Abstract
Highly active antiretroviral therapy (HAART) enables long-term suppression of plasma HIV-1 loads in infected persons, but low-level virus persists and rebounds following cessation of therapy. During HAART, this virus resides in latently infected cells, such as resting CD4+ T cells, and in other cell types that may support residual virus replication. Therapeutic eradication will require elimination of virus from all reservoirs. We report here a comprehensive analysis of these reservoirs in fluids, cells, and tissues in a rhesus macaque model that mimics HAART in HIV-infected humans. This nonhuman primate model uses RT-SHIV, a chimera of simian immunodeficiency virus containing the HIV-1 reverse transcriptase (RT). Methods were developed for extraction, preamplification, and real-time PCR analyses of viral DNA (vDNA) and viral RNA (vRNA) in tissues from RT-SHIV-infected macaques. These methods were used to identify viral reservoirs in RT-SHIV-infected macaques treated with a potent HAART regimen consisting of efavirenz, emtricitabine, and tenofovir. Plasma virus loads at necropsy ranged from 11 to 28 copies of vRNA per ml. Viral RNA and DNA were detected during HAART, in tissues from numerous anatomical locations. Additional analysis provided evidence for full-length viral RNA in tissues of animals with virus suppressed by HAART. The highest levels of vDNA and vRNA in HAART-treated macaques were in lymphoid tissues, particularly the spleen, lymph nodes, and gastrointestinal tract tissues. This study is the first comprehensive analysis of the tissue and organ distribution of a primate AIDS virus during HAART. These data demonstrate widespread persistence of residual virus in tissues during HAART.
Highly active antiretroviral therapy (HAART), consisting of potent combinations of antiretroviral drugs, has been a major advance in treatment of HIV-1 infections (22, 25, 52, 70). HAART enables long-term suppression of plasma viral RNA (vRNA) to undetectable levels for prolonged periods in many persons. This treatment results in a lower rate of emergence of drug-resistant variants, delay or prevention of disease progression, lowered mortality rates, and improved quality of life (16, 52). Nevertheless, HIV-1 inevitably reemerges following cessation of HAART (10, 50). With more sensitive reverse transcriptase PCR (RT-PCR) assays now available, it is clear that low-level viremia persists (26), even in persons on suppressive regimens for more than 7 years (11, 44). Thus, these regimens do not eradicate HIV-1, and therefore life-long treatment is required, which is costly and associated with numerous toxicities.
There are at least two mechanisms by which reservoirs of HIV-1 may be able to persist and evade HAART. The most well-characterized reservoir of HIV-1 is latently infected resting memory CD4+ T cells (10, 12, 17, 18, 47, 48). This T-cell subset is infected with stable integrated provirus that requires cellular activation to produce infectious virus. This latent form of virus is not susceptible to current antiretroviral drugs. Moreover, the pool of latently infected resting CD4+ T cells is very stable, with a half-life of nearly 4 years (17, 51, 57, 59). Latently infected resting CD4+ T cells have been detected in the peripheral blood, gastrointestinal (GI) tract, and lymph nodes of HIV-1-infected individuals and are also likely to exist in other organs containing lymphoid tissue (reviewed in reference 65). Another mechanism for evasion of HAART is residual virus replication (39, 62, 71), which can be expected in reservoirs that do not adequately transport and/or metabolically activate antiretroviral drugs. Some studies have suggested that residual viremia during HAART comes mostly from reactivation of latent virus (13, 35), whereas others have demonstrated viral evolution during HAART, which suggests ongoing virus replication (11, 62; reviewed in reference 54). These potential sources of residual viremia are not mutually exclusive, and the relative proportions of the two may vary in different patients and with different drug regimens.
In addition to resting CD4+ T cells, candidate cellular reservoirs of virus include macrophages, which may express virus for prolonged periods, follicular dendritic cells, which may hold infectious virions on their surfaces, glial cells, endothelial cells, and mast cells (15, 21, 34, 60, 68). Potential anatomical sanctuaries also include the central nervous system (CNS) and the genitourinary tract (reviewed in reference 9). Given the limitations in accessing such tissue in humans, analysis of the locations and dynamics of HIV-1 reservoirs can best be conducted with a robust animal model of AIDS. Many of the potential sanctuaries of virus are difficult to evaluate without invasive methods, which are not ethically feasible with patients during successful HAART. Accordingly, insights into the locations and dynamics of latency/persistence can best be obtained by analysis of an animal model that closely mimics HIV-1 infection and progression to AIDS in humans.
Nonhuman primate models, particularly simian immunodeficiency virus (SIV) infection of macaques, have been the most widely studied of the animal models for AIDS and have contributed significantly to the understanding of important aspects of pathogenesis and means for intervention (reviewed in reference 20). A major advantage is the similarity of macaques, anatomically, physiologically, and immunologically, to humans. Also, SIV is genetically and biologically similar to HIV-1 (reviewed in reference 28). Macaques infected with SIV exhibit a similar fatal immunodeficiency disease to that in humans infected with HIV-1, but the disease course is accelerated, permitting more rapid experimentation (reviewed in reference 20). Studies of SIV-infected macaques support the relevance of this animal model for identifying potential reservoirs of latency/persistence during HAART. Infected tissues and cellular compartments have been identified in animals with high virus loads (VLs), using immunohistochemistry, in situ hybridization, and quantitative image analysis (19, 23, 40). In addition, PCR methods have been used for sensitive detection and quantification of viral nucleic acids in cells and fluids (31, 38, 61). Collectively, these approaches have demonstrated similar tissue distributions and sites of replication in HIV-1-infected individuals (9, 23) and SIV-infected macaques (7, 29). SIV and HIV-1 exhibit similar patterns of localization in the CNS (55, 69, 72), gut-associated lymphoid tissues (27), thymus, and spleen (37). SIV, like HIV-1, infects predominantly CD4+ T cells (32, 67) and macrophages (36, 53). Brain macrophages and microglia are potential reservoirs for both of these viruses in their respective hosts (41, 69). Additional sites for SIV that could serve as reservoirs are Langerhans and dendritic cells in mucosal tissues (33). A report of SIV-infected macaques treated with two nucleoside RT inhibitors (NRTIs) provided evidence that resting CD4+ T cells in peripheral blood, lymph nodes, and spleen, but apparently not the thymus, may serve as potential viral reservoirs (56). More recently, an SIV model was reported that enables studies of reservoirs of persistent SIV during treatment with a HAART regimen (14).
A limitation to SIV as a model for HAART is that SIV is not susceptible to the nonnucleoside RT inhibitors (NNRTIs) that are widely used in AIDS therapy. This limitation has been circumvented by construction of chimeric viruses (RT-SHIVs), which consist of SIV molecular clones in which the SIV RT has been replaced with the HIV-1 RT (2, 63). Two different RT-SHIVs have been used for studies of AIDS therapy in rhesus macaques (6, 30, 43) and pigtail macaques (3). The first RT-SHIV, which is the one we used in this study, was made from the pathogenic molecular clone SIVmac239 (63). This RT-SHIV is susceptible to NRTIs and several NNRTIs, including nevirapine and efavirenz (6, 30). Moreover, RT-SHIV mutants resistant to NNRTIs selected in vitro arise as rapidly as with HIV-1 and contain similar mutations (6, 30). Additionally, the chimeric virus demonstrates high VLs and pathogenesis in rhesus macaques, comparable to those with SIVmac239 (5, 63).
This report describes a comprehensive analysis of reservoirs in RT-SHIV-infected macaques given a suppressive HAART regimen. We optimized PCR amplification methods for the sensitive quantification of both vRNA and viral DNA (vDNA) and used these to measure virus in fluids, cells, and tissues recovered from numerous anatomic sites at necropsy of drug-treated macaques. Importantly, to analyze viral reservoirs, resting CD4+ T cells were recovered from several lymphoid organs and GI tract tissues and were analyzed for vRNA and vDNA. These studies indicate that virus persists in a broad range of tissues during highly suppressive HAART.
MATERIALS AND METHODS
Virus.
Stocks of RT-SHIV were prepared as described previously (30, 43). The RT-SHIV used for these studies had the T-to-C substitution at position 8 of the SIV tRNA primer binding site which is necessary for high-level replication of RT-SHIV in vivo (58).
Animals and sample collection.
Juvenile rhesus macaques (Macaca mulatta) of 7 to 10 months of age (∼2.0 to 3.1 kg) were obtained from the retrovirus-free colony of the California National Primate Research Center (CNPRC). This facility operates according to the Guide for the Care and Use of Laboratory Animals prepared by the Committee on Care and Use of Laboratory Animals of the Institute of Laboratory Animal Resources, National Research Council. When necessary, animals were immobilized with ketamine-HCl (Parke-Davis, Morris Plains, NJ), at 10 mg/kg of body weight, injected intramuscularly. EDTA-anticoagulated blood samples were collected regularly to measure viral and immunologic parameters. Complete blood counts from these samples were done on an automated electronic cell counter (ABX Pentra 60-C+; ABX Diagnostics, Irvine, CA), and differential counts were determined manually.
Preparation and administration of antiretroviral drugs.
Efavirenz (Sustiva) was provided by Bristol-Myers Squibb (Wallingford, CT), and emtricitabine (FTC) and tenofovir (PMPA) were provided by Gilead Sciences (Foster City, CA). Efavirenz was fed at 200 mg per day by mixing the contents of a 200-mg Sustiva capsule into food such as peanut butter sandwiches. Stock solutions of FTC were prepared in phosphate-buffered saline (PBS, pH 7.4). PMPA was suspended in distilled water, with NaOH added to a final pH of 7.0. FTC and PMPA stocks were filter sterilized and stored at 4°C. These NRTIs were administered subcutaneously in the back, with a regimen of 16 mg per kg of body weight once daily for FTC and 30 mg per kg of body weight once daily for PMPA. Drug dosages were adjusted weekly according to body weight. The dose of PMPA was reduced to 15 mg/kg per day after 20 weeks of treatment to reduce any long-term renal toxicity (64). Serum chemistry panels and clinical and histopathological observations did not suggest any detectable toxicity for efavirenz or FTC at the doses used in this study.
