Abstract
A member of the sirtuin family of NAD+-dependent deacetylases, SIRT3 is identified as one of major mitochondrial deacetylase located in mammalian mitochondria responsible for deacetylation of several metabolic enzymes and components of oxidative phosphorylation. Regulation of protein deacetylation by SIRT3 is important for mitochondrial metabolism, cell survival and longevity. In this study, we identified one of the Complex II subunits, succinate dehydrogenase flavoprotein (SdhA) subunit, as a novel SIRT3 substrate in SIRT3 knock-out mice. Several acetylated Lys residues were mapped by tandem mass spectrometry and we determined the role of acetylation on Complex II activity in SIRT3 knock-out mice. In agreement with SIRT3 dependent activation of Complex I, we observed that deacetylation of SdhA subunit increased the Complex II activity in wild type mice. In addition, we treated K562 cell lines with nicotinamide and kaempferol to inhibit deacetylase activity of SIRT3 and stimulate SIRT3 expression, respectively. Stimulation of SIRT3 expression decreased acetylation of the SdhA subunit and increased Complex II activity in kaempherol-treated cells compared to control and nicotinamide treated cells. Evaluation of acetylated residues in SdhA crystal structure from porcine and chicken suggest that acetylation of the hydrophilic surface of SdhA may control the substrate entry to the active site of the protein and regulate the enzyme activity. Our findings constitute the first evidence for the regulation of Complex II activity by the reversible acetylation of the SdhA subunit as a novel substrate of the NAD+- dependent deacetylase, SIRT3.
Reversible acetylation of mitochondrial proteins is critical for regulation of many biological processes, including oxidative phosphorylation and the Krebs cycle (1–7). Flavoprotein of the succinate dehydrogenase complex (Complex II SdhA subunit) was identified as one of the acetylated proteins of mice liver mitochondria in two independent high throughput mapping of acetylated proteins by tandem mass spectrometry (1, 7). Complex II or succinate dehydrogenase (SDH) is found as an inner membrane-bound enzyme complex and it is the only enzyme that participates both in Krebs cycle and oxidative phosphorylation in mitochondria. It has four different protein subunits; hydrophilic subunits SdhA and SdhB facing the matrix side of the inner membrane and hydrophobic subunits, SdhC and SdhD, tethering the complex in the phospholipid membrane. SdhA is a 70 kDa large flavoprotein subunit containing covalently bound FAD and substrate binding site for the entry point of electrons to the Complex II. SDH plays such an important role in the mitochondria, that severe deficiency of this enzyme is incompatible with life. However, point or milder mutations in the C-terminal domain of SdhA lead to Leigh syndrome and various neurodegenerative disorders (8). Mutations of the other SDH subunits containing Fe-S cofactors have been associated with generation of reactive oxygen species causing tumor formation (9).
Post-translational modifications of SdhA by phosphorylation at Tyr residues and acetylation at lysine residues were previously reported (1, 7, 10). Interestingly, six acetylated lysine residues in SdhA were mapped in the LC-MS/MS analysis of well-fed rat mitochondria in two independent studies (1, 7). However, neither enzymes responsible for reversible acetylation and phosphorylation nor their regulatory roles of these post-translational modifications on SdhA or Complex II activity are known. Several members of the class III histone deacetylases (sirtuins) SIRT3, SIRT4, and SIRT5 have been found to reside in mitochondria (6, 11, 12). Sirtuins use NAD+ as a cosubstrate, and both SIRT3 and SIRT4 are required to maintain cell survival after genotoxic stress in a NAD+-dependent manner, and genetic variations in the human SIRT3 gene have been linked to longevity (13, 14). We have previously shown that SIRT3 expression in adipose tissue is increased by caloric restriction and cold exposure (1, 15). Mitochondrial acetyl-CoA synthetase 2 and glutamate dehydrogenase (GDH) are the two key metabolic enzymes regulated through deacetylation by SIRT3 (3, 6, 16). Thus, SIRT3 was determined to be the major deacetylase that modulates mitochondrial function in response to [NADH]/[NAD+] ratio by regulating the activity of key metabolic enzymes (6, 12, 16, 17).
In addition to metabolic enzymes, nuclear encoded subunits of the electron transport chain complexes and ribosomes responsible for the synthesis of 13 essential proteins of the oxidative phosphorylation were found to be regulated by reversible acetylation (1). In our recent studies we demonstrated that the mitochondrial ribosomal protein MRPL10 is acetylated and its deacetylation by the NAD+-dependent deacetylase SIRT3 regulates mitochondrial protein synthesis (18). Additionally, Complex I subunit NDUFA9 is also determined as a SIRT3 substrate and acetylation/deacetylation of this protein is proposed to regulate and maintain basal ATP levels in mammalian mitochondria (17). However, contribution of Complex II acetylation was overlooked on oxidative phosphorylation and ATP production in the same study (17).
Here, we confirmed that one of the subunits of Complex II, SdhA, is indeed a highly acetylated protein and it is a novel SIRT3 substrate as shown in SIRT3 knock-out mice using various proteomics techniques. We have also determined the SIRT3-dependent activation of Complex II in wild-type mice and in cells over-expressing SIRT3. Our results reported in this study suggest a more global role for SIRT3 in regulating oxidative phosphorylation by reversible acetylation of the Complex II subunit SdhA, and therefore, ATP production in mammalian mitochondria.
MATERIALS AND METHODS
Isolation of mitochondria from mice liver and enrichment of Complex II
SIRT3 knock-out mice were obtained from the Texas Institute for Genomic Medicine (Houston, TX, USA). Briefly, these mice were produced by generating embryonic stem (ES) cells (Omnibank no. OST341297) bearing a retroviral promoter trap that functionally inactivates one allele of the Sirt3 gene, as described previously (19). Liver tissue obtained from Sirt3+/+, Sirt3+/− and Sirt3−/− mice was resuspended in an isotonic mitochondrial buffer (MB) (210 mM mannitol, 70 mM sucrose, 1 mM EDTA, 10 mM HEPES-KOH, pH 7.5), supplemented with protease inhibitors (1 mM phenylmethylsulfonyl fluoride, 50 μg/ml leupeptin), and then homogenized in a Dounce homogenizer (Wheaton) on ice. The suspension was centrifuged at 400 × g on a microcentrifuge (ThermoForma) at 4 °C. This procedure was repeated twice, and supernatants were centrifuged at 10,000 × g at 4 °C for 10 min to pellet mitochondria. After lysing the mitochondrial pellets in a buffer containing 0.26 M sucrose, 20 mM Tris-HCl, pH 7.6, 40 mM KCl, 20 mM MgCl2, 0.8 mM EDTA, 0.05 mM spermine, 0.05 mM spermidine, 6 mM β-mercaptoethanol, and 1.6% Triton X-100, mitochondrial lysates were loaded on to 34% sucrose cushions and centrifuged at 100,000 × g at 4 °C for 16 h. The cushion layers enriched for acetylated proteins were acetone precipitated.
