Abstract
BACKGROUND
Human and animal studies have suggested that diet-derived flavonoids, in particular quercetin may play a beneficial role by preventing or inhibiting oncogenesis, but the underlying mechanism remains unclear. The aim of this study is to evaluate the effect(s) of quercetin on normal and malignant prostate cells and to identify the target(s) of quercetin’s action.
METHODOLOGY
We addressed this question using cells in culture and investigated whether quercetin affects key biological processes responsible for tumor cell properties such as cell proliferation and apoptosis and also studied the effect of quercetin on the proteome of prostate cancer cells using difference gel electrophoresis (DIGE) to assess changes in the expression of relevant proteins.
RESULTS
Our findings demonstrate that quercetin treatment of prostate cancer cells results in decreased cell proliferation and viability. Furthermore, we demonstrate that quercetin promotes cancer cell apoptosis by down-regulating the levels of heat shock protein (Hsp) 90. Depletion of Hsp90 by quercetin results in decreased cell viability, levels of surrogate markers of Hsp90 inhibition (intracellular and secreted), induced apoptosis and activation of caspases in cancer cells but not in normal prostate epithelial cells. Knockdown of Hsp90 by short interfering RNA also resulted in induction apoptosis similar to quercetin in cancer cells as indicated by annexin V staining.
CONCLUSION
Our results demonstrate that quercetin down-regulates the expression of Hsp90 which, in turn, induces inhibition of growth and cell death in prostate cancer cells while exerting no quantifiable effect on normal prostate epithelial cells.
Keywords: bioflavonoids, quercetin, polyphenols, anti-cancer, chaperone proteins, apoptosis
INTRODUCTION
Prostate cancer is the most frequently diagnosed, non-cutaneous malignancy of men in the industrialized world. Its incidence, which is increasing globally, varies among countries, with the US, Canada, Sweden, Australia, and France having the highest rates. The American Cancer Society estimates that in 2007 there were roughly, 218,890 new cases of prostate cancer with 27,050 deaths in the US [1]. The incidence and mortality of prostate cancer in the US is significantly higher in African-American men [1]. Despite intense efforts to develop treatments, effective agents are still not available. The profound geographic and racial variation in prostate cancer risk has stimulated investigations into how diet affects the development and progression of prostate cancer [2,3]. Epidemiological and dietary intervention studies in animals and humans have suggested that diet-derived phenolics, in particular the flavonoids, may play a beneficial role in inhibiting, reversing or retarding tumorigenesis [4]. Flavonoids are known to possess anti-inflammatory, anti-oxidant, anti-allergic, hepato-protective, anti-thrombotic, anti-viral and anti-carcinogenic activities [5]. These activities of flavonoids are thought to be mediated by interfering with a large number of mammalian enzymes, such as detoxifying enzymes that are involved in cell division and proliferation pathways [6]. Among the flavonoids, quercetin (3,3′,4′,5,7-pentahydroxyflavone) is a naturally occurring flavone found in the plant kingdom. It is a component of most edible fruits and vegetables, with the highest concentrations being found in onions, apples, and red wine [7,8]. Quercetin has many biological activities, such as anti-tumor and anti-proliferative effects on a wide range of human cancer cell lines and these effects are believed to be mediated by inhibition of glycolysis, macromolecule synthesis, and enzyme activity [9–13]. Several studies have shown that quercetin has a broad range of pharmacological properties that include selective anti-proliferative effects [14] and cell death, predominantly through an apoptotic mechanism in cancer cells but not in normal cells [15]. The anti-proliferative effect of quercetin is believed to be exerted by producing arrest in the G1 phase of the cell cycle [16] or through interaction with cell cycle-regulated proteins, like cyclin D1 and CDK-4 [17]. Quercetin also has been shown to induce release of cytochrome c and activation of caspase-9 and caspase-3 in HL-60 cells to trigger apoptosis [18]. Moreover, quercetin is believed to be a potential phosphotidylinositol-3-kinase (PI3K) inhibitor, an enzyme involved in the pivotal cell survival pathway [19]. Therefore, the ability of quercetin to prevent and/or to retard tumor growth is probably a multifunctional effect. Our previous study showed that quercetin induced gene expression and production of Th-1 derived interferon (IFN)-γ, and down-regulated Th-2 derived interleukin (IL)-4 by normal peripheral blood mononuclear cells (PBMC) [20]. Further, quercetin treatment also increased the phenotypic expression of IFN-γ positive cells and decreased IL-4 positive cells [20]. We have shown that quercetin significantly inhibits tumor necrosis factor (TNF)-α gene expression and production by PBMC [21]. Recently we showed that quercetin inhibits the colony forming ability and expression of genes related to cell cycle regulation and tumorigenesis in prostate cancer cells [13]. The present study was undertaken to determine the mechanism underlying quercetin’s actions on prostate cancer cells in an effort to evaluate the potential pharmacological use of this flavonoid for prostate cancer. We measured the anti-proliferative effect of quercetin on prostate cancer cells and identified heat shock protein 90 (Hsp90) as an important molecular target of quercetin by using difference gel electrophoresis (DIGE) to assess changes in the expression of relevant proteins.
MATERIALS AND METHODS
Cells and Culture Conditions
Normal prostate epithelial cells (NP) were obtained from Clonetics (Walkersville, MD). The human prostate cancer cell lines, PC-3, LNCaP, and DU-145 were obtained from the American Type Culture Collection (Manassas, VA) and differ significantly in their aggressive phenotypes as we already demonstrated with PC-3 > DU-145 > LNCaP [22]. DU-145 and PC-3 were isolated from metastases in brain and lumbar vertebra respectively. DU-145 and PC-3 are androgen independent. LNCaP cells are androgen responsive but their growth does not depend upon androgen. Cancer cells were cultured in RPMI 1640 medium supplemented with non-essential amino acids, l-glutamine, a twofold vitamin solution (Invitrogen Life Technologies, Grand Island, NY), sodium pyruvate, Earl’s balanced salt solution, 10% fetal bovine serum, and penicillin and streptomycin (Flow Labs, Rockville, MD). Normal prostate epithelial cells were maintained in proprietary medium as recommended by the manufacturer. Approximately 3 × 106 cells/60 mm dish were cultured at 37°C in a humidified atmosphere of 5% CO2 and 95% air for 48 hr. Cells were harvested from culture dishes after brief trypsinization. Only cell suspensions > 90% viable was used for experiments.
Treatment of Cell Cultures With Quercetin and Dihydroquercetin
Cells (1 × 105) were cultured overnight in 6-well plate. To detect cell viability, cells were transferred into 96-well plates at 1 × 104 cells per well. Quercetin or dihydroquercetin (an inactive analogue of quercetin) was dissolved in DMSO and added to the medium with final concentrations ranging from 5 to 100 µM. Cells were incubated at 37°C for 24 hr. Cells treated with DMSO alone 0.2% served as controls. Cell viability was determined by two methods including the standard trypan blue vital dye exclusion method and the Dojindo Cell Counting Kit-8 (Dojindo Molecular Technologies, Gaithersburg, MD) method as described by the manufacturer.
RNA Extraction and Real Time (RT), Quantitative (Q) Polymerase Chain Reaction (PCR)
Cytoplasmic RNA was extracted by an acid guanidinium thiocyanate–phenol–chloroform method [23] using the TRIzol® reagent (Invitrogen, Carlsbad, CA) and the final RNA pellet was dried and re-suspended in diethyl pyrocarbonate (DEPC) water and the concentration of RNA was determined using a spectrophotometer at 260 nm. DNA contamination in the RNA preparation was removed by treating the RNA preparation with DNAse (1 IU / µg of RNA, Promega, Madison, WI) for 30 min at 37°C, followed by proteinase K digestion at 37°C for 15 min and subsequent extraction with phenol/chloroform and NH4OAc/ETOH precipitation. The precipitated RNA was stored at −70°C until used. Relative abundance of Hsp90 mRNA species was assessed using the SYBR green master mix from Stratagene (La Jolla, CA) to perform Q-PCR using the ABI Prism 5700 instrument that detects and plots the increase in fluorescence versus PCR cycle number to produce a continuous measure of PCR amplification. To provide precise quantification of initial target in each PCR reaction, the amplification plot was examined at a point during the early log phase of product accumulation. This was accomplished by assigning a fluorescence threshold above background and determining the time point at which each sample’s amplification plot reaches the threshold (defined as the threshold cycle number or CT). Differences in threshold cycle number were used to quantify the relative amount of PCR target contained within each tube [24]. Relative mRNA species expression was quantitated and expressed as transcript accumulation index (TAI = 2−ΔΔ CT), calculated using the comparative CT method [25]. All data were controlled for quantity of RNA input by performing measurements on an endogenous reference gene, β-actin. In addition, results on RNA from treated samples were normalized to results obtained on RNA from the control, untreated sample.
Proteomic Analysis of the Effects of Quercetin on Prostate Cancer Cells
Proteomic studies were undertaken to analyze differential protein expression in quercetin treated and untreated PC-3 prostate cancer cells. Samples were run on 2D–DIGE (Ettan? DIGE system, GE Health Care, Piscataway, NJ.) available as a core facility at our institution.
