasmid pBUL42Δpro, which carries the HSV-1 UL42 gene under the control of the CMV IE promoter (Fig. 3A). This plasmid was originally constructed by our laboratory to test whether another ICP27-responsive HSV-1 gene, the UL42 gene (49), behaves similarly to the gC gene in a plasmid cotransfection assay. However, we found that it does not, as pBUL42Δpro expresses readily detectable UL42 mRNA in the absence of ICP27 and is stimulated only marginally, if at all, by coexpression of ICP27 (Fig. 3B, compare lanes 3 and 4). This indicates that ICP27-dependent HSV-1 genes can behave quite differently in the plasmid cotransfection assay. However, the constitutive expression of the UL42 gene and its minimal response to ICP27 made pBUL42Δpro a useful plasmid for testing whether the SE can function in a heterologous context. Therefore, the SE was inserted in the sense orientation into the 5′ half of the UL42 coding region (UL42-SE), approximating its natural position in the gC gene (Fig. 3A). The insertion was engineered to be in frame, so as not to introduce premature stop codons that could trigger the nonsense-mediated mRNA decay pathway (1). As a control, we also made a construct in which the SE was inserted in the reverse orientation (UL42-rSE). We then carried out a cotransfection experiment to see if the SE can silence the UL42 gene. In the absence of ICP27, very little, if any, mRNA was expressed from the UL42-SE gene (Fig. 3A, lane 5), indicating that the SE silences the UL42 gene. In contrast, the rSE-containing gene expressed abundant mRNA (Fig. 3A, lane 7). Strikingly, coexpression of ICP27 dramatically stimulated accumulation of UL42 mRNA from pUL42-SE (Fig. 3A, compare lanes 5 and 6). A modest stimulatory effect of ICP27 was also seen for the UL42-rSE gene. This experiment demonstrates that the SE is a transferable genetic element that can both silence a gene in the absence of ICP27 and induce it to high levels when ICP27 is present.
FIG. 3.
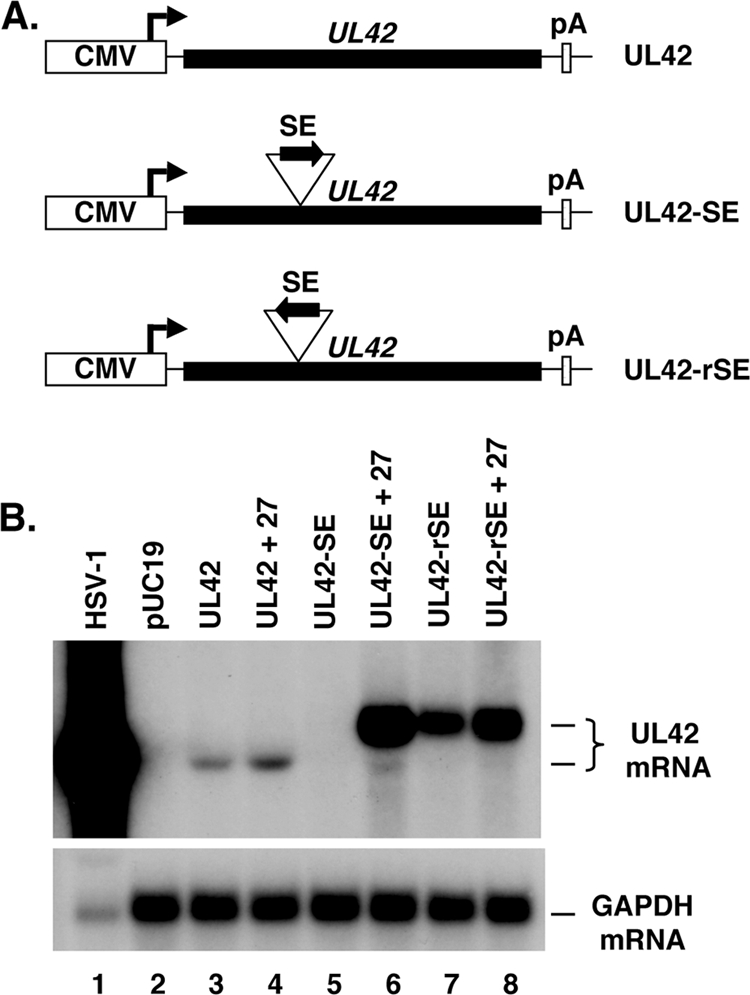
The SE confers silencing and ICP27-responsiveness when transferred to the UL42 gene. (A) Schematic diagram of the UL42 genes on pBUL42Δpro, pUL42-SE, or pUL42-rSE. (B) Cotransfection assay. The UL42 plasmids were transfected alone or with the ICP27 expression construct pC27. Total RNA was harvested at 2 d posttransfection and analyzed by Northern blotting using a UL42-specific probe.
The C-terminal half of ICP27 is important for transactivation of gC.
The above experiments demonstrate that ICP27 enhances the expression of the gC gene by overcoming the inhibitory effects of the SE. To investigate which functional domains in ICP27 are involved in this regulatory activity, we analyzed several ICP27 plasmid mutants in the pgCΔpro plasmid transfection assay (Fig. 4A). To monitor gC expression, gC protein levels at 2 days posttransfection were assessed by immunoblotting. In general, the results (Fig. 4B) suggest that mutations in the C-terminal half of ICP27 either abrogate (M11, M15, M16, and n263) or significantly weaken (n504) ICP27's ability to transactivate gC expression. In contrast, mutations in the N-terminal half of ICP27 (M4, M6, M10 d1-2, dLeu, d3-4, and d4-5) were not highly disruptive to this function of ICP27, although the M10, d1-2, d3-4, and d4-5 mutations had modest negative effects on gC expression. To ensure that comparable levels of ICP27 were expressed from the various ICP27 plasmids, we analyzed the protein samples by immunoblotting for ICP27. However, ICP27 was not efficiently detected in most samples (data not shown), likely due to immunoblotting sensitivity issues. It is thus possible that some of the mutant ICP27 polypeptides failed to transactivate gC because of their poor expression.