2D-gel electrophoresis and immunoblotting analysis
Acetone precipitated protein pellets were resuspended in Destreak rehydration buffer (Amersham Biosciences Inc.) and loaded onto the IPG strips ( pI 3–10) (Bio-Rad Laboratories, Inc.). IPG strips were rehydrated overnight and run on the Ettan IPGphor (Amersham Biosciences Inc) according to the manufacturer’s protocols. The first dimension IPG strips were equilibrated in 6 M urea, 0.375 M Tris-HCl pH 8.8, 2% SDS, 20% glycerol, and 2% (w/v) DTT for 10 min. The strips then were equilibrated in the equilibration buffer containing 2.5% (w/v) iodoacetamide and loaded onto the second dimension SDS-PAGE gel. The gels were either stained with Coomassie Blue or transferred to a PVDF membrane to be probed with N-acetyl lysine antibody at a 1:3000 dilution or SIRT3 antibody at a 1:1000 dilution (Cell Signaling Technology Inc.), a monoclonal SdhA (Complex II subunit 70 kDa Fp) antibody at a 1:5000 dilution (MitoSciences Inc.) or β-Actin Antibody at a 1:5000 dilution (Abcam Inc.). The secondary antibody was ImmunoPure Antibody Goat Anti Mouse IgG (Pierce Biochemicals Inc.) at a 1:5000 dilution or Goat Anti Rabbit IgG at a 1:1000 dilution or Affinipure Rabbit Anti-Mouse IgG, Rabbit Anti-Goat IgG, or Goat Anti-Rabbit IgG (Jackson Immuno Research) all at a 1:10,000 dilution, followed by development with the SuperSignal West Pico Chemiluminescent Substrate (Pierce Biochemicals Inc.) according to the protocol provided by the manufacturer.
Mass spectrometric identification and mapping of acetylation sites
SDS-PAGE bands and 2D-gel spots corresponding to acetylated proteins were excised and in-gel digested with trypsin prior to liquid chromatography tandem mass spectrometry (LC-MS/MS) analysis. The LC-MS/MS analyses were performed by an LTQ mass spectrometer equipped with a nano electrospray ionization source and Surveyor MS Pump Plus HPLC system and Surveyor Micro AS autosampler (ThermoFisher Co.). The in-gel tryptic digests (3–5 μL) were injected and loaded onto a peptide trap (MiChrom peptide CapTrap, C8 like resin, 0.3×1mm, 5μ) over 3 min at 10 μL/min for on-line desalting and concentration. The peptide trap was then placed in line with the analytical column, a PicoFrit column (0.075 × 150 mm) packed in-house with Supelco’s Wide Bore C18 (5μ, 300A) resin. The column was eluted at 250 nL/min using a gradient that consisted of 0.1% formic acid (Solvent A) and 0.1% formic acid in acetonitrile (Solvent B). The peptides were eluted by ramping the solvent B to 40% over 30 min. Tandem MS spectra were acquired for ions above a predetermined intensity threshold using automated data-dependent acquisition. The spectra were processed and searched against the protein sequence database Swiss-Prot using a locally maintained Mascot 2.2 (Matrix Science) and Proteome Discoverer 1.0 (ThermoFisher) search engines to identify proteins and modifications. Mass tolerance was 3 amu and 2 amu for precursor and product ions, respectively. Up to 2 missed cleavages were allowed for digestion by trypsin and methionine oxidation (+16) and lysine acetylation (+42) were considered as a variable modifications.
Cell culture
Brown preadipocytes HIB1B cells with retroviral stable expression of murine SIRT3 (78-334 aa) was previously described (15). In addition, alternative transcript of murine SIRT3 expressing a longer form of murine SIRT3 (1-334 aa)was a gift from Dr. David Sinclair of Harvard Medical School (20). The full-length of SIRT3 (1-334) cDNA was amplified by PCR with the following primers: 5′-ATAGAATTCATGGCGCTTGACCCTC-3′ and 5′-ATAGAATTCTCTGTCCTGTCCATCC-3′. The PCR product was then inserted into the EcoR I site of pBabe-puro-Flag vector (Flag-tag is inserted between EcoR I and Sal I sites of pBabe-puro). HIB1B cells with stable retroviral expression of full-length SIRT3 (1-334) were established as described (15). Mitochondria were isolated from HIB1B stable cell lines expressing truncated and full-length SIRT3 grown in Dulbecco’s Modified Eagle’s Medium (DMEM) with 10% (v/v) bovine calf serum (Hyclone, Logan, Utah), 1% (v/v) penicillin/streptomycin, and puromycin (4 μg/mL) at 37°C with 5% CO2 in a humidified atmosphere and those cells were routinely subcultured in the semi-confluent state.
Approximately, 7×107 K562 (human chronic myelogenous leukemia cell line) cells were grown in RPMI 1640 medium (Mediatech Inc.) supplemented with 10% (v/v) bovine calf serum (Hyclone, Logan, Utah) and 100 IU/ml penicillin and 100 μg/ml streptomycin, at 37°C and 5% CO2 in a humidified atmosphere. Cells were treated with nicotinamide (Calbiochem, San Diego, USA) or kaempferol (Sigma, St. Louis, MO) for 16 or 48 h at 10mM or 50 μM final concentrations, respectively. For immunoblotting, cells pellets were lysed in a buffer containing 50 mM Tris-HCl pH 7.4, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 0.5% NP-40, 0.1% SDS, supplemented with protease inhibitor cocktail (Sigma, St. Louis, MO). After incubation on ice for 10 min, soluble protein fraction was collected by centrifugation at 14,000 × g at 4°C for 15 min.
Complex II enzymatic activity assay
Mitochondria and K562 cell pellets prepared as indicated above were lysed in a buffer containing 300 mM Mannitol, 20 mM sodium phosphate, pH 7.2, 10 mM KCl, 5 mM MgCl2, and 2 mg/ml dodecyl-β-D-maltoside. Pre-incubation of varying amounts of mitochondrial or K562 cell lysates was performed in a buffer containing 300 mM Mannitol, 20 mM sodium phosphate, pH 7.2, 10 mM KCl, 5 mM MgCl2, 50 mM sodium succinate, 40 mM sodium azide, prior to the addition of 50 μM 2,6-dichloroindophenolate to fully activate the succinate dehydrogenase. The Complex II enzymatic activity was recorded by monitoring the reduction of 2,6-dichloroindophenolate at 600 nm. The rate is calculated by dividing the absorbance difference between two linear points by the time point difference (Rate = (Abs 1 − Abs 2)/(Time 2 − Time 1) (21).