Plasma viral RNA levels.
A real-time quantitative RT-PCR (TaqMan) assay with a sensitivity of 50 copies of viral RNA per ml of plasma was used to quantify RT-SHIV RNA (38). For HAART-treated macaques with VLs undetectable by this assay, virus was concentrated by centrifugation of plasma, using a modification of the method of Palmer et al. (45). This ultrasensitive vRNA assay for RT-SHIV has a sensitivity of 1 to 2 copies of vRNA per ml of the original plasma sample (our unpublished data).
Sequence analysis of rebound virus.
Nucleic acid preparation and sequence analysis of the RT-encoding region of RT-SHIV isolates of rebound virus were performed as previously described (43).
Lymphocyte phenotyping by four-color flow cytometry.
T-lymphocyte antigens were detected by direct labeling of whole blood with peridinin chlorophyll protein-conjugated anti-human CD8 (clone SK1), phycoerythrin-conjugated anti-human CD4 (clone M-T477), fluorescein-conjugated anti-human CD3 (clone SP34), and allophycocyanin-conjugated anti-human CD20 (clone L27) (all from BD, Franklin Lakes, NJ). Red blood cells were lysed and samples were fixed in paraformaldehyde by use of a Coulter Q-prep system (Coulter Corporation, Hialeah, FL). Lymphocytes were gated by forward and side light scatter and were then analyzed with a FACSCalibur flow cytometer (BD). CD4+ and CD8+ T lymphocytes were defined as CD3+ CD4+ and CD3+ CD8+, respectively. B lymphocytes were CD3− CD20+.
Necropsy and tissue collection.
Macaques were sedated, and cerebrospinal fluid (CSF) was collected. Animals were then euthanized with a barbiturate overdose, and peripheral blood was collected. Subsequently, each animal was perfused with physiological saline, and a complete necropsy was performed by staff veterinary pathologists at the CNPRC. Each necropsy included gross and microscopic examination of all tissues and organs. Tissues were collected, and separate samples were fixed in 10% neutral buffered formalin and embedded in paraffin, immersed in OCT preservative for subsequent sectioning and histopathologic analysis, snap-frozen in liquid nitrogen, and collected directly in lysis buffers for analysis of vRNA and vDNA (see below). The list of tissues is shown in Table 1. Sterile cell suspensions were prepared from lymph nodes, the spleen, and portions of the GI tract. A portion of each cell suspension was treated with lysis buffer and stored for vRNA and vDNA analysis, and another portion was cryopreserved in cell culture freezing medium, which consists of 90% fetal bovine serum (FBS) (Omega Scientific) and 10% dimethyl sulfoxide (DMSO).
TABLE 1.
Tissue virus loadsa
| Tissue and compartment | VL (copies/106 cells)b |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mmu 35389 (received HAART; plasma VL, 19; CSF VL, <50) |
Mmu 35339 (received HAART; plasma VL, 11; CSF VL, <50) |
Mmu 35343 (received HAART; plasma VL, 28; CSF VL, <50) |
Mmu 35342 (received HAART; plasma VL, 20; CSF VL, <50) |
Mmu 35349 (received HAART; plasma VL, 17; CSF VL, <50) |
Mmu 35483 (no HAART; plasma VL, 128,084; CSF VL, 18,681) |
|||||||
| DNA | RNA | DNA | RNA | DNA | RNA | DNA | RNA | DNA | RNA | DNA | RNA | |
| Lymphoid tissues | ||||||||||||
| Spleen | 290 | 3 | 5 | 3 | 750 | 420 | 370 | 67 | 540 | 33 | 4,200 | 45,000 |
| Mesenteric lymph node | 340 | 8 | 1,100 | 75 | 4,000 | 46 | 520 | 220 | 890 | 98 | 13,000 | 4,800 |
| Axial lymph node | 1,200 | 17 | 77 | 32 | 480 | 360 | 1,600 | 79 | 640 | 710 | 2,300 | 97,000 |
| Inguinal lymph node | 510 | 26 | 89 | 11 | 6,800 | 130 | 110 | 300 | 290 | <1 | 6,100 | 200,000 |
| Iliac lymph node | 440 | 8 | 87 | 46 | 2,400 | 260 | 310 | 6 | 460 | 130 | 10,000 | 67,000 |
| Cervical lymph node | 1,200 | 19 | 510 | 100 | 49 | 340 | 350 | 11 | 320 | 79 | 2,500 | 12,000 |
| Thymus | 45 | 2 | 160 | <1 | 4,800 | 140 | 31 | <1 | 74 | 94 | 140 | 430 |
| Bone marrow | 10 | <1 | <1 | <1 | 340 | <1 | 18 | <1 | <1 | <1 | 17 | 90 |
| Tonsil | 15 | <1 | 720 | 42 | 590 | 670 | 210 | 38 | 430 | 110 | 4,000 | 21,000 |
| GI tract | ||||||||||||
| Duodenum | 230 | 2 | 170 | 46 | 1,000 | 32 | <1 | 5 | 170 | 59 | 5,000 | 150,000 |
| Jejunum | 140 | 8 | 62 | <1 | 200 | 7 | 8 | 8 | 44 | 13 | 330 | 680 |
| Ileum | 88 | 3 | 120 | <1 | 390 | <1 | 140 | 23 | 3,900 | 76 | 93 | 180 |
| Cecum | 3 | <1 | <1 | <1 | 140 | 28 | 64 | 2 | 17 | 150 | 890 | 160 |
| Colon | 70 | <1 | 58 | 8 | 360 | 21 | <1 | <1 | 23 | 190 | 170 | 6,200 |
| Neurological tissues | ||||||||||||
| Cerebrum | <1 | <1 | <1 | <1 | <1 | <1 | 6 | <1 | <1 | <1 | 5 | <1 |
| Cerebellum | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | 3 | <1 | 5 |
| Choroid plexus | 15 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | 5 | <1 | 10 | 13 |
| Cervical spinal cord | 7 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | 4 |
| Lumbar spinal cord | 4 | <1 | 4 | <1 | 220 | <1 | 36 | <1 | <1 | <1 | <1 | 6 |
| Reproductive tissues | ||||||||||||
| Testes | 13 | <1 | 17 | <1 | 10 | <1 | 0 | <1 | 21 | <1 | 88 | 620 |
| Seminal vesicles | 20 | <1 | 0 | <1 | 30 | <1 | 36 | <1 | 4 | <1 | 60 | 150 |
| Prostate | 4 | <1 | 36 | <1 | 180 | <1 | 54 | <1 | 10 | 35 | 160 | 300 |
| Penis | 23 | <1 | <1 | <1 | <1 | <1 | 0 | <1 | <1 | <1 | <1 | 3 |
| Other | ||||||||||||
| Kidney | 3 | <1 | 7 | <1 | <1 | <1 | <1 | <1 | 1 | <1 | 76 | 280 |
| Liver | 6 | <1 | 9 | <1 | <1 | <1 | <1 | <1 | 25 | <1 | 7 | 250 |
| Heart | 9 | <1 | 3 | <1 | 9 | <1 | 17 | <1 | 3 | <1 | 22 | 130 |
| Lung | 3 | <1 | 1 | <1 | 18 | <1 | 14 | <1 | 2 | 9 | 340 | 3,100 |
| Bladder | 6 | <1 | 42 | <1 | <1 | <1 | 11 | <1 | 1 | <1 | 150 | 1,200 |
All tissue values are the means for two different tissue samples of each tissue. All values are normalized to 2 × 106 copies of cellular IL-2 DNA measured in each sample.
Virus loads in plasma and CSF are given as copies of RT-SHIV RNA per ml of fluid.
Enrichment of resting CD4+ T lymphocytes.
At the time of necropsy, blood and tissue samples from mesenteric lymph node (MLN), jejunum, and spleen were taken from each macaque. Single-cell suspensions were generated from MLN and spleen by liberating the lymphocytes with scalpels and passing them through a 70-μm nylon cell strainer (BD). Peripheral blood mononuclear cells (PBMC) were isolated from blood by use of Histopaque gradients following standard procedures. The single-cell suspensions from jejunum were obtained using a previously reported method (66). Briefly, 25- to 30-cm sections of jejunum were cut into approximately 0.5-cm2 pieces, using opposing scalpels. Next, the tissue pieces were transferred to medium containing Hanks buffered saline solution with 1 mM EDTA (Sigma) and 0.5 mM dithiothreitol (Sigma) and incubated at 37°C with constant agitation for 30 min. Following each of the incubations, the solution was passed through a steel mesh strainer, and the flowthrough, primarily intraepithelial lymphocytes (IELs), was collected, washed, and stored on ice in RPMI 1640 plus 5% FBS. After two or three incubations, the remaining tissue was cut into approximately 2- to 3-mm2 pieces and incubated in RPMI 1640 containing 0.5 mg/ml type II collagenase (Sigma). The flowthrough, mainly lamina propria lymphocytes (LPLs), was collected, washed, and stored on ice. After passage of the cell suspensions through 60-ml syringes loosely packed with glass wool to remove excess mucus, the IELs and LPLs were isolated by Percoll gradient centrifugation. Cells were counted, samples were set aside for flow cytometry and for PCR analysis of vDNA and vRNA in presorted cell suspensions, and remaining IELs and LPLs were then combined.
Resting CD4+ T lymphocytes were enriched from approximately 100 × 106 lymphocytes in single-cell suspensions of mesenteric lymph node, spleen, PBMC, and jejunum (IELs and LPLs), using negative selection with magnetic beads. A BD I-Mag CD4 T-lymphocyte enrichment set (BD, Franklin Lakes, NJ) was used according to the manufacturer's protocol in order to remove cells expressing the following markers: CD8, CD11b/Mac-1, CD16, CD19, CD36, CD41a, CD56, CD123, CD235a, and γδ T-cell receptor (TCR). Anti-HLA-DR and anti-CD20 microbeads (Miltenyi Biotec, Auburn, CA) were added according to the manufacturer's protocol in order to remove activated CD4+ T lymphocytes and B cells, respectively. Following resting CD4+ T-cell enrichment, separate aliquots were placed in ABI lysis buffer (Applied Biosystems, Foster City, CA) for nucleic acid analysis, stored in cell freezing medium, and analyzed by flow cytometry to estimate the purity of the cell populations.