Because of the above-described issue relating to ICP27 expression, an alternate strategy was developed to identify the ICP27 domains involved in gC transactivation. We took advantage of the fact that ICP0 is a relatively promiscuous gene transactivator (6) and thus would be expected to enhance expression of ICP27 in transfected cells, potentially enabling its detection by immunoblotting. However, by itself, ICP0 is unable to efficiently transactivate gC expression from pgCΔpro (K. Perkins and S. Rice, unpublished data). Therefore, we cotransfected pgCΔpro with pSHZ, an ICP0 expression plasmid, minus or plus WT or mutant forms of ICP27. Two days later, total cell proteins were harvested and analyzed for ICP27 and gC expression by immunoblotting (Fig. 4C). As is shown, ICP27 expression was readily apparent for the WT protein and all mutants tested, except n263. However, for some of the mutants (M11, M15, M16, and d3-4), ICP27 accumulation was reduced compared to that of the WT protein. Having verified expression of most of the mutant ICP27 polypeptides, we analyzed gC expression. In the absence of any viral transactivator, there was no detectable expression of gC from pgCΔpro (Fig. 4C, lanes 3 and 14), whereas ICP0 induced gC expression to low but detectable levels (Fig. 4C, lanes 4 and 15). As expected, when WT ICP27 was also expressed, gC accumulated to high levels (Fig. 4C, lanes 5 and 16). Among the mutants, the results were similar to those of the previous experiment, in that the C-terminal ICP27 mutants (M11, M15, M16, n263, and n504) were unable to transactivate gC, while the N-terminal mutants (M4, M6, M10, dLeu, d1-2, d3-4, and d4-5) were all competent for this activity, although the accumulation of gC was somewhat reduced in the d1-2 transfection. We conclude that the C-terminal half of ICP27 is crucial for its ability to transactivate the gC gene in transfected cells. In contrast, certain N-terminal regions, including the NES and RGG box RNA-binding domain (missing in the dLeu and d4-5 ICP27 polypeptides, respectively), are not required for this function.
The SE regulates gC gene expression during HSV-1 infection.
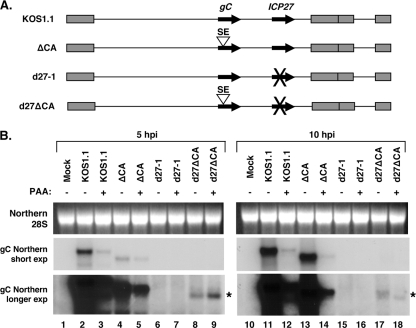
The preceding experiments demonstrate that the SE is a strong regulator of gC expression in a plasmid cotransfection assay. To investigate whether the SE also regulates gC expression during the HSV-1 infection, we constructed two HSV-1 mutants (Fig. 5A). The first, designated the ΔCA mutant, has a deletion of the 258-bp sequence in the gC coding region that we have defined as the SE. This deletion is in frame with respect to the gC open reading frame (ORF), removing codons 45 to 130. The second mutant, designated the d27ΔCA mutant, is identical to the ΔCA mutant, except that it also has the same ICP27 gene deletion found in the well-characterized ICP27-null mutant d27-1 (33).
FIG. 5.
Deletion of the SE in recombinant viruses leads to altered expression of gC mRNA during infection. (A) Schematic diagram of viral mutants engineered for this study. The ΔCA mutant has a 258-bp in-frame deletion in the gC coding region that removes the SE (gC codons 45 to 130). The d27ΔCA mutant has the same deletion in the context of the ICP27 null mutation found in d27-1. (B) Expression of gC mRNA in infected cells. Vero cells were infected with the viruses shown, either in the absence or presence of 400 μg/ml PAA. Total RNA was harvested at 5 and 10 hpi. gC mRNA expression was analyzed by Northern blotting and autoradiography. The asterisk indicates a novel gC transcript that accumulates in d27ΔCA mutant-infected cells. exp, exposure.
To study the effect of the SE on gC expression, we examined gC mRNA expression in Vero cells infected with the SE mutants or for comparison, the WT or d27-1. Because the gC gene is a true-L gene, we also wished to assess the contribution of viral DNA replication to gC expression. Therefore, the experiment included a set of infections in which PAA, an inhibitor of HSV-1 DNA replication, was added to the overlay media. At 5 and 10 h postinfection (hpi), total cell RNA was harvested, and gC mRNA levels were analyzed by Northern blotting (Fig. 5B). The results of this experiment and a repeat experiment of the same design (not shown) demonstrated that the SE deletion has at least two effects on gC mRNA expression. The first is evident when KOS1.1 and the ΔCA mutant are compared at 5 hpi (Fig. 5B, lanes 2 and 4, short exposure). It can be seen that the ΔCA mutant expresses less gC mRNA at this relatively early time point compared to WT HSV-1. However, at 10 hpi, the two viruses express comparable levels of gC mRNA (Fig. 5B, lanes 11 and 13). Thus, deletion of the SE in the ΔCA mutant results in the delayed expression of the gC gene relative to that of the WT virus.