RESULTS
Succinate dehydrogenase is acetylated and SIRT3 is responsible for its deacetylation
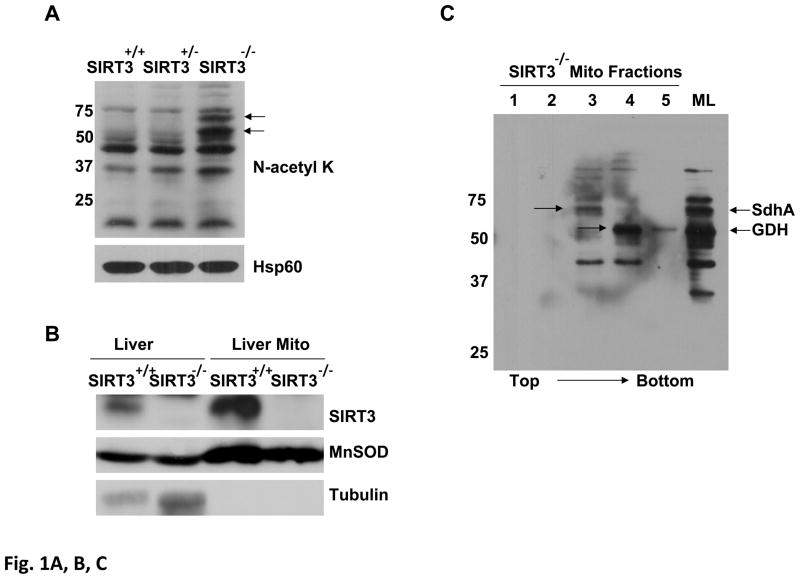
We have recently identified acetylated and phosphorylated protein(s) of mitochondrial ribosomes using a combination of immunoblotting and capillary LC-MS/MS analysis and identified NAD+-dependent SIRT3 as the deacetylase responsible for deacetylation of MRPL10 (18, 22, 23). Using a similar strategy, we identified acetylated proteins specifically deacetylated by SIRT3 in wild type and SIRT3 knock-out mice liver mitochondria to determine SIRT3 substrates. For this purpose, mitochondria were isolated from SIRT3 knock-out (Sirt3−/−), wild-type (Sirt3+/+), and heterozygote (Sirt3+/−) mouse liver mitochondria. Acetylated proteins in mitochondrial lysates were detected by immunoblotting performed with N-acetyl lysine antibody, which revealed two major protein bands at around 70 and 55 kDa with increased acetylation in SIRT3 knock-out mice mitochondrial lysate as shown by arrows (Fig. 1A). Our findings suggested that these two proteins are potential substrates of NAD+-dependent SIRT3 since they were highly acetylated in the absence of SIRT3 expression in knock-out mice (Fig. 1A). The lack of expression of SIRT3 in the whole liver or liver mitochondria from the SIRT3 knock-out mice was confirmed by immunoblot analysis (Fig. 1B)
Figure 1. Detection of SdhA as a novel SIRT3 substrate in SIRT3 knock-out mice liver mitochondria.
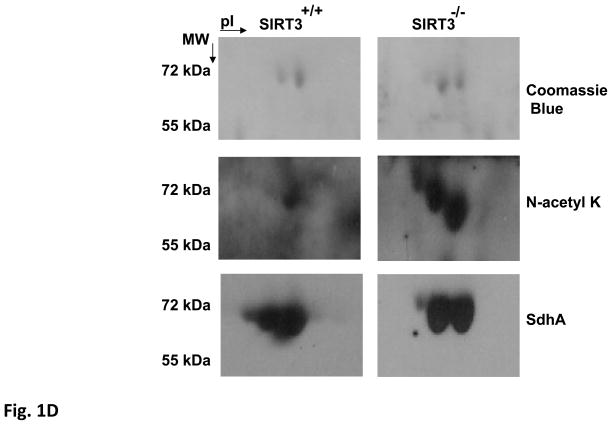
Acetylated proteins and SIRT3 expression in Sirt3+/+, Sirt3+/−, and Sirt3−/− mice liver mitochondria were evaluated by immunoblotting analysis using various antibodies. A) Mitochondrial lysates prepared as described in the Materials and Methods were separated on SDS-PAGE and increased acetylation of mitochondrial proteins was detected by immunoblotting probed with N-acetyl lysine (N-acetyl K) antibody. As a control for equal loading, protein blot was developed with Hsp60 antibody. B) SIRT3 protein levels in the liver or isolated liver mitochondria from wild type or SIRT3 deficient mice were detected by immunoblotting analysis using SIRT3 antibody. Mitochondrial protein MnSOD and cytoplasmic protein tubulin were also detected as controls. C) Approximately, 2mg of Sirt3−/− mice liver mitochondrial lysate were layered on 34% sucrose cushion and fractioned into five separate layers (the top and bottom fractions of the sucrose cushion is shown by the arrow). Equal volumes of each fraction were separated on SDS-PAGE gel and acetylated proteins in each fraction were detected by immunoblotting analysis. The total Sirt3−/− mice liver mitochondrial lysate (ML) layered on the cushion was also analyzed to locate the acetylated proteins in the fractions. Arrows show the location of SIRT3 substrates glutamate dehydrogenase (GDH) and the flavoprotein subunit of succinate dehydrogenase (SdhA). D) Approximately, 50 μl of fraction 3 from Sirt3+/+ or Sirt3−/− mice liver mitochonria was separated on 2D-gels and acetylated proteins were detected with N-acetyl lysine antibody. The acetylated 2D-gel spots corresponding to the Coomassie Blue stained gel spots were in-gel digested and identified by mass spectrometry. The protein identification determined by mass spectrometry was confirmed by immunoblotting using SdhA antibody.
To identify the proteins in these bands and simplify the protein content for 2D-gel separation, mitochondrial lysate obtained from SIRT3 knock-out mice was fractionated on a 30% sucrose cushion containing non-ionic detergent Triton-X100. Immunoblotting analysis of the fractions showed that the two major acetylated proteins at 70 and 55 kDa were in fractions 3 and 4, respectively, implying the presence of these proteins in large protein complexes (Fig. 1C). For the identification of 70 and 55 kDa proteins, 2D-gel electrophoresis was performed using fractions 3 and 4, and protein blots were probed with anti-N-acetyl lysine antibody (Fig. 1D and data not shown for fraction 4). Protein bands corresponding to acetylated proteins detected in 2D-gels were excised, in-gel digested with trypsin, and analyzed by capillary LC-MS/MS for identification. The mass spectrometric analyses of the 2D-gel spots revealed the presence of the flavoprotein subunit of succinate dehydrogenase (SdhA) and glutamate dehydrogenase (GDH-data not shown) in 70 and 55 kDa protein bands, respectively (Table I). Acetylation of glutamate dehydrogenase and the role of SIRT3 in its deacetylation was reported previously (16). Therefore, we focused our efforts on determining the acetylation and deacetylation of SdhA in mitochondria obtained from SIRT3 knockout and wild type mice. To confirm deacetylation of SdhA by SIRT3, immunoblotting and Coomassie blue stained gels of protein lysates were compared (Fig. 1D). Even though SdhA signal obtained by its specific antibody in both SIRT3 knock-out and wild type fractions were comparable, the acetylation signal significantly increased in mitochondrial fraction from SIRT3 knock-out mice (Fig. 1D). This observation supports that the deacetylation of SdhA is due to the expression of endogenous SIRT3 in wild type mice mitochondria while the absence of SIRT3 expression in knockout mice causes hyper acetylation of the SdhA subunit (Fig. 1D).
TABLE I.
