Abstract
Background: Growth factor receptor-bound protein-7 (Grb7) is an adapter-type signaling protein recruited to various tyrosine kinases, including HER2/neu. Grb7-specific inhibitors are in early development. As with other targeted therapies, response to therapy might be associated with target expression.
Materials and methods: Tissue microarrays containing 638 primary breast cancer specimens with 15-year patient follow-up were employed to assess Grb7 expression using our Automated QUantitative Analysis method; cytokeratin defines pixels as breast cancer (tumor mask) within the histospot, and Grb7 expression within the mask is measured with Cy5-conjugated antibodies.
Results: High Grb7 expression was strongly associated with decreased survival in the entire cohort and in the node-positive subset (P = 0.0034 and P = 0.0019, respectively). On multivariable analysis, it remained an independent prognostic marker (P = 0.01). High Grb7 was strongly associated with high HER2/neu, and coexpression of these molecules was associated with worse prognosis than HER2/neu overexpression alone.
Conclusions: High Grb7 defines a subset of breast cancer patients with decreased survival, indicating that Grb7 might be a valuable prognostic marker and drug target. Coexpression with HER2/neu indicates that cotargeting these molecules might be an effective approach for treating HER2/neu-positive breast cancers. Future studies using Grb7-targeting agents should include assessment of Grb7 levels.
Keywords: Grb7, HER2/neu, prognosis, therapeutic targets
introduction
Growth factor receptor-bound protein-7 (Grb7) is an adapter molecule involved in many signal transduction pathways [1, 2]. It is a member of the Grb family of proteins, which includes Grb7, Grb10 and Grb14 [3, 4]. These proteins possess no intrinsic enzymic activity and they signal through their interaction with downstream proteins [3, 4]. They participate in important cellular functions such as signal transduction, cell motility and tumor progression, as reviewed previously [3–7]. All have been found to be overexpressed in certain cancers and cancer cell lines [3–7].
Grb family proteins are differentially expressed in a variety of tissues [3–7]. They are characterized by several functional domains that include a unique and highly conserved proline-rich motif (PS/AIPNPFPEL) at the N-terminal region, a Grb and Mig region containing the Pleckstrin homology (PH) domain, a region between the PH and Src homology 2 (SH2) regions and a C-terminal SH2 domain [7, 8]. The SH2 domain binds phosphotyrosine in the context of the adjacent C-terminal amino acids and is critical for the adapter function of these proteins [9, 10]. Grb7 family proteins have been shown to interact with a variety of other proteins, including tyrosine kinase receptors and proto-oncogenes, as reviewed previously [3–7].
The human Grb7 gene is located on chromosome 17q11–21 in the immediate vicinity of the HER-2/neu and is part of the HER2/neu amplicon [11, 12]. In normal tissues, Grb7 is most abundantly expressed in the pancreas, but is also expressed in kidney, placenta, prostate, intestine, colon, liver, lung and testis [13]. The precise role of the Grb7 adapter molecule is still not clear, but studies indicate a role for Grb7 in the regulation of cell migration [2, 14–16] and tumorigenesis, as reviewed previously [3–7]. Grb7 has >20 binding partners [4], including nonreceptor tyrosine kinases and receptor tyrosine kinases including epidermal growth factor receptor (EGFR) [1] and HER-2/neu [11]. Most of these proteins interact with Grb7 through the SH2 domain of Grb7 [8, 10, 17].
Within the cell, Grb7 is present in the cytoplasm, where it interacts with upstream binding partners [14], including the members of the EGFR receptor family [11, 18]. Grb7 can also be detected in discrete regions of the plasma membrane called focal contacts, where it is bound and phosphorylated by focal adhesion kinase (FAK), which is a cytoplasmic tyrosine kinase known to play an important role in integrin-mediated signal transduction and cell migration [14]. The recruitment of Grb7 to protein complexes containing activated Ras proteins indicates that Grb7 expression may also modulate Ras signaling [19].
Grb7’s involvement in regulating cell motility is dependent on its targeting to focal contacts, its tyrosine phosphorylation by FAK and the association of its PH domain with specific phosphoinositides [2, 14]. Grb7's association with EphB1 [14] also contributes to cell migration. Grb7/FAK complex formation and increased cellular invasion have been reported in esophageal carcinoma cells, and Grb7 antisense inhibits migration in preclinical models [20, 21]. The importance of Grb7 in tumor progression and cancer cell migration has been suggested by several studies, as reviewed previously [3, 7, 22]. Grb7 has been shown to be overexpressed in a subset of esophageal and gastric cancers and in Barrett's carcinoma and has been shown to be associated with disease progression in esophageal cancer [3, 7, 22].
Several studies indicate that Grb7 is involved in breast cancer in the context of HER-2/neu amplification. Grb7 has been shown to be coamplified with HER-2/neu in most, if not all, of the breast cell lines and breast tumors with 17q11–21 amplification [11, 23–25]. This association in expression levels between Grb7 and HER2/neu has been shown in primary breast tumors both by RT-PCR [23, 26–28] and by western blot [11, 29] by several groups in small-cohort studies (<80 patients). Grb7 and HER2/neu have been found to form a complex in breast cancer cells: Grb7 coimmunoprecipitates with phosphorylated HER2/neu in breast cancer cell lines, indicating that the coexpression of these two proteins results in activation of the HER2/neu signaling pathway. Moreover, in a subgroup of human breast cancer cell lines, Grb7 associates and coexpresses with HER3 and HER4, which are known to heterodimerize with HER2/neu [18]. Knock down of Grb7 leads to decreased cell proliferation and cell cycle progression in SKBR3 and BT-474 cell lines [30]. Grb7 overexpression facilitates phosphorylation of both AKT and HER2/neu in HER2/neu-overexpressing cells [29]. In addition, Grb7 overexpression promotes tumor formation in xenograft models by HER-2/neu-expressing cells [29].