The second effect of the SE deletion on gC mRNA expression is most evident when d27-1- and d27ΔCA mutant-infected cells are compared at 5 hpi (Fig. 5B, lanes 6 and 8, long exposure). It can be seen that the d27ΔCA mutant expresses a gC transcript (Fig. 5B, lane 8, asterisk) that is not detected with the d27-1 infection. The failure of d27-1 to express a gC transcript is consistent with prior work that shows that gC mRNA expression is heavily dependent on ICP27 (33). Interestingly, accumulation of the novel gC transcript in d27ΔCA mutant-infected cells was resistant to PAA (Fig. 5B, lane 9), suggesting that viral DNA replication is not required for its expression. Although this result could be explained if the PAA was not effective at inhibiting HSV-1 DNA synthesis, this does not appear to be the case, as PAA effectively reduced gC mRNA expression in the KOS1.1 and ΔCA mutant infections (e.g., see Fig. 5B, 10 hpi, lanes 12 and 14). Low levels of gC transcript were also seen with the d27ΔCA mutant infection at 10 hpi (Fig. 5 B, lanes 17 and 18). Together, these results suggest that in the absence of ICP27, the SE blocks expression of the gC transcript. This is consistent with the results of our transfection studies, which indicate that the SE functions as a gene silencing element.
Deletion of the SE leads to a general delay in HSV-1 gene expression.
We next carried out experiments to explore the first effect of the SE deletion described above, i.e., the delay in gC mRNA expression in the ΔCA mutant. To see if the delayed expression phenotype is specific for the gC gene, as expected, we analyzed the expression of another true-L gene, US11 (18). We were surprised to find that expression of US11 mRNA was also delayed in the ΔCA mutant (data not shown). Given this unexpected effect on another L gene, we wished to ensure that this phenotype is due to the SE deletion, rather than to an unrelated secondary mutation. Thus, we isolated a viral rescuant of the ΔCA mutant, designated the rΔCA mutant, in which the SE deletion was restored to the WT sequence by in vivo recombination. We then used Northern blotting to examine gC and US11 mRNA expression in Vero cells infected with the ΔCA mutant (two independent stocks were analyzed), KOS1.1, or the rΔCA mutant. The results are shown in Fig. 6A. At both 4 and 5 hpi, it can be seen that the ΔCA mutant expresses significantly less gC and US11 mRNA than does the WT or the rΔCA mutant. Note that the US11 probe also detects the overlapping US12 (ICP47) IE transcript, as well as the US10 L transcript. These transcripts, particularly the US10 one, also appeared to be reduced at 4 and 5 hpi in the CA mutant. However, by 7 hpi, the ΔCA mutant expressed levels of gC, US11, US10, and US12 transcripts that were comparable to those of KOS1.1 and the rΔCA mutant. This analysis shows that the ΔCA mutant exhibits a delay in the expression of at least three L genes and that this phenotype is due to the SE deletion.
To see if the SE deletion in the ΔCA mutant also affects IE and DE genes, we used immunoblotting to look at the expression of IE proteins ICP0, ICP27, and ICP4 and the DE protein ICP8. Both 2.5- and 5-hpi time points were examined. We also included a gC deletion mutant, d44 (31), in our analysis. The results, shown in Fig. 6B, demonstrate that both IE and DE protein expression levels are delayed in the ΔCA mutant compared to KOS1.1, the rΔCA mutant, or d44. This indicates that deletion of the SE results in a general delay in viral gene expression. It is notable that the gC deletion mutant d44 does not share this phenotype, since this mutant also lacks the SE (as well as most of the rest of the gC coding region). This suggests that the delay in gene expression exhibited by the ΔCA mutant is not due to a loss of a cis-acting regulatory sequence. Rather, given that the ΔCA mutant encodes a mutant gC polypeptide lacking residues 45 to 130, the most probable explanation of these results is that the mutant gC polypeptide is present on the viral envelope and confers a delay in viral entry. This possibility is further explored in Discussion.
The SE blocks an ICP27- and DNA replication-independent pathway of gC gene expression.
We next explored the second effect of the SE deletion, which is that when present in the context of an ICP27 null mutation in the d27ΔCA mutant, it leads to expression of a gC transcript (Fig. 5B, asterisks) that is not seen in the control d27-1 infection. As ICP27 promotes retention of the gC intron during infection (42), it might be expected that this novel transcript is predominantly spliced. To investigate this, we used an RT-PCR assay (42) (Fig. 7A) to determine the level of gC mRNA splicing, using RNA samples that were isolated at 5 hpi. The results indicate that the gC transcript made in d27ΔCA mutant-infected cells is largely spliced, as predicted (Fig. 7B, lanes 8 and 9). In contrast, the gC transcript produced by the WT and ΔCA infections is mostly unspliced (lanes 2 to 5), consistent with ICP27's ability to promote intron retention. Note that a spliced gC transcript can also be seen with the d27-1 samples, despite the fact that no gC transcript is detected when 5-hpi RNA is examined by Northern blotting (Fig. 5B). This is likely due to the semiquantitative nature of the RT-PCR assay and the fact that it can amplify low-level transcripts.