Peptides Detected from Tryptic Digests of Acetylated Band Detected in SIRT3 −/− Mice Mitochondria by LC-MS/MS Analysis
| Peptide Sequence | m/z | Mascot Score |
|---|---|---|
| ISQLYGDLK | 1036.6 | 38 |
| HTLSYVDIK | 1075.6 | 41 |
| WHFYDTVK | 1095.6 | 48 |
| NTVIATGGYGR | 555.4 | 83 |
| SMQNHAAVFR | 589.2 | 66 |
| AFGGQSLKAcetFGK | 592.2 | 74 |
| KHTLSYVDIK | 602.8 | 57 |
| AKNTVIATGGYGR | 655.2 | 82 |
| GEGGILINSQGER | 666.2 | 84 |
| VTLEYRPVIDK | 667.3 | 63 |
| TGHSLLHTLYGR | 678.2 | 88 |
| ANAGEESVMNLDK | 698.2 | 98 |
| VDEYDYSKPIQGQQK | 899.8 | 82 |
| VRVDEYDYSKPIQGQQK | 1027.4 | 102 |
| NTVIATGGYGRTYFSCTSAHTSTGDGTAMVTR | 1102.1 | 41 |
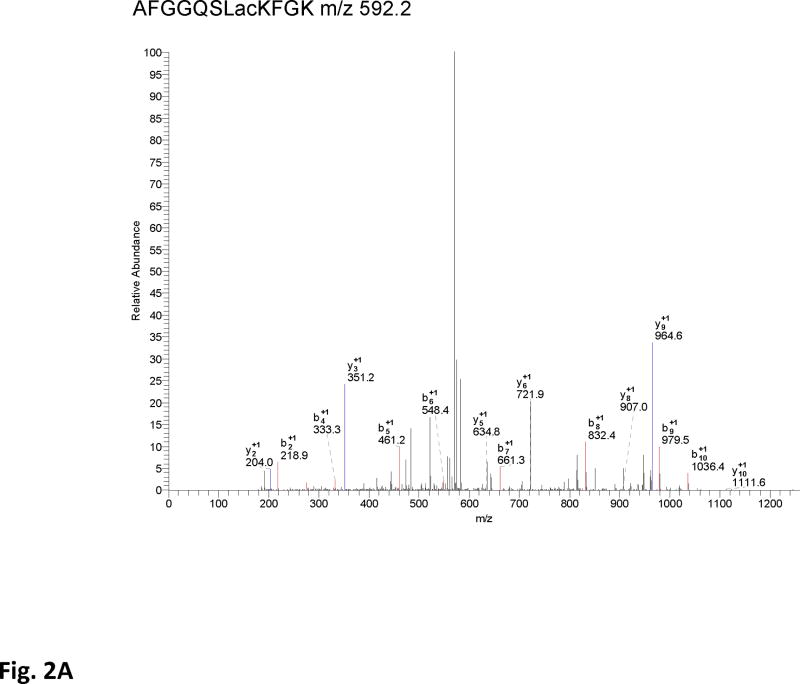
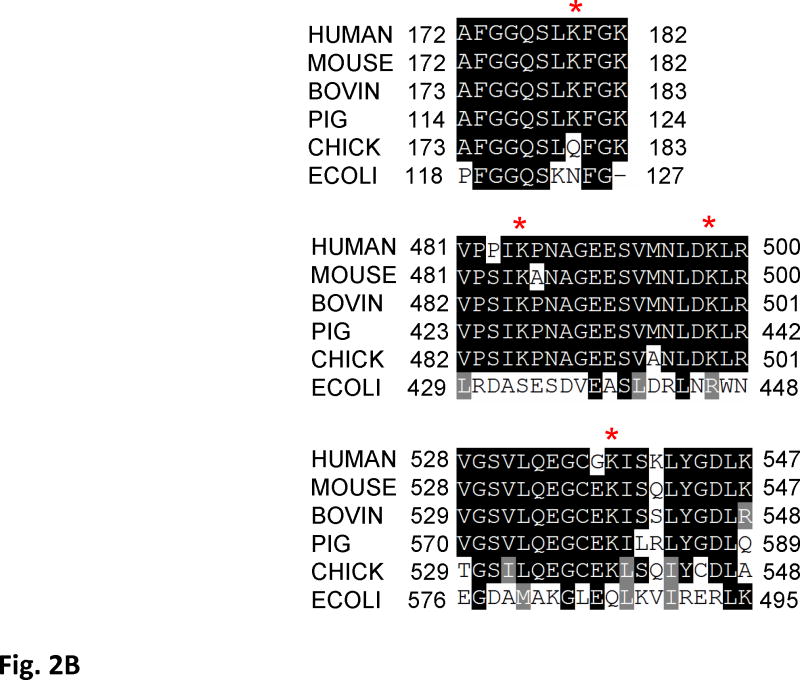
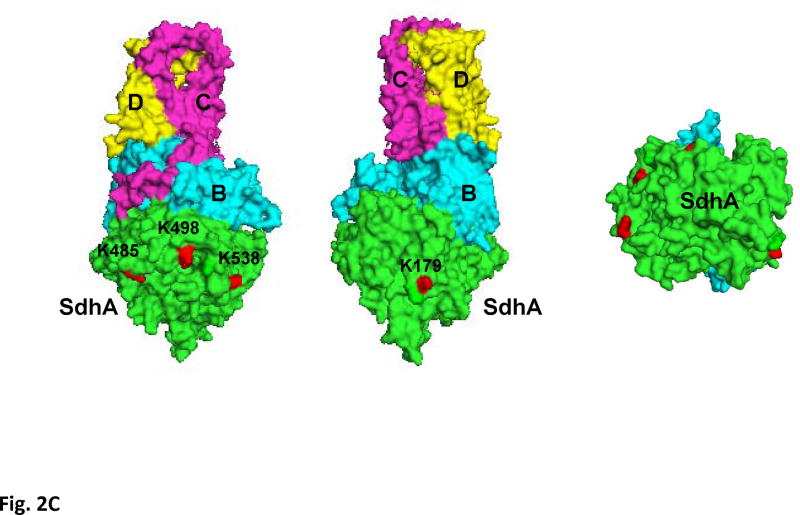
In addition to confirming the acetylation of the SdhA subunit by immunoblotting, one of the acetylated tryptic peptides was also identified with a Mascot score of 74 in the LC-MS/MS analysis of the 2D-gel spots that was previously detected. The CID spectrum of the acetylated peptide AFGGQSLacKFGK is given in Fig. 2A. In high throughput analysis of acetylated proteins from well-fed rat liver mitochondria, several other acetylated lysines were previously identified (1, 7) Alignment of these acetylated peptides with the conserved regions in several other mammalian and chicken mitochondrial, and E. coli SdhA shows that the acetylated lysines are highly conserved in these proteins (Fig. 2B). To demonstrate the location of acetylated lysines in the SdhA subunit, we modeled Complex II structure using the coordinates of the chicken mitochondrial Complex II (Fig 2C) (24). In this structure, conserved acetylated lysine residues (K179, K485, K498, and K538) in the mouse sequence were labeled in red surfaces in the SdhA subunit (Fig. 2). All these residues are located on the hydrophilic surface of the subunit supporting the reversible acetylation of these residues by changes in [NADH]/[NAD] ratios.