DIGE
After treatment with quercetin the PC-3 cells were washed twice with 1X PBS (Invitrogen Life Technologies). Total protein was extracted using standard cell lysis buffer (30 mM Tris–HCl; 8 M urea; 4% w / v) CHAPS, adjusted to pH 8.5 for 10 min on ice. Cell lysate was centrifuged at 4°C for 10 min at 12,000g and was further purified by precipitation with chloroform/methanol as described [26]. Samples were re-suspended in standard cell lysis buffer. Final cell lysate protein concentrations were determined using Coomassie Protein Reagent (Bio-Rad, Hercules, CA) and used for DIGE analysis. PC-3 cells cultured identically without quercetin served as the control. The Ettan DIGE technique was used to detect differences in protein abundance between normal and experimental samples. The Ettan DIGE system uses three CyDye DIGE fluors (Cy2, Cy3, Cy5), each with a unique fluorescent wavelength and matched for mass and charge. CyDyes form a covalent bond with the free epsilon amino group on lysine residues of the sample proteins. CyDyes label approximately 2% of the lysine residues. This system allows for two experimental samples and an internal standard to be simultaneously separated on the same gel. The internal standard is a pool of equal amount of all the experimental samples labeled with Cy2 dye. The use of an internal standard facilitates accurate inter-gel matching of spots and allows for data normalization between gels to minimize gel-to-gel experimental variability [27]. Cell lysates were labeled with CyDye per the manufacturer. All reagents used were from GE Healthcare (Piscataway, NJ). Briefly, 50 µg of cell lysate was labeled with 400 pmol of either Cy3 or Cy5 or Cy2 (Cy2 was used to label the internal standard) on ice for 30 min and then quenched with a 50-fold molar excess of free lysine. Cy3, Cy5, and Cy2 labeled samples and unlabeled protein (500–800 µg) were then pooled. An equal volume of 2X sample buffer was added incubated on ice for 10 min. The total volume of sample was adjusted to 450 µl with re-hydration buffer [4% (w / v) Pharmalytes 3–10 nonlinear (NL); 13 mM DTT]. Samples were applied to immobilized nonlinear pH gradient (IPG) strips (24 cm, pH 3–10) and absorbed by active re-hydration at 30 V for 13 hr. Iso-electric focusing (IEF) was carried out using an IPGphor IEF system with a three phase program; first phase at 500 V for 1 hr, second phase at 1,000 V for 1 hr and third phase (linear gradient) 8,000–64,000 V for 2 hr (50 µA maximum per strip). Prior to separation in the second dimension, strips were equilibrated for 15 min in equilibration buffer I [50 mM Tris–HCl, 6 M urea, 30%(v / v) glycerol, 2% (w / v) SDS, 0.5% (w / v) DTT]. The strips were again equilibrated for 15 min in the equilibration buffer II [50 mM Tris–HCl, 6 M urea 30% (v / v) glycerol, 2% (w / v) SDS, 4.5% (w / v) iodoacetamide] and the equilibrated IPG strips were then transferred onto 18 cm × 20 cm, 12.5% uniform polyacrylamide gels poured between low fluorescence glass plates. Gels were bonded to inner plates using Bind-Silane solution (GE Health Care) according to the manufacturer supplied instruction. Strips were overlaid with 0.9% agarose in 1X running buffer containing bromophenol blue and were run for 16 hr (1.8 W per gel, overnight) at 15°C, in an Ettan DALT electrophoresis system. After the run was completed, the 2D gels were scanned three times with a Typhoon 9410 imager, each time at different excitation wavelengths [Cy3, 580 BP 30 / green (532 nm); Cy5, 670 BP 30 / red (633 nm); Cy2, 520 BP 40 / blue (488 nm)]. Images were cropped in ImageQuant v5.2 software and then imported into DeCyder Differential In-gel Analysis (DIA) software v 5.0 from GE Healthcare for spot identification and normalization of spot intensities within each gel. Gels were fixed in 30% (v / v) methanol, 7.5% (v / v) acetic acid for 3 hr and stained with SYPRO-Ruby dye (Molecular Probes, Eugene, OR) overnight at room temperature. Gels were de-stained in water and scanned using the Typhoon 9410 scanner. Spots of interest were excised from the gel using the Ettan Spot Picker. DeCyder software (GE Healthcare) has been specifically developed for use with Ettan DIGE. DeCyder software allows for automatic detection of spots, background subtraction, quantitation, normalization, internal standardization and integral matching. DeCyder DIA draws boundaries around spots in a composite gel image obtained from the intra-gel overlap of the Cy2, Cy3, and Cy5 scanned images and normalizes the data from each CyDye to account for differences in dye fluorescence intensity, scanner sensitivity. To avoid artifacts due to gel heterogeneity at the edges of the images, we define an area of interest within the gel. An exclusion filter using peak-slope, area, height and volume is defined to eliminate non-protein spots from further analysis. The abundance difference between samples run on the same gel is then analyzed. The biological variation analysis (BVA) module of DeCyder is used to match all image comparisons from in-gel analysis for a cross-gel statistical analysis. DeCyder BVA initially calculates normalized intensities (standard abundance) for all spots by comparison to the internal standard and from this an average volume ratio and a Student’s paired t-test derived P-value are calculated for each spot. A paired t-test derived P-value of ≤0.05 was considered statistically significant [27].
Nano high performance liquid chromatography and tandem mass spectroscopy (HPLC-MS/MS)
Excised spots were sent to the Proteomic Analysis Laboratory at the University of Arizona for analysis. In-gel digestion and HPLC-MS/MS were performed as described by Breci et al. [28]. Briefly, gel slices were de-stained [28–30] and digested with trypsin [31]. The tryptic peptides were extracted with 5% formic acid/50% CH3CN. HPLC was performed using a microbore system (Surveyor, Thermo Finnigan, San Jose, CA). The HPLC column eluate was directed into a Thermo Finnigan LCQ Deca XP Plus ion trap mass spectrometer. Automated peak recognition, dynamic exclusion and daughter ion scanning of the two most intense ions were performed using X calibur software [32,33]. Spectra were scanned over the range 400–1,400 mass units. MS/MS data were analyzed using SEQUEST software and searched against the latest version of the National Center for Biotechnology’s public non-redundant protein database [27]. Statistical significance was determined using Student’s t-test (Sigmastat, SPSS, Inc.) P < 0.05 was considered significant.
Measurement of Hsp90 and Hsp90 Client Protein Levels by Western Blotting
Cell lysates were prepared by re-suspending cells in lysis buffer (65 mmol/L Tris–HCl (pH 7.4), 150 mmol/L NaCl, 1 mmol/L EDTA, 1% NP40, 1% sodium deoxycholate, 1 µg/ml aprotinin, 100 µg/ml phenylmethylsulfonyl fluoride) for 30 min at 4°C and cleared by centrifugation for 30 min at 13,000g. Supernates were collected and stored at −80°C until used. Total protein concentration was determined using Coomassie Protein Reagent (Bio-Rad). Other client proteins have been used as indicators of Hsp90 inhibition in clinical trials of Hsp90 inhibitors [34]. Raf-1 and cyclin-dependent kinase (cdk) 4 are destabilized by Hsp90 inhibition and are used in this study as Hsp90 client proteins. Western blotting for Hsp90 and Hsp90 client proteins was done using the antibodies from (Santa Cruz Biotechnology, Inc., Santa Cruz, CA). β-actin expression was used as an internal control.
Immunolocalization of Hsp90
Prostate tumor cells and normal prostate epithelial cells were plated on chamber slides and incubated overnight followed by fixation in an ice-cold methanol/acetone (1:1) mixture. Cells were stained with Hsp90 primary antibody (Santa Cruz Biotechnology, Inc.) at 4°C overnight followed by a fluorescein isothiocyanate (FITC)-labeled secondary antibody for detection by immunofluorescent microscopy. Appropriate internal controls were used during all immuno-staining procedures. Imaging was done using a Leica Confocal Laser Scanning Microscope (TCS SP2 AOBS, Leica Micro-sysytems, Heidelgergh, GmbH) with an oil immersion objective lens and an argon ion laser was applied to excite the FITC.
Transfection of Hsp90 Double-Stranded siRNA
The Hsp90 siRNA pre-designed and validated by the manufacturer, Hsp90 control siRNA and scrambled siRNA bearing no homology with any relevant human gene which served as a control were all from Ambion (Austin, TX). The siRNA duplex was dissolved in RNAase free sterile water supplied by the manufacturer to a final concentration of 20 mM. For transfection, 1 × 105 cells were plated into 6-well plates and incubated overnight. Lipofectamine? 2000 (Invitrogen) reagent (10 µl) was added to 250 µl Opti-MEM medium (Invitrogen Life Technologies) and incubated at room temperature for 5 min. Hsp90, Hsp90 control siRNA or scrambled siRNA (40 nM) was added and incubated for another 20 min. A third set of transfections was done without any siRNA (mock transfection). The transfection reagent/siRNA complex was added to the wells containing 2 ml RPMI with 10% FBS and incubated for 4 hr at 37°C until medium was removed and replaced with fresh medium. Assays were done at 24, 48, and 72 hr after transfection.
Determination of Apoptosis Using the Annexin V Assay
Apoptosis was quantified by detecting surface exposure of phosphatidylserine in apoptotic cells using the Annexin V-FITC/PI (propidium iodide) apoptosis detection kit (BD Biosciences, CA). Briefly, cells were seeded in 3.5 cm dishes in 1 ml of medium and incubated with either quercetin or dihydroquercetin at doses of 5, 10, 25, 50, 100 µM for 24 hr respectively. Adherent and floating cells were combined and treated according to the manufacturer’s instructions and measured with FITC/PI staining using flow cytometry (Becton Dickinson, San Jose, CA). Apoptotic cells (annexin V+PI−) were differentiated from necrotic cells (annexin V+PI+, including apoptotic cells at late stage).
Terminal Deoxynucleotidyl Transferase-Mediated d UTP Nick End Labeling (TUNNEL) Assay for Measurement of in Situ Apoptosis
Prostate tumor cells were cultured on chamber slides. Cells were fixed with 4% paraformaldehyde in PBS (pH 7.4) for 1 hr at 25°C. Permeabilization of cells was achieved by incubation with 0.1% Triton X-100 in 0.1% sodium citrate for 2 min on ice. The TUNEL assay was then carried out according to the manufacturer’s instructions (Roche). Signal conversion was achieved using an anti-fluorescein antibody conjugated with alkaline phosphatase, and methyl green was used as a counterstain.
Caspase-3 and -9 Activity Assays
Caspase-9 and caspase-3 activities were measured by the Apo Target? assay kit (Invitrogen) per manufacturer’s instruction. Briefly, following treatment with the appropriate concentration of either quercetin, dihydroquercetin (0, 5, 10, 25, 50, 100 µM) or 0.2% DMSO for 24 hr, cells were trypsinized, harvested and lyzed in 50 µl of ice-cold lysis buffer and incubated on ice for 10 min. The samples where centrifuged at 10,000g for 2 min to collect the cytosol extract. To determine caspase 3 or 9 activity, identical amounts of cytosolic protein in 50 µl volume were aliquoted into a 96-well plate and 50 µl of 2X reaction buffer containing 10 mM of DTT was added to each well. The reaction was initiated by adding 5 µl of a 4 mM substrate (a synthetic tetra peptide specific for each caspase) solution containing p-nitroanilide (pNA) and the plate was incubated for 2 hr at 37°C. At the end of incubation the plate was read at 400 nm in a spectrophotometer. The increase or decrease in OD was recorded and was used to calculate the units of caspase activity per mg protein.
Insulin Like Growth Factor Binding Protein (IGFBP)-2 and Human Epidermal Growth Factor (HER)-2 ELISA
The level of IGFBP-2 and the secreted extracellular domain of HER-2 were measured using ELISA kits from R&D Systems (Minneapolis, MN) and Calbiochem (San Diego, CA) respectively. The assay was done according to the manufacturer’s protocol.
Statistical Analysis
All experiments with cells were repeated at least three times. Values are expressed as the mean ± SE. The significance of the difference between the control and each experimental test condition was analyzed by unpaired t-test, and a value of P < 0.05 was considered statistically significant.