Detection of vDNA and vRNA in cells and tissues by PCR.
Levels of vRNA in HAART-treated animals are below the level of detection by conventional methods (immunohistochemistry and in situ hybridization). Accordingly, we developed methods for real-time PCR, using a preamplification procedure similar to that reported for detection of low-copy-number RNA transcripts (8). Preamplification of RT-SHIV DNA and RNA and of cellular interleukin-2 (IL-2) DNA and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) RNA was validated as described previously (8). Real-time PCR and RT-PCR, for RT-SHIV DNA and RNA, respectively, were performed as described previously (38). The primers used in these assays amplify a region of gag that is present in viral RNA and in full-length RT-SHIV transcripts; this region is not present in multiply spliced mRNA or short transcripts that are hypothesized to be made during viral latency (1). Thus, this assay provides a preliminary test to identify sites of virus replication.
Extraction and preamplification of viral DNA and RNA were performed at the UC Davis Lucy Whittier Molecular and Diagnostics Core Facility. Briefly, tissue samples in ABI lysis buffer in 96-well plates were treated with proteinase K, and tissues were disrupted by addition of two metal beads to each well and processed through an ABI 2000 GenoGrinder (Applied Biosystems, Foster City, CA). RNA and DNA were prepared from these lysates, using an Applied Biosystems model 6100 nucleic acid prep station. For RNA analysis, 100 μl of lysate was transferred to a 1.5-ml microcentrifuge tube on ice containing 6 μl of 5 M NaCl and 10 μl of glycogen (5 mg/ml). After mixing, 300 μl of absolute ethanol was added to each tube, followed by gentle mixing. Tubes were then stored at −20°C overnight. The ethanol precipitate was pelleted by centrifugation, and the pellet was washed with 70% ethanol. Ethanol was removed and residual ethanol allowed to evaporate. Pellets were resuspended in 20 μl of water, treated with DNase (10 U, 37°C for 15 min), and heated to 85°C for 5 min to inactivate DNase. Aliquots were tested for DNA contamination by PCR (no RT), using primer pairs for cellular IL-2 DNA. The RNA was used to synthesize cDNA in reaction mixtures with RT (SuperScript III), random hexamer primers, and RNase inhibitor. DNA was extracted from 100 μl of lysate and precipitated with cold ethanol.
Due to extremely low levels of vRNA and vDNA in tissues from HAART-treated humans or animals, we used a sensitive assay that includes a preamplification step prior to real-time PCR. Preamplification of DNA or the cDNA made from RNA was conducted as described by Baumgarth et al. (8), with conditions optimized for our primers for SIV and cellular genes. Preamplification was performed for 20 cycles with Advantage 2 DNA polymerase. For analysis of cDNA, PCR amplification was done with primer pairs that detect the cellular GAPDH gene and SIV gag. For analysis of DNA, primers for the cellular IL-2 gene and SIV gag were used. All of these reactions were validated with known standards. The threshold cycle (CT) values obtained by TaqMan RT-PCR analysis of the preamplified material were linear with respect to cycle number.
Real-time PCR was performed, and levels of DNA or RNA were determined with standard curves as previously described (38, 43). For samples that were quantified after preamplification, real-time PCR of IL-2 DNA was performed both before and after preamplification to determine the preamplification factor. After real-time PCR, values were divided by this preamplification factor to calculate levels of viral and cellular DNA in the tissue samples.
For detection of RT-SHIV gag, pol, and env sequences, nested PCR amplifications were performed with cDNAs obtained from tissues as described above. The two primer sets for gag were the same as those for the preamplification step and the real-time PCR assay (38). The first round of nested PCR amplification of gag was done with primers SIV 456F and SIV 756R, and the second round used primers SIV 510F and SIV 592R (38). The primer sets for pol were for the HIV-1 RT region of RT-SHIV. The first round of nested PCR amplification for pol used primers HXB2-EcoRV and HXB2-3341R, and the second round used HXB2-3145 and HXB2-3253R (43). For env, the first-round nested primers were 239-8961 (5′-CTC TTG ACT TGG CTA TTC AGC AAC) and 239-9266R (5′-CTC CAT GGA GTA TTC ATA TAC TG), and the second-round primers were 239-8997 (5′-ATC GAG AGT ATA CCA GAT CCT C) and 239-9064R (5′-CCA CCC ATA TTG TAG GTA GGT). Nested PCR was performed using GoTaq DNA polymerase (Promega, Madison, WI) with green buffer under the manufacturer's recommended conditions. Reaction mixtures contained primers at a concentration of 0.4 μM and a 200 μM concentration of each of the four deoxynucleoside triphosphates. A 2-μl aliquot of cDNA was used for the first round of PCR amplification, and 2 μl of the first-round product was used in the second round. To test for potential DNA contamination, nested PCR amplification was done on samples of the no-RT controls from the cDNA synthesis reaction. First-round PCR amplification conditions were 95°C for 2 min, followed by 40 cycles of 95°C for 30 s, 50°C for 30 s, and 72°C for 23 s and ending with an extension step at 72°C for 5 min. Conditions for the second round were 95°C for 2 min, followed by 30 cycles of 95°C for 20 s, 50°C for 20 s, and 72°C for 7 s and ending with an extension step at 72°C for 5 min. Second-round PCR amplification products were analyzed by electrophoresis on a 2% agarose gel and were imaged with a Gel Logic 100 imaging system using Kodak 1D software (Eastman Kodak Company, Rochester, NY). Size markers were All Purpose Lo DNA markers (Bionexus, Oakland, CA).
RESULTS
Efficacy of HAART regimen in macaques.
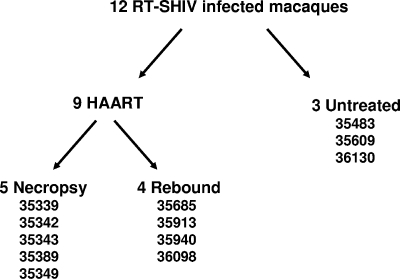
The experimental design for this study and specific animals in each group are shown in Fig. 1. Twelve juvenile rhesus macaques were each inoculated intravenously with cell-free RT-SHIV as described previously (43), and all became persistently infected, with VLs reaching a peak between 2 and 4 weeks postinfection (Fig. 2). At 6 weeks postinfection, nine animals were started on HAART consisting of efavirenz, tenofovir, and emtricitabine. The other three RT-SHIV-infected macaques were maintained as untreated controls (no drugs). Data from an additional 10 control (no drugs), RT-SHIV-infected animals from two other studies were included in this study. The mean VL for this group of 13 RT-SHIV-infected control animals is shown in Fig. 2. The three untreated control animals in this study had VLs within the range reported in previous studies (30, 43).
FIG. 1.
Experimental design for treatment and analysis of RT-SHIV-infected macaques.
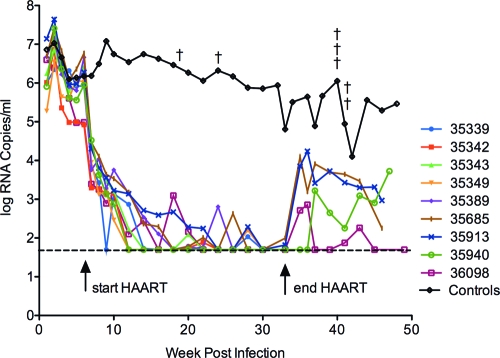
FIG. 2.
Plasma virus RNA in RT-SHIV-infected rhesus macaques treated with efavirenz plus emtricitabine plus tenofovir. Plasma VLs in the nine individual animals of the drug-treated group are shown. The arrow at 6 weeks indicates the time that drug therapy was initiated, and the arrow at 32 weeks indicates the time at which drug therapy was stopped. The mean VL for the control group was obtained from a total of 13 RT-SHIV-infected macaques that received no HAART. A cross indicates the necropsy of a control animal and a reduction of the total number of animals in the group from that point forward. The dotted line indicates the lower limit of detection of the VL assay (50 copies of viral RNA per ml of plasma).
Plasma VLs in the HAART-treated macaques rapidly declined, with a pattern similar to the decline in HIV-1 loads observed with HAART in human patients (46) and to that in our previous report of RT-SHIV-infected macaques treated with efavirenz plus lamivudine plus tenofovir (43). By 24 weeks postinoculation (18 weeks of therapy), plasma VLs were below the level of detection with our standard assay (<50 copies per ml) in all drug-treated animals, and VLs remained suppressed for the duration of therapy, with only occasional blips (Fig. 2). Linear regression analysis of the VL decline to fewer than 50 copies per ml was determined to be biphasic (P = 0.02), using the Joinpoint regression program, version 3.4.1 (Statistical Research and Applications Branch, National Cancer Institute). Sufficient amounts of plasma for measurement of VLs below 50 copies per ml of plasma were not obtained from these animals during the course of infection, except at necropsy. However, a longitudinal analysis of this further suppression will be reported for another group of HAART-treated macaques (our unpublished data).
Although plasma vRNA was not detected with our standard VL assay, it was detected with a more sensitive VL assay and larger volumes of plasma available at necropsy. The five HAART-treated animals that were analyzed at necropsy had VLs ranging from 11 to 28 copies of vRNA per ml of plasma (Table 1).
After 26 weeks of HAART, drug therapy was stopped in four of the treated macaques (Fig. 1), and these animals were maintained for a 12-week period of observation. After cessation of HAART, VLs rebounded in all four animals (Fig. 2). Consensus sequence analysis (43) of the rebound virus did not reveal detectable levels of mutations in RT that are known to confer resistance to efavirenz, emtricitabine, or tenofovir (data not shown). These results are in agreement with our previous study of viral rebound after cessation of HAART (43) and indicate that the rebound virus is not primarily drug resistant. The other five HAART-treated animals were euthanized, and necropsies were performed (Fig. 1) for pathological and histopathologic analysis and for collection of fluids, tissues, and cells for analysis of vRNA and vDNA.