Figure 2. Acetylation of SdhA at conserved K179, K485, K498, and K538 residues.
A) The CID spectrum of acetylated peptide detected in LC-MS/MS analysis of 2D-gel spot of SdhA from SIRT3 knock-out mice mitochondria. B) Primary sequence alignment of acetylated peptides from mice SdhA and its homologs from different species. The human, bovine, pig, chicken, and E. coli SdhA were aligned with acetylated peptides of mouse SdhA. (*) denotes the acetylated Lys residues detected in the LC-MS/MS analysis. The alignment was created with CLUSTALW program in Biology Workbench and displayed in BOXSHADE. C) Crystal structure model of the chicken SdhA (PDB# 1YQ3) representing the all four subunits SdhA (green), SdhB (cyan), SdhC, and SdhD (yellow and pink, respectively). The conserved Lys residues found to be acetylated in mouse SdhA (shown by asterisks in B) were colored in red.
Role of hyper-acetylation of SdhA on Complex II activity
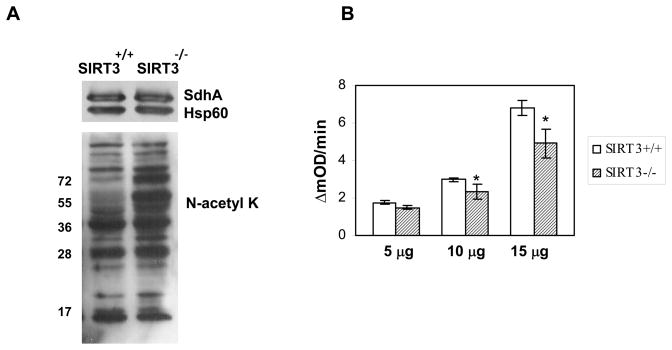
To determine the effect of acetylation on oxidation of succinate to fumarate by Complex II activity, we measured the oxidation of 2,6-dichloroindophenolate (DCIP) in mitochondrial suspensions obtained from SIRT3 knock-out and wild type mice. First, mitochondrial suspensions obtained from these mice were separated on a 12% SDS-PAGE and evaluated for the SdhA, Hsp60, and acetylation levels by immunoblotting of the same gel probed with specific antibodies. Although the same amount of SdhA and Hsp60 were loaded in the gels, the degree of acetylation was much higher in mitochondrial suspension from SIRT3 knock-out mice compared to wild type mice (Fig. 3A). After confirming the presence of equal amounts of SdhA in these samples, we performed the Complex II activity assays at several different amounts of mitochondrial suspensions obtained from SIRT3 knock-out and wild type mice (Fig. 3B). In these assays, the activity of complex II was followed by the transfer of electrons from succinate to DCIP at 600 nm (Fig. 3B). As plotted in Fig. 3B, rate of reactions were measured as changes in absorbance at 600 nm over time as a function of amount of mitochondrial suspension used in the assays. At 15 μg of mitochondria suspension, the difference between the rate of Complex II activity from SIRT3 knock-out mice and wild type mice was about 30% (Fig. 3B). To demonstrate the linearity of the % inhibition detected by the assay, different amounts of mitochondrial lysate was used; however, % inhibition did not change significantly above 15 μg of mitochondria suspension. Here, the reduction of DCIP was directly related to SdhA activity since electrons from succinate are first transferred to enzyme bound cofactor, FAD, in SdhA subunit. For this reason, the decrease in Complex II activity can be attributed to increased acetylation of SdhA in mitochondria from the SIRT3 knock-out mice (Fig. 3B).
Figure 3. Regulation of Succinate Dehydrogenase (Complex II) activity by acetylation of SdhA.
Hyper acetylation of SdhA decreases Complex II activity in SIRT3 knock-out mice. A) Equal amounts of lysates obtained from Sirt3+/+, Sirt3+/−, and Sirt3−/− mice liver mitochondria were separated on 12% SDS-PAGE and probed with N-acetyl lysine (N-acetyl K), SdhA and Hsp60 antibodies. B) Complex II activity was measured as the rate of DCIP reduction, monitored at 600 nm using different amounts of mitochondrial lysates from Sirt3+/+ and Sirt3−/− mice liver mitochondria. Asterisks denote p<0.005.
Role of increased SIRT3 expression on deaceylation of SdhA and Complex II activity
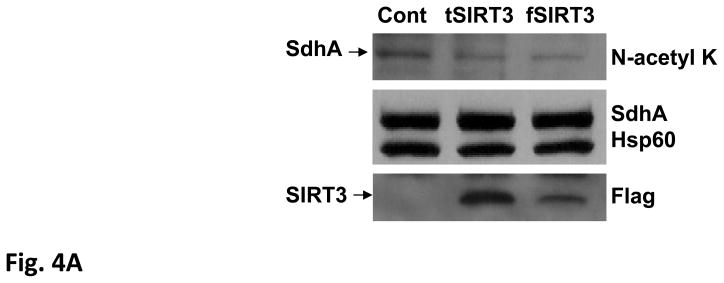
The significant increase in acetylation of several proteins in SIRT3 knock-out mice mitochondria (Fig. 1A and 3A) prompted us to determine the effect of SIRT3 over-expression. For this purpose, we used brown preadipocyte HIB1B cells with retroviral stable expression of murine SIRT3 (78-334 aa) as described before (15). In addition, alternative transcripts of murine SIRT3 were found recently to express proteins with extension at the N-terminus (20). Accordingly, we have generated HIB1B cells with retroviral expression of the long form of SIRT3 (1-334 aa). To determine the role of SIRT3-dependent deacetylation of mitochondrial proteins, mitochondria were isolated from HIB1B control and stable cells expressing two different forms of the SIRT3 gene. In the immunoblotting analysis performed with N-acetyl lysine antibody, we observed a general decrease in acetylation of some of the acetylated protein bands and a protein at around 70 kDa in mitochondrial lysates obtained from SIRT3 over-expression cells. This 70 kDa band overlapped with the SdhA signal in the reprobing of the blot with the SdhA antibody (Fig. 4A).
Figure 4. Role of SIRT3 over-expression on SdhA deacetylation and Complex II activity.
