Abstract
Aims
To elucidate the expression and regulation of survivin in normal tissues.
Methods and results
A novel monoclonal antibody (12C4) to survivin was generated. Application of this antibody to determine survivin expression in human normal adult tissues revealed that most adult tissues do not express survivin and, where it is present, survivin is largely restricted to a small subset of epithelial cells and cells with proliferative potential such as thymus. Survivin expression among positive tissues showed individual variations, ranging from zero to <5% positive cells in epithelial cell populations. Testis is the only human adult tissue highly expressing survivin, with 60–70% positivity in the nuclei of spermatogonia. Consistent with deregulated expression of survivin associated with oncogenesis, we found that certain ligands and transcription factors differentially modulate survivin promoter activity in cancer cells versus normal/untransformed cells.
Conclusion
Deregulation of survivin transcription controls in individual epithelial cells may contribute to oncogenesis in various human adult tissues.
Keywords: cancer tissues, human adult tissues, survivin expression
Introduction
Survivin is a novel member of the inhibitor of apoptosis (IAP1) protein family, which has been implicated in both the inhibition of apoptosis and the promotion of cell division.1,2 Survivin is readily detected in multiple cancer types and its role in tumour cell growth and survival is established.3,4 Although survivin is expressed during normal development, only a limited number of normal adult tissues have been reported to express this protein. These include thymus,5 endometrium, 6 bone marrow CD34+ cells7 and colonic mucosa.8 In contrast to these limited examples of survivin-positive tissues, survivin expression has not been detected in other human adult tissues tested, including peripheral blood, lymph node, kidney, skeletal muscle, liver, lung, brain and heart.5 These observations suggest that survivin has a limited role in healthy adult tissues. Consistently, recent studies have revealed a role for survivin in normal physiology in a highly regulated manner. For example, survivin plays a role in the maturation and/or development of T cells,9,10 neutrophils11 and erythroid cells/megakaryocytes. 12 Thus, elucidation of survivin expression in human adult tissues is important for further investigation of the role and regulation of survivin in normal organs, tissues or cells. However, the expression of survivin in many human adult tissues, such as thyroid, pituitary, ovary, adrenal, tonsil, cervix, etc., has not been determined. Moreover, the expression profile and localization of survivin in the known survivin-positive adult tissues remain unclear. Elucidation of these points would not only shed light on the potential role of survivin in normal adult tissues but also aid our understanding of its role and regulation during oncogenesis
In this study, we have characterized a novel survivin monoclonal antibody (mAb) (12C4), which recognizes survivin and survivin variants. Using this newly developed 12C4 antibody, we determined the expression profile and localization of survivin in various human adult tissues (including many that have not been previously tested). These observations, together with the known roles of survivin in oncogenesis and cancer progression,13,14 provide insight into the possible pathogenesis of tumours from individual epithelial cells with deregulated expression of survivin. This, in turn, may lead to new strategies in cancer prevention and therapeutics.
Materials and methods
GENERATION OF VARIOUS GLUTATHIONINE-S-TRANSFERASE–SURVIVIN FUSION PROTEIN CONSTRUCTS
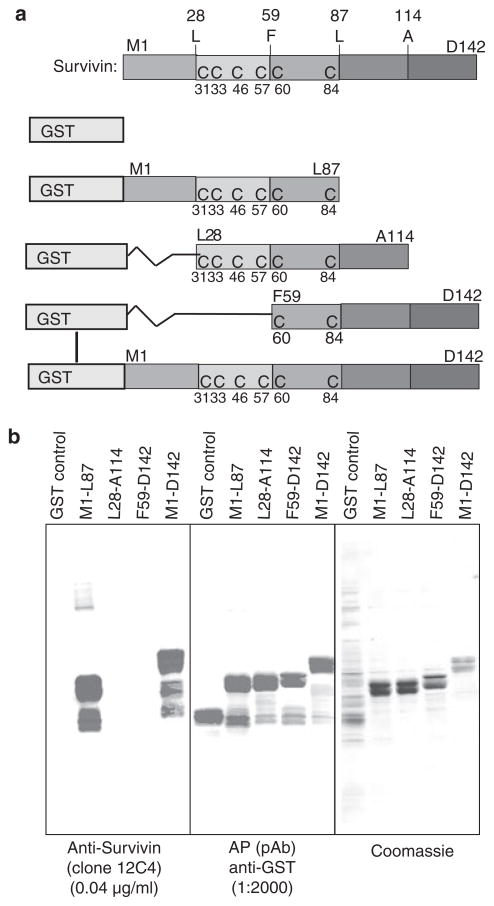
Various truncated survivin cDNAs were amplified by polymerase chain reaction (PCR) using a full-length survivin cDNA as a template and subcloned into the pIVEX-GST expression vector downstream of the glutathionine-S-transferase (GST) gene in frame (Figure 1a) at Nde1 and Xho1 sites. The primers used for the survivin cDNA amplification were as follows: M1 (5′-TTA CCT CAT ATG GGT GCC CCG ACG TTG CCC CCT-3′), L28 (5′-TTA CCT CAT ATG TTG GAG GGC TGC GCC TGC ACC C-3′), F59 (5′-TTA CCT CAT ATG TTC TGC TTC AAG GAG CTG GAA GGC T-3′), L87 (5′-TCC TAA CTC GAG AAG GAA AGC GCA ACC GGA CGA ATG CT-3′), A114 (5′-TCC TAA CTC GAG TGC AAT TTT GTT CTT GGC TCT TTC TCT-3′) and D142 (5′-TCC TAA CTC GAG ATC CAT GGC AGC CAG CTG CTC GAT-3′), which were synthesized from DNA-Technologies (Århus, Denmark). All of the obtained constructs were confirmed by sequencing.
Figure 1.
Characterization of the anti-survivin monoclonal antibody, 12C4. a, Diagram of the glutathionine-S-transferase (GST)-fused survivin proteins with and without various truncations. Generation of the corresponding vectors for expressing these proteins is described in Materials and methods. b, The first 27 amino acids of the survivin protein sequence are essential for recognition by clone 12C4. The 12C4 antibody was characterized by Western blot analysis (left panel; note that top bands are survivin and low bands are degraded) using various truncated survivin proteins as indicated at the top. Protein expression was confirmed by Western blotting with anti-GST antibodies (middle panel), and protein loading was visualized by Coomassie blue staining (right panel).