In the live phase, all of the HAART-treated macaques in this report showed normal weight gains and did not present with significant clinical findings. Hematologic values, including numbers and ratios of CD4+ and CD8+ T cells, remained in the normal reference range for all of these animals (data not shown).
Necropsy analysis included both gross pathology and histopathology. One of the three RT-SHIV-infected, untreated macaques, Mmu 35609, exhibited signs of the lymphoproliferative phase of simian AIDS, including prominent lymphoid tissue in several anatomic sites and secondary follicles in spleen, lymph nodes, and bone marrow. Cryptosporidium and Trichomonas in the GI tract provided evidence of an immunocompromised state in Mmu 35609. The other two animals (Mmu 35483 and Mmu 36130) in this group of RT-SHIV-infected macaques had not developed signs of progression to simian AIDS at the time of necropsy. Also, all other organs examined appeared normal.
For the five RT-SHIV-infected macaques that were euthanized for necropsy during HAART, pathological analysis revealed no opportunistic infections and histologic analysis showed that lymphoid organs were normal. Three of the four RT-SHIV-infected macaques that received HAART for 26 weeks followed by removal of this drug regimen for a 12-week period (Mmu 35685, Mmu 35913, and Mmu 36098) showed no pathological signs of immunodeficiency and no abnormalities of major organs. However, Mmu 35940, a member of this group, entered the lymphodepletion stage of simian AIDS, as revealed by depletion of lymphocytes in the lymph nodes, spleen, and GI tract (data not shown).
Tissue distribution of vRNA and vDNA.
Extraction procedures and PCR/RT-PCR assays for detection and quantification of RT-SHIV DNA and RNA in tissues from infected macaques were developed to measure tissue levels of vDNA and vRNA (see Materials and Methods). IL-2 DNA levels were used to normalize vDNA and vRNA to a per cell basis, and GAPDH RNA provided a measure of RNA yield and integrity. In preliminary studies, we determined that vRNA and vDNA levels in HAART-treated animals were below the level of detection with our standard assays (data not shown). Therefore, a preamplification step was introduced for detection of low-copy-number RNAs (8). Preamplification efficiencies for SIV gag and cellular IL-2 DNA were nearly identical, as determined from the slopes of the lines for CT values versus cycles of preamplification (Fig. 3). Linear regression analysis determined that the slopes of the two lines were not significantly different (P > 0.77). To assess the reproducibility of preamplification and TaqMan assays, we performed duplicate determinations for vDNA, cellular IL-2 DNA, vRNA, and cellular GAPDH RNA with a preparation of PBMC from a control (no drug) RT-SHIV-infected macaque. In all cases, variation was <2.5-fold (data not shown). The number of target cells for virus relative to connective tissue and nontarget cells may vary considerably within an organ. Accordingly, to address the issue of sampling variation, we collected four random pieces from each of three different organs from an RT-SHIV-infected macaque that was not treated with HAART. Nucleic acids from these independent samples were extracted, preamplified, and analyzed for vDNA and vRNA by real-time PCR. The amount of variation in random samples from the same organ was <3.5-fold for vDNA (Fig. 4A) or vRNA (Fig. 4B). One-way analysis of variance (ANOVA) determined no significant difference within each organ sampled (P > 0.05).
FIG. 3.
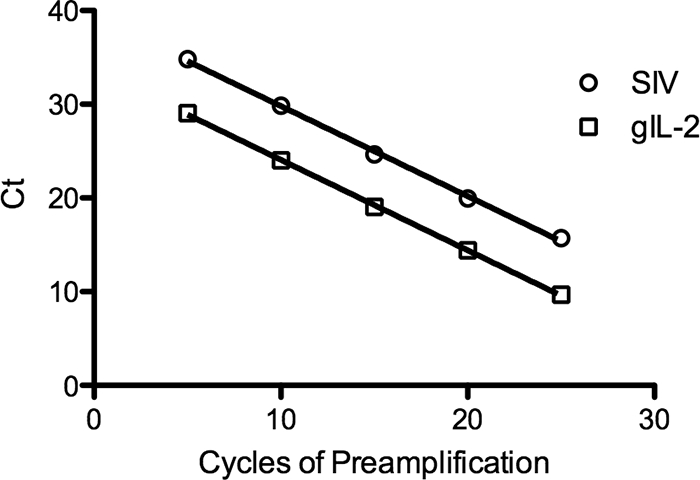
Preamplification of SIV and cellular IL-2 DNAs. SIV gag and genomic IL-2 (gIL-2) DNAs were preamplified by PCR for 5, 10, 15, 20, and 25 cycles. Quantitative PCR analysis was then performed on these samples. Each point represents the average CT value for three replicates. The greatest standard deviation of any point was 0.28. Error bars are too small to be visible.
FIG. 4.
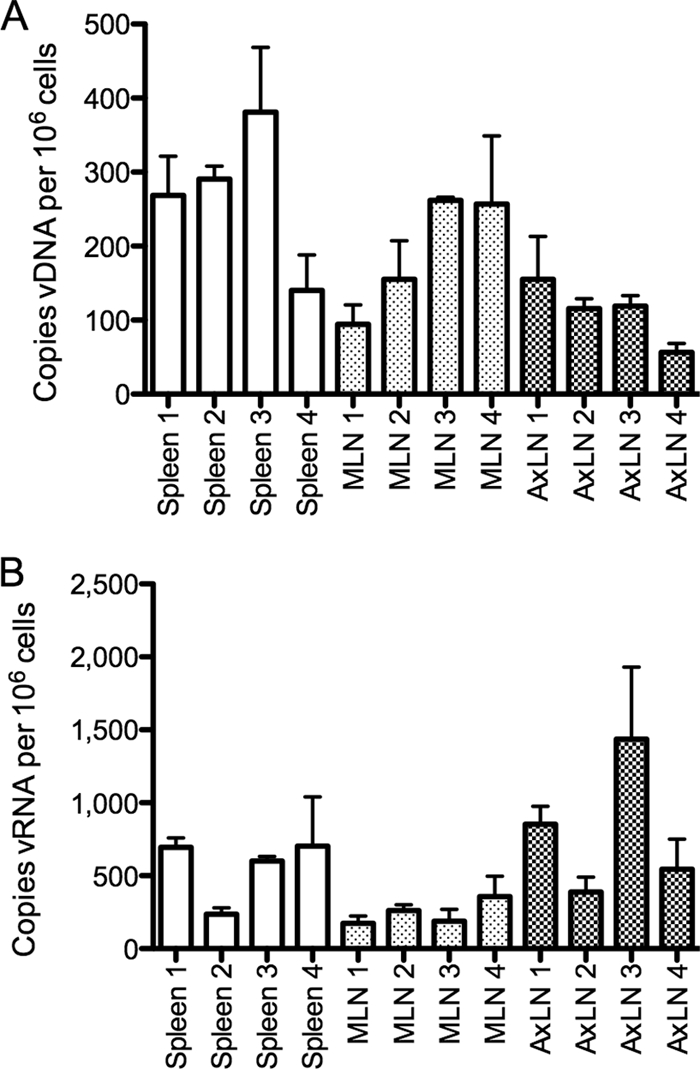
Biological variation of vDNA and vRNA levels in multiple samples of the same tissues. (A) vDNA; (B) vRNA. Four samples were taken from each of the following tissues of a single RT-SHIV-infected macaque (Mmu 36606; plasma VL, 4,300 copies vRNA per ml): spleen, a mesenteric lymph node (MLN), and an axillary lymph node (AxLN). Nucleic acid extraction, preamplification, and real-time PCR for measuring levels of vRNA and vDNA were performed independently for each tissue sample, in duplicate. Each bar represents the mean for two replicates, with an error bar for the standard deviation.
One of the control RT-SHIV-infected macaques (not receiving HAART) was randomly chosen for analysis of vDNA and vRNA in tissues (Table 1). In this animal (Mmu 35483), the highest levels of vDNA and vRNA were in lymphoid tissues, namely, the spleen, lymph nodes, tonsils, and GI tract tissues. In most of these tissues, vRNA levels were substantially higher than levels of vDNA. Lower levels of vDNA and vRNA were detected in neural and reproductive tissues and in nontarget tissues (kidney, liver, heart, lung, and bladder). Virus detected in these nontarget tissues may be due to trace amounts of blood remaining in the tissues from necropsy.
In HAART-treated macaques, tissue levels of vDNA and vRNA were much lower than those in tissues from the animal that received no drugs (Table 1). The highest levels of vDNA and vRNA in HAART-treated macaques were in lymphoid tissues, particularly the spleen and lymph nodes. Lower levels of vDNA and vRNA (per 106 cells) were measured in GI tract tissues. This may be due, at least in part, to a lower percentage of CD4+ T cells in GI tract tissues (Table 2). The spleen and all lymph nodes tested from each of the HAART-treated macaques showed detectable levels of vRNA (Table 1). Some animals also contained vRNA in the thymus (3 of 5 animals) and tonsils (4 of 5 animals). None of the HAART-treated macaques had detectable vRNA in bone marrow. All of the lymphoid tissues from HAART-treated animals had vDNA, except for the bone marrow from two macaques. Most GI tract tissues of the HAART-treated macaques also contained detectable vRNA and vDNA, but the levels were generally lower on a per cell basis than those in the lymph nodes and spleen. The exception to this was one of the HAART-treated macaques, Mmu 35349, which had higher levels of vRNA and vDNA in GI tract tissues than the other members of the HAART-treated group. In contrast to the case for the control macaque (no HAART), levels of vDNA were considerably higher than vRNA levels in the lymphoid and GI tract tissues. Therefore, these tissues probably harbor transcriptionally inactive provirus, such as latent or defective proviruses.
TABLE 2.