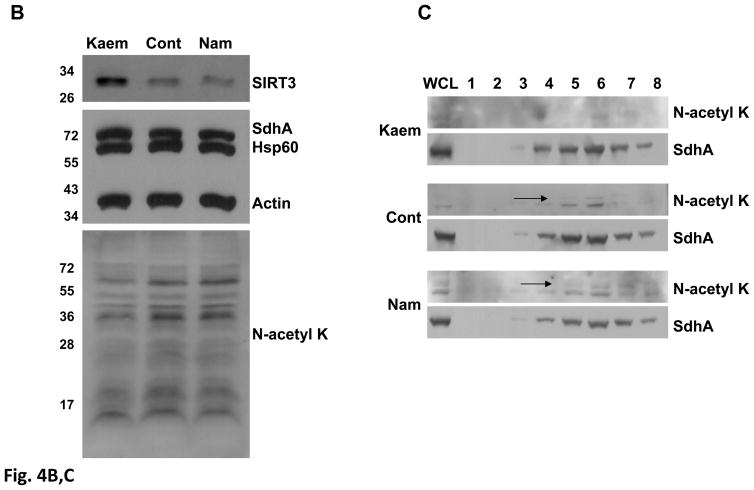
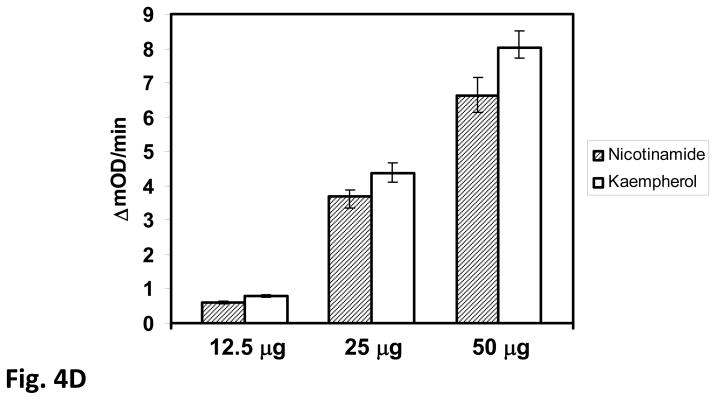
Over-expression of SIRT3 in HIB1B cells increases Complex II activity by deacetylation of SdhA. A) Mitochondria from control and HIB1B cells stably expressing Flag-tagged truncated (tSIRT3) and full-length (fSIRT3) were isolated and about 20 μg of the mitochondrial lysate from each cell line was separated on 12% SDS-PAGE. Immunoblotting analyses were performed with antibodies described in Fig. 3A and Flag-tag antibody. B) Immunoblotting analysis of K562 cell lysates obtained from control (Cont), nicotinamide (Nam), and kaempferol (Kaem) treated cells. Approximately, 20 μg of control and treated K562 cell lysates from each sample was loaded onto 12% SDS-PAGE and immunoblotting was performed as described above. Actin and Hsp60 blots were shown to ensure equal loading in the protein lanes. C) Equal amounts (about 2mg) of control and treated K562 cells lysates were layered on 34% sucrose cushion and fractionated into 8-1 mL aliquots after high speed centrifugation. Equal volumes of each fraction (1–8) were acetone precipitated and loaded on 12% SDS-PAGE gels for immunoblotting analysis. Arrows show the location of acetylated protein overlapping with the SdhA signal. D) Complex II activity was monitored using different amounts of kaempferol and nicotinamide treated K562 cell lysates. The analysis was done in triplicate and values shown are the mean ±SD.
Stimulation of sirtuins, class III histone deacetylases, by several polyphenolic compounds such as resveratrol and kaempferol has been suggested recently (25–27). Specifically, kaempferol treatment of the chronic myelogenous leukemia, K562, cell line has been shown to increase SIRT3 expression in these cell lines (27). Moreover, nicotinamide is a general sirtuin inhibitor and has been shown to inhibit SIRT3-dependent deacetylation of GDH and NDUFA9 (17, 28). To demonstrate the effect of SIRT3 expression on Complex II activity, we treated K562 cells with 50 μM of kaempferol or 10 mM nicotinamide for either 16 or 48 h and, monitored the changes in acetylation and expression of SIRT3 by immunoblotting analysis using whole cell lysates (Fig 4B). Reprobing of the membranes was performed with SdhA and Hsp60 antibodies to ensure equal amount of protein loading in the SDS-PAGE. Consistent with the increased expression of SIRT3 in kaempherol treated cells, the overall acetylation level of proteins decreased compared to the control and nicotinamide treated cells (Fig. 4B). In addition to the detection of overall changes in acetylation of proteins in K562 cells, we fractionated the cell lysates treated with kaempferol and nicotinamide along with untreated cells on 34% sucrose cushion containing 1.6% Triton-X100 to enrich for SdhA protein. Similar to the pattern obtained in fractionation of mice liver mitochondria (Fig. 1C), SdhA remained associated and sedimented with the rest of the Complex II subunits in fractionation of kaempferol and nicotinamide treated cells as confirmed by immunoblotting analyses (Fig. 4C). Especially in the nicotinamide treated and the control cells, acetylated protein signal (indicated by arrows) overlapped with the SdhA signal in the reprobing of the membranes with the specific SdhA antibody. On the other hand, acetylation of SdhA was significantly reduced in kaempferol treated cells, despite the strong SdhA signal obtained with the Sdh antibody in the reprobing. Interestingly, the acetylation signal coming from the lower band was also affected by kaempferol and nicotinamide treatments (Fig. 4C).
Again, to determine the role of SdhA acetylation on Complex II activity, we performed Complex II enzyme activity assays using whole cell lysates obtained from nicotinamide and kaempferol treated K562 cells, which revealed that the Complex II was about 20% more active in kaempferol treated cells compared to the Complex II activity from nicotinamide treated cells ( Fig 4D). The Complex II activity in control cells was not similar to activity of nicotinamide treated cells (data not shown).
DISCUSSION
Mitochondria are required for the production of more than 90% of the ATP required for survival of eukaryotic cells in oxidative phosphorylation. Regulation of oxidative phosphorylation and Krebs cycle components by post-translational modifications has already been established (1, 7, 29, 30). ADP/ATP and [NADH]/[NAD+] ratios are important for regulation of these pathways either by post-translational modifications such as phosphorylation and acetylation or by allosteric regulation. Regulation of mitochondrial function by phosphorylation is known for a long time; however, the recent progress in identification of mitochondria specific NAD-dependent sirtuins such as SIRT3, SIRT4, and SIRT5, revealed the importance of [NADH]/[NAD+] ratio in regulation of protein/enzyme function in post-translational modifications by reversible acetylation (14, 28). One of the best characterized mitochondrial NAD-dependent deacetylase, SIRT3, has been known to regulate activities of several metabolic enzymes and the Complex I subunit NDUFA9 by deacetylation (17). Moreover, we have recently discovered its pivotal role in regulation of mitochondrially encoded proteins of oxidative phosphorylation by mitochondrial protein synthesis by specific deacetylation of a ribosomal protein MRPL10 (18).
In this study, comparison of acetylated proteins in wild type and SIRT3 knock-out mice mitochondria has led us to a novel substrate for SIRT3, the flavoprotein of succinate dehydrogenase complex (SdhA), along with a known substrate, glutamate dehydrogenase. SdhA is one of the hydrophilic subunits of the succinate dehydrogenase involved in Krebs cycle and oxidative phosphorylation in mammalian mitochondria. Previously, in two independent high throughput surveys of the acetylated proteins of rat liver, several acetylated peptides were mapped from SdhA (1, 7) while it was reported as an unacetylated protein in a comprehensive study of SIRT3-dependent deacetylation of Complex I subunit NDUFA9 (17). However, the role of acetylation in the enzyme activity and the deacetylase responsible for this modification were not determined previously.