We next wished to characterize gC protein expression in the d27ΔCA mutant infection. However, initial experiments indicated that neither the gC-specific monoclonal antibody (H1104) that we commonly use to detect gC nor a commercial gC antibody (Abcam) reacted with the gC expressed by the d27ΔCA mutant (or the ΔCA mutant), likely because the SE deletion removes or alters the epitopes. As an alternate strategy, we took advantage of the fact that the novel gC transcript expressed by the d27ΔCA mutant is predominantly spliced. We previously showed that the gC splice leads to a variant form of gC, designated gCsec, that has an alternate C-terminal sequence. This form of gC can be detected using a rabbit antisera raised against this unique C terminus (42). Thus, Vero cells were mock infected or infected with KOS1.1, the ΔCA mutant, d27-1, or the d27ΔCA mutant. At 16 hpi, total infected proteins were collected and analyzed by immunoblotting for gCsec expression. Consistent with our past work, KOS1.1. expressed readily detectable gCsec (Fig. 7C, lane 2) as a result of low-level expression of the spliced transcript (42). As expected, given the true-L nature of the gC gene, gCsec expression was completely blocked in the WT infection by PAA (Fig. 7C, lane 3). The ΔCA mutant expressed a gCsec polypeptide of lower apparent molecular weight (Fig. 7C, lane 4). The reduced size is consistent with the deletion of residues 45 to 130 as a result of the SE mutation. Unlike with the WT virus, however, expression of the gCsec-related polypeptide by the ΔCA mutant was not completely blocked by PAA (Fig. 7C, lane 5). No gCsec was detected in the d27-1 infection, a result that is consistent with our inability to detect gC mRNA by Northern blotting. However, the deleted form of gCsec was readily detectable in the d27ΔCA mutant infection (lane 8), and the levels were completely unaffected by PAA (Fig. 7C, lane 9). This result is consistent with the Northern analysis shown in Fig. 5B. To ensure that this effect was specific for gC, we also examined expression of the US11 protein by immunoblotting. No expression was seen in either the d27-1 or the d27ΔCA mutant infection. Moreover, expression of US11 in the KOS1.1 and the ΔCA mutant infections was completely blocked by PAA. Thus, the results of our RNA and protein expression analyses of d27ΔCA mutant-infected cells demonstrate that when the SE is absent from the HSV-1 genome, the gC gene is expressed in an ICP27- and DNA replication-independent manner. This confirms that the SE functions during infection to genetically silence the gC gene.
DISCUSSION
Identification of an ICP27-responsive element in the HSV-1 gC gene.
In this study, we have used both plasmid cotransfection assays and recombinant virus analysis to investigate the cis-acting sequences that mediate the ICP27-dependent expression of the gC gene. Using these approaches, we identified a 258-bp sequence in the coding region of the gC gene, termed the SE, that functions to silence expression of the gC gene in the absence of ICP27 and to confer expression when ICP27 is present. Moreover, this sequence can be transferred to a heterologous viral gene, where it functions similarly. Thus, for the first time, an ICP27-responsive element has been identified in a physiologically relevant ICP27 target gene.
It is important to point out that we have not yet determined the minimal extent of the SE or defined its important sequence determinants. It is possible that the element is significantly smaller than 258 bp and/or that it is composed of multiple independent elements. We are thus continuing to map this regulatory sequence in more detail. Another point worth emphasizing is that although our study shows that the SE is critical in conferring ICP27-dependent expression, our results do not imply that the SE is the only sequence in the gC gene that mediates ICP27-dependent expression. In fact, our transfection studies strongly indicate that there are other ICP27-responsive sequences, since a gC construct which lacks the SE (pProgC5d+ in Fig. 1A) is still strongly transactivated by ICP27 (Fig. 1B). Thus, the SE-deleted gC gene in the ΔCA and d27ΔCA mutants should not be viewed as a gene that is devoid of all ICP27-dependent regulation.
In both our transfection and virus infection experiments, we found that deletion of the SE from the gC gene allows ICP27-independent accumulation of gC mRNA (Fig. 1 and 5). However, it is evident that this effect is more dramatic in the transfection assays than in the infected-cell experiments. That is, during d27ΔCA mutant infection, gC gene expression is only a small fraction of that seen with the WT infection. There are several possible reasons for the relatively modest expression of gC in d27ΔCA mutant-infected cells. First, as mentioned above, the SE-deleted gC gene of the d27ΔCA mutant still likely retains ICP27-responsive sequences that could limit its expression in the absence of ICP27. Second, the gC gene is a true-L gene whose expression is highly dependent on viral DNA replication. Thus, active viral DNA replication will greatly amplify gC expression in the WT HSV-1 infection, while no similar amplification will occur in the d27ΔCA mutant infection because efficient viral DNA replication requires ICP27 (23, 33). Of course, this effect is not relevant in the transfection studies. Third, the infection experiments utilize the endogenous and likely relatively weak gC promoter, whereas the transfection experiments utilize the powerful CMV IE promoter. We suspect that expression of the gC gene in d27ΔCA mutant-infected cells would be significantly higher if the gene's transcription were driven by the CMV IE promoter or a similarly strong promoter.
Possible mechanisms by which the SE confers ICP27 response.
Our results indicate that the SE inhibits gC gene expression at the level of mRNA accumulation and that ICP27 in some way alleviates this inhibition to enable efficient expression. We will first discuss possible mechanisms by which the SE might inhibit gC expression. Relevant to this is our past study of ICP27's effects on the transfected gC gene (31). This work included nuclear run-on transcription assays that provided evidence that although ICP27 has a profound stimulatory effect on gC mRNA accumulation, it does not affect the gene's transcription rate. This suggests that pre-mRNA is transcribed from the transfected gC gene in the absence of ICP27 but that it is rapidly degraded, likely in the nucleus. We thus hypothesize that the SE represents an mRNA instability determinant that triggers pre-mRNA decay in the nucleus. However, other mechanisms are also feasible. For example, it is possible that the SE inhibits mRNA 3′-end formation and/or polyadenylation, which would also lead to reduced transcript levels.