Virus loads in single-cell suspensions and resting CD4+ T cellsa
| Monkey (treatment) | Tissue | Single-cell suspensiond |
Resting CD4+ T cellsd |
||||||
|---|---|---|---|---|---|---|---|---|---|
| % CD4+ cellsb | % HLA-DR+ cellsc | DNA level | RNA level | % CD4+ cellsb | % HLA-DR+ cellsc | DNA level | RNA level | ||
| Mmu 35339 (HAART) | PBMC | 30.2 | 12.4 | 78 | <1 | 90.5 | 0.92 | 92 | <2 |
| MLN | 60 | 14.6 | 47 | <1 | 97.5 | 0.12 | 340 | <1 | |
| Spleen | 30.9 | 26.3 | 31 | <1 | 90.4 | 1.3 | 91 | <1 | |
| Jejunum | 36.3 | 2.8 | * | * | 92.2 | 1.1 | * | <4 | |
| Mmu 35342 (HAART) | PBMC | 45.6 | 4.3 | 86 | 150 | 93.6 | 0.59 | 170 | <1 |
| MLN | 50.8 | 16.4 | 90 | <1 | 96.7 | 0.3 | 230 | 40 | |
| Spleen | 30.7 | 14.9 | 92 | <1 | 90.6 | 0.78 | * | * | |
| Jejunum | 18.5 | 4 | <2 | <1 | 84.1 | ND | 53 | * | |
| Mmu 35343 (HAART) | PBMC | 53.4 | 7.2 | 71 | <2 | 93.8 | 0.47 | 160 | 4 |
| MLN | 58.4 | 23.5 | 130 | <2 | 97.6 | 0.26 | 230 | 12 | |
| Spleen | 35 | 36.5 | 95 | <2 | 96 | 0.28 | 190 | 4 | |
| Jejunum | ND | ND | * | * | 79.6 | 0.22 | 750 | 14 | |
| Mmu 35349 (HAART) | PBMC | 47.6 | 10.5 | 180 | <2 | 92.4 | 1.12 | 170 | <1 |
| MLN | 37.8 | 30.4 | 230 | 10 | 94.4 | 0.37 | 460 | <2 | |
| Spleen | 26.2 | 37.1 | 87 | <2 | 94.9 | 0.065 | 470 | * | |
| Jejunum | 16.4 | 4.25 | 100 | * | 81.4 | 0.24 | 490 | * | |
| Mmu 35389 (HAART) | PBMC | 41.9 | 13.7 | 200 | <4 | 92.5 | 2.8 | 79 | 10 |
| MLN | 41.9 | 23.9 | 82 | <1 | 91.5 | 1 | 170 | <3 | |
| Spleen | 28.4 | 43.6 | 34 | <1 | 89 | 4.9 | 310 | <5 | |
| Jejunum | 35.7 | 4.3 | * | * | 92.3 | 1.35 | 29 | <3 | |
| Mmu 35483 (no HAART) | PBMC | ND | ND | 370 | 1040 | ND | ND | 470 | 390 |
| MLN | ND | ND | 375 | 390 | ND | ND | 550 | 870 | |
| Spleen | ND | ND | 290 | 1000 | ND | ND | 2400 | 9000 | |
| Jejunum | ND | ND | * | * | ND | ND | * | * | |
All tissue values are the means for two different tissue samples. All VL values are normalized to 2 × 106 copies of cellular IL-2 DNA measured in each sample.
Percentage of CD3+ cells expressing CD4 and negative for CD8.
Percentage of lymphocytes expressing HLA-DR.
*, no sample or low nucleic acid level in sample; ND, not determined.
Low levels of vDNA were consistently detected in tissues from the reproductive tract of HAART-treated macaques (these were sexually immature males), but vRNA was detected in only one prostate sample (Table 1). There was little or no vRNA or vDNA detected in CSF or neural tissues from these HAART-treated macaques (Table 1).
With the method of nested RT-PCR amplification, we were able to detect gag, pol, and env sequences in tissue samples from HAART-treated animals (Fig. 5). Thus, the vRNA detected was most likely full length, implying the synthesis of viral genomes.
FIG. 5.
Analysis of viral transcripts in tissues. Axillary lymph nodes were obtained at necropsy from one macaque after rebound from HAART (Mmu 35913; plasma viral load, 930 copies/ml) and from another macaque during HAART (Mmu 35343; plasma viral load, 28 copies/ml). PCR amplification was performed to detect gag, pol, and env RNAs. A control reaction without RT (mock cDNA sample) was included for each animal. Lane M, Bionexus All Purpose Lo marker.
Analysis of vRNA and vDNA in resting CD4+ T cells.
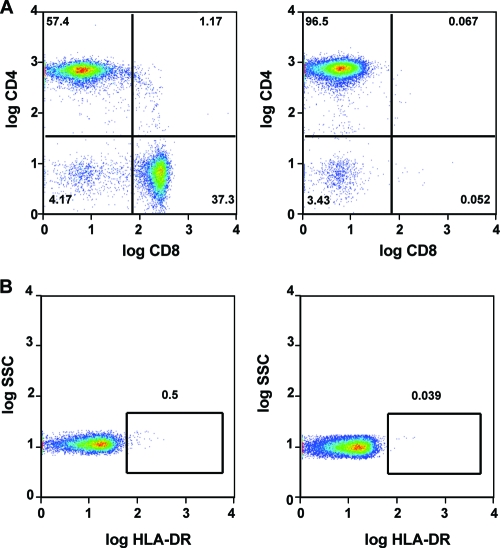
Single-cell suspensions were isolated from necropsy tissues of mesenteric lymph node, spleen, jejunum, and PBMC from the HAART-treated macaques and a control (no drug treatment) RT-SHIV-infected macaque. Resting CD4+ T lymphocytes were enriched from the single-cell suspensions by negative selection to prevent cell activation during the process. Magnetic beads were used to remove cells expressing the following markers: CD8, CD11b/Mac-1, CD16, CD19, CD36, CD41a, CD56, CD123, CD235a, and γδ TCR. Activated CD4+ T lymphocytes and B cells were removed with anti-HLA-DR and anti-CD20 microbeads, as described in Materials and Methods. Although the original depletion cocktail included CD19 beads, B-cell removal was significantly enhanced when CD20 beads were added. Flow cytometry analysis showed that this enrichment produced populations that were predominantly resting CD4+ T cells (Fig. 6 and Table 2) and had greatly reduced levels of activated HLA-DR+ T cells (Table 2). Resting CD4+ T-cell populations in PBMC, spleen, MLN, and jejunum ranged from 18 to 60% of pre-enrichment cell suspensions. Postenrichment suspensions contained 80 to 97% resting CD4+ T cells.
FIG. 6.
Enrichment of resting CD4+ T cells. MLN lymphocytes from a representative animal (Mmu 35389) were enriched for CD4+ T cells by negative magnetic bead selection. Additional HLA-DR beads were included to remove activated T cells. CD20 beads were added to remove B cells, as the original bead cocktail containing CD19 beads was ineffective. (A) Removal of CD8+ T cells after enrichment. (B) Removal of HLA-DR+ CD4+ T cells. Left panels show samples before enrichment; right panels show samples after enrichment.
RT-SHIV DNA was detected in both the single-cell suspensions and the enriched resting CD4+ T-cell populations from all macaques, but at a lower level in the HAART-treated group than in the control (Table 2). Levels of RT-SHIV DNA were generally higher (per 106 total cells) in all of the enriched resting CD4+ cell populations than in the single-cell suspensions from which they were derived. However, there was little or no enrichment relative to numbers of CD4+ T cells. Surprisingly, there was little or no vRNA detected in either the cell suspensions or the enriched populations of resting CD4+ T lymphocytes from the HAART-treated group. Most of these tissues had been positive for vRNA in assays of extracts obtained directly from lysis of the tissue pieces (Table 1); accordingly, these data suggest that vRNA was lost during preparation of the cell suspensions. There was not a general loss of intracellular RNA in these samples, because the cell suspensions and enriched populations had normal ratios of cellular GAPDH RNA to IL-2 DNA. However, extracellular vRNA (such as RNA in virions) or vRNA associated with dendritic cells could be lost in the Histopaque gradients and by the washing steps used to prepare the single-cell suspensions. Therefore, it is likely that most of the vRNA detected in tissue pieces is either extracellular virus or is contained in cells that are lost during preparation of cell suspensions with Histopaque gradients.
DISCUSSION
This study is the first comprehensive analysis of the tissue and organ distribution of a primate AIDS virus during HAART. The HAART regimen studied in these RT-SHIV-infected macaques, efavirenz, emtricitabine, and tenofovir, is commonly used in humans and is one of the most suppressive and durable combinations for AIDS therapy (24, 49). As in the majority of humans receiving this HAART regimen, all of the macaques in this study remained healthy, with no adverse clinical effects during the treatment period. Virus loads in plasmas of treated macaques declined rapidly, with a biphasic curve similar to that for HIV-1 levels in humans initiating HAART (46), and low-level viremia (<50 copies per ml) persisted in plasmas of all treated macaques. Importantly, these drugs were given up to and including the day of necropsy for the animals used to analyze distribution of virus. Therefore, the results represent viral distribution during HAART. Levels of vRNA in these HAART-treated animals were highest (on a per cell basis) in the spleen and lymph nodes. In general, levels of vRNA in these tissues were suppressed less by HAART than the plasma VLs (relative to tissue and plasma VLs, respectively, of the control RT-SHIV-infected macaque that did not receive HAART). Lower levels of vRNA remained in tissues from the GI tract and in the CSF. Viral DNA was most prevalent in lymphoid and GI tract tissues but was also detected in the CNS and the reproductive tract. Viral RNA was detected by real-time RT-PCR of a gag region, but gag, pol, and env sequences were found in similar proportions by nested PCR of tissue samples from HAART-treated animals. Thus, the vRNA detected was most likely full length.