RECOMBINANT GST–SURVIVIN FUSED PROTEIN EXPRESSION
Escherichia coli Rosetta (DE3) (Novagen, VWR International, Aps, Denmark) was used for the expression of the pIVEX-GST and the four GST–survivin derivative constructs. Escherichia coli was grown at 37°C in Luria–Bertani broth in the presence of Cabennicilin (100 μg/ml), Chloramphenicol (40 μg/ml), glucose and isopropylthio-β-D-galactoside induction. The cultures were precipitated, resuspended in water and stored in aliquots at − 20°C. Prior to Western blotting, the E. coli lysates containing the individual recombinant protein were normalized according to the intensities observed on Coomassie-stained sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS–PAGE).
ANTI-SURVIVIN MONOCLONAL ANTIBODY DEVELOPMENT
Mouse anti-survivin mAb clone 12C4 (DakoCytomation; M3624, Carpinteria, CA, USA) was developed using E. coli DE3-derived recombinant survivin as antigen. Female NMRCF1 mice (Charles River Laboratoriums, Les Oncins, France) were immunized and boosted two to four times, intradermally, at 2-week intervals with E. coli-derived recombinant survivin in 0.9% saline combined with alum (State Serum Institute, Copenhagen, Denmark) 1 mg per immunization. Four days after the last injection, spleen cells were fused with P3-X63-Ag 8.653 using standard techniques. 15 Antibodies in plasma and hybridoma culture supernatants were detected by their reaction with survivin using enzyme-linked immunosorbent assay. Positive hybridomas were screened by immunohistochemistry on formalin-fixed paraffin-embedded tissues.
WESTERN BLOT ANALYSIS
Recombinant GST and GST–survivin derivatives, GST-M1-L87, GST-L28-A114, GST-F59-D142 and GST-M1-D142 or various cell lysates were separated on the SDS–PAGE and blotted onto an Immuno-Blot polyvinylidene fluoride membrane (Bio-Rad, Hercules, CA, USA) with the blotting buffer (10 mM 3-[cyclohexylamino]-1-propanesulphonic acid, pH 11.0 and 15% ethanol). A total of 1.5–1.8 A hours were applied in the blotting process. Membranes were then blocked in 1× Tris-buffered NaCl solution with Tween 20 (TBS–T) buffer (50 mM Tris, 0.5 M NaCl, 0.5% Tween 20), followed by incubation of the membrane with the anti-survivin mAb (clone 12C4, 0.04 μg/ml), the survivin polyclonal antibody (1: 500; Santa Cruz, Santa Cruz, CA, USA), actin antibody (1: 5000; Sigma, St Louis, MO, USA) or the anti-GST antibody (1: 2000; Amersham GE-Healthcare, Piscataway, NJ, USA), which was dependent on individual experiments, for 1 h at room temperature or overnight at 2–8°C in 1× TBS–T buffer. After washing with 1× TBS–T buffer, the membranes were incubated with either the goat antimouse horseradish peroxidase (HRP)-conjugated secondary antibody (1: 2000; DakoCytomation; P0447) or goat peroxidase-conjugated antirabbit IgG (1: 50 000; Sigma) for detection of bound anti-survivin and the rabbit antigoat HRP-conjugated secondary antibody (1: 2000; DakoCytomation; P0449) for detection of bound anti-GST, which was dependent on the individual experiments. After washing in 1× TBS–T buffer, antibodies on the membrane were visualized using 3,3′-diaminobenzidene (DAB)+ substrate chromogen (DakoCytomation; K3468) or the enhanced chemiluminescence protein detection kit (Perkin Elmer, Shelton, CT, USA). Membranes prepared in parallel were routinely stained with Coomassie blue to verify equal protein transfer to the membrane in some experiments.
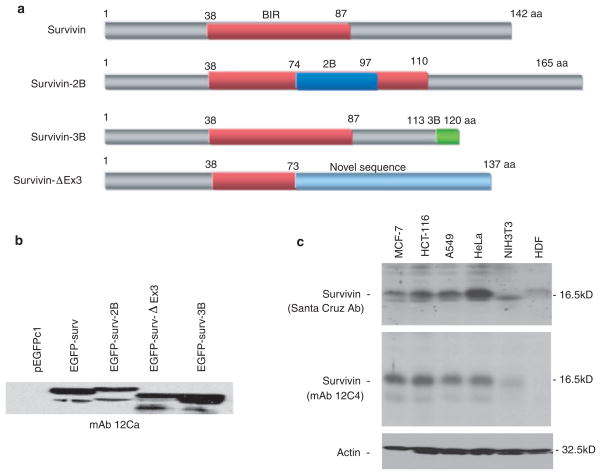
MOLECULAR CLONING OF SURVIVIN VARIANTS AND CELL TRANSFECTION
Expression vectors for survivin,1,16 survivin-2B and survivin-ΔEx317 have been previously characterized. For cloning of the pEGFPc1-survivin-3B expression vector, survivin-3B cDNA was amplified with a pair of primers of HsE1ATG-5′ (forward primer, 5′-GGA ATT CCA TGG GTG CCC CGA CGT TG-3′, EcoR I site underlined) and 3B-3′P1 (reverse primer, 5′-GCT CTA GAT CAT TCT GCT AAC AGA GCT CTC TCA ATT TTG TTC TTG GCT C-3′, Xba I site underlined) using survivin cDNA as a template. After digestion with EcoR I and Xba I, the PCR DNA fragment was subsequently cloned into the EcoR I and Xba I sites into the pEGFPc1 vector in frame. The resulting pEGFPc1-survivin-3B vector was confirmed by sequencing. Transfection of pEGFPc1-survivin, pEGFPc1-survivin-2B, pEGFPc1-survivin-ΔEx3 and pEGFPc1-survivin-3B into HeLa cells was performed using Lipofectamine™ 2000 as previous described,17 followed by Western blot analysis using anti-survivin mAb 12C4.