Small-molecule and peptidomimetic inhibitors of Grb7 are currently being assessed in laboratory models, as reviewed previously [7]. One of the promising anti-Grb7 peptides, G7-18NATE, binds selectively to the SH2 domain of Grb7 (with no detectable binding to other related family members) [31]. It inhibits the binding of Grb7 to various tyrosine kinases, including the ErbB family [31]. This drug has been shown to be well tolerated in mice [32] and has been shown to inhibit breast cancer cell proliferation with no effect on nonmalignant cells [22]. Specifically, G7-18NATE inhibits proliferation of SKBR3, ZR-7530, MDA-MB-361 and MDA-MB-231 breast cancer cells and has no significant effects on the non-HER2/neu-expressing MCF-7cells, and nonmalignant MCF-10A or NIH3T3 cells [22]. G7-18NATE is synergistic with trastuzumab in inhibiting SKBR3 cells, which overexpress HER2/neu [22, 33] and is also synergistic with adriamycin in inhibiting SKBR3 cells [22].
A number of studies have assessed the importance of Grb7 in predicting prognosis in breast cancer at the messenger RNA (mRNA) level. Grb7 is included in the 21-gene set (Oncotype DX assay) used for predicting prognosis in early-stage breast cancer based on mRNA levels [34]. Cobleigh et al. [28] and Vinatzer et al. [27] studied the expression levels of Grb7 mRNA in 78 and 85 breast cancer patients, respectively, and found that higher expression of Grb7 mRNA was associated with worse prognosis. Given the potential importance of Grb7 as a drug target and the potential discordance between mRNA and protein levels, we assessed expression of this protein in a large cohort of breast cancer specimens and studied the association with survival and other clinical variables. To the best of our knowledge, no large-scale quantitative studies have been conducted on Grb7 protein expression in clinical breast cancer specimens. To obtain more accurate, objective expression measures, we used our newly developed method of Automated QUantitative Analysis (AQUA) of in situ protein expression. This method has been validated and has proven to be more accurate than pathologist-based scoring of 3,3′-diaminobenzidine stain. As with some other targeted therapies, it is possible that response to Grb7-targeting drugs might be associated with expression levels of the target in tumors, and quantitative assays need to be developed to predict response. Other markers that have both prognostic and predictive value, such as HER2/neu and hormone receptors, have significantly impacted our ability to appropriately select therapeutic regimens for breast cancer. We found that Grb7 expression is variable and that high expression is associated with decreased survival and HER2/neu overexpression.
materials and methods
cell lines and western blots
MDA-MB-435, MDA-MB-231, MCF-7, T47D, ZR-7510, BT-20, BT-474 and SKBR3 human breast cancer cell lines were purchased from ATCC (Manassas, VA). Western blotting of protein extracts was carried out using standard methods. Grb7 expression was detected by overnight incubation with rabbit anti-Grb7 immunoglobulin G H-70 (Santa Cruz Biotechnology, Santa Cruz, CA) at 1:1000. Protein loading was assessed using mouse anti-β-actin (Sigma-Aldrich, St Louis, MO) at 1:5000.
cell fractionation
BT-474 cells were grown to 80% confluence, collected and incubated in 400-μl hypotonic buffer for 30 min on ice. A 2-μl sample was removed to verify that 80%–90% of cells were stained by Trypan blue. The remaining cells were spun at 1200×g for 15 min and the cytoplasmic fraction was collected from the supernatant. The pellet was rinsed, spun down for 30 seconds at 1000×g, suspended in 400-μl hypertonic buffer (10 mM NaPO4, 150 mM NaCl, 5 mM ethylenediaminetetraacetic acid), sonicated for 1 min and incubated on ice for 5 min. Sonication was repeated, and the pellet was incubated for 30 min on ice and spun down for 10 min at 360×g. Supernatant containing the nuclear fraction was collected. Purity of fractions was confirmed by western blot and determined by the presence of α-tubulin (cytoplasmic marker) and laminB1 (nuclear marker) using anti-laminB1 ab16048 (Abcam, Cambridge, UK) and anti-α-tubulin (Sigma-Aldrich) antibodies.
tissue microarray construction
Tissue microarrays (TMAs) were constructed as previously described [35]. A total of 319 node-negative and 319 node-positive breast cancer cores, each measuring 0.6 mm in diameter, were spaced 0.8 mm apart. Specimens and clinical information were collected with approval of a Yale University Institutional Review Board. The cohort has been described and validated in previous publications [35]. Estrogen receptor staining was positive in 52%, progesterone receptor in 46% and HER2/neu in 14%. Nuclear grade 3 (on a 1–3 scale) was seen in 28% of the specimens, and 59% were larger than 2 cm. The histological subtypes included 72% invasive ductal carcinomas and 14% lobular carcinomas and 14% had mixed or other histology. The specimens were resected between 1962 and 1980, with a follow-up range between 4 months and 53 years, and a mean follow-up time of 12.6 years. Age at diagnosis ranged from 24 to 88 years (mean age 58 years). Complete treatment history was not available for the entire cohort. Most patients were treated with local irradiation. None of the node-negative patients were given adjuvant systemic therapy. A minority of the node-positive patients (∼15%) received chemotherapy, and ∼27% of those treated after 1978 received tamoxifen. The time between tumor resection and tissue fixation was not available. A pathologist reviewed slides from all of the blocks to select representative areas of invasive tumor to be cored. The cores were placed on the TMA using a Tissue Microarrayer (Beecher Instruments, Silver Spring, MD). The TMAs were then cut to 5-μm sections and placed on glass slides using an adhesive tape-transfer system (Instrumedics, Inc., Hackensack, NJ) with UV cross-linking.