One important point is that the SE obstructs gC expression in uninfected cells. Thus, the machinery involved in inhibiting gene expression must be cellular. If we are correct in our hypothesis that the mRNA degradation machinery is involved, then suspect pathways would be host systems involved in degrading nuclear pre-mRNAs. However, at present, not a great deal is known of how Pol II transcripts are degraded in the eukaryotic cell nucleus. Such degradation appears to be mediated primarily by RNA quality control systems that destroy pre-mRNAs if they are improperly processed or in some way deemed aberrant (for recent reviews, see references 13 and 50). Although the SE is extremely rich in C/A nucleotides and thus has an unusual nucleotide composition, it is not obvious that it would be perceived as aberrant by the cell. This raises the possibility that an as yet unidentified cellular system is responsible for degrading SE-containing transcripts. If so, then the gC gene may provide a useful tool to identify and characterize this system.
How does ICP27 overcome the silencing effect? Since the silencing mechanism is unknown, ICP27's role is, not surprisingly, somewhat obscure at present. ICP27 could either directly interfere with the silencing machinery, or alternately, it could block access of the machinery to the gC transcript. Fortunately, we did obtain some information about the functional domains of ICP27 that are involved in this activity. Our results indicate that the C-terminal half of ICP27, which is conserved among other mammalian herpesviruses, is critical for this function. However, neither the N-terminal leucine-rich NES nor the RGG box RNA-binding domain of ICP27 is required. These domains are critical for ICP27's mRNA export function (17), suggesting that the rescue of SE-containing transcripts occurs by a mechanism that is distinct from ICP27's effects on mRNA transport. Furthermore, the fact that ICP27's RNA-binding domain is dispensable strongly supports the argument that the mechanism by which ICP27 overcomes silencing does not involve its direct binding to the gC transcript. This is also consistent with ICP27's known RNA-binding specificity: it appears to bind preferentially to G-rich sequences (28, 43), whereas the SE is relatively low in G residues (17.9%). One sequence feature in the SE of note is an abundance of short runs of 3 to 7 C residues. This is potentially of interest since ICP27 is known to associate with hnRNP K (51), a cellular RNA-binding protein that is capable of binding to short C runs (48).
One important finding is that the SE can still function when transferred to the HSV-1 UL42 gene. Thus, the SE can be considered a discrete genetic element that both silences gene expression and confers ICP27 response. However, in other experiments, we found that the SE did not function to confer either silencing or ICP27 response when inserted into the coding region of the E. coli β-galactosidase gene (data not shown). Moreover, a single insertion of the SE into the 3′ UTR of a CMV IE promoter-driven enhanced green fluorescent protein gene had little effect on mRNA levels, whereas two head-to-tail insertions conferred modest silencing and response to ICP27 (data not shown). Thus, it appears that gene context is quite critical to the function of the SE. Sorting out the parameters involved in these context effects will likely aid in our understanding of the mechanisms involved.
Another aspect of the UL42 experiments that deserves comment is the fact that ICP27 induces high-level expression of the UL42-SE gene, at least in comparison to the much lower expression levels seen for the parental UL42 construct (Fig. 3B, compare lane 6 to 2 and 3). One interpretation of this result is that ICP27 not only overcomes the silencing effect but that it also actively enhances expression of the SE-containing gene over and above the basal level. If this is true, then it is conceivable that the SE contains separable positive and negative elements. However, other interpretations are possible. One relevant point in this regard is that inserting the “reverse” SE into the same position of the UL42 gene also led to high basal levels of UL42 mRNA (Fig. 3B, compare lane 7 and 3). A similar result is seen when an HSV-1 gB gene sequence is inserted at this position (C. Xu, L. Sedlackova, and S. Rice, unpublished data). However, neither the reverse SE nor the gB sequence silences UL42 expression or confers a strong ICP27 response. These results suggest that the sequence of the UL42 gene at the insertion site (which corresponds to an AfeI restriction site) may somehow limit UL42 expression and that disruption of this sequence by any insertion will lead to enhanced gene expression. If this is correct, then ICP27's apparent ability to enhance mRNA accumulation from the UL42-SE gene may simply reflect the disruption of the putative UL42 inhibitory sequence. Further experiments involving other SE-containing gene constructs should allow us to resolve this issue.
The SE plays a role in the replication-dependent expression of gC.
One very surprising finding was that the ICP27-independent expression of gC in a d27ΔCA mutant was almost completely resistant to an inhibitor of viral DNA replication. These results were unexpected, because the gC gene is a true-L gene known to be highly dependent on viral DNA replication. Our results suggest that there is an active pathway for gC gene expression that is both ICP27- and viral DNA replication-independent. However, this pathway appears to be blocked by the SE, likely at a posttranscriptional level. These results are consistent with a recent study that showed that the TATA-binding protein and Pol II assemble on the gC promoter in the presence of a viral DNA replication inhibitor (39). We suggest that this reflects the active transcription of the gC gene during the early phase of infection. However, the transcripts made may be rapidly degraded in an SE-dependent manner. Interestingly, a past study using nuclear run-on transcription assays reported that there is active transcription of the gC gene under conditions of DNA replication inhibition (53). On the other hand, a similar analysis by a different group reported the opposite result (8). In light of our results, it may be worth revisiting the question of whether posttranscriptional mechanisms contribute to the replication dependence of gC and possibly other HSV-1 true-L genes.