For HIV-1-infected patients receiving HAART, there is evidence that plasma viremia does not originate from blood CD4+ T cells (4). Therefore, it is critical to evaluate effects of HAART and other therapeutic strategies on tissue virus levels. Within tissues, it is also essential to evaluate effects on specific cell types (macrophages, microglia, dendritic cells, and mast cells) that may harbor virus. Some of these cells may be less accessible to antiviral drugs. The RT-SHIV-macaque model will enable future studies on simultaneous measurements of both VL and drug levels in the same tissue or population of cells. Importantly, this approach will identify tissue sites of poor drug penetration and facilitate strategies to eliminate residual virus replication.
Persistently infected resting CD4+ T cells have been detected in the peripheral blood, GI tract, and lymph nodes of HIV-1-infected individuals and are also likely to reside in other organs containing lymphoid tissue (reviewed in reference 65). Reservoirs of SIV were also reported for resting CD4+ T cells in the peripheral blood, lymph nodes, and spleen, but apparently not the thymus, of macaques experimentally infected with a brain-passaged derivative of the pathogenic SIVmac239 strain (56). Our results demonstrate that RT-SHIV DNA levels were substantially higher than vRNA levels in most of the lymphoid and GI tract samples and that RT-SHIV DNA, but little or no RNA, was detected in resting CD4+ T cells obtained from the PBMC, mesenteric lymph nodes, spleens, and jejunums of HAART-treated macaques. These findings are consistent with a widespread distribution of latent viral reservoirs in RT-SHIV-infected macaques.
A major impediment to therapeutic eradication of HIV-1 is the persistence of virus in reservoirs that are not eliminated by the best current HAART regimens. It is generally believed that eradication of HIV-1 from infected individuals will require simultaneous inhibition of residual virus replication and the purging or elimination of latent reservoirs of virus. RT-SHIV-infected rhesus macaques provide a model that can directly address issues pertaining to viral reservoirs that evade HAART, and this model can be used to fully test novel eradication strategies. Advantages of the model are the ability to perform controlled experiments in which all animals are infected at the same time with the same virus and animals in each group receive the same drug therapy; the controlled experimental conditions permit studies to address critical issues about the importance of the timing of initiation of intense antiviral drug therapy. In addition, invasive experiments can be performed during successful antiviral therapy for analyses of reservoirs, latency, and residual virus replication. Another advantage of the model is that an important, clinically meaningful end point, virus rebound after cessation of therapy, can be monitored in this experimental system. Successful eradication strategies must reduce, delay, or eliminate rebound virus after therapy is stopped. Eradication strategies, particularly those based on reactivation of latent virus from resting CD4+ T cells (42), may require approaches that are too risky to test in humans. This nonhuman primate model may be particularly important for testing regimens for “induction” therapy to activate latent virus as well as for evaluating novel antiviral drugs for distribution (penetration) in tissue and cellular reservoirs of virus.
Acknowledgments
This work was supported by NIH program project grant P01-AI058708. This work was also supported by NIH grant 1R01-RR025996 (T.W.N. and R.F.S.). J. D. Deere was supported by NIH training grant T32-AI60555. R.F.S. is also supported by NIH CFAR grant 2P30-AI-050409 and by the Department of Veterans Affairs.
We thank the other members of the NIH program project (E. Verdin, W. Greene, and M. Peterlin) for helpful discussions. We thank E. Cannavo, N. Pedersen, and M. Gardner for helpful discussions and critical reviews of the manuscript. We thank the UC Davis Lucy Whittier Molecular and Diagnostic Core Facility for RNA/DNA extractions and TaqMan assays. We also thank K. Van Rompay, L. Hirst, T. Dearman, A. Spinner, R. Tarara, D. Canfield, and others in the veterinary staff, colony services, and clinical laboratory of the California National Primate Research Center (base grant RR-00169 from NIH) for expert technical assistance.
Footnotes
Published ahead of print on 23 December 2009.
REFERENCES
- 1.Adams, M., L. Sharmeen, J. Kimpton, J. M. Romeo, J. V. Garcia, B. M. Peterlin, M. Groudine, and M. Emerman. 1994. Cellular latency in human immunodeficiency virus-infected individuals with high CD4 levels can be detected by the presence of promoter-proximal transcripts. Proc. Natl. Acad. Sci. USA 91:3862-3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ambrose, Z., V. Boltz, S. Palmer, J. M. Coffin, S. H. Hughes, and V. N. Kewalramani. 2004. In vitro characterization of a simian immunodeficiency virus-human immunodeficiency virus (HIV) chimera expressing HIV type 1 reverse transcriptase to study antiviral resistance in pigtail macaques. J. Virol. 78:13553-13561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ambrose, Z., S. Palmer, V. F. Boltz, M. Kearney, K. Larsen, P. Polacino, L. Flanary, K. Oswald, M. Piatak, Jr., J. Smedley, W. Shao, N. Bischofberger, F. Maldarelli, J. T. Kimata, J. W. Mellors, S. L. Hu, J. M. Coffin, J. D. Lifson, and V. N. KewalRamani. 2007. Suppression of viremia and evolution of human immunodeficiency virus type 1 drug resistance in a macaque model for antiretroviral therapy. J. Virol. 81:12145-12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bailey, J. R., A. R. Sedaghat, T. Kieffer, T. Brennan, P. K. Lee, M. Wind-Rotolo, C. M. Haggerty, A. R. Kamireddi, Y. Liu, J. Lee, D. Persaud, J. E. Gallant, J. Cofrancesco, Jr., T. C. Quinn, C. O. Wilke, S. C. Ray, J. D. Siliciano, R. E. Nettles, and R. F. Siliciano. 2006. Residual human immunodeficiency virus type 1 viremia in some patients on antiretroviral therapy is dominated by a small number of invariant clones rarely found in circulating CD4+ T cells. J. Virol. 80:6441-6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balzarini, J., E. De Clercq, and K. Uberla. 1997. SIV/HIV-1 hybrid virus expressing the reverse transcriptase gene of HIV-1 remains sensitive to HIV-1-specific reverse transcriptase inhibitors after passage in rhesus macaques. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 15:1-4. [DOI] [PubMed] [Google Scholar]
- 6.Balzarini, J., M. Weeger, M. J. Camarasa, E. De Clercq, and K. Uberla. 1995. Sensitivity/resistance profile of a simian immunodeficiency virus containing the reverse transcriptase gene of human immunodeficiency virus type 1 (HIV-1) toward the HIV-1-specific non-nucleoside reverse transcriptase inhibitors. Biochem. Biophys. Res. Commun. 211:850-856. [DOI] [PubMed] [Google Scholar]
- 7.Baskin, G. B., L. N. Martin, M. Murphey-Corb, F. S. Hu, D. Kuebler, and B. Davison. 1995. Distribution of SIV in lymph nodes of serially sacrificed rhesus monkeys. AIDS Res. Hum. Retrovir. 11:273-285. [DOI] [PubMed] [Google Scholar]
- 8.Baumgarth, N., R. Szubin, G. M. Dolganov, M. R. Watnik, D. Greenspan, M. Da Costa, J. M. Palefsky, R. Jordan, M. Roederer, and J. S. Greenspan. 2004. Highly tissue substructure-specific effects of human papilloma virus in mucosa of HIV-infected patients revealed by laser-dissection microscopy-assisted gene expression profiling. Am. J. Pathol. 165:707-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blankson, J. N., D. Persaud, and R. F. Siliciano. 2002. The challenge of viral reservoirs in HIV-1 infection. Annu. Rev. Med. 53:557-593. [DOI] [PubMed] [Google Scholar]
- 10.Chun, T. W., and A. S. Fauci. 1999. Latent reservoirs of HIV: obstacles to the eradication of virus. Proc. Natl. Acad. Sci. USA 96:10958-10961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chun, T. W., D. C. Nickle, J. S. Justement, J. H. Meyers, G. Roby, C. W. Hallahan, S. Kottilil, S. Moir, J. M. Mican, J. I. Mullins, D. J. Ward, J. A. Kovacs, P. J. Mannon, and A. S. Fauci. 2008. Persistence of HIV in gut-associated lymphoid tissue despite long-term antiretroviral therapy. J. Infect. Dis. 197:714-720. [DOI] [PubMed] [Google Scholar]
- 12.Chun, T. W., L. Stuyver, S. B. Mizell, L. A. Ehler, J. A. Mican, M. Baseler, A. L. Lloyd, M. A. Nowak, and A. S. Fauci. 1997. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc. Natl. Acad. Sci. USA 94:13193-13197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dinoso, J. B., S. Y. Kim, A. M. Wiegand, S. E. Palmer, S. J. Gange, L. Cranmer, A. O'Shea, M. Callender, A. Spivak, T. Brennan, M. F. Kearney, M. A. Proschan, J. M. Mican, C. A. Rehm, J. M. Coffin, J. W. Mellors, R. F. Siliciano, and F. Maldarelli. 2009. Treatment intensification does not reduce residual HIV-1 viremia in patients on highly active antiretroviral therapy. Proc. Natl. Acad. Sci. USA 106:9403-9408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dinoso, J. B., S. A. Rabi, J. N. Blankson, L. Gama, J. L. Mankowski, R. F. Siliciano, M. C. Zink, and J. E. Clements. 2009. A simian immunodeficiency virus-infected macaque model to study viral reservoirs that persist during highly active antiretroviral therapy. J. Virol. 83:9247-9257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Embretson, J., M. Zupancic, J. L. Ribas, A. Burke, P. Racz, K. Tenner-Racz, and A. T. Haase. 1993. Massive covert infection of helper T lymphocytes and macrophages by HIV during the incubation period of AIDS. Nature 362:359-362. [DOI] [PubMed] [Google Scholar]
- 16.Fauci, A. S. 1999. The AIDS epidemic—considerations for the 21st century. N. Engl. J. Med. 341:1046-1050. [DOI] [PubMed] [Google Scholar]
- 17.Finzi, D., J. Blankson, J. D. Siliciano, J. B. Margolick, K. Chadwick, T. Pierson, K. Smith, J. Lisziewicz, F. Lori, C. Flexner, T. C. Quinn, R. E. Chaisson, E. Rosenberg, B. Walker, S. Gange, J. Gallant, and R. F. Siliciano. 1999. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat. Med. 5:512-517. [DOI] [PubMed] [Google Scholar]
- 18.Finzi, D., M. Hermankova, T. Pierson, L. M. Carruth, C. Buck, R. E. Chaisson, T. C. Quinn, K. Chadwick, J. Margolick, R. Brookmeyer, J. Gallant, M. Markowitz, D. D. Ho, D. D. Richman, and R. F. Siliciano. 1997. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 278:1295-1300. [DOI] [PubMed] [Google Scholar]
- 19.Fuller, C. L., Y. K. Choi, B. A. Fallert, S. Capuano III, P. Rajakumar, M. Murphey-Corb, and T. A. Reinhart. 2002. Restricted SIV replication in rhesus macaque lung tissues during the acute phase of infection. Am. J. Pathol. 161:969-978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gardner, M. B., M. P. Carlos, and P. A. Luciw. 2004. Simian retroviruses, p. 195-262. In G. P. Wormser (ed.), AIDS and other manifestations of HIV infection, 4th ed. Raven Press, New York, NY.