immunoflourescence
One set of two slides (each containing a core from different areas of the tumor for the same patient) was stained for Grb7. Staining was carried out for AQUA as described [36]. Briefly, slides were deparaffinized in xylene and transferred through two changes of 100% ethanol. For antigen retrieval, the slides were boiled in a pressure cooker containing 6.5 mM sodium citrate (pH 6.0). Endogenous peroxidase activity was blocked in a mixture of methanol and 2.5% hydrogen peroxide for 30 min at room temperature. To reduce nonspecific background staining, slides were incubated at room temperature for 30 min in 0.3% bovine serum albumin (BSA) in 1X Tris-buffered saline. Slides were incubated at 4°C overnight in a humidity tray with rabbit anti-Grb7 antibody (at 1:100) diluted in Tris-buffered saline containing 0.3% BSA. Goat anti-rabbit HRP-b polymer backbone (Envision; Dako, Carpinteria, CA) was used as a secondary reagent and Cy5-tyramide (Perkin Elmer Life Science, Massachusetts) was used to visualize the target. To create a tumor mask, slides were simultaneously incubated with a mouse anti-human cytokeratin antibody (Dako) at 1:200 and were visualized with a secondary Alexa 488-conjugated goat anti-mouse antibody (Molecular Probes, Inc. Eugene,OR). Coverslips were mounted with ProLong Gold antifade reagent with 4′,6-diamidino-2-phenylindole (DAPI; Invitrogen, Carlsbad, CA). Staining for estrogen receptor, progesterone receptor and HER2/neu was carried out as previously described [37].
automated image acquisition and analysis
Images were analyzed using algorithms that have been extensively described [36]. Multiple monochromatic, high-resolution (1024 × 1024 pixel, 0.5-μm) grayscale images were obtained for each histospot, using the ×10 objective of an Olympus AX-51 epifluorescence microscope (Olympus, Melville, NY) with an automated microscope stage and digital image acquisition driven by custom program and macro-based interfaces with IPLabs software (Scanalytics Inc., Fairfax, VA). Areas of tumor were distinguished from stroma by creating a mask with the cytokeratin signal tagged with Alexa 488. Coalescence of cytokeratin at the cell surface was used to identify the membrane–cytoplasm compartment within the tumor mask, while DAPI was used to identify the nuclear compartment within the tumor mask. Grb7 was visualized with Cy5 (red). Cy5 was used because its emission peak is outside the color spectrum of tissue autoflourescence. The target signal was scored on a scale of 0–255 (AQUA score) for the nuclear and cytoplasmic compartments. Two images (one in focus and one out of focus) were taken of the compartment-specific tags and the target marker. A percentage of the out-of-focus image was subtracted from the in-focus image for each pixel, representing the signal-to-noise ratio of the image. An algorithm described as Rapid Exponential Subtraction Algorithm was used to subtract the out-of-focus information in a uniform fashion for the entire microarray. Subsequently, the PLACE algorithm (Pixel Locale Assignment for Compartmentalization of Expression) was used to assign each pixel in the image to a specific subcellular compartment and the signal in each location is calculated. Pixels that cannot accurately be assigned to a compartment were discarded. The AQUA scores were calculated as the average signal intensity per unit of compartment area and expressed on a scale of 0–255.
data analysis
JMP version 5.0 software was used (SAS Institute, Cary, NC). The prognostic significance of parameters was assessed using the Cox proportional hazards model with survival as an end point. Associations between continuous AQUA scores of target expression and clinical and pathological parameters were assessed by unpaired Student's t-tests. Survival curves were generated using the Kaplan–Meier method, with significance evaluated using the Mantel–Cox long-rank test.
results
western blots
Western blots using the polyclonal H-70 antibody specific for Grb7 on a panel of lysates from breast cancer cell lines are shown in supplemental Figure 1 (available at Annals of Oncology online). We identified a single 57-kDa band. Expression was very high in the SKBR3 and BT-474 cell lines and lower in the other cell lines, consistent with reports in the literature [11, 29, 30].
immunoflourescent staining
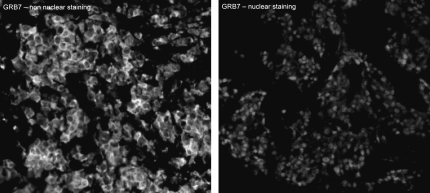
Grb7 staining was either nuclear or cytoplasmic, and many specimens had both nuclear and cytoplasmic staining. Examples of Grb7 staining patterns in breast tumor samples are shown in Figure 1. Details of the masking and compartmentalization of cells are given in supplemental Figure 2 (available at Annals of Oncology online). Given that previous studies report that Grb7 expression is cytoplasmic, we carried out cell fractionation on BT-474 cells and demonstrated Grb7 presence in both the nuclear and cytoplasmic fractions (supplemental Figure 3, available at Annals of Oncology online). To account for intratumor heterogeneity, two separate sets of slides—each containing a core from a different area of the tumor for each patient—were used to evaluate the expression of Grb7. The same was done for ER, PR and HER2/neu as described in our previous work [37]. In the case of Grb7, both nuclear and nonnuclear compartments were analyzed. HER2/neu did not have significant amounts of nuclear staining, and only the membranous/cytoplasmic compartments were analyzed, and vice versa, for ER and PR staining. We carried out log-regression analyses to assess the correlation between Grb7 expression in the two slides, as demonstrated in supplemental Figure 4 (available at Annals of Oncology online). The matching spots on each array were highly correlated for all four markers (R = 0.7 for Grb7, R = 0.74 for the ER, R = 0.79 for the PR and R = 0.92 for HER2/neu). AQUA scores ranged from 2.8 to 157.5 for the total Grb7 score (median 27.1), from 2.34 to 94.097 for PR (median 6.58), from 1.085 to 61.164 for 300 HER2/neu (median 2.03) and from 2.29 to 105.39 for ER (median 13.46). For each of the markers, the AQUA scores from both sets of slides were combined to give a single dataset. Tumor spots were deemed uninterpretable if they had insufficient tumor cells, loss of tissue in the spot or an abundance of necrotic tissue. For patients who had two interpretable histospots, a composite score was formed by taking the average of the two scores. For patients with only one interpretable core, the single score was used. The combined dataset for Grb7 had scores for 570 patients. We obtained scores for HER2/neu, ER and PR for 608 patients, 601 patients and 595 patients, respectively.