Although we were not able to look at the accumulation of the standard form of the gC protein due to a lack of a reactive antibody, we were able to monitor the expression of gCsec, a secreted variant of gC (42). The data presented in Fig. 7C show that accumulation of gCsec in d27ΔCA mutant-infected cells is completely independent of viral DNA replication. In contrast, gCsec expression is still quite dependent on replication in ΔCA mutant-infected cells, when ICP27 is present. This difference may reflect the effect of DNA replication, which would be highly active in the ΔCA mutant infection but not the d27ΔCA mutant infection. Interestingly, we have consistently observed a small amount of replication-independent gCsec expression in the ΔCA mutant infections (Fig. 7C, lane 5), whereas gCsec expression in the WT infection is completely dependent upon viral DNA replication (Fig. 7C, compare lanes 2 and 1), as expected. This is further evidence that the SE plays a role in the replication-dependent expression of gC.
Deletion of the SE from the gC coding sequence appears to delay viral entry.
We were surprised to find that all viral gene expression is delayed in the ΔCA mutant relative to WT HSV-1 or a genetic rescuant. This effect does not appear to be due to deletion of a cis-acting regulatory sequence, since a gC deletion mutant that also lacks the SE (as well as most of the rest of the gC gene) did not show the same phenotype. We believe that the most likely explanation for the delayed gene expression of the ΔCA mutant is that it has an entry delay as a result of its altered gC polypeptide. The ΔCA mutant form of gC lacks residues 45 to 130 of the gC polypeptide. However, it retains the gC signal sequence and transmembrane domain and is thus likely to be present on the viral envelope (see reference 45 for a review of gC structure and function). Although gC is not required for viral entry in cell culture, it is an important cell attachment protein by virtue of its interaction with heparan sulfate-containing proteoglycans. Moreover, it forms hetero-oligomers on the viral particle with the viral glycoproteins that mediate membrane penetration (10), those being gB, gD, and gH/gL. Interestingly, a recent study indicated that a mutant form of gC increases the sensitivity of HSV-1 virions to neutralization with anti-gB, anti-gD, and anti-gH/gL antibodies, providing further evidence that gC interacts with these proteins on the surface of virions (12). It is thus possible that the ΔCA mutant form of gC, by virtue of altered interactions with the entry glycoproteins, interferes with and hence delays entry. Further experiments will be required to confirm this hypothesis.
The apparent delay in entry exhibited by the ΔCA mutant, and likely the d27ΔCA mutant as well (although we have not examined this mutant for early gene expression), somewhat complicates the interpretation of their phenotypes. Unfortunately, because the SE maps to the gC coding region, it is impossible to delete this cis element without at the same time disrupting the structure of the gC polypeptide. This points to the need to better map the SE and define its essential sequence determinants. With this information in hand, it should be possible to design SE-inactivating mutations that only minimally affect the encoded polypeptide, or ideally, do not alter it all.
Do elements similar to the SE regulate other HSV-1 ICP27-responsive genes?
It is interesting to speculate that other ICP27-dependent DE and L genes may have elements that are similar to the SE. At present, the most striking sequence characteristic of the SE is its unusually high C+A content. To see if other HSV-1 genes possess similar sequences, we used a DNA base composition program (http://molbiol-tools.ca/Jie_Zheng/dna.html) to survey the entire HSV-1 strain 17 genome. The average C+A content of the genome was found to be 49.7%. Using a window size of 100 nucleotides, we identified 41 sequences, including the SE, that have a C+A content of >70%. Several of these are located in the coding strand of known ICP27-dependent transcripts and thus have the potential to affect gene expression similarly to SE. We are currently using our plasmid cotransfection system to test whether some of these sequences function as silencing or ICP27-responsive elements in our assays.
Acknowledgments
We thank Joy Lengyel for help with plasmid constructions, J. J. Diaz for the gift of US11 antiserum, and Bob Geraghty for helpful discussions. We also appreciate continuing suggestions and support from the members of the Rice, Schiff, and Bresnahan laboratories. Finally, Linse Lahti, Megan Ahl, and Lindsey Foran provided valuable technical help.
This research was supported by NIH grant R01-AI42737.
Footnotes
Published ahead of print on 30 December 2009.
