Abstract
The Ras GTPases act as binary switches for signal transduction pathways that are important for growth regulation and tumorigenesis. Despite the biochemical simplicity of this switch, Ras proteins control multiple pathways, and the functions of the four mammalian Ras proteins are not overlapping. This raises an important question—how does a Ras protein selectively regulate a particular activity? One recently emerging model suggests that a single Ras protein can control different functions by acting in distinct cellular compartments. A critical test of this model is to identify pathways that are selectively controlled by Ras when it is localized to a particular compartment. A recent study has examined Ras signaling in the fission yeast Schizosaccharomyces pombe, which expresses only one Ras protein that controls two separate evolutionarily conserved pathways. This study demonstrates that whereas Ras localized to the plasma membrane selectively regulates a MAP kinase pathway to mediate mating pheromone signaling, Ras localized to the endomembrane activates a Cdc42 pathway to mediate cell polarity and protein trafficking. This study has provided unambiguous evidence for compartmentalized signaling of Ras.
Keywords: signal transduction, oncogene, cancer, Rho, GTPase, lipid rafts, S. pombe, Prenylation
The mystery of cancer started to be unveiled in the early 1960s when the retroviral transfection assay was used to systematically identify retrovirus that caused transformation of animal cells.1 These studies led to the isolation of many “oncogenes.” However, it was not known whether these oncogenes were “authentic” viral genes or whether they were, in fact, mutated forms of human genes that had been hijacked by the virus. The latter possibility was proved in the early 1980s by the isolation from a human tumor of a mutated form of the H-ras gene that carried the same mutation as the transforming v-ras found in the Harvey Sarcoma Virus.2 Mutations in ras were found to be frequent in human tumors, and before p53 was discovered, ras mutations were the most prevalent gene alteration found in human cancer. Enthusiasm for the study of the ras oncogene was enhanced when it was appreciated that the gene product was a GTP binding protein, a class of regulatory molecules whose biochemical properties were becoming widely known. These two seminal observations combined to raise early, and perhaps premature, optimism that oncogenesis operates by simple rules.
Ras proteins, like most G-proteins, have rather simple biochemical properties that are well understood. They bind GDP and GTP with similar affinity. Whereas Ras proteins are biologically inactive in the GDP-bound state, the GTP-bound forms are active and can stimulate the activities of effectors. Ras proteins are kept in the GDP-bound state through their intrinsic but weak GTPase activity, which is enhanced by GTPase activating proteins (GAPs).3 Activation requires guanine nucleotide exchange factors (GEFs) that induce a conformational change in Ras to open the nucleotide binding pocket, allowing the exchange of the bound GDP for the free GTP present in tenfold greater abundance than GDP in the cytoplasm.4
Whereas on a biochemical level Ras operates as a simple binary switch, Ras signaling pathways in cells are complex. There are three ras genes in mammals that encode four proteins, H-Ras, N-Ras, K-Ras-4A and K-Ras-4B. All but K-Ras4A are ubiquitously expressed. These four Ras proteins are nearly identical in amino acid sequences at the N-terminus, which includes the domain that binds to GEFs and effectors. In vitro, each Ras isoform can be activated by any one of several GEFs, and the human genome encodes many such proteins (http://www.gdb.org/gdb/). Furthermore, each activated Ras can, in turn, regulate a long (and growing) list of effectors.5 Despite the fact that these structurally similar Ras proteins possess the same intrinsic ability to interact with many GEFs and effectors, they appear to have distinct functions in vivo. For example, whereas mice lacking either N-ras or H-ras or both are viable, K-ras is essential for embryonic development,6,7 and, in human tumors, mutations in K-ras are far more frequent than those of H and N-ras.8 How a given Ras protein is coupled to a particular set of GEFs and effectors in order to selectively regulate a specific signaling pathway is of major interest to Ras biologists.