We believe that the data presented here convincingly clarifies the discrepancy reported in the literature and demonstrates that SIRT3 is indeed the major mitochondrial deacetylase controlling the oxidative phosphorylation by reversible lysine acetylation (16, 17). In the comparison of 2D-gel immunoblotting of SIRT3−/− and SIRT3+/+ mice liver mitochondria, SdhA was found to be hyperacetylated in the absence of SIRT3; however, it is possible that the degree of acetylation in wild-type mice is regulated by availability of acetyl-coA and/or [NADH] levels in the mitochondria. For this reason, we have not observed complete deacetylation of SdhA in the wild-type mice liver mitochondria lysates (Fig. 1D and 3A). More importantly, we have shown the effect of hyper-acetylation on Complex II activity in SIRT3−/− liver mitochondria (Fig. 3B). Interestingly, the Complex II activity in SIRT3 knock-out mice was about 30% lower than that of the wild-type, possibly due to incomplete deacetylation of SdhA in the wild-type mice (Fig. 3). Previously, none of the Complex II subunit proteins was reported as acetylated proteins for the immunocaptured Complex II components in SIRT3 knock-out mice (17). This discrepancy could be due to the sample preparation used by Ahn et al. as they determined the acetylation of Complex II components after immunocapturing of the complex (17). In addition to changes in SdhA acetylation and Complex II activity in SIRT3−/− and SIRT3+/+ mice mitochondria, we have shown a decrease in SdhA activity while increased acetylation was observed in cells treated with a general deacetylase inhibitor, nicotinamide. In contrast, kaempferol treatment of the same cell line caused an increase in expression of SIRT3 and deacetylation of SdhA accompanied by a 20% increase in Complex II activity possibly due to SIRT3-dependent deacetylation of SdhA. Surprisingly, the changes in acetylation of SdhA did not completely inhibit the Complex II activity. As proposed previously, it is likely that only a minor proportion of the protein is acetylated or acetylation only partially regulates the enzyme activity even though mitochondrial protein hyper-acetylation is dramatic in SIRT3 knock-out mice (16). Additionally, conserved acetylated lysine residues in mammalian SdhA are located on the surface of the protein, away from the active site of the enzyme. Therefore, it is feasible to expect that acetylation of the positively charged residues on the surface of the enzyme might either slightly change affinity of the enzyme for its negatively charged substrate, succinate, or induce conformational changes to reduce the activity of the enzyme (Fig. 2B).
Regulation of Complex II activity by reversible acetylation of SdhA subunit relates how oxidative phosphorylation and Krebs cycle components are regulated by metabolite levels in mammalian mitochondria. In the case of high levels of reduced cofactors such as NADH and FADH2 present in the mitochondria, there is no need for further oxidation of acetyl-coA in the Krebs cycle for generation of these cofactors to support oxidative phosphorylation. Thus, it would be reasonable to suggest that acetylation of SdhA just slows down the Krebs cycle, as this process will also cause accumulation of acetyl-coA in the mitochondria. On the other hand, when NAD+ level increases in the mitochondria, SIRT3 and other NAD+ dependent deacetylases will be activated and deacetylate SdhA and other acetylated components of the Krebs cycle. In agreement with stimulation of catalytic activities of metabolic enzymes such as glutamate dehydrogenase and acetyl-coA synthetase 2 by deacetylation, deacetylation of SdhA also stimulates Complex II or succinate dehydrogenase activity to promote Krebs cycle for the generation of reduced NADH and FADH2, as they are the electron donors for ATP synthesis in oxidative phosphorylation. Another potential regulation of Complex II activity is by phosphorylation of the SdhA subunit as it was found to be phosphorylated by Fgr tyrosine kinase in vitro (10). Given its importance in oxidative phosphorylation, it could be suggested that this enzyme can be regulated through cooperation or interplay between these two different post-translational modifications at varying metabolite levels. Moreover, in the case of complete inhibition of the complex, succinate accumulation resulting from the decreased SdhA activity may cause deleterious effects in the cell due to the absence of additional mitochondrial metabolic enzymes those can metabolize succinate (8, 9).
Acknowledgments
We thank Dr. David Sinclair for providing the full-length murine SIRT3 cDNA.
This work was supported by a NIH grant to Q.T (R01DK075978) and E.C.K (R01GM071034).
Abbreviations
- 2D-gel
two dimensional-gel electrophoresis
- FAD
flavin adenine dinucleotide
- K562
human chronic myelogenous leukemia cells
- LC-MSMS/MS
liquid chromatography-tandem mass spectrometry
- NAD+
nicotinamide adenine dinucleotide
- SdhA
succinate dehydrogenase subunit A
- sirtuin or SIRT
silent information regulator two (Sir2) homolog
References
- 1.Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, Cheng T, Kho Y, Xiao H, Xiao L, Grishin NV, White M, Yang XJ, Zhao Y. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell. 2006;23:607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 2.Jackson PJ, Harris DA. Sites of protein-protein interaction on the mitochondrial F1-ATPase inhibitor protein. Biochem J. 1986;235:577–583. doi: 10.1042/bj2350577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hallows WC, Lee S, Denu JM. Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc Natl Acad Sci U S A. 2006;103:10230–10235. doi: 10.1073/pnas.0604392103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dinardo MM, Musicco C, Fracasso F, Milella F, Gadaleta MN, Gadaleta G, Cantatore P. Acetylation and level of mitochondrial transcription factor A in several organs of young and old rats. Biochem Biophys Res Commun. 2003;301:187–191. doi: 10.1016/s0006-291x(02)03008-5. [DOI] [PubMed] [Google Scholar]
- 5.Gerhart-Hines Z, Rodgers JT, Bare O, Lerin C, Kim SH, Mostoslavsky R, Alt FW, Wu Z, Puigserver P. Metabolic control of muscle mitochondrial function and fatty acid oxidation through SIRT1/PGC-1alpha. EMBO J. 2007;26:1913–1923. doi: 10.1038/sj.emboj.7601633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schwer B, Bunkenborg J, Verdin RO, Andersen JS, Verdin E. Reversible lysine acetylation controls the activity of the mitochondrial enzyme acetyl-CoA synthetase 2. Proc Natl Acad Sci U S A. 2006;103:10224–10229. doi: 10.1073/pnas.0603968103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 8.Briere JJ, Favier J, El Ghouzzi V, Djouadi F, Benit P, Gimenez AP, Rustin P. Succinate dehydrogenase deficiency in human. Cell Mol Life Sci. 2005;62:2317–2324. doi: 10.1007/s00018-005-5237-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.King A, Selak MA, Gottlieb E. Succinate dehydrogenase and fumarate hydratase: linking mitochondrial dysfunction and cancer. Oncogene. 2006;25:4675–4682. doi: 10.1038/sj.onc.1209594. [DOI] [PubMed] [Google Scholar]