REFERENCES
- 1.Chang, Y. F., J. S. Imam, and M. F. Wilkinson. 2007. The nonsense-mediated decay RNA surveillance pathway. Annu. Rev. Biochem. 76:51-74. [DOI] [PubMed] [Google Scholar]
- 2.Chen, I. H., L. Li, L. Silva, and R. M. Sandri-Goldin. 2005. ICP27 recruits Aly/REF but not TAP/NXF1 to herpes simplex virus type 1 transcription sites although TAP/NXF1 is required for ICP27 export. J. Virol. 79:3949-3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen, I. H., K. S. Sciabica, and R. M. Sandri-Goldin. 2002. ICP27 interacts with the RNA export factor Aly/REF to direct herpes simplex virus type 1 intronless mRNAs to the TAP export pathway. J. Virol. 76:12877-12889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dai-Ju, J. Q., L. Li, L. A. Johnson, and R. M. Sandri-Goldin. 2006. ICP27 interacts with the C-terminal domain of RNA polymerase II and facilitates its recruitment to herpes simplex virus 1 transcription sites, where it undergoes proteasomal degradation during infection. J. Virol. 80:3567-3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DeLuca, N. A., and P. A. Schaffer. 1988. Physical and functional domains of the herpes simplex virus transcriptional regulatory protein ICP4. J. Virol. 62:732-743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Everett, R. D. 2000. ICP0, a regulator of herpes simplex virus during lytic and latent infection. Bioessays 22:761-770. [DOI] [PubMed] [Google Scholar]
- 7.Frink, R. J., R. Eisenberg, G. Cohen, and E. K. Wagner. 1983. Detailed analysis of the portion of the herpes simplex virus type 1 genome encoding glycoprotein C. J. Virol. 45:634-647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Godowski, P. J., and D. M. Knipe. 1986. Transcriptional control of herpesvirus gene expression: gene functions required for positive and negative regulation. Proc. Natl. Acad. Sci. U. S. A. 83:256-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Graham, F. L., and A. J. van der Eb. 1973. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology 52:456-467. [DOI] [PubMed] [Google Scholar]
- 10.Handler, C. G., R. J. Eisenberg, and G. H. Cohen. 1996. Oligomeric structure of glycoproteins in herpes simplex virus type 1. J. Virol. 70:6067-6070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Homa, F. L., T. M. Otal, J. C. Glorioso, and M. Levine. 1986. Transcriptional control signals of a herpes simplex virus type 1 late (gamma 2) gene lie within bases −34 to +124 relative to the 5′ terminus of the mRNA. Mol. Cell. Biol. 6:3652-3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hook, L. M., J. Huang, M. Jiang, R. Hodinka, and H. M. Friedman. 2008. Blocking antibody access to neutralizing domains on glycoproteins involved in entry as a novel mechanism of immune evasion by herpes simplex virus type 1 glycoproteins C and E. J. Virol. 82:6935-6941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Houseley, J., J. LaCava, and D. Tollervey. 2006. RNA-quality control by the exosome. Nat. Rev. Mol. Cell Biol. 7:529-539. [DOI] [PubMed] [Google Scholar]
- 14.Hughes, R. G., Jr., and W. H. Munyon. 1975. Temperature-sensitive mutants of herpes simplex virus type 1 defective in lysis but not in transformation. J. Virol. 16:275-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ingram, A., A. Phelan, J. Dunlop, and J. B. Clements. 1996. Immediate early protein IE63 of herpes simplex virus type 1 binds RNA directly. J. Gen. Virol. 77:1847-1851. [DOI] [PubMed] [Google Scholar]
- 16.Jean, S., K. M. LeVan, B. Song, M. Levine, and D. M. Knipe. 2001. Herpes simplex virus 1 ICP27 is required for transcription of two viral late (gamma2) genes in infected cells. Virology 283:273-284. [DOI] [PubMed] [Google Scholar]
- 17.Johnson, L. A., and R. M. Sandri-Goldin. 2009. Efficient nuclear export of herpes simplex virus 1 transcripts requires both RNA binding by ICP27 and ICP27 interaction with TAP/NXF1. J. Virol. 83:1184-1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson, P. A., C. MacLean, H. S. Marsden, R. G. Dalziel, and R. D. Everett. 1986. The product of gene US11 of herpes simplex virus type 1 is expressed as a true late gene. J. Gen. Virol. 67:871-883. [DOI] [PubMed] [Google Scholar]
- 19.Koffa, M. D., J. B. Clements, E. Izaurralde, S. Wadd, S. A. Wilson, I. W. Mattaj, and S. Kuersten. 2001. Herpes simplex virus ICP27 protein provides viral mRNAs with access to the cellular mRNA export pathway. EMBO J. 20:5769-5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kohler, A., and E. Hurt. 2007. Exporting RNA from the nucleus to the cytoplasm. Nat. Rev. Mol. Cell Biol. 8:761-773. [DOI] [PubMed] [Google Scholar]
- 21.Lengyel, J., C. Guy, V. Leong, S. Borge, and S. A. Rice. 2002. Mapping of functional regions in the amino-terminal portion of the herpes simplex virus ICP27 regulatory protein: importance of the leucine-rich nuclear export signal and RGG box RNA-binding domain. J. Virol. 76:11866-11879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li, L., L. A. Johnson, J. Q. Dai-Ju, and R. M. Sandri-Goldin. 2008. Hsc70 focus formation at the periphery of HSV-1 transcription sites requires ICP27. PLoS One 3:e1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McCarthy, A. M., L. McMahan, and P. A. Schaffer. 1989. Herpes simplex virus type 1 ICP27 deletion mutants exhibit altered patterns of transcription and are DNA deficient. J. Virol. 63:18-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McLauchlan, J., A. Phelan, C. Loney, R. M. Sandri-Goldin, and J. B. Clements. 1992. Herpes simplex virus IE63 acts at the posttranscriptional level to stimulate viral mRNA 3′ processing. J. Virol. 66:6939-6945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McLauchlan, J., S. Simpson, and J. B. Clements. 1989. Herpes simplex virus induces a processing factor that stimulates poly(A) site usage. Cell 59:1093-1105. [DOI] [PubMed] [Google Scholar]
- 26.Mears, W. E., V. Lam, and S. A. Rice. 1995. Identification of nuclear and nucleolar localization signals in the herpes simplex virus regulatory protein ICP27. J. Virol. 69:935-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mears, W. E., and S. A. Rice. 1998. The herpes simplex virus immediate-early protein ICP27 shuttles between nucleus and cytoplasm. Virology 242:128-137. [DOI] [PubMed] [Google Scholar]