- 21.Gavegnano, C., and R. F. Schinazi. 2009. Antiretroviral therapy in macrophages: implication for HIV eradication. Antivir. Chem. Chemother. 20:63-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gulick, R. M., J. W. Mellors, D. Havlir, J. J. Eron, C. Gonzalez, D. McMahon, D. D. Richman, F. T. Valentine, L. Jonas, A. Meibohm, E. A. Emini, and J. A. Chodakewitz. 1997. Treatment with indinavir, zidovudine, and lamivudine in adults with human immunodeficiency virus infection and prior antiretroviral therapy. N. Engl. J. Med. 337:734-739. [DOI] [PubMed] [Google Scholar]
- 23.Haase, A. T., K. Henry, M. Zupancic, G. Sedgewick, R. A. Faust, H. Melroe, W. Cavert, K. Gebhard, K. Staskus, Z. Q. Zhang, P. J. Dailey, H. H. Balfour, Jr., A. Erice, and A. S. Perelson. 1996. Quantitative image analysis of HIV-1 infection in lymphoid tissue. Science 274:985-989. [DOI] [PubMed] [Google Scholar]
- 24.Hammer, S. M., J. J. Eron, Jr., P. Reiss, R. T. Schooley, M. A. Thompson, S. Walmsley, P. Cahn, M. A. Fischl, J. M. Gatell, M. S. Hirsch, D. M. Jacobsen, J. S. Montaner, D. D. Richman, P. G. Yeni, and P. A. Volberding. 2008. Antiretroviral treatment of adult HIV infection: 2008 recommendations of the International AIDS Society-USA panel. JAMA 300:555-570. [DOI] [PubMed] [Google Scholar]
- 25.Hammer, S. M., K. E. Squires, M. D. Hughes, J. M. Grimes, L. M. Demeter, J. S. Currier, J. J. Eron, Jr., J. E. Feinberg, H. H. Balfour, Jr., L. R. Deyton, J. A. Chodakewitz, and M. A. Fischl. 1997. A controlled trial of two nucleoside analogues plus indinavir in persons with human immunodeficiency virus infection and CD4 cell counts of 200 per cubic millimeter or less. AIDS Clinical Trials Group 320 Study Team. N. Engl. J. Med. 337:725-733. [DOI] [PubMed] [Google Scholar]
- 26.Havlir, D. V., M. C. Strain, M. Clerici, C. Ignacio, D. Trabattoni, P. Ferrante, and J. K. Wong. 2003. Productive infection maintains a dynamic steady state of residual viremia in human immunodeficiency virus type 1-infected persons treated with suppressive antiretroviral therapy for five years. J. Virol. 77:11212-11219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heise, C., P. Vogel, C. J. Miller, A. Lackner, and S. Dandekar. 1993. Distribution of SIV infection in the gastrointestinal tract of rhesus macaques at early and terminal stages of AIDS. J. Med. Primatol. 22:187-193. [PubMed] [Google Scholar]
- 28.Hirsch, V. M., and J. D. Lifson. 2000. Simian immunodeficiency virus infection of monkeys as a model system for the study of AIDS pathogenesis, treatment, and prevention. Adv. Pharmacol. 49:437-477. [DOI] [PubMed] [Google Scholar]
- 29.Hirsch, V. M., P. M. Zack, A. P. Vogel, and P. R. Johnson. 1991. Simian immunodeficiency virus infection of macaques: end-stage disease is characterized by widespread distribution of proviral DNA in tissues. J. Infect. Dis. 163:976-988. [DOI] [PubMed] [Google Scholar]
- 30.Hofman, M. J., J. Higgins, T. B. Matthews, N. C. Pedersen, C. Tan, R. F. Schinazi, and T. W. North. 2004. Efavirenz therapy in rhesus macaques infected with a chimera of simian immunodeficiency virus containing reverse transcriptase from human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 48:3483-3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hofmann-Lehmann, R., R. K. Swenerton, V. Liska, C. M. Leutenegger, H. Lutz, H. M. McClure, and R. M. Ruprecht. 2000. Sensitive and robust one-tube real-time reverse transcriptase-polymerase chain reaction to quantify SIV RNA load: comparison of one- versus two-enzyme systems. AIDS Res. Hum. Retrovir. 16:1247-1257. [DOI] [PubMed] [Google Scholar]
- 32.Hoxie, J. A., B. S. Haggarty, S. E. Bonser, J. L. Rackowski, H. Shan, and P. J. Kanki. 1988. Biological characterization of a simian immunodeficiency virus-like retrovirus (HTLV-IV): evidence for CD4-associated molecules required for infection. J. Virol. 62:2557-2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu, J., M. Pope, C. Brown, U. O'Doherty, and C. J. Miller. 1998. Immunophenotypic characterization of simian immunodeficiency virus-infected dendritic cells in cervix, vagina, and draining lymph nodes of rhesus monkeys. Lab. Invest. 78:435-451. [PubMed] [Google Scholar]
- 34.Joling, P., L. J. Bakker, J. A. Van Strijp, T. Meerloo, L. de Graaf, M. E. Dekker, J. Goudsmit, J. Verhoef, and H. J. Schuurman. 1993. Binding of human immunodeficiency virus type-1 to follicular dendritic cells in vitro is complement dependent. J. Immunol. 150:1065-1073. [PubMed] [Google Scholar]
- 35.Joos, B., M. Fischer, H. Kuster, S. K. Pillai, J. K. Wong, J. Boni, B. Hirschel, R. Weber, A. Trkola, and H. F. Gunthard. 2008. HIV rebounds from latently infected cells, rather than from continuing low-level replication. Proc. Natl. Acad. Sci. USA 105:16725-16730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kitagawa, M., A. A. Lackner, D. J. Martfeld, M. B. Gardner, and S. Dandekar. 1991. Simian immunodeficiency virus infection of macaque bone marrow macrophages correlates with disease progression in vivo. Am. J. Pathol. 138:921-930. [PMC free article] [PubMed] [Google Scholar]
- 37.Lackner, A. A., P. Vogel, R. A. Ramos, J. D. Kluge, and M. Marthas. 1994. Early events in tissues during infection with pathogenic (SIVmac239) and nonpathogenic (SIVmac1A11) molecular clones of simian immunodeficiency virus. Am. J. Pathol. 145:428-439. [PMC free article] [PubMed] [Google Scholar]
- 38.Leutenegger, C. M., J. Higgins, T. B. Matthews, A. F. Tarantal, P. A. Luciw, N. C. Pedersen, and T. W. North. 2001. Real-time TaqMan PCR as a specific and more sensitive alternative to the branched-chain DNA assay for quantitation of simian immunodeficiency virus RNA. AIDS Res. Hum. Retrovir. 17:243-251. [DOI] [PubMed] [Google Scholar]
- 39.Lewin, S. R., M. Vesanen, L. Kostrikis, A. Hurley, M. Duran, L. Zhang, D. D. Ho, and M. Markowitz. 1999. Use of real-time PCR and molecular beacons to detect virus replication in human immunodeficiency virus type 1-infected individuals on prolonged effective antiretroviral therapy. J. Virol. 73:6099-6103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li, Q., L. Duan, J. D. Estes, Z. M. Ma, T. Rourke, Y. Wang, C. Reilly, J. Carlis, C. J. Miller, and A. T. Haase. 2005. Peak SIV replication in resting memory CD4+ T cells depletes gut lamina propria CD4+ T cells. Nature 434:1148-1152. [DOI] [PubMed] [Google Scholar]
- 41.Liu, Z. Q., S. Muhkerjee, M. Sahni, C. McCormick-Davis, K. Leung, Z. Li, V. H. Gattone II, C. Tian, R. W. Doms, T. L. Hoffman, R. Raghavan, O. Narayan, and E. B. Stephens. 1999. Derivation and biological characterization of a molecular clone of SHIV(KU-2) that causes AIDS, neurological disease, and renal disease in rhesus macaques. Virology 260:295-307. [DOI] [PubMed] [Google Scholar]
- 42.North, T. W., and P. A. Luciw. 2007. Nonhuman primate model for viral latency/persistence during highly active antiretroviral therapy (HAART) for AIDS, p. 83-110. In A. M. Brown (ed.), Persistent and latent infection by HIV-1 and related lentiviruses. Research Signpost, Kerala, India.