Figure 1.
Immunoflourescent growth factor receptor-bound protein-7 (Grb7) staining patterns: Predominantly cytoplasmic Grb7 (left panel) and nuclear Grb7 (right panel) staining in two breast cancer histospots at ×60 magnification.
Given that the nodal status often determines the standard clinical approach to patients, we assessed the prognostic value of Grb7 in the entire cohort, as well as within the node-positive and node-negative subsets of patients. Using the Cox univariate survival analysis of AQUA scores, we found that Grb7 expression (cytoplasmic and total) was associated with survival in the entire cohort and in the node-positive subset (P = 0.01 and P = 0.007, respectively, for total Grb7), as shown in Table 1. Since the nonnuclear Grb7 scores seemed to be the best predictors of survival, since Grb7 is known to be functional in the cytoplasm and since most histospots had mainly nonnuclear Grb7 staining, the remainder of the analyses will focus on the nonnuclear Grb7 scores.
Table 1.
Association between Grb7 and breast cancer-specific survival by Cox univariate analysis with 15-year follow-up
| Patient group | Grb7 total |
Grb7 nonnuclear |
Grb7 nuclear |
|||
| P value | n | P value | n | P value | n | |
| Node negative | 0.7 | 254 | 0.93 | 252 | 0.54 | 249 |
| Node positive | 0.007 | 262 | 0.0019 | 261 | 0.036 | 259 |
| All | 0.013 | 516 | 0.0034 | 513 | 0.063 | 508 |
Significant P-value (<0.05) are in bold. Grb7, growth factor receptor-bound protein-7.
Using the Cox proportional hazards model, we carried out multivariable analysis to assess the independent prognostic value of Grb7 expression. High Grb7 expression remained an independent prognostic marker, as shown in Table 2, as were tumor size, PR and nodal involvement. Interestingly, once we included Grb7 in our multivariable model, HER2/neu no longer remained an independent prognostic variable.
Table 2.
Multivariable Cox proportional hazards analysis of Grb7 and other commonly used clinical and pathological variables with 15-year follow-up
| Variable | 95% Confidence interval | P value |
| Grb7a | 1.00–1.01 | 0.01 |
| HER2/neua | 0.96–0.99 | 0.41 |
| PRa | 0.97–0.98 | 0.01 |
| ERa | 0.98–0.99 | 0.06 |
| Nodal status | 0.54–0.63 | <0.0001 |
| Tumor size | 0.6–0.72 | <0.0001 |
| Menopausal status (postmenopausal) | 0.66–0.80 | 0.01 |
| Nuclear grade (1 versus 2 and 3) | 0.87–1.06 | 0.56 |
Continuous Automated QUantitative Analysis scores.
Grb7, growth factor receptor-bound protein-7; PR, progesterone receptor; ER, estrogen receptor.
In order to assess the association between Grb7 expression and other commonly used clinical and pathological parameters, we carried out analyses of variance, and as shown in Table 3. For patients who had AQUA scores for all four biomarkers, we assessed the associations between continuous biomarker scores by Spearman's rho test, as demonstrated in Table 3. High Grb7 expression was associated with high nuclear grade and high HER2/neu.
Table 3.
Association between growth factor receptor-bound protein-7 scores and commonly used clinical and pathological variables
| One way ANOVA t-test | ||
| Variable | t-test | P value |
| Tumor size >2cm | 0.27 | 0.78 |
| Nodal status (node positive) | 1.65 | 0.1 |
| Age at diagnosis >50 | 1.09 | 0.27 |
| Nuclear grade (2–3 versus 1) | 2.5 | 0.012 |
| Spearman's rank correlation | ||
| Variable | Rho | P value |
| ERa | −0.022 | 0.6 |
| PRa | −0.05 | 0.25 |
| HER2/neua | 0.410 | <0.0001 |
Continuous Automated QUantitative Analysis scores were used.
ANOVA, analysis of variance; ER, estrogen receptor; PR, progesterone receptor.