- 28.Mears, W. E., and S. A. Rice. 1996. The RGG box motif of the herpes simplex virus ICP27 protein mediates an RNA-binding activity and determines in vivo methylation. J. Virol. 70:7445-7453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nabel, G. J., S. A. Rice, D. M. Knipe, and D. Baltimore. 1988. Alternative mechanisms for activation of human immunodeficiency virus enhancer in T cells. Science 239:1299-1302. [DOI] [PubMed] [Google Scholar]
- 30.Pearson, A., D. M. Knipe, and D. M. Coen. 2004. ICP27 selectively regulates the cytoplasmic localization of a subset of viral transcripts in herpes simplex virus type 1-infected cells. J. Virol. 78:23-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perkins, K. D., J. Gregonis, S. Borge, and S. A. Rice. 2003. Transactivation of a viral target gene by herpes simplex virus ICP27 is posttranscriptional and does not require the endogenous promoter or polyadenylation site. J. Virol. 77:9872-9884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rice, S. A., and D. M. Knipe. 1988. Gene-specific transactivation by herpes simplex virus type 1 alpha protein ICP27. J. Virol. 62:3814-3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rice, S. A., and D. M. Knipe. 1990. Genetic evidence for two distinct transactivation functions of the herpes simplex virus alpha protein ICP27. J. Virol. 64:1704-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rice, S. A., and V. Lam. 1994. Amino acid substitution mutations in the herpes simplex virus ICP27 protein define an essential gene regulation function. J. Virol. 68:823-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rice, S. A., V. Lam, and D. M. Knipe. 1993. The acidic amino-terminal region of herpes simplex virus type 1 alpha protein ICP27 is required for an essential lytic function. J. Virol. 67:1778-1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rice, S. A., L. S. Su, and D. M. Knipe. 1989. Herpes simplex virus alpha protein ICP27 possesses separable positive and negative regulatory activities. J. Virol. 63:3399-3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roizman, B., D. M. Knipe, and R. J. Whitley. 2007. Herpes simplex viruses, p. 2501-2601. In D. M. Knipe and P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 38.Sacks, W. R., C. C. Greene, D. P. Aschman, and P. A. Schaffer. 1985. Herpes simplex virus type 1 ICP27 is an essential regulatory protein. J. Virol. 55:796-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sampath, P., and N. A. Deluca. 2008. Binding of ICP4, TATA-binding protein, and RNA polymerase II to herpes simplex virus type 1 immediate-early, early, and late promoters in virus-infected cells. J. Virol. 82:2339-2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sandri-Goldin, R. M. 1998. ICP27 mediates HSV RNA export by shuttling through a leucine-rich nuclear export signal and binding viral intronless RNAs through an RGG motif. Genes Dev. 12:868-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sandri-Goldin, R. M. 2004. Viral regulation of mRNA export. J. Virol. 78:4389-4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sedlackova, L., K. D. Perkins, J. Lengyel, A. K. Strain, V. L. van Santen, and S. A. Rice. 2008. Herpes simplex virus type 1 ICP27 regulates expression of a variant, secreted form of glycoprotein C by an intron retention mechanism. J. Virol. 82:7443-7455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sokolowski, M., J. E. Scott, R. P. Heaney, A. H. Patel, and J. B. Clements. 2003. Identification of herpes simplex virus RNAs that interact specifically with regulatory protein ICP27 in vivo. J. Biol. Chem. 278:33540-33549. [DOI] [PubMed] [Google Scholar]
- 44.Soliman, T. M., R. M. Sandri-Goldin, and S. J. Silverstein. 1997. Shuttling of the herpes simplex virus type 1 regulatory protein ICP27 between the nucleus and cytoplasm mediates the expression of late proteins. J. Virol. 71:9188-9197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spear, P. G. 2004. Herpes simplex virus: receptors and ligands for cell entry. Cell Microbiol. 6:401-410. [DOI] [PubMed] [Google Scholar]
- 46.Steiner, I., P. G. Kennedy, and A. R. Pachner. 2007. The neurotropic herpes viruses: herpes simplex and varicella-zoster. Lancet Neurol. 6:1015-1028. [DOI] [PubMed] [Google Scholar]
- 47.Stingley, S. W., J. J. Ramirez, S. A. Aguilar, K. Simmen, R. M. Sandri-Goldin, P. Ghazal, and E. K. Wagner. 2000. Global analysis of herpes simplex virus type 1 transcription using an oligonucleotide-based DNA microarray. J. Virol. 74:9916-9927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thisted, T., D. L. Lyakhov, and S. A. Liebhaber. 2001. Optimized RNA targets of two closely related triple KH domain proteins, heterogeneous nuclear ribonucleoprotein K and alphaCP-2KL, suggest distinct modes of RNA recognition. J. Biol. Chem. 276:17484-17496. [DOI] [PubMed] [Google Scholar]
- 49.Uprichard, S. L., and D. M. Knipe. 1996. Herpes simplex ICP27 mutant viruses exhibit reduced expression of specific DNA replication genes. J. Virol. 70:1969-1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vanacova, S., and R. Stefl. 2007. The exosome and RNA quality control in the nucleus. EMBO Rep. 8:651-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wadd, S., H. Bryant, O. Filhol, J. E. Scott, T. Y. Hsieh, R. D. Everett, and J. B. Clements. 1999. The multifunctional herpes simplex virus IE63 protein interacts with heterogeneous ribonucleoprotein K and with casein kinase 2. J. Biol. Chem. 274:28991-28998. [DOI] [PubMed] [Google Scholar]
- 52.Watson, R. J., and J. B. Clements. 1980. A herpes simplex virus type 1 function continuously required for early and late virus RNA synthesis. Nature 285:329-330. [DOI] [PubMed] [Google Scholar]
- 53.Weinheimer, S. P., and S. L. McKnight. 1987. Transcriptional and post-transcriptional controls establish the cascade of herpes simplex virus protein synthesis. J. Mol. Biol. 195:819-833. [DOI] [PubMed] [Google Scholar]
- 54.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]