IMMUNOHISTOCHEMISTRY
Immunohistochemistry was performed using the Catalysed Signal Amplification system II (DakoCytomation; K1497). Formalin-fixed paraffin-embedded specimens of various normal and neoplastic human tissues were assembled into tissue microarrays. A subset of the tissue samples was provided by the Cooperative Human Tissue Network, which is funded by the National Cancer Institute. Multiple cores for each tissue type were mounted on positively charged microscope slides. Single tissue blocks of colorectal and breast adenocarcinoma specimens were also procured for testing. The mounted tissue sections were deparaffinized in Histo-Clear (National Diagnostics, Atlanta, GA, USA) and rehydrated in graded alcohols. After the tissue sections were pretreated in a citrate-based target retrieval solution (DakoCytomation; S1700) at 95–99°C in a water bath for 40 min, the tissue sections were cooled for 20 min at room temperature. The tissue sections were then incubated with a hydrogen peroxide solution for 5 min to quench endogenous peroxidase activity. After rinsing and incubation of the tissue sections in two fresh TBS–T buffer baths for 3 min each, the sections were incubated with protein block for 5 min. Anti-survivin mAb (clone 12C4; DakoCytomation; M3624) was applied next at a concentration of 1.0 μg/ml and incubated for 15 min. An IgG2a subclass-matched mouse mAb to Aspergillus niger glucose oxidase (DakoCytomation; X0943) was used as the negative control reagent. The sections were rinsed and incubated in two fresh TBS–T buffer baths for 3 min each. The HRP-conjugated antimouse immunoglobulin antibody was applied to the sections and incubated for 15 min. Following TBS–T washes, the sections were incubated with a fluorescyl-tyramide amplification reagent for 15 min. After washing in TBS–T, antifluorescein–HRP was added to slides and allowed to incubate for 15 min. Finally, the DAB+ substrate chromogen reagent was prepared and applied to the sections for 5 min. The sections were washed in dH2O, counterstained and mounted. All reagent incubations were conducted at room temperature.
CELL CULTURE AND VITAMIN D3 TREATMENT
MCF-7, HeLa, HCT116 or NIH3T3 cell lines were grown in Dulbecco’s Modified Eagle’s Medium (DMEM) medium supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin and 0.1 μg/ml streptomycin in a 5% CO2 incubator. MCF-10A cells were grown in DMEM/F12 medium (v/v 50: 50) supplemented with 20 ng/ml epidermal growth factor, 500 ng/ml hydrocortisone, 100 ng/ml cholera toxin, 10 μg/ml insulin, 2 mM L-glutamine and 5% FBS. Cells grown in normal conditions were treated with vitamin D3 for 48 h after transfection, followed by luciferase activity assay (see below).
TRANSFECTION AND LUCIFERASE REPORTER ASSAY
MCF-7, HCT116, HeLa, NIH3T3 or MCF-10A cells were seeded in 48-well plates (0.5–1 × 105/well) 1 day before transfection. Cells in each well were transfected by the survivin promoter-luciferase construct pLuc-284018 and pRL-TK [TK promoter-driven Renilla-luciferase construct (Promega, Madison, WI, USA), internal control vector for normalization] along with or without (VD3 treatment only) one of AP-2 member expression vectors (α, β, γ)19 or the control empty vector (pRSV). Briefly, 300 ng of a pLuc-2840 and 5–10 ng of pRL-TK together with or without (VD3 treatment only) 20–100 ng of an AP-2 vector or pRSV empty vector were in 30 μl serum-free DMEM (making master mix when possible). The above DNA solution was then mixed with Lipofectamine™ 2000 in 30 μl serum-free DMEM (1.5 μl of LipofectamineTM 2000 for 1 μg of DNA) in a 1.5-ml tube for each well of 48-well plates. After incubation at room temperature for 20–25 min, the DNA/Lipofectamine 2000 mixture (60 μl) was added to each well containing 250 μl of complete cell growth medium and incubated in a CO2 incubator for 36–48 h before processing the luciferase assay [Dual-Luciferase Reporter Assay System (Promega)]. The transfected cells in the 48-well plates were washed with PBS and lysed with 1× passive lysis buffer (60–100 μl/well) on a shaker for up to 1 h at 4°C. Cell lysate (10–20 μl/well) was used for measurement of luciferase activities in a luminometer by first mixing the cell lysate (10–20 μl) with 10–20 μl of luciferase assay reagent for measuring Firefly luciferase activity, and subsequently adding 10–20 μl of Stop & Glo reagent for measuring Renilla luciferase activity. Data were normalized to Renilla luciferase activity (internal control) as arbitrary units.
Results
THE PEPTIDE EPITOPE THAT REACTS WITH THE ANTI-SURVIVIN MONOCLONAL ANTIBODY IS MAPPED TO THE N-TERMINAL REGION OF SURVIVIN
Anti-survivin mAb (clone 12C4) was developed using purified survivin protein as an antigen. To map the epitope recognized by clone 12C4, bacterially expressed full-length (M1-D142) and truncated GST–survivin fusion proteins were employed as shown in Figure 1a. Immunoblotting analysis of these fusion proteins revealed that 12C4 recognized an epitope within full-length survivin and in the fusion protein containing its N-terminal 87 amino acids (Figure 1b, lanes 2 and 5). However, deletion of either 27 or 57 amino acids from the N-terminus of survivin abrogated the ability of this antibody to recognize survivin (Figure 1b, lanes 3 and 4). Collectively, these data demonstrate that the 12C4 antibody recognizes an epitope close to the N-terminus of survivin which is at least partially within the first 27 amino acids of the protein.
THE 12C4 SURVIVIN MONOCLONAL ANTIBODY EVENLY RECOGNIZES SURVIVIN AND ITS KNOWN SPLICE VARIANTS
Many commercial survivin antibodies preferentially detect only certain variants of survivin. For example, the survivin polyclonal antibody from Santa Cruz (sc-10811) could not effectively pick up survivin-ΔEx3 (not shown). Since the N-terminal region is common to the known variants of survivin (survivin, survivin-2B, survivin-3B and survivin-ΔEx3) (Figure 2a), we suspected that 12C4 would have similar affinity to all splice variants of survivin. Consistent with its recognition of an epitope in a common region, immunoblotting analysis of lysates from cells transfected with survivin variants demonstrated that 12C4 recognizes all of the variants with approximately equal affinity (Figure 2b). Thus, this antibody should prove useful for detecting the overall expression of the survivin gene.
Figure 2.