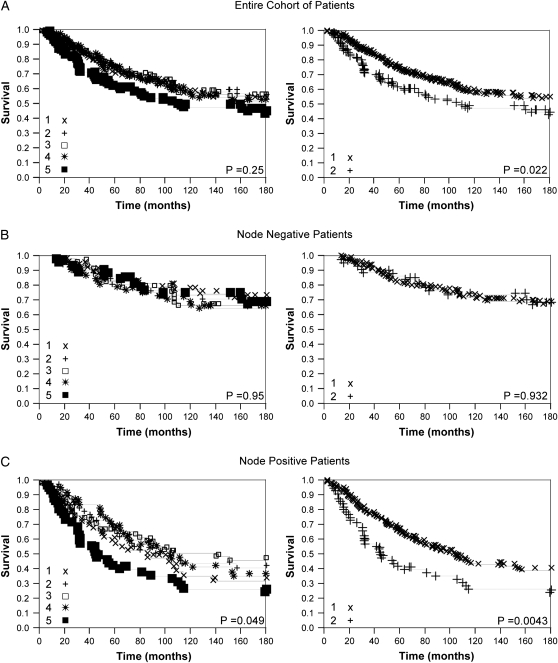
For the purpose of visual demonstration, continuous nonnuclear Grb7 AQUA scores were divided arbitrarily into quintiles, reflecting the use of routine statistical divisions in the absence of a commonly used justification for division of expression levels. Kaplan–Meier survival curves were generated for the Grb7 scores as shown in Figure 2 (left panel). Using these divisions, log-rank analysis revealed an association between quintiles of Grb7 expression and survival in the node-positive subset only (P = 0.049). The figure demonstrates clear convergence of the lower four quintiles of Grb7 expression, while the highest quintile is clearly associated with decreased survival, providing clear rationale and justification for division of the population into high expressers (top quintile) and low expressers (lower 80% of Grb7 expressers). Using this cut point we found a strong association between high Grb7 expression and decreased survival in the entire cohort and the node-positive subset by log-rank analysis (P = 0.02 and P = 0.0043, respectively), as shown in Figure 2 (right panel). To provide further justification for this cut point, we used the nonparametric Kolmogorov–Smirnov test to compare the empirical Grb7 distributions of the subset of individuals who lived at least 15 years and uncensored individuals who lived <15 years. Based on this test these two distributions are significantly different (P < 0.046) and the largest difference between the corresponding cumulative distribution functions occurs when binarizing Grb7 expression levels at an AQUA score of 31.81 (73rd percentile), which is close to the cut point implemented in the Kaplan–Meier curves.
Figure 2.
Kaplan–Meier survival curves for growth factor receptor-bound protein-7. Automated QUantitative Analysis scores divided into quintiles (left) and dichotomized by the top quintile versus the rest (right) for the entire cohort of patients (A), node-negative patients (B) and node-positive patients (C). Left panel: 1 = first (lowest) quintile, 2 = second quintile, 3 = third quintile, 4 = fourth quintile, 5 = fifth quintile. Right panel: 1 = low expressers (first four quintiles), 2 = high expressers (fifth quintile).
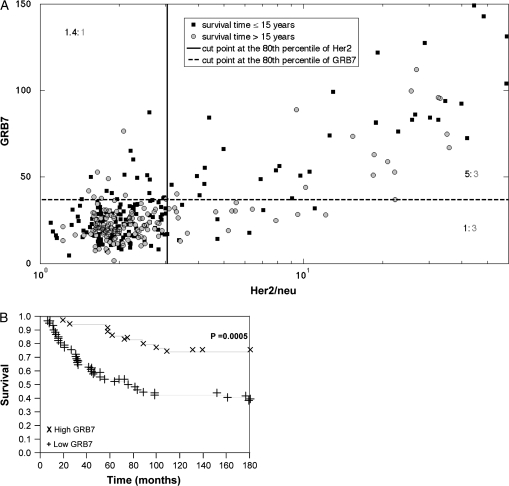
Despite the clear association between Grb7 and HER2/neu expression, many cases have different expression ratios between these two markers. Therefore, we further analyzed the joint distribution of these markers in the context of survival, as visually demonstrated in the two-dimensional plot in Figure 3A. Grb7 AQUA scores were divided into two groups using the 80th percentile cut point described above, with the top quintile of continuous AQUA scores in one group, and the lower 80% of scores in the second group. HER2/neu scores were similarly binarized by the 80th percentile, reflecting the approximate designation of HER2/neu scores into ‘high’ and ‘low’ categories in clinical practice. To further assess cut points, we used a method developed by Peacock [38], detailed in the supplemental materials (available at Annals of Oncology online). There is a clear abundance of nonsurvivors (62.5%, black squares) in the high Grb7/high HER2/neu group (top right quadrant) when compared with the patients who have high expression of HER2/neu, but low expression of Grb7 (37.5%). To further demonstrate the difference between Grb7 high and low expressers within the HER2/neu-overexpressing subset, we generated Kaplan–Meier survival curves in Figure 3B. By log-rank analysis at 15 years, we demonstrate that patients who are high expressers for both Grb7 and HER2/neu have decreased survival when compared with patients who only have high HER2/neu expression (P = 0.0005).
Figure 3.
Visualization of coexpression patterns of HER2/neu and growth factor receptor-bound protein-7 (Grb7) and the association with survival. (A) A two-dimensional plot divided into four quadrants with high Grb7 (top 20%) expressers in the top quadrants and high HER2/neu expressers (top 20%) in the two right-hand quadrants. Patients who were dead of disease at 15 years are depicted by black squares, while those who were alive at 15 years are depicted by gray circles. The ratios shown in the different quadrants are corrected for relative frequencies of survivors versus nonsurvivors in the entire population. The top right quadrant includes patients who have high expression of both markers and contains an abundance of nonsurvivors when compared with the lower right quadrant containing high HER2/neu, low Grb7 expressers. This is further demonstrated in the Kaplan–Meier survival curves in panel B, which show a significant difference in survival between high and low Grb7 expressers among patients with HER2/neu-overexpressing tumors.
discussion
In this work we assessed expression of Grb7 protein levels in a quantitative fashion on a large cohort of primary breast cancer specimens and evaluated the association with breast cancer-specific survival. The method of analysis used gives objective and continuous measures of expression, rather than routinely used pathologist-based divisions of staining into nominal scores or ‘positive/negative’. Our results were reproducible when using a second set of array with different cores from tumors of the same patients.