RESULTS
Effect of Quercetin on the Viability of Prostate Cancer Cells
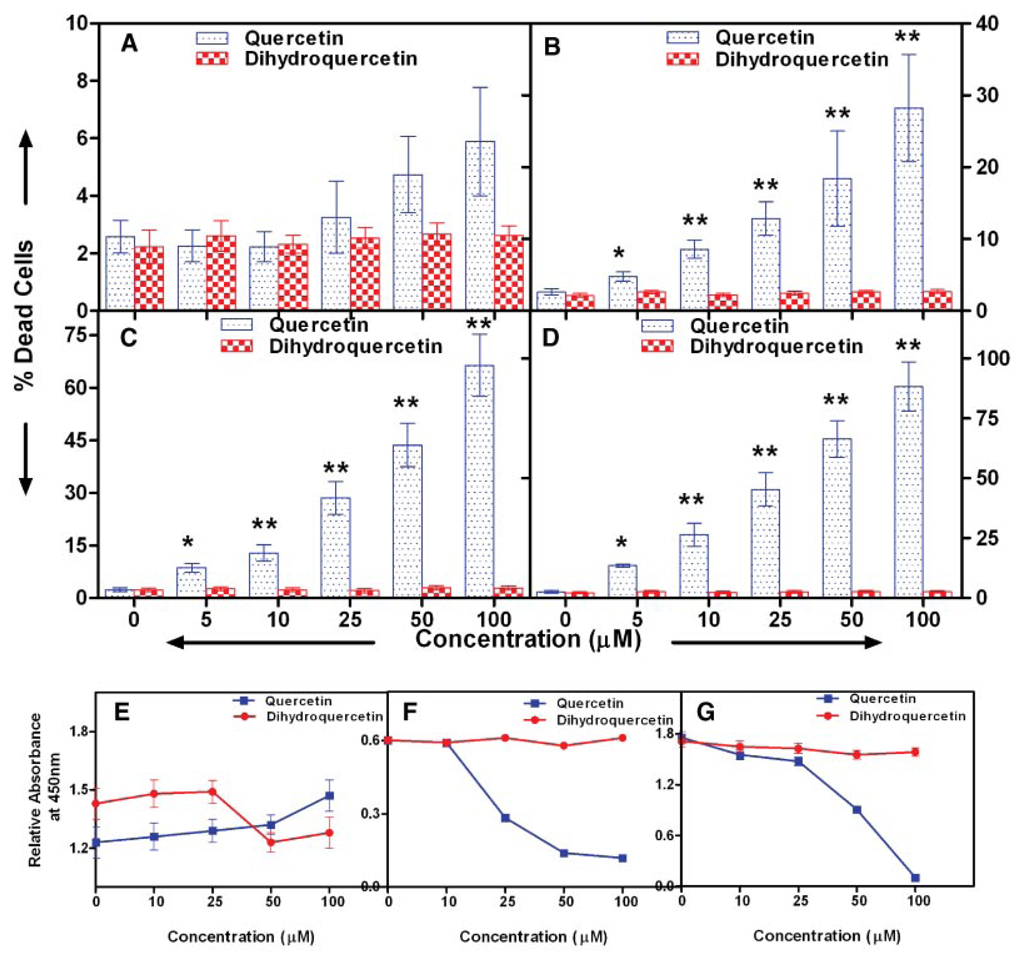
To determine if quercetin is cytotoxic to human prostate cancer cells we used the trypan blue exclusion method to assay cell viability. Cells were treated with 0–100 µM of quercetin or dihydroquercetin for 24 hr. Our results show that quercetin induced death in all three prostate cancer cell lines in a dose-dependent manner after 24 hr of incubation (Fig. 1). The effect of quercetin on viability was most impressive with the two aggressive prostate cancer cell lines. A concentration of 100 µM of quercetin produced 88% and 66% mortality in PC-3 and DU-145 cells respectively. The dose of quercetin yielding 50% mortality (IC50) was 30.2 µM for PC-3 cells and 64.2 µM for DU-145 cells. The extent of cytotoxicity induced in LNCaP cells was not as dramatic (Fig. 1B). Also treatment of prostate cancer cells or normal prostate epithelial cells with dihydo-quercetin had no significant effect on viability at all concentration studied (Fig. 1A). The above data were confirmed by incubating prostate cancer cells with different concentrations of quercetin and assessing the residual numbers of cells by measuring the absorbance of cultures at 450 nm. As shown in Figure 1E–G quercetin decreased the numbers of residual prostate cancer cells, LNCaP and PC-3, in a dose-dependent manner while having no effect on normal prostate epithelial cell numbers at equal concentration of quercetin. The inactive analog, dihydroquercetin, had no effect on either normal or malignant prostate cells as did a 0.2% DMSO vehicle control.
Fig. 1.
Effect of quercetin on the viability of prostate cancer cells. A: Normal prostate epithelial cells, (B) LNCaP, (C)DU-145, and (D) PC-3 prostate cancer cells were treated with increasing concentrations of quercetin for 24 hr and viability was determined by trypan blue vital dye exclusion. Viability was confirmed by a proprietary tetrazolium dye method using absorbance at 450 nm as described in the text: (E) normal prostate epithelial cells, (F) LNCaP, and (G) PC-3 cells. Values are mean ± SE of six experiments; *P < 0.05 and **P < 0.001 compared to treatment with dihydroquercetin which did not affect the viability of any of the cells in this study. [Color figure can be viewed in the online issue, which is available at http://www.interscience.wiley.com.]
Proteomic Analysis of the Effects of Quercetin on Prostate Cancer Cells
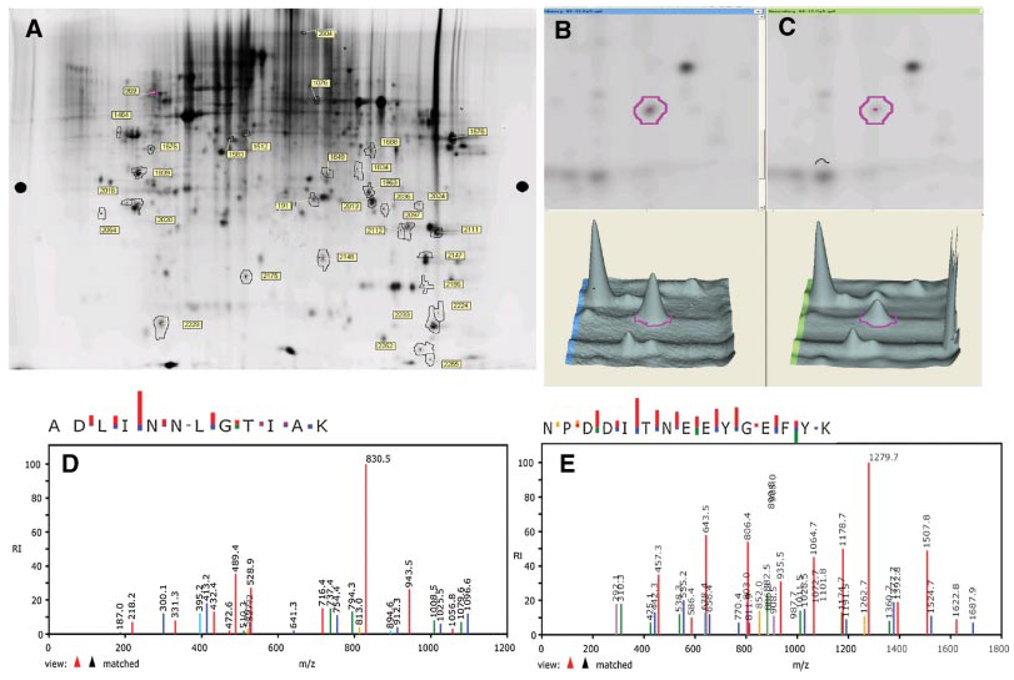
For these experiments we used PC-3 cells because they are the most aggressive phenotype and were most sensitive to treatment with quercetin. PC-3 cells were washed with PBS and lysates were prepared using a standard cell lysis buffer. Proteins from lysates of untreated PC-3 cells were labeled with Cy3 dye whereas proteins from lysates of quercetin treated cells were labeled with Cy5 dye and were subjected to two-dimensional (2D) gel electrophoresis. The images of the Cy3 and Cy5 stained gels were superimposed to reveal unique proteins and/or significant differences in the intensities of protein expression. After 2D gel electrophoresis, the analysis of the Cy2, Cy3, and Cy5 stained images was done using the BVA module of DeCyder software. Approximately 2,500 protein spots were visualized and several differences were observed after quercetin treatment. Of the ~2,500 protein spots selected by DeCyder software from an average of three experiments, 67% (1,675 spots) showed no significant change, 16% (400 spots) showed decreased abundance, and 18% (450 spots) revealed an increase in abundance. Statistically significant (Student’s t-test P < 0.05) changes were observed in 32 different proteins, as measured by spot volume ratios shown in the sypro-ruby stained 2D gel showing all protein spots (Fig. 2A). Twenty-five spots were selected in the order of the significance of their respective spot volume ratios for further analysis by LC-MS/MS. Proteins from 10 gel spots were successfully identified. The significantly and differentially expressed, selected spots were indicated by numbered outlines. Determination of tryptic peptide masses permitted identification of the specific parent proteins in most cases with multiple matching in a few instances (Table I). Coincidently the expression of all identified proteins was significantly down-regulated by treatment of PC-3 cells with 50µM quercetin; down-regulation of spot # 191 was highest among the identified proteins (−1.75-fold, P < 0.001). Further examination of the differentially expressed proteins showing significant changes in expression did not suggest that they were either associated or co-regulated with the exception of the enzymes of oxidative metabolism and, more significantly, members of the stress/chaperone family of proteins and receptor membrane component. Spot # 191 was subsequently identified as Hsp90. The spot intensity of spot # 191 is compared for untreated (Fig. 2B) and quercetin treated (Fig. 2C) PC-3 cells. Figure 2D, E shows the elution profiles of two representative Hsp90 peptides from HPLC-MS/MS. Other significantly down-regulated proteins include tumor protein D52 (−1.69-fold, P = 0.032), tropomyosin 3 (−1.66-fold, P = 0.035) and progesterone receptor membrane component (−1.66-fold, P = 0.028). Table I is a list of identified proteins from PC-3 cells arranged on the basis of the significance (P-value) of their fold-decrease in response to treatment with quercetin.
Fig. 2.
Proteomic analysis on the effects of quercetin on prostate cancer cells showing that Hsp90 is maximally modulated (down-regulated) by treatment of PC-3 cells with quercetin. A: Representative 2D-Sypro-Ruby stained proteome image of quercetin treated PC-3 cells.PC-3 cells (1 × 106) were cultured with 50µM of quercetin for 24hr. Total protein was extracted, subjected to DIGE analysis, and stained with SYPRO Ruby protein stain as described in the text. The pH increases from left to right and the molecular mass decreases from the top to the bottom of the gels. Identified protein spots are outlined and numbered, three separate experiments yielded similar results. Abundance of spot # 191 from (B) untreated and (C) quercetin treated PC-3 cells as determined by DeCyder software. Of all proteins, spot # 191 showed the greatest change (−1.75-fold) after treatment with quercetin. D,E: MS/MS spectra of ion fragments from two tryptic peptides, ADLINNLGTIAK and NPDDITNEEYGEFYK respectively, obtained from spot # 191. The spectra represent the ion fragments that matched Hsp90 in the Sequest empirical database (noted as “view ▲▲ matched” at the lower left of each spectrum). [Color figure can be viewed in the online issue, which is available at http://www.interscience.wiley.com.]