Specificity of mAb clone 12C4 to various survivin variants/isoforms. a, Diagram of protein structures for the different survivin variants. b, Clone 12C4 recognizes all survivin isoforms with similar affinity. HeLa cells were transfected with pEGFPc1 empty vector or with various survivin variants as shown. Expression of enhanced green fluorescence protein (EGFP)-fused survivin variants was determined by Western blots using the clone 12C4 (note: top bands are survivin variants, and low bands are degraded). c, Clone 12C4 specifically recognizes endogenous survivin expression in various cancer cell lines. Survivin expression in lysates from various normal and cancer cell lines was determined using the 12C4 survivin mAb and the survivin polyclonal antibody from Santa Cruz (sc-10811) by Western blots, respectively. Actin is an internal control for protein loading. MCF-7, a breast cancer cell line; HCT-116, a colon cancer cell line; A549, a lung cancer cell line; HeLa, a cervical carcinoma cell line; NIH3T3, a mouse embryo fibroblast cell line which is known with very low or no survivin expression; HDF, a primary normal human dermal fibroblast cell line, which was also shown to be survivin negative.29
To test further the sensitivity of the 12C4 mAb for recognition of endogenous survivin expression, cell lysates isolated from various normal and cancer cell lines were used to test the expression of survivin by Western blots using 12C4 as well as the survivin antibody from Santa Cruz (sc-10811). As shown (Figure 2c), 12C4 effectively recognized survivin expression in various cancer cell types but not normal (NIH3T3 and HDF) cells (Figure 2c).
EXPRESSION OF SURVIVIN IN NORMAL HUMAN ADULT TISSUES IS LARGELY RESTRICTED TO A SMALL SUBSET OF EPITHELIAL CELLS AND CELLS WITH PROLIFERATIVE POTENTIAL
Having established the specificity of the 12C4 antibody, this antibody was used to analyse the expression and cellular/subcellular distribution of survivin in various adult human tissues by immunohistochemistry. To avoid possible variation among individuals, analysis was performed on two to four samples from different individuals. Furthermore, staining protocols were carefully controlled to avoid potential ‘false-positive’ staining of nuclei.20 Our data revealed that most normal human adult tissues do not express survivin (Table 1), which is consistent with previous observations. 13 However, survivin was detected in multiple epithelial tissues, where it was largely restricted to a very small subset (< 5%) of epithelial cells (Table 2). Further analysis revealed that survivin-positive cells were restricted to the intestinal crypts and basal layers of squamous epithelia, areas of epithelial cell proliferation (Table 2 and Figure 3). In addition, survivin was also detected in lymphatic/haematopoietic cells, where its expression was also restricted to sites of cell division, such as the spleen and thymus (Table 2). Restriction of expression to proliferating compartments in normal adult tissues points to a link between survivin expression and cell division in non-transformed adult tissue, an idea consistent with the known M-phase functions of survivin. Interestingly, our data revealed that testis is the only human adult tissue that highly expresses survivin, with 60–70% positivity in the nuclei of spermatogonia (Table 2, Figure 3a). Since spermatogonia undergo rapid cell division during spermatogenesis, these data reinforce the link between survivin expression and proliferation in non-transformed adult tissues. The restriction of survivin expression to proliferating compartments of adult tissues, together with the reported requirement for survivin in epithelial renewal of intestine,8 not only points to a critical role for survivin in proliferation of non-transformed adult cells, but also indicates that the expression and function of survivin is strictly regulated in these cells. These findings, together with the role of survivin in oncogenesis,14 suggest that deregulated survivin expression in individual cells is a critical step in the oncogenic process.
Table 1.
Survivin-negative normal human adult tissues
| Human adult tissues (specimen no. tested) | Survivin positivity |
|---|---|
| Adrenal (3) | 0/3 |
| Breast (3) | 0/3 |
| Brain, cerebellum (3) | 0/3 |
| Brain, cerebrum (3) | 0/3 |
| Heart (3) | 0/3 |
| Kidney (3) | 0/3 |
| Liver (3) | 0/3 |
| Lung (3) | 0/3 |
| Mesothelial cells (3) | 0/3 |
| Nerve (2) | 0/2 |
| Ovary (3) | 0/3 |
| Parathyroid (3) | 0/3 |
| Pituitary (3) | 0/3 |
| Salivary gland (3) | 0/3 |
| Skeletal muscle (3) | 0/3 |
| Thyroid (2) | 0/2 |
Table 2.
Survivin-positive normal human adult tissues
| Normal tissue type (specimen no. tested) | Description of specific staining profile and localization |
|---|---|
| Bone marrow (3) | 2/3 Progenitor cells (< 5%), nuclear; 1/3 histiocytes (< 5%), cytoplasmic |
| Cervix (3) | 3/3 Basal epithelial cells (< 5%), nuclear |
| Colon (2) | 2/2 Deep glandular epithelial cells (< 5%), nuclear; 1/2 lymphocytes (< 5%), nuclear |
| Oesophagus (2) | 1/2 Basal epithelial cells (< 5%), nuclear |
| Pancreas (3) | 1/3 Acinar cells (20%), granular cytoplasmic |
| Prostate (3) | 2/3 Glandular epithelial cells (< 5%), nuclear and/or cytoplasmic |
| Skin (3) | 2/3 Keratinocytes (5–10%), nuclear; 1/3 keratinocytes (< 5%), nuclear |
| Small intestine (3) | 3/3 Epithelial cells (< 5%), nuclear; 2/3 lymphocytes (< 5%), nuclear |
| Spleen (3) | 3/3 Lymphocytes (< 5%), nuclear |
| Stomach (3) | 2/3 Glandular epithelium (< 5%), nuclear; 1/3 lymphocytes (< 5%), nuclear |
| Testis (3) | 3/3 Spermatogonia (60–70%), nuclear |
| Thymus (3) | 3/3 Lymphocytes (< 5%), nuclear |
| Tonsil (3) | 3/3 Basal epithelial cells (< 5%), nuclear; lymphocytes (< 5%), nuclear |
| Uterus (3) | 2/3 Glandular epithelial cells (< 5%), nuclear; embryonic stromal cells (< 5%), nuclear; myometrial cells (< 5%), nuclear |
Figure 3.