Consistent with published reports regarding prognostic value of Grb7 mRNA levels [27, 28], we demonstrated that high Grb7 protein expression is associated with decreased survival. In our study high Grb7 protein expression was strongly associated with decreased survival in the entire cohort (P = 0.0034) and in the node-positive subset (P = 0.0019) by Cox univariate analysis of continuous scores. The association with survival in the node-negative subset was not significant. High Grb7 expression retained its independence as a prognostic factor on multivariable analysis (P = 0.01). Thus, high Grb7 expression defines a subset of breast cancer patients with decreased survival. To the best of our knowledge, this is the first large cohort study assessing Grb7 protein expression and its association with survival.
Staging of primary breast cancer is an important step in selecting patients for adjuvant chemotherapy, which reduces the likelihood of disease recurrence and death. Standard prognostic variables (tumor size, grade, stage, hormone receptor expression and lymph node involvement) are routinely used to select the course of therapy. Molecular markers in the primary specimen that are highly associated with survival, such as Grb7, could supplement standard staging information and enable us to identify breast cancer patients at high risk for disease recurrence, thus avoiding the toxicity associated with chemotherapy for the vast majority of patients who are cured by local therapy alone [39, 40].
In addition to supplementing staging information, expression of molecular markers such as Grb7 can assist us in improving selection of specific therapies. We found a strong association between expression of Grb7 and HER2/neu. These two genes are both located on chromosome 17q11–21, which is a commonly amplified region in breast carcinomas, particularly those that have been shown to be driven by HER2/neu [11, 12, 23]. We demonstrated that coexpression of these molecules was associated with worse prognosis than high HER2/neu alone, indicating that Grb7 coexpression might be necessary for HER2/neu function. The importance of HER2/neu in breast cancer is well established [41]; HER2/neu is overexpressed in approximately 20%–30% of invasive breast cancers and is associated with poor prognosis, as reviewed previously [42]. Drugs that target HER2/neu have been shown to prolong survival, as reviewed previously [43–45]. Given that Grb7 strongly binds to HER2/neu via its SH2 domain, the improved survival among HER2/neu overexpressers who have low Grb7 compared with those with high levels as seen in Figure 3 further supports the notion that Grb7 is essential for signaling through HER2/neu. This finding has very important clinical implications for the treatment of breast cancer: only a subset of HER2/neu-overexpressing breast cancers are sensitive to trastuzumab [43, 46, 47], and it is possible that in the trastuzumab-resistant cases, signaling through the HER2/neu pathway is not driving tumor growth due to low levels of Grb7. Conversely, in patients with overexpression of both proteins, trastuzumab efficacy might be augmented by concurrent targeting of Grb7.
As detailed in the introduction, inhibitors of Grb7 are currently in preclinical development, as reviewed previously [7]. One of the promising anti-Grb7 peptides, G7-18NATE, inhibits proliferation of HER2/neu-overexpressing cells and has no significant effects on the non-HER2/neu-expressing MCF-7cells and nonmalignant MCF-10A or NIH3T3 cells [22]. G7-18NATE is synergistic with trastuzumab in vitro in HER2/neu-overexpressing cells [22, 33]. Since Grb7 also binds to other oncogenic receptor tyrosine kinases, such as EGFR [1], ErbB3 [18] and ErbB4 [18], combining Grb7 inhibitors with inhibitors of these kinases might be beneficial in breast cancers that are driven by these receptors or in other diseases.
In summary, we have demonstrated a strong association between high Grb7 expression and decreased survival in primary breast cancer, both in the entire patient cohort and among the node-positive patients. Grb7 expression could supplement current standard immunohistochemical analyses in identifying patients at the highest risk for relapse, who might benefit from aggressive adjuvant therapy. The association between Grb7 expression and sensitivity to Grb7-targeting agents remains to be determined, yet our data indicate that Grb7 might be a good drug target in a subset of breast cancer patients, particularly HER2/neu-positive breast cancers. Future studies using Grb7-targeting agents should include assessment of Grb7 levels.
funding
National Institutes of Health (R33 CA 106709, R33 CA 110511 to D.L.R.); Breast Cancer Alliance to H.M.K; Susan G. Komen Foundation (BCTR88406) to Y.K.
disclosures
DLR and RLC are founders, stockholders and consultants of HistoRx, a private corporation to which Yale University has given exclusive rights to produce and distribute software and technologies embedded in AQUA. Yale University retains patent rights for the AQUA technology. The other authors have no competing interests.