TABLE I.
List of Proteins Modulated in PC-3 Cells on Treatment With Quercetin, Identified by 2D-DIGE Coupled With LC-MS/MS Analysis
| Master spot # |
Identified proteins | Accession number |
Fold Change |
P-value | Theoretical Mass |
Theoretical pI |
Biological function |
|---|---|---|---|---|---|---|---|
| 191 | Heat shock protein Hsp90-alpha and -beta |
gi123678 | −1.75 | 0.001 | 84.6 | 4.94 | Involved in cellular stress associated with malignancy, essential for the functions of a range of oncogenic proteins, including Raf-1, Akt/PKB, ErbB2, mutant p53 |
| 2,016 | Tumor protein D52 isoform 3 | gi 4827038 | −1.69 | 0.032 | 19.9 | 4.94 | Tumor-associated antigen, is over expressed in the majority of breast, prostate and ovarian cancers, involved in activation of cell division protein |
| 1,839 | Tropomyosin 3 | gi24119203 | −1.66 | 0.035 | 29.0 | 4.75 | Is an actin-binding protein the isoform expression of which involves multiple genes and alternative processing of RNA |
| 2,020 | Tumor protein D52 isoform 3 | gi4827038 | −1.66 | 0.05 | 19.9 | Tumor-associated antigen, is overexpressed in the majority of breast, prostate and ovarian cancers, involved in activation of cell division protein |
|
| 2,054 | Progesterone receptor membrane component 1 |
gi5729875 | −1.64 | 0.028 | 21.7 | 4.56 | Interacts with plasminogen activator inhibitor RNA binding protein-1 (PAIRBP1), a membrane-associated protein involved in the anti-apoptotic action of progesterone |
| 2,229 | Glyceraldehyde-3-phosphate dehydrogenase |
gi7669492 | −1.55 | 0.038 | 36.0 | 8.57 | Metabolic enzyme, involved in activation of colony stimulating factor |
| 1,676 | Elongation factor 1 delta isoform 2 |
gi25453472 | −1.44 | 0.024 | 31.1 | 4.90 | Transcription factor |
| 2,148 | Cofilin 1 (non-muscle) | gi5031635 | −1.39 | 0.016 | 18.5 | 8.22 | Cytoskeletal protection, involved in actin depolymerization, cell motility, and apoptosis |
| 2,175 | Heat shock protein 70 | gi16507237 | −1.21 | 0.023 | 72.3 | 4.97 | Stress protein |
| 969 | Human platelet profilin | gi3891601 | −1.2 | 0.044 | 36.0 | 8.47 | Main regulator of the transition of globular actin (G-actin) to filamentous actin (F-actin) |
PC-3 cells were cultured with and without quercetin (50 µmol/L) for 24 hr (n = 3 independent experiments using three different PC-3 culture). Protein was extracted and subjected to DIGE as described in Materials and Methods Section. Data represent statistically significant down-regulated proteins (Student’s t-test) that were identified using HPLC-MS/MS. Data are represented as protein name, gene accession number (gi No), theoretical mass and function, theoretical iso-electric point.
Hsp90 Is Elevated in Prostate Cancer Cells
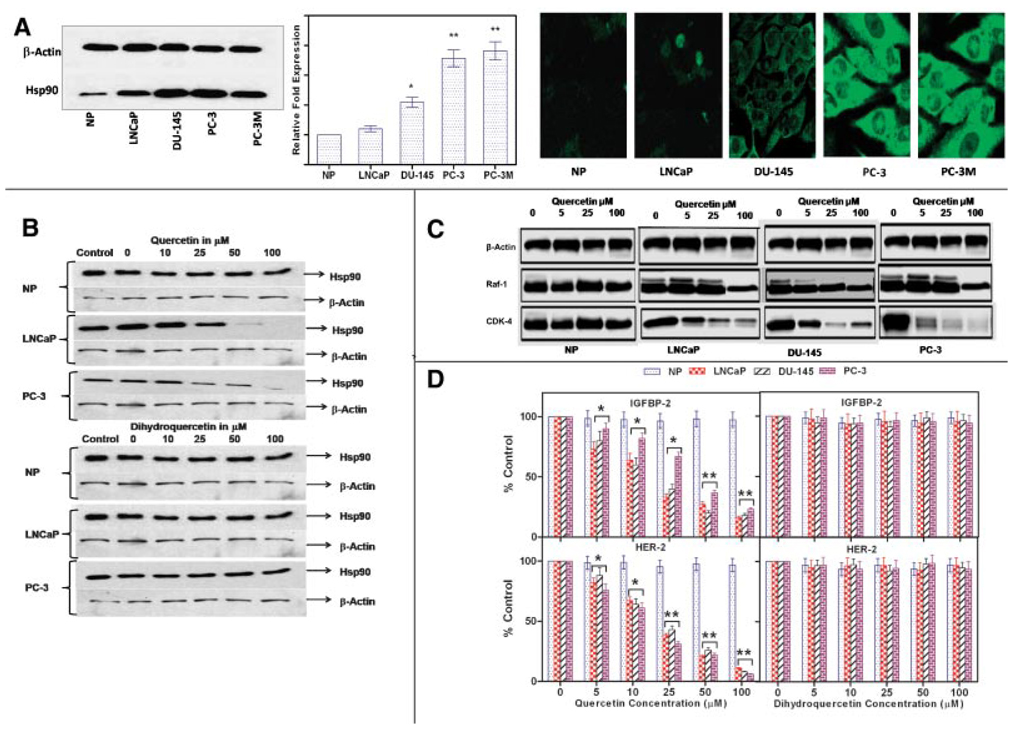
Having identified Hsp90 as a protein whose expression is significantly modulated on treatment of prostate cancer cells with quercetin we then compared Hsp90 levels in prostate cancer cells to normal prostate epithelial cells using Western blots. Compared with non-malignant, prostate epithelial cells, Hsp90 levels in 3 of 4 prostate cancer cell lines examined were higher (Fig. 3A first panel). Hsp90 levels correlated with the aggressive phenotype of prostate cancer cell lines as we previously reported [22]. Q-PCR studies confirmed our Western blot findings (data not shown). The above data were supported by the observation that gene expression of Hsp90 was significantly higher in human prostate cancer cells grown in immuno-deficient nude mice compared with normal prostate epithelial cells as determined by Q-PCR (Fig.3A second panel).Immuno-cytochemical staining of PC-3 and normal prostate epithelial cells revealed higher cytosolic expression of Hsp90 in the tumor cell line (Fig. 3A third panel). Similar results were obtained with other prostate cancer cell lines DU-145, PC-3, and PC-3M. No significant staining was detected in healthy prostate epithelial cells or negative controls.
Fig. 3.
Effect of quercetin on the expression of Hsp90 and Hsp90 client proteins by prostate cancer cells. A, Panel1: a representative Western blot of prostate cancer cell lines showing increased Hsp90 levels compared with normal prostate epithelial cells; Panel2: Hsp90 gene expression by human prostate tumors grown sub-cutaneously in nude mice as determined by quantitative PCR and Panel 3: Hsp90 expression in situ as shown by immunofluoresence in normal prostate epithelial cells and prostate cancer cells. (*P < 0.05, **P < 0.001; n = 6.) Expression of Hsp90 was normalized to the housekeeping gene, β-Actin. B: Representative Western blots showing quercetin treatment decreases Hsp90 expression in prostate cancer cells. LNCaP, PC-3, and normal prostate epithelial cells were treated with various concentrations of quercetin or dihydroquercetin as described in the text. Cell lysates were prepared and 10 µg aliquots of protein were loaded per lane on 10% Tris gels and blotted with an antibody specific for Hsp90. Membranes were stripped and re-probed for β-actin. DMSO served as a vehicle for quercetin and dihydroquercetin. Final concentrations of DMSO never exceeded 0.2%. C: Representative Western blots showing the degradation of intracellular Hsp90 client proteins in normal and malignant prostate cancer cells in response to quercetin. Cells were treated with varying concentrations of quercetin as indicated in the text. Cell lysates were prepared and 10 µg aliquots of protein were loaded per lane on 10% Tris gels and blotted with an antibody specific for Cdk-4 and Raf-1. D: ELISA data showing dose-dependent suppression of the levels of the secreted client proteins IGFBP-2 and HER-2, whose expression correlate directly with the expression of Hsp90, normalized to untreated control in normal and malignant prostate cancer cells. Cells were treated with varying concentrations of quercetin or dihydroquercetin as indicated in the text, culture medium was collected at 24 hr and levels of the secreted client proteins IGFBP-2 and HER-2, whose expression correlates directly with the expression of Hsp90 were measured as described in the text. Values are mean ± SE of six experiments; *P < 0.05 and **P < 0.001 compared to untreated cells. Quercetin inhibited IGFBP-2 and HER-2 secretion in a dose-dependent manner. [Color figure can be viewed in the online issue, which is available at http://www.interscience.wiley.com.]
Treatment of Prostate Cancer Cells With Quercetin Down-Regulates the Expression of Hsp90 Client Proteins
To directly determine the effects of quercetin on Hsp90 expression prostate cancer cells were incubated with various concentrations of quercetin. After 24 hr cells were lysed, protein extracted and assayed for Hsp90 by Western blots. Our results (Fig. 3B) demonstrate a dose-dependent reduction of Hsp90 expression by LNCaP and PC-3 caused by quercetin. However quercetin treatment had no effect on normal prostate epithelial cells. The inactive analog, dihydroquercetin, and 0.2% DMSO used as a vehicle control similarly had no effects on Hsp90 by both prostate cancer cells and normal prostate epithelial cells. These results demonstrate that quercetin specifically inhibits Hsp90 expression by prostate cancer cells. To further confirm that the selective cytotoxic effect of quercetin on prostate cancer cells is mediated via down-regulation of Hsp90 protein we measured the expression of the two most commonly used, well established client proteins for Hsp90 inhibition, Raf-1 and cdk-4, using Western blots. Treatment of prostate cancer cell lines with quercetin for 24 hr decreased Raf-1 and cdk-4 levels in all three prostate cancer cell lines in a dose-dependent manner but had no measurable effect on the levels of these proteins in normal prostate epithelial cells (Fig. 3C).
The expression of both insulin like growth factor binding protein (IGFBP)-2 and human epidermal growth factor receptor (HER)-2 extracellular domains are also regulated by Hsp90. We further tested whether quercetin down-regulates the secretion of these two additional markers of Hsp90. Prostate cancer cells were cultured for 24 hr with quercetin or its inactive analog, dihydroquercetin, medium was collected and assayed for IGFBP-2 and HER-2 extra cellular domain by ELISA. As shown in Figure 3D both proteins were suppressed in a dose-dependent manner by quercetin in all the three prostate cancer cell lines but not by the inactive analog, dihydroquercetin. Treatment with quercetin or dihydroquercetin had no effects on normal prostate epithelial cells. To summarize treatment of prostate cancer cells with quercetin specifically and selectively inhibits their expression of Hsp90 and its surrogate markers in a dose-dependent manner. Normal prostate epithelial cells were unaffected by treatment with quercetin.