- 10.Salvi M, Morrice NA, Brunati AM, Toninello A. Identification of the flavoprotein of succinate dehydrogenase and aconitase as in vitro mitochondrial substrates of Fgr tyrosine kinase. FEBS Lett. 2007;581:5579–5585. doi: 10.1016/j.febslet.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 11.Michishita E, Park JY, Burneskis JM, Barrett JC, Horikawa I. Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol Biol Cell. 2005;16:4623–4635. doi: 10.1091/mbc.E05-01-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Onyango P, Celic I, McCaffery JM, Boeke JD, Feinberg AP. SIRT3, a human SIR2 homologue, is an NAD-dependent deacetylase localized to mitochondria. Proc Natl Acad Sci U S A. 2002;99:13653–13658. doi: 10.1073/pnas.222538099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rose G, Dato S, Altomare K, Bellizzi D, Garasto S, Greco V, Passarino G, Feraco E, Mari V, Barbi C, BonaFe M, Franceschi C, Tan Q, Boiko S, Yashin AI, De Benedictis G. Variability of the SIRT3 gene, human silent information regulator Sir2 homologue, and survivorship in the elderly. Exp Gerontol. 2003;38:1065–1070. doi: 10.1016/s0531-5565(03)00209-2. [DOI] [PubMed] [Google Scholar]
- 14.Yang H, Yang T, Baur JA, Perez E, Matsui T, Carmona JJ, Lamming DW, Souza-Pinto NC, Bohr VA, Rosenzweig A, de Cabo R, Sauve AA, Sinclair DA. Nutrient-Sensitive Mitochondrial NAD(+) Levels Dictate Cell Survival. Cell. 2007;130:1095–1107. doi: 10.1016/j.cell.2007.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi T, Wang F, Stieren E, Tong Q. SIRT3, a mitochondrial sirtuin deacetylase, regulates mitochondrial function and thermogenesis in brown adipocytes. J Biol Chem. 2005;280:13560–13567. doi: 10.1074/jbc.M414670200. [DOI] [PubMed] [Google Scholar]
- 16.Lombard DB, Alt FW, Cheng HL, Bunkenborg J, Streeper RS, Mostoslavsky R, Kim J, Yancopoulos G, Valenzuela D, Murphy A, Yang Y, Chen Y, Hirschey MD, Bronson RT, Haigis M, Guarente LP, Farese RV, Jr, Weissman S, Verdin E, Schwer B. Mammalian Sir2 Homolog SIRT3 Regulates Global Mitochondrial Lysine Acetylation. Mol Cell Biol. 2007;27:8807–8814. doi: 10.1128/MCB.01636-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ahn BH, Kim HS, Song S, Lee IH, Liu J, Vassilopoulos A, Deng CX, Finkel T. A role for the mitochondrial deacetylase Sirt3 in regulating energy homeostasis. Proc Natl Acad Sci USA. 2008;105:14447–14452. doi: 10.1073/pnas.0803790105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang Y, Cimen H, Shi T, Han M-J, Deng J-H, Koc H, Palacios OM, Bai Y, Tong Q, Koc EC. NAD+-dependent deacetylase, SIRT3, regulates mitochondrial protein synthesis by deacetylation of mitochondrial ribosomal protein MRP-L10. Revision requested. 2009 doi: 10.1074/jbc.M109.053421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Orsolya MP, Carmona JJ, Shaday M, Chen KY, Manabe Y, Ward JL, III, Goodyear LJ, Tong Q. Diet and exercise signals regulate SIRT3 and activate AMPK and PGC-1α in skeletal muscle. Aging. 2009;392:608–611. doi: 10.18632/aging.100075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jin L, Galonek H, Israelian K, Choy W, Morrison M, Xia Y, Wang X, Xu Y, Yang Y, Smith JJ, Hoffmann E, Carney DP, Perni RB, Jirousek MR, Bemis JE, Milne JC, Sinclair DA, Westphal CH. Biochemical characterization, localization, and tissue distribution of the longer form of mouse SIRT3. Protein Sci. 2009;18:514–525. doi: 10.1002/pro.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Birch-Machin MA, Turnbull DM. Assaying mitochondrial respiratory complex activity in mitochondria isolated from human cells and tissues. Methods Cell Biol. 2001;65:97–117. doi: 10.1016/s0091-679x(01)65006-4. [DOI] [PubMed] [Google Scholar]
- 22.Miller JL, Koc H, Koc EC. Identification of phosphorylation sites in mammalian mitochondrial ribosomal protein DAP3. Protein Sci. 2008;17:251–260. doi: 10.1110/ps.073185608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller JL, Cimen H, Koc H, Koc EC. Phosphorylated proteins of the mammalian mitochondrial ribosome: implications in protein synthesis. J Proteome Res. 2009;8:4789–4798. doi: 10.1021/pr9004844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang LS, Sun G, Cobessi D, Wang AC, Shen JT, Tung EY, Anderson VE, Berry EA. 3-nitropropionic acid is a suicide inhibitor of mitochondrial respiration that, upon oxidation by complex II, forms a covalent adduct with a catalytic base arginine in the active site of the enzyme. J Biol Chem. 2006;281:5965–5972. doi: 10.1074/jbc.M511270200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lagouge M, Argmann C, Gerhart-Hines Z, Meziane H, Lerin C, Daussin F, Messadeq N, Milne J, Lambert P, Elliott P, Geny B, Laakso M, Puigserver P, Auwerx J. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell. 2006;127:1109–1122. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 26.Baur JA, Pearson KJ, Price NL, Jamieson HA, Lerin C, Kalra A, Prabhu VV, Allard JS, Lopez-Lluch G, Lewis K, Pistell PJ, Poosala S, Becker KG, Boss O, Gwinn D, Wang M, Ramaswamy S, Fishbein KW, Spencer RG, Lakatta EG, Le Couteur D, Shaw RJ, Navas P, Puigserver P, Ingram DK, de Cabo R, Sinclair DA. Resveratrol improves health and survival of mice on a high-calorie diet. Nature. 2006;444:337–342. doi: 10.1038/nature05354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marfe G, Tafani M, Indelicato M, Sinibaldi-Salimei P, Reali V, Pucci B, Fini M, Russo MA. Kaempferol induces apoptosis in two different cell lines via Akt inactivation, Bax and SIRT3 activation, and mitochondrial dysfunction. J Cell Biochem. 2009;106:643–650. doi: 10.1002/jcb.22044. [DOI] [PubMed] [Google Scholar]
- 28.Haigis MC, Mostoslavsky R, Haigis KM, Fahie K, Christodoulou DC, Murphy AJ, Valenzuela DM, Yancopoulos GD, Karow M, Blander G, Wolberger C, Prolla TA, Weindruch R, Alt FW, Guarente L. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic beta cells. Cell. 2006;126:941–54. doi: 10.1016/j.cell.2006.06.057. [DOI] [PubMed] [Google Scholar]
- 29.Distler AM, Kerner J, Hoppel CL. Post-translational modifications of rat liver mitochondrial outer membrane proteins identified by mass spectrometry. Biochim Biophys Acta. 2007;1774:628–636. doi: 10.1016/j.bbapap.2007.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gottlieb RA. Identification of targets of phosphorylation in heart mitochondria. Methods Mol Biol. 2007;357:127–137. doi: 10.1385/1-59745-214-9:127. [DOI] [PubMed] [Google Scholar]