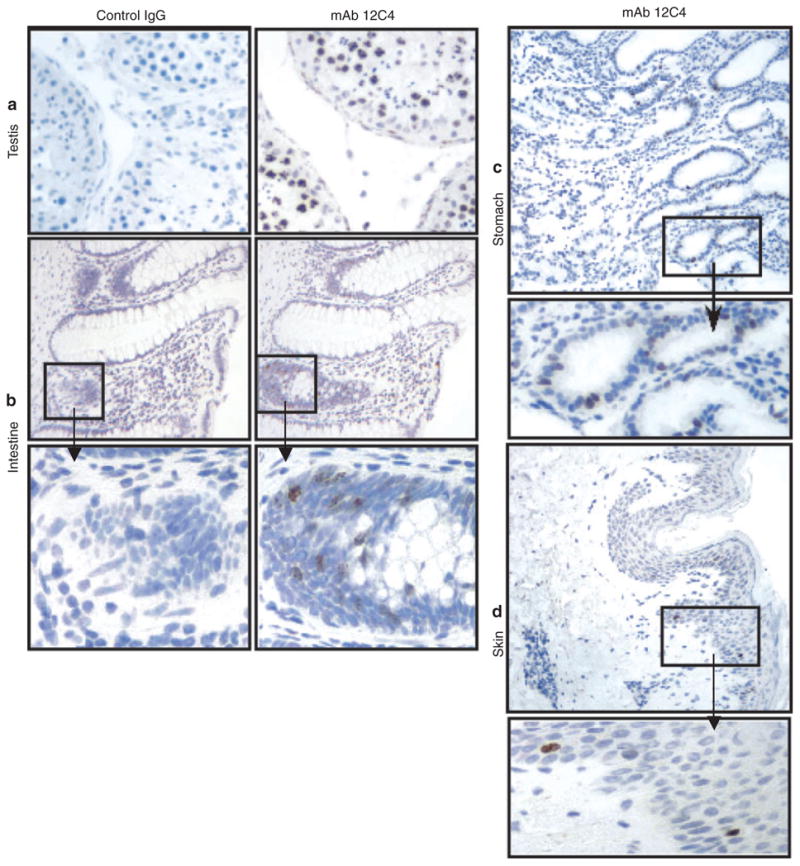
Images from the immunohistochemical study of survivin expression in human adult tissue. Survivin is highly expressed in human testis (a). Survivin is expressed in a limited subset of epithelial cells in the intestinal crypts (b) and basal layers of squamous epithelia (c,d). Left panels in a and b are negative controls processed with an IgG2a subclass mouse mAb (negative control reagent).
EXPRESSION OF SURVIVIN IN HUMAN NEOPLASTIC TISSUES
Survivin expression was also restricted to a subset of the tumour cell population among the neoplastic tissues tested. The majority of tumour types tested on the neoplastic tissue array (16/20) were found to express survivin, but only a subset of the specimens from each tumour type were immunoreactive. Of the 72 specimens evaluated, 35 exhibited positive staining with anti-survivin. Within the positive specimens, immunoreactivity was confined to < 5% of tumour cells present in 18/35 cases. The tumours, which exhibited positivity in ≥ 5% of tumour cells, included colon adenocarcinoma, leiomyomas, melanoma and thyroid carcinoma. Anti-survivin was also tested on a series of single tissue blocks of colorectal and breast adenocarcinoma and the majority of these specimens (11/11 colorectal carcinomas and 12/14 breast carcinomas) exhibited survivin positivity, although expression was focal (< 5%) in 11/25 cases tested. In the remaining 12 positive tumours the percent positive tumour cells ranged from 5 to 40%. These data are consistent with the idea that the majority of cells in tumours are quiescent and that only a subpopulation are proliferative at any one time.
DEREGULATION OF SURVIVIN PROMOTER ACTIVITY IN TUMOUR-DERIVED CELLS VERSUS NON-TRANSFORMED CELLS
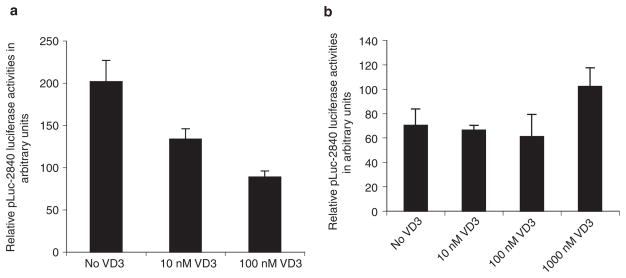
Growing evidence suggests that deregulated expression of survivin contributes to oncogenesis.14 We reasoned that if deregulation of survivin expression in individual cells plays a role in the oncogenic process, we should be able to find certain ligands and/or transcription factors that are involved in survivin transcription controls, showing different behaviour to modulate survivin promoter activity in cancer cells versus normal/untransformed cells. Previously, we have shown that inhibition of survivin expression by vitamin D3 (VD3) is essential for VD3-mediated growth arrest and apoptosis induction in MCF-7E breast cancer cells.21 Consistent with this finding, VD3 down-regulated survivin promoter activity in MCF-7E breast cancer cells (Figure 4a). In contrast, VD3 treatment did not show inhibitory effects on survivin promoter activity in normal fibroblast cells (Figure 4b). This finding implies that oncogenesis may involve changes in the regulation of survivin in response to certain ligands.
Figure 4.
Different modulation of survivin promoter activity by vitamin D3 (VD3). a, VD3 down-regulates survivin promoter activity in MCF-7E breast cancer cells (charcoal dextrin-stripped medium). MCF-7E cells were transfected with the survivin promoter-luciferase reporter construct, pLuc-2840, and treated with and without VD3 for 48 h before the luciferase assay. Luciferase activity was normalized to Renilla luciferase (internal control) activity as arbitrary units and plotted as histograms. Each bar represents the mean ± SD derived from three independent assays. b, VD3 has no inhibitory effect on survivin promoter activity in NIH3T3 fibroblast cells. The experimental conditions were the same as in a. Each bar represents the mean ± SD derived from three independent assays.