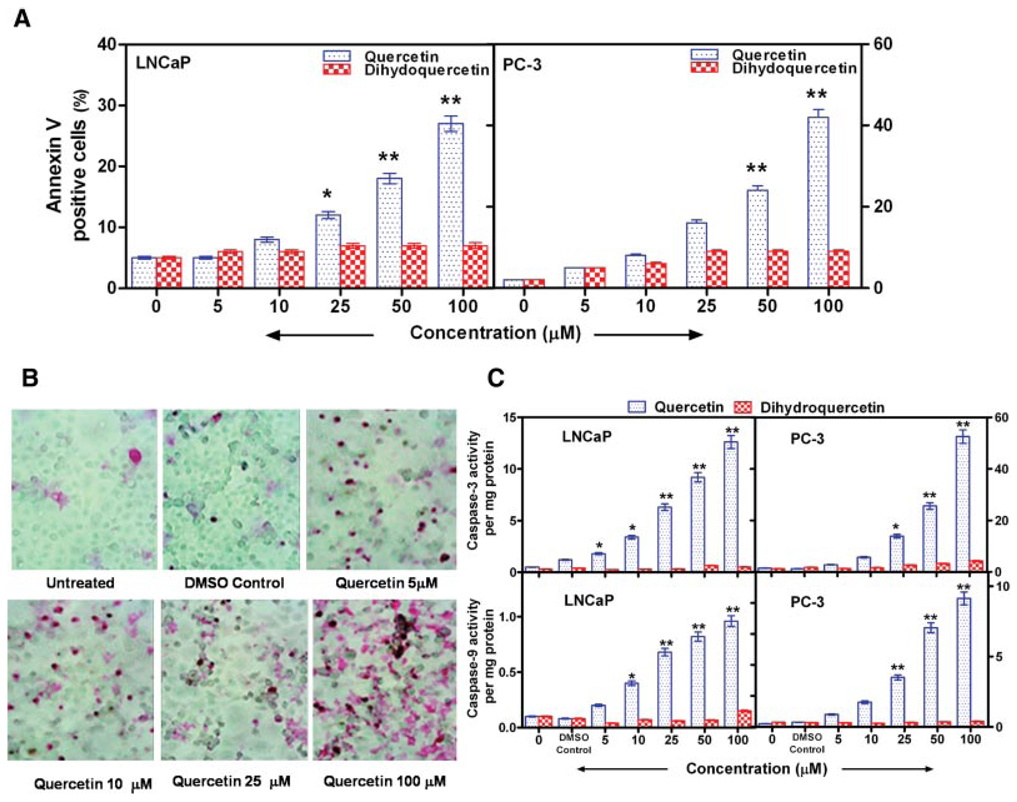
Quercetin Mediated Inhibition of Hsp90 Stimulates Apoptosis of Prostate Cancer Cells
The effect of the Hsp90 inhibitor quercetin, on the induction of apoptosis of prostate cancer cells was determined after 24 hr of incubation using annexin V staining. A dose-dependent response in both LNCaP and PC-3 cells was observed (Fig. 4A). Quercetin treatment of LNCaP cells resulted in a three- to five fold increase in annexin V–positive cells compared with the control group treated with dihydroquercetin. However, with the more aggressive PC-3 cells the increase ranged from 4- to 20-fold. Apoptosis also was measured in situ using the TUNEL stain. PC-3 cells incubated with quercetin show a dose-dependent increase in TUNEL staining compared with control cells incubated with the vehicle alone (Fig. 4B). Similar results were obtained with LNCaP and DU-145 cells (data not shown). The effect of quercetin and dihydroquercetin on caspase-3 (effector caspase) and caspase-9 (initiator caspase) activation was examined. Quercetin increased caspase-3 and caspase-9 activities in both LNCaP and PC-3 cells, whereas dihydroquercetin had no effect on caspases (Fig. 4C). Treatment of LNCaP cells with 50 µM quercetin for 24 hr led to a >7-fold increase in caspase-3 and a 4-fold increase in caspase-9 activities. PC-3 cells incubated with 50 µM quercetin resulted in an 8.9-fold elevation of caspase-3 and a 6.3-fold increase in caspase-9 compared with incubation with dihydroquercetin. Elevated caspase levels were apparent with lower concentrations of quercetin. The above data demonstrate that quercetin, which decreases Hsp90 expression, also initiates apoptosis of prostate cancer cells.
Fig. 4.
Quercetin mediated inhibition of Hsp90 stimulates apoptosis of prostate cancer cells. A: Increase in annexin V−positive cells (assayed by flow cytometry) following treatment of LNCaP and PC-3 prostate cancer cells with varying concentrations of quercetin for 24 hr. Incubation with dihydroquercetin or 0.2% DMSO did not induce apoptosis. B: Increased apoptosis of PC-3 cells treated with varying doses of quercetin for 24 hr as determined by in situ TUNEL staining as described in the text. C: Increased caspase-3 and -9 activities in prostate cancer cells, LNCaP and PC-3. Cells were incubated with or without varying concentrations of quercetin or dihydroquercetin for 24 hr. Caspase activities were measured by a colorimetric assay as described in the text. DMSO (0.2%) did not have any effect on caspase activation. Values are mean ± SE from six experiments; *P < 0.05 and **P < 0.001 compared to treatment with dihydroquercetin. [Color figure can be viewed in the online issue, which is available at http://www.interscience.wiley.com.]
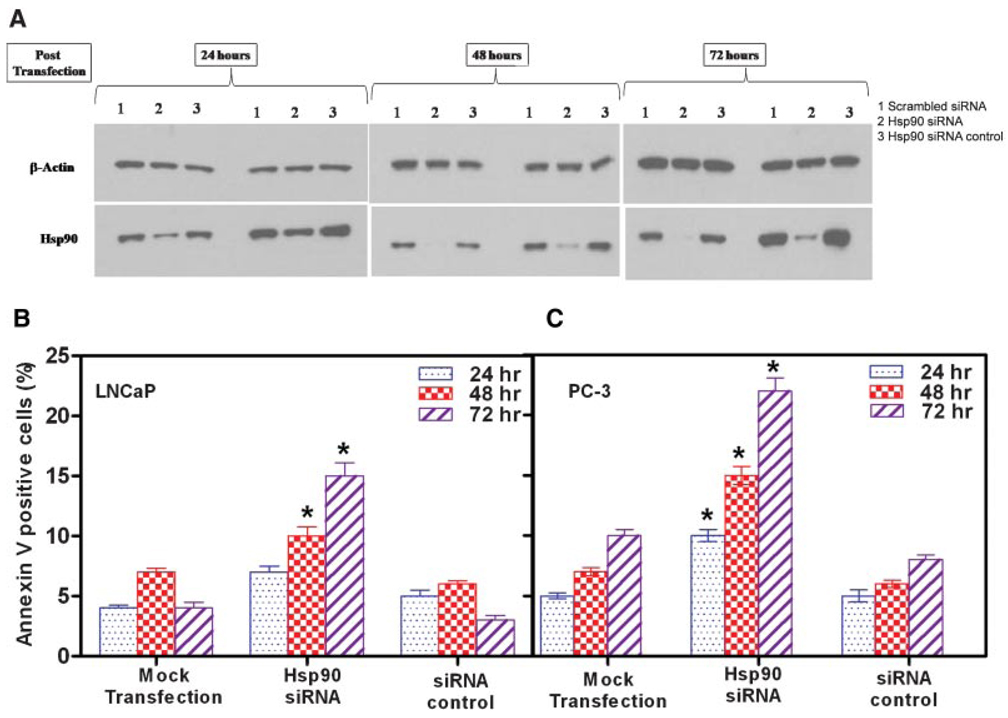
Knockdown of Hsp90 Gene Expression by siRNA Promotes Apoptosis of Prostate Cancer Cells
To confirm that apoptosis of prostate cancer cells is mediated via Hsp90 and to exclude any non-Hsp90-related effects of flavonoids, we transfected prostate cancer cells with double-stranded siRNA oligonucleotides. Cells were harvested at 24, 48, and 72 hr after transfection, and Hsp90 expression was analyzed by Western blot (Fig. 5A). Whereas Hsp90-specific siRNA suppressed Hsp90 synthesis almost completely, control (scrambled) siRNA had no effect on Hsp90 expression. A set of mock transfected cells showed no decrease in Hsp90 synthesis. Actin expression was unaffected by treatment with either Hsp90-specific or scrambled control siRNA. Next, we assessed the effect of Hsp90-specific siRNA on cell viability. Inhibition of Hsp90 using siRNA significantly decreased cell viability in both LNCaP (37.4 ± 2.3% decrease compared to mock) and PC-3 (65.1 ± 3.1% decrease compared to mock) cells. We then determined the extent of apoptosis at 24, 48, and 72 hr after incubation with siRNA using annexin V staining. Transfection of LNCaP and PC-3 cells with Hsp90 siRNA after 48 and 72 hr significantly increased annexin V–positive cells (Fig. 5B). In contrast, only a slight enhancement in the number of annexin V–positive cells was detected after the first 24 hr after transfection, a time when Hsp90 expression had decreased only slightly. A 3.2-fold increase in annexin V–positive cells occurred after 72 hr of treatment with Hsp90-specific siRNA in both LNCaP and PC-3 cells. Transfections with scrambled siRNA serving as a control did not show any alterations in annexin V–positive cells. Control scrambled siRNA altered neither activity compared with mock-transfected cells. Hsp90-specific siRNA and annexin V stained profiles showed that Hsp90-specific oligonucleotides were able to initiate apoptosis in prostate tumor cancer cells in vitro.
Fig. 5.
Knockdown of Hsp90 gene expression by siRNA promotes apoptosis of prostate cancer cells. Cells were transfected with 40 nM Hsp90 siRNA, 40 nM scrambled control siRNA, or mock transfection using the transfection reagent without siRNA. Annexin V−positive cells were measured by flow cytometry after 24, 48, and 72 hr. A: Representative Western blots from three separate experiments which yielded similar results. Hsp90 siRNA induced marked suppression of Hsp90 expression in LNCaP and PC-3 cells, whereas β-actin expression was unaffected. Hsp90 siRNA enhances apoptosis of (B) LNCaP and PC-3 prostate cancer cells as measured by annexin V-positive cells. Results are mean ± SE of six experiments; *P < 0.05 and **P < 0.001, compared with control siRNA. [Color figure can be viewed in the online issue, which is available at http://www.interscience.wiley.com.]
In summary direct inhibition of Hsp90 expression by prostate cancer cells using siRNA specific for Hsp90 selectively induced apoptosis of these cells confirming our hypothesis that the mechanism underlying the specific and selective cytotoxic effects of quercetin on prostate cancer cells is due to the induction of apoptosis by inhibiting the expression of Hsp90.