- 43.North, T. W., K. K. Van Rompay, J. Higgins, T. B. Matthews, D. A. Wadford, N. C. Pedersen, and R. F. Schinazi. 2005. Suppression of virus load by highly active antiretroviral therapy in rhesus macaques infected with a recombinant simian immunodeficiency virus containing reverse transcriptase from human immunodeficiency virus type 1. J. Virol. 79:7349-7354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Palmer, S., F. Maldarelli, A. Wiegand, B. Bernstein, G. J. Hanna, S. C. Brun, D. J. Kempf, J. W. Mellors, J. M. Coffin, and M. S. King. 2008. Low-level viremia persists for at least 7 years in patients on suppressive antiretroviral therapy. Proc. Natl. Acad. Sci. USA 105:3879-3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Palmer, S., A. P. Wiegand, F. Maldarelli, H. Bazmi, J. M. Mican, M. Polis, R. L. Dewar, A. Planta, S. Liu, J. A. Metcalf, J. W. Mellors, and J. M. Coffin. 2003. New real-time reverse transcriptase-initiated PCR assay with single-copy sensitivity for human immunodeficiency virus type 1 RNA in plasma. J. Clin. Microbiol. 41:4531-4536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perelson, A. S., P. Essunger, Y. Cao, M. Vesanen, A. Hurley, K. Saksela, M. Markowitz, and D. D. Ho. 1997. Decay characteristics of HIV-1-infected compartments during combination therapy. Nature 387:188-191. [DOI] [PubMed] [Google Scholar]
- 47.Perelson, A. S., P. Essunger, and D. D. Ho. 1997. Dynamics of HIV-1 and CD4+ lymphocytes in vivo. AIDS 11(Suppl. A):S17-S24. [PubMed] [Google Scholar]
- 48.Persaud, D., T. Pierson, C. Ruff, D. Finzi, K. R. Chadwick, J. B. Margolick, A. Ruff, N. Hutton, S. Ray, and R. F. Siliciano. 2000. A stable latent reservoir for HIV-1 in resting CD4(+) T lymphocytes in infected children. J. Clin. Invest. 105:995-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pozniak, A. L., J. E. Gallant, E. DeJesus, J. R. Arribas, B. Gazzard, R. E. Campo, S. S. Chen, D. McColl, J. Enejosa, J. J. Toole, and A. K. Cheng. 2006. Tenofovir disoproxil fumarate, emtricitabine, and efavirenz versus fixed-dose zidovudine/lamivudine and efavirenz in antiretroviral-naive patients: virologic, immunologic, and morphologic changes—a 96-week analysis. J. Acquir. Immune Defic. Syndr. 43:535-540. [DOI] [PubMed] [Google Scholar]
- 50.Press, N., M. W. Tyndall, E. Wood, R. S. Hogg, and J. S. Montaner. 2002. Virologic and immunologic response, clinical progression, and highly active antiretroviral therapy adherence. J. Acquir. Immune Defic. Syndr. 31(Suppl. 3):S112-S117. [DOI] [PubMed] [Google Scholar]
- 51.Ramratnam, B., J. E. Mittler, L. Zhang, D. Boden, A. Hurley, F. Fang, C. A. Macken, A. S. Perelson, M. Markowitz, and D. D. Ho. 2000. The decay of the latent reservoir of replication-competent HIV-1 is inversely correlated with the extent of residual viral replication during prolonged anti-retroviral therapy. Nat. Med. 6:82-85. [DOI] [PubMed] [Google Scholar]
- 52.Richman, D. D. 2001. HIV chemotherapy. Nature 410:995-1001. [DOI] [PubMed] [Google Scholar]
- 53.Ringler, D. J., M. S. Wyand, D. G. Walsh, J. J. MacKey, P. K. Sehgal, M. D. Daniel, R. C. Desrosiers, and N. W. King. 1989. The productive infection of alveolar macrophages by simian immunodeficiency virus. J. Med. Primatol. 18:217-226. [PubMed] [Google Scholar]
- 54.Rong, L., and A. S. Perelson. 2009. Modeling HIV persistence, the latent reservoir, and viral blips. J. Theor. Biol. 260:308-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ryzhova, E. V., P. Crino, L. Shawver, S. V. Westmoreland, A. A. Lackner, and F. Gonzalez-Scarano. 2002. Simian immunodeficiency virus encephalitis: analysis of envelope sequences from individual brain multinucleated giant cells and tissue samples. Virology 297:57-67. [DOI] [PubMed] [Google Scholar]
- 56.Shen, A., M. C. Zink, J. L. Mankowski, K. Chadwick, J. B. Margolick, L. M. Carruth, M. Li, J. E. Clements, and R. F. Siliciano. 2003. Resting CD4+ T lymphocytes but not thymocytes provide a latent viral reservoir in a simian immunodeficiency virus-Macaca nemestrina model of human immunodeficiency virus type 1-infected patients on highly active antiretroviral therapy. J. Virol. 77:4938-4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Siliciano, J. D., J. Kajdas, D. Finzi, T. C. Quinn, K. Chadwick, J. B. Margolick, C. Kovacs, S. J. Gange, and R. F. Siliciano. 2003. Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat. Med. 9:727-728. [DOI] [PubMed] [Google Scholar]
- 58.Soderberg, K., L. Denekamp, S. Nikiforow, K. Sautter, R. C. Desrosiers, and L. Alexander. 2002. A nucleotide substitution in the tRNA(Lys) primer binding site dramatically increases replication of recombinant simian immunodeficiency virus containing a human immunodeficiency virus type 1 reverse transcriptase. J. Virol. 76:5803-5806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Strain, M. C., H. F. Gunthard, D. V. Havlir, C. C. Ignacio, D. M. Smith, A. J. Leigh-Brown, T. R. Macaranas, R. Y. Lam, O. A. Daly, M. Fischer, M. Opravil, H. Levine, L. Bacheler, C. A. Spina, D. D. Richman, and J. K. Wong. 2003. Heterogeneous clearance rates of long-lived lymphocytes infected with HIV: intrinsic stability predicts lifelong persistence. Proc. Natl. Acad. Sci. USA 100:4819-4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sundstrom, J. B., J. E. Ellis, G. A. Hair, A. S. Kirshenbaum, D. D. Metcalfe, H. Yi, A. C. Cardona, M. K. Lindsay, and A. A. Ansari. 2007. Human tissue mast cells are an inducible reservoir of persistent HIV infection. Blood 109:5293-5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Suryanarayana, K., T. A. Wiltrout, G. M. Vasquez, V. M. Hirsch, and J. D. Lifson. 1998. Plasma SIV RNA viral load determination by real-time quantification of product generation in reverse transcriptase-polymerase chain reaction. AIDS Res. Hum. Retrovir. 14:183-189. [DOI] [PubMed] [Google Scholar]
- 62.Tobin, N. H., G. H. Learn, S. E. Holte, Y. Wang, A. J. Melvin, J. L. McKernan, D. M. Pawluk, K. M. Mohan, P. F. Lewis, J. I. Mullins, and L. M. Frenkel. 2005. Evidence that low-level viremias during effective highly active antiretroviral therapy result from two processes: expression of archival virus and replication of virus. J. Virol. 79:9625-9634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Uberla, K., C. Stahl-Hennig, D. Bottiger, K. Matz-Rensing, F. J. Kaup, J. Li, W. A. Haseltine, B. Fleckenstein, G. Hunsmann, B. Oberg, and J. Sodroski. 1995. Animal model for the therapy of acquired immunodeficiency syndrome with reverse transcriptase inhibitors. Proc. Natl. Acad. Sci. USA 92:8210-8214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Van Rompay, K. K., R. P. Singh, L. L. Brignolo, J. R. Lawson, K. A. Schmidt, B. Pahar, D. R. Canfield, R. P. Tarara, D. L. Sodora, N. Bischofberger, and M. L. Marthas. 2004. The clinical benefits of tenofovir for simian immunodeficiency virus-infected macaques are larger than predicted by its effects on standard viral and immunologic parameters. J. Acquir. Immune Defic. Syndr. 36:900-914. [DOI] [PubMed] [Google Scholar]
- 65.Veazey, R. S., and A. A. Lackner. 2004. Getting to the guts of HIV pathogenesis. J. Exp. Med. 200:697-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Veazey, R. S., M. Rosenzweig, D. E. Shvetz, D. R. Pauley, M. DeMaria, L. V. Chalifoux, R. P. Johnson, and A. A. Lackner. 1997. Characterization of gut-associated lymphoid tissue (GALT) of normal rhesus macaques. Clin. Immunol. Immunopathol. 82:230-242. [DOI] [PubMed] [Google Scholar]
- 67.Veazey, R. S., I. C. Tham, K. G. Mansfield, M. DeMaria, A. E. Forand, D. E. Shvetz, L. V. Chalifoux, P. K. Sehgal, and A. A. Lackner. 2000. Identifying the target cell in primary simian immunodeficiency virus (SIV) infection: highly activated memory CD4(+) T cells are rapidly eliminated in early SIV infection in vivo. J. Virol. 74:57-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ward, J. M., T. J. O'Leary, G. B. Baskin, R. Benveniste, C. A. Harris, P. L. Nara, and R. H. Rhodes. 1987. Immunohistochemical localization of human and simian immunodeficiency viral antigens in fixed tissue sections. Am. J. Pathol. 127:199-205. [PMC free article] [PubMed] [Google Scholar]
- 69.Williams, K. C., S. Corey, S. V. Westmoreland, D. Pauley, H. Knight, C. deBakker, X. Alvarez, and A. A. Lackner. 2001. Perivascular macrophages are the primary cell type productively infected by simian immunodeficiency virus in the brains of macaques: implications for the neuropathogenesis of AIDS. J. Exp. Med. 193:905-915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wong, J. K., H. F. Gunthard, D. V. Havlir, Z. Q. Zhang, A. T. Haase, C. C. Ignacio, S. Kwok, E. Emini, and D. D. Richman. 1997. Reduction of HIV-1 in blood and lymph nodes following potent antiretroviral therapy and the virologic correlates of treatment failure. Proc. Natl. Acad. Sci. USA 94:12574-12579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang, L., C. Chung, B. S. Hu, T. He, Y. Guo, A. J. Kim, E. Skulsky, X. Jin, A. Hurley, B. Ramratnam, M. Markowitz, and D. D. Ho. 2000. Genetic characterization of rebounding HIV-1 after cessation of highly active antiretroviral therapy. J. Clin. Invest. 106:839-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zink, M. C., V. A. Laast, K. L. Helke, A. K. Brice, S. A. Barber, J. E. Clements, and J. L. Mankowski. 2006. From mice to macaques—animal models of HIV nervous system disease. Curr. HIV Res. 4:293-305. [DOI] [PubMed] [Google Scholar]