Supplementary Material
References
- 1.Margolis B, Silvennoinen O, Comoglio F, et al. High-efficiency expression/cloning of epidermal growth factor-receptor-binding proteins with Src homology 2 domains. Proc Natl Acad Sci U S A. 1992;89:8894–8898. doi: 10.1073/pnas.89.19.8894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shen TL, Han DC, Guan JL. Association of Grb7 with phosphoinositides and its role in the regulation of cell migration. J Biol Chem. 2002;277:29069–29077. doi: 10.1074/jbc.M203085200. [DOI] [PubMed] [Google Scholar]
- 3.Daly RJ. The Grb7 family of signalling proteins. Cell Signal. 1998;10:613–618. doi: 10.1016/s0898-6568(98)00022-9. [DOI] [PubMed] [Google Scholar]
- 4.Han DC, Shen TL, Guan JL. The Grb7 family proteins: structure, interactions with other signaling molecules and potential cellular functions. Oncogene. 2001;20:6315–6321. doi: 10.1038/sj.onc.1204775. [DOI] [PubMed] [Google Scholar]
- 5.Holt LJ, Daly RJ. Adapter protein connections: the MRL and Grb7 protein families. Growth Factors. 2005;23:193–201. doi: 10.1080/08977190500196267. [DOI] [PubMed] [Google Scholar]
- 6.Shen TL, Guan JL. Grb7 in intracellular signaling and its role in cell regulation. Front Biosci. 2004;9:192–200. doi: 10.2741/1229. [DOI] [PubMed] [Google Scholar]
- 7.Pero SC, Daly RJ, Krag DN. Grb7-based molecular therapeutics in cancer. Expert Rev Mol Med. 2003;5:1–11. doi: 10.1017/S1462399403006227. [DOI] [PubMed] [Google Scholar]
- 8.Margolis B. The GRB family of SH2 domain proteins. Prog Biophys Mol Biol. 1994;62:223–244. doi: 10.1016/0079-6107(94)90013-2. [DOI] [PubMed] [Google Scholar]
- 9.Cohen GB, Ren R, Baltimore D. Modular binding domains in signal transduction proteins. Cell. 1995;80:237–248. doi: 10.1016/0092-8674(95)90406-9. [DOI] [PubMed] [Google Scholar]
- 10.Pawson T, Gish GD, Nash P. SH2 domains, interaction modules and cellular wiring. Trends Cell Biol. 2001;11:504–511. doi: 10.1016/s0962-8924(01)02154-7. [DOI] [PubMed] [Google Scholar]
- 11.Stein D, Wu J, Fuqua SA, et al. The SH2 domain protein GRB-7 is co-amplified, overexpressed and in a tight complex with HER2 in breast cancer. Embo J. 1994;13:1331–1340. doi: 10.1002/j.1460-2075.1994.tb06386.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gullick WJ. The role of the epidermal growth factor receptor and the c-erbB-2 protein in breast cancer. Int J Cancer Suppl. 1990;5:55–61. doi: 10.1002/ijc.2910460708. [DOI] [PubMed] [Google Scholar]
- 13.Frantz JD, Giorgetti-Peraldi S, Ottinger EA, Shoelson SE. Human GRB-IRbeta/GRB10. Splice variants of an insulin and growth factor receptor-binding protein with PH and SH2 domains. J Biol Chem. 1997;272:2659–2667. doi: 10.1074/jbc.272.5.2659. [DOI] [PubMed] [Google Scholar]
- 14.Han DC, Shen TL, Miao H, et al. EphB1 associates with Grb7 and regulates cell migration. J Biol Chem. 2002;277:45655–45661. doi: 10.1074/jbc.M203165200. [DOI] [PubMed] [Google Scholar]
- 15.Han DC, Shen TL, Guan JL. Role of Grb7 targeting to focal contacts and its phosphorylation by focal adhesion kinase in regulation of cell migration. J Biol Chem. 2000;275:28911–28917. doi: 10.1074/jbc.M001997200. [DOI] [PubMed] [Google Scholar]
- 16.Han DC, Guan JL. Association of focal adhesion kinase with Grb7 and its role in cell migration. J Biol Chem. 1999;274:24425–24430. doi: 10.1074/jbc.274.34.24425. [DOI] [PubMed] [Google Scholar]
- 17.Pawson T. SH2 and SH3 domains in signal transduction. Adv Cancer Res. 1994;64:87–110. doi: 10.1016/s0065-230x(08)60835-0. [DOI] [PubMed] [Google Scholar]
- 18.Fiddes RJ, Campbell DH, Janes PW, et al. Analysis of Grb7 recruitment by heregulin-activated erbB receptors reveals a novel target selectivity for erbB3. J Biol Chem. 1998;273:7717–7724. doi: 10.1074/jbc.273.13.7717. [DOI] [PubMed] [Google Scholar]
- 19.Rodriguez-Viciana P, Sabatier C, McCormick F. Signaling specificity by Ras family GTPases is determined by the full spectrum of effectors they regulate. Mol Cell Biol. 2004;24:4943–4954. doi: 10.1128/MCB.24.11.4943-4954.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tanaka S, Sugimachi K, Kawaguchi H, et al. Grb7 signal transduction protein mediates metastatic progression of esophageal carcinoma. J Cell Physiol. 2000;183:411–415. doi: 10.1002/(SICI)1097-4652(200006)183:3<411::AID-JCP14>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 21.Tanaka S, Mori M, Akiyoshi T, et al. A novel variant of human Grb7 is associated with invasive esophageal carcinoma. J Clin Invest. 1998;102:821–827. doi: 10.1172/JCI2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pero SC, Shukla GS, Cookson MM, et al. Combination treatment with Grb7 peptide and Doxorubicin or Trastuzumab (Herceptin) results in cooperative cell growth inhibition in breast cancer cells. Br J Cancer. 2007;96:1520–1525. doi: 10.1038/sj.bjc.6603732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kauraniemi P, Barlund M, Monni O, Kallioniemi A. New amplified and highly expressed genes discovered in the ERBB2 amplicon in breast cancer by cDNA microarrays. Cancer Res. 2001;61:8235–8240. [PubMed] [Google Scholar]
- 24.Kauraniemi P, Kuukasjarvi T, Sauter G, Kallioniemi A. Amplification of a 280-kilobase core region at the ERBB2 locus leads to activation of two hypothetical proteins in breast cancer. Am J Pathol. 2003;163:1979–1984. doi: 10.1016/S0002-9440(10)63556-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luoh SW. Amplification and expression of genes from the 17q11 approximately q12 amplicon in breast cancer cells. Cancer Genet Cytogenet. 2002;136:43–47. doi: 10.1016/s0165-4608(01)00657-4. [DOI] [PubMed] [Google Scholar]