DISCUSSION
Epidemiological and statistical data indicate that about 35% of human cancer mortality is attributable to diet [35]. It is well accepted that the consumption of food products containing high amounts of flavonoids, a diet rich in fruits and vegetables, is associated with a reduced risk of cancer [36,37]. The mechanisms underlying the cancer-protective effects of these naturally occurring polyphenolic compounds, however, remain elusive. We previously reported that quercetin, a major plant polyphenols, inhibited the colony forming ability of prostate cancer cells [13]. In the same study we showed that 25 µM quercetin inhibited expression of a variety of genes involved in the cell cycle and oncogenesis in prostate cancer cells. Therefore we undertook the present investigation to determine the mechanisms underlying the effect of quercetin on prostate cancer cells. We observed that quercetin is selectively cytotoxic to prostate cancer cells by inducing apoptosis while sparing normal prostate epithelial cells. Our results concur with a report that quercetin inhibits the growth of other tumor cell lines [38]. In our previous report [13] 25 µM quercetin inhibited colony formation by prostate cancer cells, comparable to the concentrations of quercetin (50–100 µM) shown in the present study to be cytotoxic to these cells. These doses of quercetin are comparable to concentrations used by others in cell culture studies [39]. Whether these concentrations can be achieved in vivo in humans is unknown. A recent study [17] indicates that >50 µM of quercetin might be achieved in circulation when quercetin at 150 mg/kg body weight was given orally to rats. Moreover, quercetin at doses of 40–1,900 mg/kg/day in rats showed no signs of toxicity or death [40]. This suggests that quercetin at 100 µM is safe and could be used at 100 µM for therapeutic application and is consistent with the doses used in our study.
Previous studies suggested multiple different targets underlying the effects of quercetin on cancer cells [16,18,19,41]. In an effort to identify the molecular targets of quercetin in prostate cancer cells we used the proteomic approach to help us explore this aspect of our study. The data presented here demonstrate the utility of proteomics-based analysis to identify the mode of action of agents like quercetin. Prior to this study, quercetin was characterized as a potent anti-proliferative agent capable of inhibiting tumor cell growth in vitro and in animal models. Attempts to identify the molecular mechanisms underlying the anti-tumor activity of quercetin have been unsuccessful to date. We therefore undertook a proteomic analysis to identify changes in cellular proteins as candidate targets of quercetin treatment of prostate cancer cells. Recently proteomics was used to identify the molecular targets of another plant natural product, bengamides, found to be a new class of methonine aminopeptidase inhibitors [42]. Our data on changes in the proteome of PC-3 cells treated with quercetin, demonstrate that Hsp90 is the protein that was maximally modulated (down-regulated) among all the proteins studied. Hsp90, a molecular chaperone, is required for the stability and function of mutated, chimeric, and overexpressed proteins. These proteins include mutated p53, Bcr–Abl, Cdk4, mitogen-activated protein kinase kinase (MEK1/2), Raf-1, Akt, HER2, and hypoxia-inducible factor 1[alpha] (HIF-1[alpha]) from various signaling pathways [43]. The mechanism of action of Hsp90 is thought to include the stepwise recruitment of co-chaperones, including Hsp70, p50cdc37, p60hop, and p23 [44]. Hsp90 also is believed to be involved in adaptation of cancer cells to many environmental stresses, such as hypoxia and nutrient deprivation [45], and is commonly overexpressed in cancer cells. Consequently, cancer cells may be dependent on Hsp90 for their growth and survival [45]. It is conceivable that the down-regulation of Hsp90 by quercetin can lead to modulation of its chaperone function, thereby accelerating degradation of numerous oncogenic client proteins that participate in the regulation of cell proliferation, survival, and angiogenesis [46]. In this study we showed that Hsp90 levels are elevated in prostate cancer cells compared to normal prostate epithelial cells and treatment of prostate cancer cells down-regulates the expression of Hsp90 and its client proteins. Suppression of the client proteins modulated by Hsp90 likely is secondary to down-regulation of the expression of Hsp90. Similar observations were described by Zhang et al. [34] who reported that treatment of breast and colon cancer cells with inhibitors of Hsp90 also down-regulated the expression of Hsp90 client proteins. We also measured the effect of quercetin on two widely accepted biomarkers of Hsp90 inhibition, IGFBP-2 and HER-2 extracellular domain [34]. These two secreted proteins also are indicators of tumor presence and progression. Cancer cells control cell proliferation and tumor growth by producing IGFBPs that enhance the function of insulin like growth factor (IGF)-I and IGF-II [47–49]. IGFBP-2 is activated by a variety of pathways, such as the role of IGFR in estrogen receptor signaling all of which are regulated by Hsp90 [43]. Circulating HER-2 comprises the extracellular domain of HER-2. Full-length HER-2 is cleaved by a sheddase near the transmembrane domain to generate a 110-kDa fragment that is detectable in serum of most individuals and in the culture medium of HER-2-overexpressing cells [50,51]. Elevated HER-2 extracellular domain can be detected in the serum of patients with high HER-2-overexpressing tumors and is correlated with poor prognosis and reduced response to chemotherapy [41,52,53]. Decreased levels of HER-2 have been observed in cultured cancer cells and in the serum of mice treated with Hsp90 inhibitors [34]. The present study shows that treatment of prostate cancer cells with quercetin resulted in a dose-dependent decrease in the levels of these two secreted biomarkers of Hsp90 inhibition which confirms our conclusion that Hsp90 protein is a primary target for quercetin action.
Our data show that prostate cancer cells overexpress Hsp90 compared to normal prostate epithelial cells, supporting our premise that Hsp90 increases the resistance of tumor cells to apoptosis. This observation is consistent with reports by others showing increased heat shock protein mRNA levels in human prostate carcinomas [54,55]. High levels of Hsp90 expression also correlate with increased drug resistance in human glioma [56] and multiple myeloma [57] cells and in other human cancers [58].We also showed that levels of Hsp90 protein in prostate cancer cell lines is in the following order LNCaP < DU-145 < PC-3. This is particularly interesting as this is the order of increasing aggressive phenotype of these cells lines as we reported [22]. Thus increasing expression of Hsp90 correlates with the aggressive phenotype of prostate cancer cells. As a control, the inactive analog, dihydroquercetin, which does not inhibit Hsp90 [59] also did not alter cell viability of any prostate cancer cell line. Therefore, the effect observed with quercetin can be ascribed to the reduction in Hsp90. Measurements of lactate dehydrogenase activity in cell culture medium after 12 and 24 hr of treatment with quercetin or dihydroquercetin did not show any significant increase, indicating that necrosis was not the underlying mechanism decreasing cell viability with quercetin (data not shown). In contrast, both caspase-3 and caspase-9 activities in LNCaP and PC-3 cells were significantly increased following incubation with quercetin, strongly supporting that apoptosis was responsible for quercetin-induced cell death. To confirm this observation, annexin V labeling and TUNEL staining (markers of apoptosis) also were significantly increased following quercetin induced inhibition of Hsp90 expression in prostate cancer cell lines. Our data agree with a study by Brusselmans et al. [60] showing that quercetin increased apoptosis in LNCaP cells. Apoptosis can occur via two major pathways. The extrinsic pathway is characterized by activation of death receptors on the cell surface belonging to the TNF/nerve growth factor receptor superfamily and recruitment of the initiator caspases, 8 and 10. The intrinsic or mitochondrial pathway is independent of death receptor signaling and involves the release of cytochrome c from the mitochondria with formation of a complex of cytochrome c, the cytoplasmic apoptotic protease-activating factor (Apaf)-1, and procaspase-9. Both pathways converge to the activation of downstream effector caspase-3, caspase-6, and caspase-7 [61–63]. Hsps are highly conserved proteins that play a role in fundamental cellular processes. Hsp90 is one of the main Hsps consisting of both constitutively expressed and stress-inducible members, and it is a molecular chaperone that protects the organism from stress-induced cell injury. Secondly, it has been reported to act on several steps of the apoptotic cascade. The detailed mechanism of the anti-apoptotic effect of Hsp90 is yet to be determined. However, it seems to be cell type specific. A study done by Cohen-Saidon et al. [64] demonstrated that the anti-apoptotic activity of Bcl-2 in mast cells is dependent on its association with Hsp90beta. Dissociation of these 2 proteins inhibits the anti-apoptotic activity of Bcl-2 on mast cells by initiating the release of cytochrome c from mitochondria into the cytosol and increasing the activity of caspase-3 and caspase-7. This effect was significantly enhanced by specifically knocking-out Hsp90 be using RNA interference. In another study Georgakis et al. [65] report that inhibition of Hsp90 function by the specific inhibitor, 17-allylamino-17-demethoxy-geldanamycin (17-AAG), resulted in induction of apoptosis in Hodgkin’s lymphoma cell lines that was mediated by caspase-dependent and caspase-independent mechanisms. In the present study, we showed that quercetin-mediated inhibition of Hsp90 expression in the prostate cancer cell lines, PC-3 and LNCaP, induced apoptosis and caused activation of caspase-3 and -9. These results are consistent with the observations that Hsp90 inhibition can prevent caspase-9 recruitment by its ability to interfere with Apaf-1 to block apoptosome formation. From our data, we conclude that, in prostate cancer cell lines, Hsp90 also plays a major role in caspase-dependent apoptosis. To rule out any effects quercetin unrelated to Hsp90 inhibition, we used RNA interference to selectively knockout of the Hsp90 gene and assessed apoptosis. We showed that Hsp90 siRNA resulted in increased apoptosis of prostate cancer cell lines. These results clearly support the fact that inhibition of Hsp90 expression results in apoptotic cell death of prostate cancer cells and that quercetin-induced cell death is mediated via its effect on Hsp90. However, because the extent of apoptosis was some-what greater with quercetin than with Hsp90 siRNA, it is possible that in addition to promoting apoptosis, quercetin may affect viability by other mechanisms to be determined. Nevertheless the major effect of quercetin appears to be mediated by inhibition of Hsp90 expression based on the following effects on all prostate cancer cell lines tested without affecting normal prostate epithelial cells: (a) quercetin significantly decreased Hsp90 expression, (b) quercetin induced apoptosis, and (c) the inactive analog, dihydroquercetin, did not inhibit Hsp90 and did not induce apoptosis.
In summary, the present study demonstrates that quercetin has anti-proliferative and pro-apoptotic effects on prostate cancer cells and these effects are mediated via down-regulation of the expression of Hsp90. Interestingly, in all three prostate cancer cell lines studied, a remarkable dose-response correlation was observed between quercetin-induced suppression of Hsp90 expression and inhibition of cell growth and induction of apoptosis. The effect of quercetin is specific for malignant cells and has no apparent effects on normal prostate epithelial cells. Moreover the effect of quercetin on the viability of prostate cancer cell lines corresponded to the aggressive phenotype of the cells with the greatest effect on PC-3 cells and the lowest effect on LNCaP cells; the effect on DU-145 cells was intermediate between the other two cell lines. Our results suggest that quercetin may be a potential candidate for both the prevention and treatment of prostate cancer without the undesirable side-effects of conventional chemotherapy. Further in vivo investigations to determine the doses of quercetin required for optimal cancer chemoprevention and therapy are warranted. In conclusion our findings demonstrate that the ability of quercetin to induce apoptosis in cancer cells is directly associated with its Hsp90 inhibitory activity, thereby providing a new mechanism by which polyphenolic dietary plant compounds may exert their anti-cancer effects.