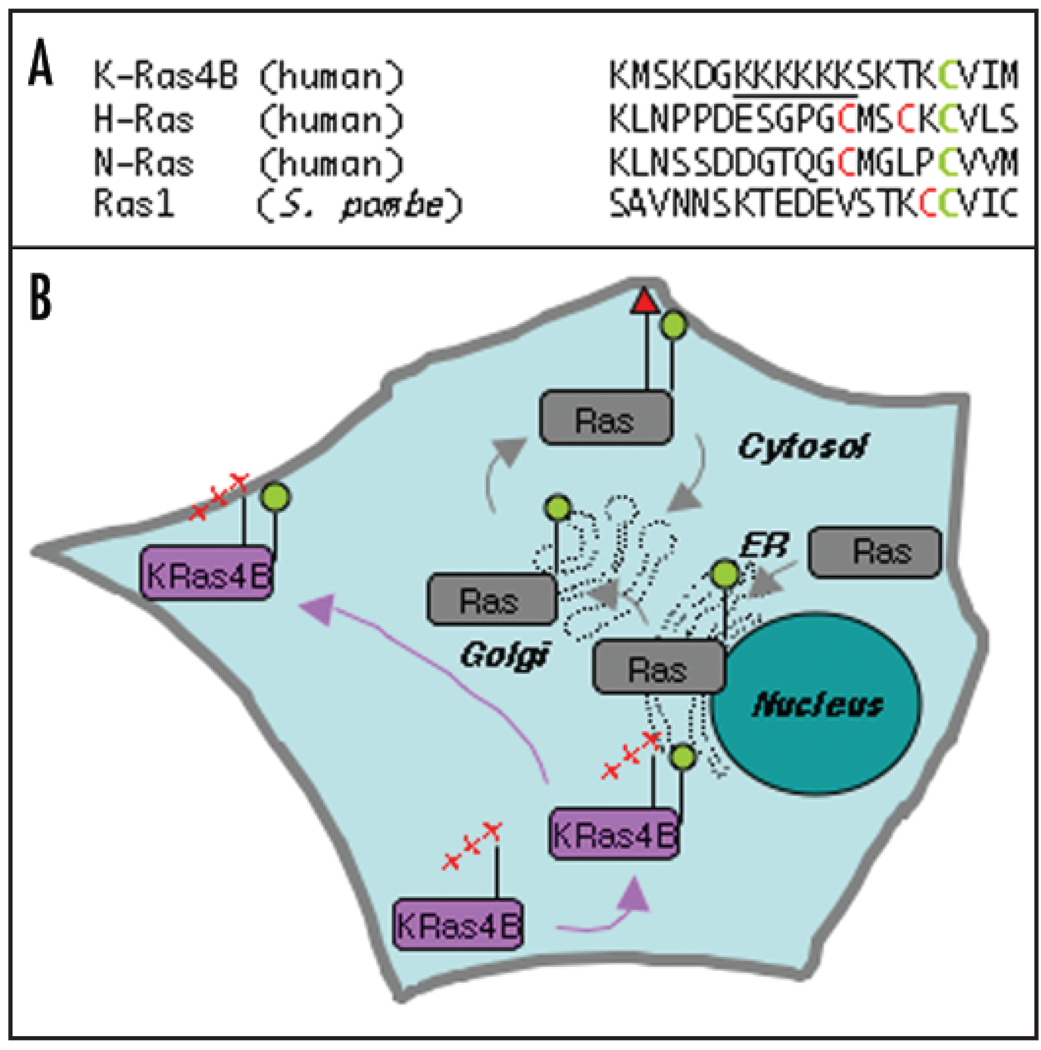
Although Ras proteins are nearly identical at their N-termini, they differ substantially at the C-termini, where the final 10–20 amino acids are called the hypervariable region (Fig. 1A). Thus, it seems reasonable to assume that the hypervariable region can determine signaling selectivity. The hypervariable region contains amino acid residues that are the targets of the covalent modifications that influence where Ras proteins localize in the cell. All Ras proteins contain a CAAX motif at the C-terminus (where C is cysteine, A is an aliphatic amino acid, and X is any amino acid) whose cysteine is farnesylated, a reaction catalyzed in the cytosol by farnesyl transferase. Farnesylation facilitates the localization of Ras to the endoplastic reticulum (ER, Fig. 1B), where the AAX amino acids of the CAAX sequence are proteolytically removed and the farnesylated cysteine is then carboxyl methylated. Immediately upstream of the CAAX box, all Ras proteins except K-Ras4B contain either one (N and K-Ras4A) or two (H-Ras) additional cysteines which are palmitoylated on the Golgi. Palmitoylation targets these Ras isoforms to the plasma membrane (PM). K-Ras4B, in contrast, does not have cysteines that can undergo palmitoylation, but rather is targeted to the PM by a polylysine sequence whose net positive charge is thought to interact with the negatively charged lipid head groups of the inner leaflet of the PM.