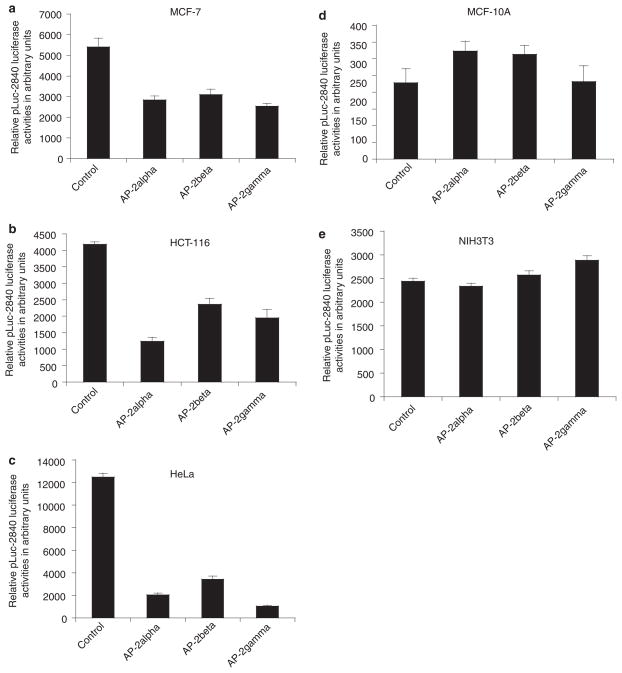
We have shown that survivin promoter differentially interacts with nuclear proteins isolated from cancerous and non-cancerous cells13 and data in Figure 4 also point to differences in the regulation of survivin in normal and cancerous cells. Therefore, we examined the effects of various transcription factors on survivin transcription in various cell types. These studies led to the finding that Ap-2 family transcription factors (α, β, γ) strongly inhibit survivin promoter activity in various tumour-derived cancer cells (Figure 5a–c), but have no significant effect on survivin transcription controls in non-transformed MCF10A breast epithelial cells and NIH3T3 fibroblast cells (Figure 5d,e). Thus, deregulation of survivin transcription controls in individual cells may contribute to oncogenesis and cancer progression.
Figure 5.
Different modulation of survivin promoter activity in cancerous versus non-transformed cells by AP-2 family transcriptional factors. The survivin promoter-luciferase construct, pLuc-2840, was transfected into various cell lines together with pRSV empty vector or one of the AP-2 expression vectors (α, β, γ) as shown. Cells were lysed for luciferase reporter assay 36–48 h after transfection. Luciferase activity was normalized to Renilla luciferase (internal control) activity as arbitrary units and plotted as histograms. Representative experiments are shown. Each bar represent the means ± SD from two independent experiments.
Discussion
Previous studies have shown that deregulated expression of survivin is involved in oncogenesis14 and cancer progression.13 While the expression, subcellular localization, regulation and function of survivin in cancerous tissues and/or cells have been extensively studied, knowledge of its expression and localization in normal human adult tissues is incomplete. Existing studies indicate that survivin is expressed in thymus and placenta,5 endometrium,6 cord blood and bone marrow CD34+ cells7 and colonic mucosa,8 but not in peripheral blood, lymph node, pancreas, kidney, skeletal muscle, liver, lung, brain or heart.5 However, it is not clear whether survivin is expressed in other human adult tissues such as thyroid, pituitary, ovary, adrenal, tonsil, cervix, etc. This information is essential not only for a better understanding of the role of survivin in normal tissue homeostasis, but also to shed light on oncogenic processes and to aid the development of new strategies for cancer prevention and therapeutics.
In the current study, we have characterized a novel survivin mAb (12C4), which evenly recognizes all known splice variants of survivin. Using this newly characterized novel antibody (12C4), the expression and localization of survivin were examined by immunohistochemistry in various tissue types, including tissues not previously examined (Tables 1 and 2). Survivin was found to be largely restricted to a small subset (< 5%) of normal epithelial cells. This observation suggests that survivin expression among epithelial cells of human adult tissues is highly regulated and only a very small subset of these cells express survivin at a time. Notably, these cells were limited to proliferative compartments of positive tissues, reinforcing the link between proliferation and the highly controlled expression of survivin in non-transformed cells. The expression of survivin was also evaluated in a range of tumour types (Tables 3 and 4). In these samples, survivin expression was also found to be limited to a subset of cancer cells, arguing that survivin is also tightly regulated in tumour cells, although it may use different mechanisms. This is consistent with the notion that only a subset of tumour cells is in a proliferative state at any one time and most tumour cells are quiescent. Moreover, we have provided evidence that certain ligands and transcription factors differentially modulate survivin promoter activity in various cancer cells in contrast to normal/untransformed cells. Many mechanisms may result in the deregulation of survivin expression, including activation, inactivation or mutation of survivin transcription factors. For example, it has been demonstrated that wild-type p53 tumour suppressor protein transcriptionally down-regulates survivin expression.22–24 Thus, disruption of p53 function could be one possible mechanism for survivin deregulation during carcinogenesis in certain tissues. The findings of this study not only support the notion that deregulation of survivin expression in individual epithelial cells may contribute to oncogenic processes, but also provide new perspectives for cancer prevention and therapeutics.
Table 3.
Survivin expression in neoplastic human tissue array specimens
| Neoplastic tissue type (no. tested) | Specific staining |
|---|---|
| Lung adenoca (3/3) | 0 |
| Hepatocellular Ca (4/4) | 0 |
| Renal cell Ca (4/4) | 0 |
| Colonic adenoca (2/4) | 0 |
| Colonic adenoca (2/4) | Tumour cells (30–40%), nuclear |
| Ovarian Ca (3/3) | Tumour cells (< 5%), nuclear |
| Breast Ca (2/2) | Tumour cells (< 5%), nuclear |
| Thyroid Ca (2/4) | Tumour cells (90%), cytoplasmic |
| Thyroid Ca (1/4) | 0 |
| Thyroid Ca (1/4) | Tumour cells (< 5%), nuclear |
| Gastric Ca (3/4) | 0 |
| Gastric Ca (1/4) | Tumour cells (5%), nuclear |
| Pancreatic Ca (2/3) | Tumour cells (< 5%), nuclear |
| Pancreatic Ca (1/3) | 0 |
| Prostatic Ca (2/2) | Tumour cells (< 5%), nuclear |
| Astrocytoma (3/4) | 0 |
| Astrocytoma (1/4) | Tumour cells (< 5%), nuclear |
| Mesothelioma (2/4) | 0 |
| Mesothelioma (2/4) | Tumour cells (< 5%), nuclear |
| Undifferentiated Ca (1/4) | Tumour cells (5%), nuclear |
| Undifferentiated Ca (2/4) | Tumour cells (< 5%), nuclear |
| Undifferentiated Ca (1/4) | 0 |
| Lymphoma (2/4) | Tumour cells (5%), nuclear |
| Lymphoma (2/4) | 0 |
| Melanoma (2/4) | 0 |
| Melanoma (1/4) | Tumour cells (5%), nuclear |
| Melanoma (1/4) | Tumour cells (20%), nuclear and cytoplasmic |
| Rhabdomyosarcoma (1/4) | Tumour cells (< 5%), nuclear |
| Rhabdomyosarcoma (3/4) | 0 |
| Malignant fibrous histocytoma (1/4) | Tumour cells (< 5%), nuclear |
| Malignant fibrous histocytoma (3/4) | Tumour cells (5%), nuclear |
| Ewing’s sarcoma (1/3) | Tumour cells (< 5%), nuclear |
| Ewing’s sarcoma (2/3) | Tumour cells (10%), nuclear |
| Leiomyoma (2/4) | Tumour fasicles (91%), cytoplasmic |
| Leiomyoma (2/4) | 0 |
| Carcinoid (4/4) | 0 |
Plasma cell and vascular endothelial cell staining (cytoplasmic) was observed in many tissues
Table 4.