ACKNOWLEDGMENTS
The authors thank Ramnik Chawda, Division of Allergy Immunology and Rheumatology, Department of Medicine, State University of New York at Buffalo, Buffalo, NY 14203 for technical assistance. This work was supported by the Margaret Duffy and Robert Cameron Troup Memorial Fund for Cancer Research of the Kaleida Health System, Buffalo, NY.
REFERENCES
- 1.Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics, 2007. CA Cancer J Clin. 2007;57(1):43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- 2.Heinonen OP, Albanes D, Virtamo J, Taylor PR, Huttunen JK, Hartman AM, Haapakoski J, Malila N, Rautalahti M, Ripatti S, Maenpaa H, Teerenhovi L, Koss L, Virolainen M, Edwards BK. Prostate cancer and supplementation with alpha-tocopherol and beta-carotene: Incidence and mortality in a controlled trial. J Natl Cancer Inst. 1998;90(6):440–446. doi: 10.1093/jnci/90.6.440. [DOI] [PubMed] [Google Scholar]
- 3.Kolonel LN. Fat, meat, and prostate cancer. Epidemiol Rev. 2001;23(1):72–81. doi: 10.1093/oxfordjournals.epirev.a000798. [DOI] [PubMed] [Google Scholar]
- 4.Kris-Etherton PM, Hecker KD, Bonanome A, Coval SM, Binkoski AE, Hilpert KF, Griel AE, Etherton TD. Bioactive compounds in foods: Their role in the prevention of cardiovascular disease and cancer. Am J Med. 2002;113 Suppl 9B:71S–88S. doi: 10.1016/s0002-9343(01)00995-0. [DOI] [PubMed] [Google Scholar]
- 5.Middleton E, Jr, Kandaswami C, Theoharides TC. The effects of plant flavonoids on mammalian cells: Implications for inflammation, heart disease, and cancer. Pharmacol Rev. 2000;52(4):673–751. [PubMed] [Google Scholar]
- 6.Chen C, Kong AN. Dietary chemopreventive compounds and ARE/EpRE signaling. Free Radic Biol Med. 2004;36(12):1505–1516. doi: 10.1016/j.freeradbiomed.2004.03.015. [DOI] [PubMed] [Google Scholar]
- 7.Hakkinen SH, Karenlampi SO, Heinonen IM, Mykkanen HM, Torronen AR. Content of the flavonols quercetin, myricetin, and kaempferol in 25 edible berries. J Agric Food Chem. 1999;47(6):2274–2279. doi: 10.1021/jf9811065. [DOI] [PubMed] [Google Scholar]
- 8.Formica JV, Regelson W. Review of the biology of Quercetin and related bioflavonoids. Food Chem Toxicol. 1995;33(12):1061–1080. doi: 10.1016/0278-6915(95)00077-1. [DOI] [PubMed] [Google Scholar]
- 9.Castillo MH, Perkins E, Campbell JH, Doerr R, Hassett JM, Kandaswami C, Middleton E., Jr The effects of the bioflavonoid quercetin on squamous cell carcinoma of head and neck origin. Am J Surg. 1989;158(4):351–355. doi: 10.1016/0002-9610(89)90132-3. [DOI] [PubMed] [Google Scholar]
- 10.Scambia G, Ranelletti FO, Benedetti Panici P, Piantelli M, Bonanno G, De Vincenzo R, Ferrandina G, Pierelli L, Capelli A, Mancuso S. Quercetin inhibits the growth of a multidrug-resistant estrogen-receptor-negative MCF-7 human breast-cancer cell line expressing type II estrogen-binding sites. Cancer Chemother Pharmacol. 1991;28(4):255–258. doi: 10.1007/BF00685531. [DOI] [PubMed] [Google Scholar]
- 11.Ranelletti FO, Ricci R, Larocca LM, Maggiano N, Capelli A, Scambia G, Benedetti-Panici P, Mancuso S, Rumi C, Piantelli M. Growth-inhibitory effect of quercetin and presence of type-II estrogen-binding sites in human colon-cancer cell lines and primary colorectal tumors. Int J Cancer. 1992;50(3):486–492. doi: 10.1002/ijc.2910500326. [DOI] [PubMed] [Google Scholar]
- 12.Elattar TM, Virji AS. The inhibitory effect of curcumin, genistein, quercetin and cisplatin on the growth of oral cancer cells in vitro. Anticancer Res. 2000;20(3A):1733–1738. [PubMed] [Google Scholar]
- 13.Nair HK, Rao KV, Aalinkeel R, Mahajan S, Chawda R, Schwartz SA. Inhibition of prostate cancer cell colony formation by the flavonoid quercetin correlates with modulation of specific regulatory genes. Clin Diagn Lab Immunol. 2004;11(1):63–69. doi: 10.1128/CDLI.11.1.63-69.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Csokay B, Prajda N, Weber G, Olah E. Molecular mechanisms in the antiproliferative action of quercetin. Life Sci. 1997;60(24):2157–2163. doi: 10.1016/s0024-3205(97)00230-0. [DOI] [PubMed] [Google Scholar]
- 15.Wei YQ, Zhao X, Kariya Y, Fukata H, Teshigawara K, Uchida A. Induction of apoptosis by quercetin: Involvement of heat shock protein. Cancer Res. 1994;54(18):4952–4957. [PubMed] [Google Scholar]
- 16.Yoshida M, Yamamoto M, Nikaido T. Quercetin arrests human leukemic T-cells in late G1 phase of the cell cycle. Cancer Res. 1992;52(23):6676–6681. [PubMed] [Google Scholar]
- 17.Ma Z, Hung Nguyen T, Hoa Huynh T, Tien Do P, Huynh H. Reduction of rat prostate weight by combined quercetin-finasteride treatment is associated with cell cycle deregulation. J Endocrinol. 2004;181(3):493–507. doi: 10.1677/joe.0.1810493. [DOI] [PubMed] [Google Scholar]
- 18.Wang IK, Lin-Shiau SY, Lin JK. Induction of apoptosis by apigenin and related flavonoids through cytochrome c release and activation of caspase-9 and caspase-3 in leukaemia HL-60 cells. Eur J Cancer. 1999;35(10):1517–1525. [PubMed] [Google Scholar]
- 19.Datta SR, Brunet A, Greenberg ME. Cellular survival A play in three Akts. Genes Dev. 1999;13(22):2905–2927. doi: 10.1101/gad.13.22.2905. [DOI] [PubMed] [Google Scholar]
- 20.Nair MP, Kandaswami C, Mahajan S, Chadha KC, Chawda R, Nair H, Kumar N, Nair RE, Schwartz SA. The flavonoid, quercetin, differentially regulates Th-1 (IFNgamma) and Th-2 (IL4) cytokine gene expression by normal peripheral blood mononuclear cells. Biochim Biophys Acta. 2002;1593(1):29–36. doi: 10.1016/s0167-4889(02)00328-2. [DOI] [PubMed] [Google Scholar]
- 21.Nair MP, Mahajan S, Reynolds JL, Aalinkeel R, Nair H, Schwartz SA, Kandaswami C. The flavonoid quercetin inhibits proinflammatory cytokine (tumor necrosis factor alpha) gene expression in normal peripheral blood mononuclear cells via modulation of the NF-kappa beta system. Clin Vaccine Immunol. 2006;13(3):319–328. doi: 10.1128/CVI.13.3.319-328.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aalinkeel R, Nair MP, Sufrin G, Mahajan SD, Chadha KC, Chawda RP, Schwartz SA. Gene expression of angiogenic factors correlates with metastatic potential of prostate cancer cells. Cancer Res. 2004;64(15):5311–5321. doi: 10.1158/0008-5472.CAN-2506-2. [DOI] [PubMed] [Google Scholar]
- 23.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162(1):156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 24.Shively L, Chang L, LeBon JM, Liu Q, Riggs AD, Singer-Sam J. Real-time PCR assay for quantitative mismatch detection. Biotechniques. 2003;34(3):498–502. doi: 10.2144/03343st01. 504. [DOI] [PubMed] [Google Scholar]
- 25.Mahajan SD, Schwartz SA, Nair MP. Immunological assays for chemokine detection in in-vitro culture of CNS cells. Biol Proced Online. 2003;5:90–102. doi: 10.1251/bpo50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wessel D, Flugge UI. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal Biochem. 1984;138(1):141–143. doi: 10.1016/0003-2697(84)90782-6. [DOI] [PubMed] [Google Scholar]
- 27.Tonge R, Shaw J, Middleton B, Rowlinson R, Rayner S, Young J, Pognan F, Hawkins E, Currie I, Davison M. Validation and development of fluorescence two-dimensional differential gel electrophoresis proteomics technology. Proteomics. 2001;1(3):377–396. doi: 10.1002/1615-9861(200103)1:3<377::AID-PROT377>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 28.Breci L, Hattrup E, Keeler M, Letarte J, Johnson R, Haynes PA. Comprehensive proteomics in yeast using chromatographic fractionation, gas phase fractionation, protein gel electrophoresis, and isoelectric focusing. Proteomics. 2005;5(8):2018–2028. doi: 10.1002/pmic.200401103. [DOI] [PubMed] [Google Scholar]
- 29.Gharahdaghi F, Weinberg CR, Meagher DA, Imai BS, Mische SM. Mass spectrometric identification of proteins from silver-stained polyacrylamide gel: A method for the removal of silver ions to enhance sensitivity. Electrophoresis. 1999;20(3):601–605. doi: 10.1002/(SICI)1522-2683(19990301)20:3<601::AID-ELPS601>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 30.Cooper B, Eckert D, Andon NL, Yates JR, Haynes PA. Investigative proteomics: Identification of an unknown plant virus from infected plants using mass spectrometry. J Am Soc Mass Spectrom. 2003;14(7):736–741. doi: 10.1016/S1044-0305(03)00125-9. [DOI] [PubMed] [Google Scholar]
- 31.Wilm M, Shevchenko A, Houthaeve T, Breit S, Schweigerer L, Fotsis T, Mann M. Femtomole sequencing of proteins from polyacrylamide gels by nano-electrospray mass spectrometry. Nature. 1996;379(6564):466–469. doi: 10.1038/379466a0. [DOI] [PubMed] [Google Scholar]
- 32.Haynes PA, Gygi SP, Figeys D, Aebersold R. Proteome analysis: Biological assay or data archive? Electrophoresis. 1998;19(11):1862–1871. doi: 10.1002/elps.1150191104. [DOI] [PubMed] [Google Scholar]