- 26.Bieche I, Onody P, Tozlu S, et al. Prognostic value of ERBB family mRNA expression in breast carcinomas. Int J Cancer. 2003;106:758–765. doi: 10.1002/ijc.11273. [DOI] [PubMed] [Google Scholar]
- 27.Vinatzer U, Dampier B, Streubel B, et al. Expression of HER2 and the coamplified genes Grb7 and MLN64 in human breast cancer: quantitative real-time reverse transcription-PCR as a diagnostic alternative to immunohistochemistry and fluorescence in situ hybridization. Clin Cancer Res. 2005;11:8348–8357. doi: 10.1158/1078-0432.CCR-05-0841. [DOI] [PubMed] [Google Scholar]
- 28.Cobleigh MA, Tabesh B, Bitterman P, et al. Tumor gene expression and prognosis in breast cancer patients with 10 or more positive lymph nodes. Clin Cancer Res. 2005;11:8623–8631. doi: 10.1158/1078-0432.CCR-05-0735. [DOI] [PubMed] [Google Scholar]
- 29.Bai T, Luoh SW. GRB-7 facilitates HER-2/neu-mediated signal transduction and tumor formation. Carcinogenesis. 2008;29:473–479. doi: 10.1093/carcin/bgm221. [DOI] [PubMed] [Google Scholar]
- 30.Kao J, Pollack JR. RNA interference-based functional dissection of the 17q12 amplicon in breast cancer reveals contribution of coamplified genes. Genes Chromosomes Cancer. 2006;45:761–769. doi: 10.1002/gcc.20339. [DOI] [PubMed] [Google Scholar]
- 31.Pero SC, Oligino L, Daly RJ, et al. Identification of novel non-phosphorylated ligands, which bind selectively to the SH2 domain of Grb7. J Biol Chem. 2002;277:11918–11926. doi: 10.1074/jbc.M111816200. [DOI] [PubMed] [Google Scholar]
- 32.Tanaka S, Pero SC, Taguchi K, et al. Specific peptide ligand for Grb7 signal transduction protein and pancreatic cancer metastasis. J Natl Cancer Inst. 2006;98:491–498. doi: 10.1093/jnci/djj105. [DOI] [PubMed] [Google Scholar]
- 33.Yakes FM, Chinratanalab W, Ritter CA, et al. Herceptin-induced inhibition of phosphatidylinositol-3 kinase and Akt is required for antibody-mediated effects on p27, cyclin D1, and antitumor action. Cancer Res. 2002;62:4132–4141. [PubMed] [Google Scholar]
- 34.Paik S, Shak S, Tang G, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 35.Dolled-Filhart M, McCabe A, Giltnane J, et al. Quantitative in situ analysis of beta-catenin expression in breast cancer shows decreased expression is associated with poor outcome. Cancer Res. 2006;66:5487–5494. doi: 10.1158/0008-5472.CAN-06-0100. [DOI] [PubMed] [Google Scholar]
- 36.Camp RL, Chung GG, Rimm DL. Automated subcellular localization and quantification of protein expression in tissue microarrays. Nat Med. 2002;8:1323–1327. doi: 10.1038/nm791. [DOI] [PubMed] [Google Scholar]
- 37.Nadler Y, Camp RL, Giltnane JM, et al. Expression patterns and prognostic value of Bag-1 and Bcl-2 in breast cancer. Breast Cancer Res. 2008;10:R35. doi: 10.1186/bcr1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peacock J. Two-dimensional goodness-of-fit testing in astronomy. Mon Not R Astr Soc. 1983;202:615–627. [Google Scholar]
- 39.Fisher B, Dignam J, Wolmark N, et al. Tamoxifen and chemotherapy for lymph node-negative, estrogen receptor-positive breast cancer. J Natl Cancer Inst. 1997;89:1673–1682. doi: 10.1093/jnci/89.22.1673. [DOI] [PubMed] [Google Scholar]
- 40.Fisher B, Anderson S, Tan-Chiu E, et al. Tamoxifen and chemotherapy for axillary node-negative, estrogen receptor-negative breast cancer: findings from National Surgical Adjuvant Breast and Bowel Project B-23. J Clin Oncol. 2001;19:931–942. doi: 10.1200/JCO.2001.19.4.931. [DOI] [PubMed] [Google Scholar]
- 41.Ross JS, Fletcher JA. The HER-2/neu oncogene in breast cancer: prognostic factor, predictive factor, and target for therapy. Oncologist. 1998;3:237–252. [PubMed] [Google Scholar]
- 42.Carney WP, Leitzel K, Ali S, et al. HER-2/neu diagnostics in breast cancer. Breast Cancer Res. 2007;9:207. doi: 10.1186/bcr1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Demonty G, Bernard-Marty C, Puglisi F, et al. Progress and new standards of care in the management of HER-2 positive breast cancer. Eur J Cancer. 2007;43:497–509. doi: 10.1016/j.ejca.2006.10.020. [DOI] [PubMed] [Google Scholar]
- 44.Slamon DJ, Clark GM, Wong SG, et al. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235:177–182. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- 45.Sunami E, Shinozaki M, Sim MS, et al. Estrogen receptor and HER2/neu status affect epigenetic differences of tumor-related genes in primary breast tumors. Breast Cancer Res. 2008;10:R46. doi: 10.1186/bcr2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cobleigh MA, Vogel CL, Tripathy D, et al. Multinational study of the efficacy and safety of humanized anti-HER2 monoclonal antibody in women who have HER2-overexpressing metastatic breast cancer that has progressed after chemotherapy for metastatic disease. J Clin Oncol. 1999;17:2639–2648. doi: 10.1200/JCO.1999.17.9.2639. [DOI] [PubMed] [Google Scholar]
- 47.Piccart-Gebhart MJ, Procter M, Leyland-Jones B, et al. Trastuzumab after adjuvant chemotherapy in HER2-positive breast cancer. N Engl J Med. 2005;353:1659–1672. doi: 10.1056/NEJMoa052306. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.