Survivin expression in human colorectal and breast adenocarcinoma tissues
| Neoplastic tissue type (no. tested) | Specific staining |
|---|---|
| Colorectal carcinoma | Tumour cells (< 5%), nuclear |
| Colorectal carcinoma | Tumour cells (< 5%), nuclear |
| Colorectal carcinoma | Tumour cells (40%), nuclear |
| Colorectal carcinoma | Tumour cells (5%), nuclear |
| Colorectal carcinoma | Tumour cells (10%), nuclear |
| Colorectal carcinoma | Tumour cells (< 5%), nuclear |
| Colorectal carcinoma | Tumour cells (< 5%), nuclear |
| Colorectal carcinoma | Tumour cells (40%), nuclear |
| Colorectal carcinoma | Tumour cells (30%), nuclear |
| Colorectal carcinoma | Tumour cells (< 5%), nuclear |
| Colorectal carcinoma | Tumour cells (20%), nuclear |
| Breast carcinoma | 0 |
| Breast carcinoma | 0 |
| Breast carcinoma | Tumour cells (< 5%), nuclear |
| Breast carcinoma | Tumour cells (< 5%), nuclear |
| Breast carcinoma | Tumour cells (< 5%), nuclear |
| Breast carcinoma | Tumour cells (< 5%), nuclear |
| Breast carcinoma | Tumour cells (< 5%), nuclear |
| Breast carcinoma | Tumour cells (10%), nuclear |
| Breast carcinoma | Tumour cells (10%), nuclear |
| Breast carcinoma | Tumour cells (10%), nuclear |
| Breast carcinoma | Tumour cells (< 5%), nuclear |
| Breast carcinoma | Tumour cells (30%), nuclear |
| Breast carcinoma | Tumour cells (5%), nuclear |
| Breast carcinoma | Tumour cells (30%), nuclear |
In addition to the above findings, several other observations made in this study are worthy of mention. Consistent with the previous report that survivin mRNA is highly expressed in mouse testis,25 we found that approximately 60–70% of spermatogonia of human testis express survivin (Table 2 and Figure 3a), although the potential role for survivin in sperm maturation remains to be investigated. Some of the results on survivin expression obtained in this study differ from results in previous reports. For example, in previous studies5,25 survivin mRNA was not detected by Northern blots in the spleen of human and mouse. In contrast, our study revealed that a small percentage (< 5%) of the lymphocytes from spleen do express survivin. In addition, a high level of survivin mRNA was detected in thymus of both human and mouse,5,25 whereas our studies indicate that < 5% of lymphocytes in human thymus tissue express survivin (Table 2). Thus, survivin mRNA levels may not give an accurate reflection of the extent of survivin protein expression in individual cells within a tissue. Furthermore, the finding that one out of three pancreatic samples tested showed 20% positivity in acinar cells is in conflict with reports that normal pancreatic tissue is survivin negative. Nevertheless, lack of survivin expression in the majority of adult pancreas samples is consistent with the fact that this tissue is not generally a site of active cell division.26 However, pancreatitis is associated with acinar cell proliferation and induction of survivin expression.27,28 Current studies are directed at further investigating possible links between pancreatic diseases and survivin expression.
In summary, our studies reveal that the expression of survivin in various human adult tissues is, for the most part, restricted to a very small subset of epithelial cells and that certain ligands and transcription factors differentially modulate survivin promoter activity in various cancer cells versus normal/untransformed cells. These findings are consistent with a role of deregulated expression of survivin in oncogenesis14 and suggest that deregulation of survivin expression in individual epithelial cells contributes to oncogenesis in various human adult tissues.
Acknowledgments
We thank Dr Lily Yang (Emory University, Atlanta, GA, USA) for MCF-10A and HDF cells. This work was sponsored in part by a NIH R01 Grant (CA109481) and a grant from Concern Foundation (Beverly Hill, CA) to F.L., and by shared resources supported by NIH Cancer Center Support Grant CA16056 to Roswell Park Cancer Institute.