- 33.Andon NL, Hollingworth S, Koller A, Greenland AJ, Yates JR, III, Haynes PA. Proteomic characterization of wheat amyloplasts using identification of proteins by tandem mass spectrometry. Proteomics. 2002;2(9):1156–1168. doi: 10.1002/1615-9861(200209)2:9<1156::AID-PROT1156>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 34.Zhang H, Chung D, Yang YC, Neely L, Tsurumoto S, Fan J, Zhang L, Biamonte M, Brekken J, Lundgren K, Burrows F. Identification of new biomarkers for clinical trials of Hsp90 inhibitors. Mol Cancer Ther. 2006;5(5):1256–1264. doi: 10.1158/1535-7163.MCT-05-0537. [DOI] [PubMed] [Google Scholar]
- 35.Doll R, Peto R. The causes of cancer: Quantitative estimates of avoidable risks of cancer in the United States today. J Natl Cancer Inst. 1981;66(6):1191–1308. [PubMed] [Google Scholar]
- 36.Riboli E, Norat T. Epidemiologic evidence of the protective effect of fruit and vegetables on cancer risk. Am J Clin Nutr. 2003;78(3 Suppl):559S–569S. doi: 10.1093/ajcn/78.3.559S. [DOI] [PubMed] [Google Scholar]
- 37.Tadjalli-Mehr K, Becker N, Rahu M, Stengrevics A, Kurtinaitis J, Hakama M. Randomized trial with fruits and vegetables in prevention of cancer. Acta Oncol. 2003;42(4):287–293. doi: 10.1080/02841860310011852. [DOI] [PubMed] [Google Scholar]
- 38.Williams RJ, Spencer JP, Rice-Evans C. Flavonoids: Antioxidants or signalling molecules? Free Radic Biol Med. 2004;36(7):838–849. doi: 10.1016/j.freeradbiomed.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 39.Aghdassi A, Phillips P, Dudeja V, Dhaulakhandi D, Sharif R, Dawra R, Lerch MM, Saluja A. Heat shock protein 70 increases tumorigenicity and inhibits apoptosis in pancreatic adenocarcinoma. Cancer Res. 2007;67(2):616–625. doi: 10.1158/0008-5472.CAN-06-1567. [DOI] [PubMed] [Google Scholar]
- 40.Dunnick JK, Hailey JR. Toxicity and carcinogenicity studies of quercetin, a natural component of foods. Fundam Appl Toxicol. 1992;19(3):423–431. doi: 10.1016/0272-0590(92)90181-g. [DOI] [PubMed] [Google Scholar]
- 41.Carney WP, Neumann R, Lipton A, Leitzel K, Ali S, Price CP. Monitoring the circulating levels of the HER2/neu oncoprotein in breast cancer. Clin Breast Cancer. 2004;5(2):105–116. doi: 10.3816/cbc.2004.n.014. [DOI] [PubMed] [Google Scholar]
- 42.Towbin H, Bair KW, DeCaprio JA, Eck MJ, Kim S, Kinder FR, Morollo A, Mueller DR, Schindler P, Song HK, van Oostrum J, Versace RW, Voshol H, Wood J, Zabludoff S, Phillips PE. Proteomics-based target identification: Bengamides as a new class of methionine aminopeptidase inhibitors. J Biol Chem. 2003;278(52):52964–52971. doi: 10.1074/jbc.M309039200. [DOI] [PubMed] [Google Scholar]
- 43.Neckers L, Ivy SP. Heat shock protein 90. Curr Opin Oncol. 2003;15(6):419–424. doi: 10.1097/00001622-200311000-00003. [DOI] [PubMed] [Google Scholar]
- 44.Pearl LH, Prodromou C. Structure and in vivo function of Hsp90. Curr Opin Struct Biol. 2000;10(1):46–51. doi: 10.1016/s0959-440x(99)00047-0. [DOI] [PubMed] [Google Scholar]
- 45.Gabai VL, Kabakov AE. Induction of heat-shock protein synthesis and thermotolerance in EL-4 ascites tumor cells by transient ATP depletion after ischemic stress. Exp Mol Pathol. 1994;60(2):88–99. doi: 10.1006/exmp.1994.1008. [DOI] [PubMed] [Google Scholar]
- 46.Sharp S, Workman P. Inhibitors of the HSP90 molecular chaperone: Current status. Adv Cancer Res. 2006;95:323–348. doi: 10.1016/S0065-230X(06)95009-X. [DOI] [PubMed] [Google Scholar]
- 47.Maxwell P, van den Berg HW. Changes in the secretion of insulin-like growth factor binding proteins -2 and -4 associated with the development of tamoxifen resistance and estrogen independence in human breast cancer cell lines. Cancer Lett. 1999;139(2):121–127. doi: 10.1016/s0304-3835(99)00009-9. [DOI] [PubMed] [Google Scholar]
- 48.Dubois V, Couissi D, Schonne E, Remacle C, Trouet A. Intracellular levels and secretion of insulin-like-growth-factor-binding proteins in MCF-7/6, MCF-7/AZ and MDA-MB-231 breast cancer cells. Differential modulation by estrogens in serum-free medium. Eur J Biochem. 1995;232(1):47–53. doi: 10.1111/j.1432-1033.1995.tb20779.x. [DOI] [PubMed] [Google Scholar]
- 49.Bhat RV, Engber TM, Zhu Y, Miller MS, Contreras PC. Identification of insulin-like growth factor binding protein-2 as a biochemical surrogate marker for the in vivo effects of recombinant human insulin-like growth factor-1 in mice. J Pharmacol Exp Ther. 1997;281(1):522–530. [PubMed] [Google Scholar]
- 50.Codony-Servat J, Albanell J, Lopez-Talavera JC, Arribas J, Baselga J. Cleavage of the HER2 ectodomain is a pervanadate-activable process that is inhibited by the tissue inhibitor of metalloproteases-1 in breast cancer cells. Cancer Res. 1999;59(6):1196–1201. [PubMed] [Google Scholar]
- 51.Zabrecky JR, Lam T, McKenzie SJ, Carney W. The extracellular domain of p185/neu is released from the surface of human breast carcinoma cells, SK-BR-3. J Biol Chem. 1991;266(3):1716–1720. [PubMed] [Google Scholar]
- 52.Sandri MT, Johansson H, Colleoni M, Zorzino L, Passerini R, Orlando L, Viale G. Serum levels of HER2 ECD can determine the response rate to low dose oral cyclophosphamide and methotrexate in patients with advanced stage breast carcinoma. Anticancer Res. 2004;24(2C):1261–1266. [PubMed] [Google Scholar]
- 53.Hayes DF, Yamauchi H, Broadwater G, Cirrincione CT, Rodrigue SP, Berry DA, Younger J, Panasci LL, Millard F, Duggan DB, Norton L, Henderson IC. Circulating HER-2/erbB-2/c-neu (HER-2) extracellular domain as a prognostic factor in patients with metastatic breast cancer: Cancer and Leukemia Group B Study 8662. Clin Cancer Res. 2001;7(9):2703–2711. [PubMed] [Google Scholar]
- 54.Williams CR, Tabios R, Linehan WM, Neckers L. Intratumor injection of the Hsp90 inhibitor 17AAG decreases tumor growth and induces apoptosis in a prostate cancer xenograft model. J Urol. 2007;178(4 Pt 1):1528–1532. doi: 10.1016/j.juro.2007.05.120. [DOI] [PubMed] [Google Scholar]
- 55.Gabai VL, Budagova KR, Sherman MY. Increased expression of the major heat shock protein Hsp72 in human prostate carcinoma cells is dispensable for their viability but confers resistance to a variety of anticancer agents. Oncogene. 2005;24(20):3328–3338. doi: 10.1038/sj.onc.1208495. [DOI] [PubMed] [Google Scholar]
- 56.Panner A, Murray JC, Berger MS, Pieper RO. Heat shock protein 90alpha recruits FLIPS to the death-inducing signaling complex and contributes to TRAIL resistance in human glioma. Cancer Res. 2007;67(19):9482–9489. doi: 10.1158/0008-5472.CAN-07-0569. [DOI] [PubMed] [Google Scholar]
- 57.Chatterjee M, Jain S, Stuhmer T, Andrulis M, Ungethum U, Kuban RJ, Lorentz H, Bommert K, Topp M, Kramer D, Muller-Hermelink HK, Einsele H, Greiner A, Bargou RC. STAT3 and MAPK signaling maintain overexpression of heat shock proteins 90alpha and beta in multiple myeloma cells, which critically contribute to tumor-cell survivalx. Blood. 2007;109(2):720–728. doi: 10.1182/blood-2006-05-024372. [DOI] [PubMed] [Google Scholar]
- 58.Whitesell L, Lindquist SL. HSP90 and the chaperoning of cancer. Nat Rev Cancer. 2005;5(10):761–772. doi: 10.1038/nrc1716. [DOI] [PubMed] [Google Scholar]
- 59.Budagova KR, Zhmaeva SV, Grigor’ev AN, Goncharova AY, Kabakov AE. Flavonoid dihydroquercetin, unlike quercetin, fails to inhibit expression of heat shock proteins under conditions of cellular stress. Biochemistry (Mosc) 2003;68(9):1055–1061. doi: 10.1023/a:1026081016923. [DOI] [PubMed] [Google Scholar]
- 60.Brusselmans K, Vrolix R, Verhoeven G, Swinnen JV. Induction of cancer cell apoptosis by flavonoids is associated with their ability to inhibit fatty acid synthase activity. J Biol Chem. 2005;280(7):5636–5645. doi: 10.1074/jbc.M408177200. [DOI] [PubMed] [Google Scholar]
- 61.Denecker G, Vercammen D, Declercq W, Vandenabeele P. Apoptotic and necrotic cell death induced by death domain receptors. Cell Mol Life Sci. 2001;58(3):356–370. doi: 10.1007/PL00000863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hengartner MO. The biochemistry of apoptosis. Nature. 2000;407(6805):770–776. doi: 10.1038/35037710. [DOI] [PubMed] [Google Scholar]
- 63.Hajra KM, Liu JR. Apoptosome dysfunction in human cancer. Apoptosis. 2004;9(6):691–704. doi: 10.1023/B:APPT.0000045786.98031.1d. [DOI] [PubMed] [Google Scholar]
- 64.Cohen-Saidon C, Carmi I, Keren A, Razin E. Antiapoptotic function of Bcl-2 in mast cells is dependent on its association with heat shock protein 90beta. Blood. 2006;107(4):1413–1420. doi: 10.1182/blood-2005-07-2648. [DOI] [PubMed] [Google Scholar]
- 65.Georgakis GV, Li Y, Rassidakis GZ, Martinez-Valdez H, Medeiros LJ, Younes A. Inhibition of heat shock protein 90 function by 17-allylamino-17-demethoxy-geldanamycin in Hodgkin’s lymphoma cells down-regulates Akt kinase, dephosphorylates extracellular signal-regulated kinase, and induces cell cycle arrest and cell death. Clin Cancer Res. 2006;12(2):584–590. doi: 10.1158/1078-0432.CCR-05-1194. [DOI] [PubMed] [Google Scholar]