Abbreviations
- DAB
3,3′-diaminobenzidene
- DMEM
Dulbecco’s Modified Eagle’s Medium
- EGFP
enhanced green fluorescence protein
- FBS
fetal bovine serum
- GST
glutathionine-S-transferase
- HRP
horseradish peroxidase
- IAP
inhibitor of apoptosis
- mAb
monoclonal antibody
- SDS–PAGE
sodium dodecyl sulphate–polyacrylamide gel electrophoresis
- TBS–T
Tris-buffered NaCl solution with Tween 20
References
- 1.Li F, Ambrosini G, Chu EY, et al. Control of apoptosis and mitotic spindle checkpoint by survivin. Nature. 1998;396 (6711):580–584. doi: 10.1038/25141. [DOI] [PubMed] [Google Scholar]
- 2.Li F, Ackermann EJ, Bennett CF, et al. Pleiotropic cell-division defects and apoptosis induced by interference with survivin function. Nat Cell Biol. 1999;1:461–466. doi: 10.1038/70242. [DOI] [PubMed] [Google Scholar]
- 3.Altieri DC. Survivin, versatile modulation of cell division and apoptosis in cancer. Oncogene. 2003;22:8581–8589. doi: 10.1038/sj.onc.1207113. [DOI] [PubMed] [Google Scholar]
- 4.Altieri DC. Molecular circuits of apoptosis regulation and cell division control: the survivin paradigm. J Cell Biochem. 2004;92:656–663. doi: 10.1002/jcb.20140. [DOI] [PubMed] [Google Scholar]
- 5.Ambrosini G, Adida C, Altieri DC. A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nat Med. 1997;3:917–921. doi: 10.1038/nm0897-917. [DOI] [PubMed] [Google Scholar]
- 6.Konno R, Yamakawa H, Utsunomiya H, Ito K, Sato S, Yajima A. Expression of survivin and Bcl-2 in the normal human endometrium. Mol Hum Reprod. 2000;6:529–534. doi: 10.1093/molehr/6.6.529. [DOI] [PubMed] [Google Scholar]
- 7.Fukuda S, Pelus LM. Regulation of the inhibitor-of-apoptosis family member survivin in normal cord blood and bone marrow CD34(+) cells by hematopoietic growth factors: implication of survivin expression in normal hematopoiesis. Blood. 2001;98:2091–2100. doi: 10.1182/blood.v98.7.2091. [DOI] [PubMed] [Google Scholar]
- 8.Zhang T, Otevrel T, Gao Z, Ehrlich SM, Fields JZ, Boman BM. Evidence that APC regulates survivin expression: a possible mechanism contributing to the stem cell origin of colon cancer. Cancer Res. 2001;61:8664–8667. [PubMed] [Google Scholar]
- 9.Xing Z, Conway EM, Kang C, Winoto A. Essential role of survivin, an inhibitor of apoptosis protein, in T cell development, maturation, and homeostasis. J Exp Med. 2004;199:69–80. doi: 10.1084/jem.20031588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Okada H, Bakal C, Shahinian A, et al. Survivin loss in thymocytes triggers p53-mediated growth arrest and p53-independent cell death. J Exp Med. 2004;199:399–410. doi: 10.1084/jem.20032092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Altznauer F, Martinelli S, Yousefi S, et al. Inflammation-associated cell cycle-independent block of apoptosis by survivin in terminally differentiated neutrophils. J Exp Med. 2004;199:1343–1354. doi: 10.1084/jem.20032033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gurbuxani S, Xu Y, Keerthivasan G, Wickrema A, Crispino JD. Differential requirements for survivin in hematopoietic cell development. Proc Natl Acad Sci USA. 2005;102:11480–11485. doi: 10.1073/pnas.0500303102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li F. Survivin study: what is the next wave? J Cell Physiol. 2003;197:8–29. doi: 10.1002/jcp.10327. [DOI] [PubMed] [Google Scholar]
- 14.Li F. Role of survivin and its splice variants in tumorigenesis. Br J Cancer. 2005;92:212–216. doi: 10.1038/sj.bjc.6602340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Galfre G, Milstein C. Preparation of monoclonal antibodies: strategies and procedures. Methods Enzymol. 1981;73 (Part B):3–46. doi: 10.1016/0076-6879(81)73054-4. [DOI] [PubMed] [Google Scholar]
- 16.Grossman D, McNiff JM, Li F, Altieri DC. Expression and targeting of the apoptosis inhibitor, survivin, in human melanoma. J Invest Dermatol. 1999;113:1076–1081. doi: 10.1046/j.1523-1747.1999.00776.x. [DOI] [PubMed] [Google Scholar]
- 17.Ling X, Yang J, Tan D, et al. Differential expression of survivin–2B and survivin–DeltaEx3 is inversely associated with disease relapse and patient survival in non-small-cell lung cancer (NSCLC) Lung Cancer. 2005 Jun 3;49:353–361. doi: 10.1016/j.lungcan.2005.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li F, Altieri DC. Transcriptional analysis of human survivin gene expression. Biochem J. 1999;344 (Part 2):305–311. doi: 10.1042/0264-6021:3440305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bosher JM, Totty NF, Hsuan JJ, Williams T, Hurst HC. A family of AP-2 proteins regulates c-erbB-2 expression in mammary carcinoma. Oncogene. 1996;13:1701–1707. [PubMed] [Google Scholar]
- 20.Li F, Yang J, Ramnath N, Javle MM, Tan D. Nuclear or cytoplasmic expression of survivin: what is the significance? Int J Cancer. 2005;114:509–512. doi: 10.1002/ijc.20768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li F, Ling X, Huang H, et al. Differential regulation of survivin expression and apoptosis by vitamin D(3) compounds in two isogenic MCF-7 breast cancer cell sublines. Oncogene. 2005;24:1385–1395. doi: 10.1038/sj.onc.1208330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoffman WH, Biade S, Zilfou JT, Chen J, Murphy M. Transcriptional repression of the anti-apoptotic survivin gene by wild type p53. J Biol Chem. 2002;277:3247–3257. doi: 10.1074/jbc.M106643200. [DOI] [PubMed] [Google Scholar]
- 23.Mirza A, McGuirk M, Hockenberry TN, et al. Human survivin is negatively regulated by wild-type p53 and participates in p53-dependent apoptotic pathway. Oncogene. 2002;21:2613–2622. doi: 10.1038/sj.onc.1205353. [DOI] [PubMed] [Google Scholar]
- 24.Zhou M, Gu L, Li F, Zhu Y, Woods WG, Findley HW. DNA damage induces a novel p53-survivin signaling pathway regulating cell cycle and apoptosis in acute lymphoblastic leukemia cells. J Pharmacol Exp Ther. 2002;303:124–131. doi: 10.1124/jpet.102.037192. [DOI] [PubMed] [Google Scholar]
- 25.Kobayashi K, Hatano M, Otaki M, Ogasawara T, Tokuhisa T. Expression of a murine homologue of the inhibitor of apoptosis protein is related to cell proliferation. Proc Natl Acad Sci USA. 1999;96:1457–1462. doi: 10.1073/pnas.96.4.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kami K, Doi R, Koizumi M, et al. Survivin expression is a prognostic marker in pancreatic cancer patients. Surgery. 2004;136:443–448. doi: 10.1016/j.surg.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 27.Bateman AC, Turner SM, Thomas KS, et al. Apoptosis and proliferation of acinar and islet cells in chronic pancreatitis: evidence for differential cell loss mediating preservation of islet function. Gut. 2002;50:542–548. doi: 10.1136/gut.50.4.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tashiro M, Nakamura H, Taguchi M, Yoshikawa H, Otsuki M. Expression of survivin after acute necrohemorrhagic pancreatitis in rats. Pancreas. 2003;26:160–165. doi: 10.1097/00006676-200303000-00012. [DOI] [PubMed] [Google Scholar]
- 29.Yang L, Cao Z, Li F, et al. Tumor-specific gene expression using the survivin promoter is further increased by hypoxia. Gene Ther. 2004;11:1215–1223. doi: 10.1038/sj.gt.3302280. [DOI] [PMC free article] [PubMed] [Google Scholar]