Figure 1.
The hypervariable regions of Ras proteins influence where they localize in the cell. (A) The membrane targeting sequences within the hyper-variable regions of indicated Ras proteins from humans and S. pombe are shown. The cysteine residue that is the target for farnesylation is marked in green while that for palmitoylation is marked in red. The polylysine region in K-Ras-4B is underlined. (B) In human cells, newly synthesized Ras proteins localize in the cytoplasm and are devoid of lipidation. N, H, and K-Ras4A (represented by a grey box) are first farnesylated (green circle) to associate with the endomembrane. These proteins are further palmitoylated (red triangle) in the Golgi to localize to the PM, and these proteins can return to the Golgi by depalmitoylation. The polylysine region (red + signs) in K-Ras-4B provides an alternative mode of association with the PM because it interacts with the negatively charged lipids on the inner leaflet of the PM.
The differences in the mode by which Ras proteins associate with various cellular membranes appear to affect where they concentrate in the cell (Fig. 1B). At steady state, H and N-Ras can be found on both the PM and Golgi, but K-Ras-4B is only detectable on the PM.9 This difference in the localization of Ras proteins leads to a model whereby Ras proteins can localize to different cell compartments in order to control different functions. Major support for this idea came from a study by Chiu et al, in which H-Ras was engineered such that it could be specifically targeted to the endomembrane (either Golgi or ER) of fibroblasts.10 These ectopically expressed mutant H-Ras proteins could be activated on the endomembrane and from this location stimulate three well-characterized Ras pathways (Erk, Akt and Jnk). Moreover, oncogenic Ras similarly targeted to the endomembrane was able to transform cells. Endogenous H-Ras is also activated on the endomembrane, as demonstrated by “bystander” fluorescence resonance energy transfer.10
The compartmentalized Ras model predicts that Ras in different subcellular locations can regulate distinct functions, but this idea was not unambiguously validated by the studies of Chiu et al. because the ectopically expressed, endomembrane-targeted Ras proteins activated, to some degree, all Ras pathways studied without absolute selectivity. Compelling evidence supporting compartment-specific signaling came from studies of the budding yeast Saccharomyces cerevisiae, which contains two Ras proteins, Ras1 and Ras2.11 Adenylyl cyclase was the first Ras effector identified in this (or any) organism,12 and was presumed to act on the PM. Sobering et al have more recently identified an additional Ras effector in yeast called Endoplasmic Reticulum Ras Inhibitor 1 (Eri1),13 which functions in the biosynthesis of the glycosylphosphatidylinositol anchor, a process that occurs in the ER. Furthermore, Ras2, when overexpressed, cofractionated with Eri1 in the ER in a GTP-dependent manner.14 Since Ras2 is presumed to activate adenylyl cyclase on the PM, this study substantially strengthened the compartmentalization model by showing that Ras2 can interact with a second effector on a distinct compartment. However, in addition to Ras2, S. cerevisiae also expresses Ras1. Therefore, as in mammalian cells, although one can alter one Ras pathway, one cannot be certain that distinct signaling outcomes are regulated by the same molecule in different locations. Thus a simpler system limited to one Ras isoform would offer a unique opportunity to definitively test the compartmentalization model.
In contrast to budding yeast and mammalian cells, the fission yeast Schizosaccharomyces pombe contains only one Ras ortholog, Ras1.15 Whereas wild type cells are elongated in cell shape and can mate, ras1 cells are abnormally round and sterile. The sterility and round cell shape are two phenotypes that are easily scored and they facilitated the identification of two evolutionarily conserved Ras pathways.16–18 Ras1 regulates Byr2, a MAP kinase kinase, to control mating through a MAPK regulated transcriptional program. Ras1 also regulates Scd1/Ral1, a GEF for Cdc42, to control morphogenesis through the regulation of the cytoskeleton. Because S. pombe contains a single Ras protein that regulates two effectors, it presents an ideal system to study the mechanisms of Ras signaling selectivity.
In a study that was just published, Onken et al.19 tested the hypothesis that the two pathways in S. pombe regulated by Ras1 are controlled from distinct subcellular compartments. The authors constructed a pair of Ras1 mutants that are specifically targeted to the PM and endomembrane, respectively. Ras1-C215S is restricted to the endomembrane due to a cysteine-to-serine substitution that abolishes palmitoylation (Fig. 1A). Ras1-Rit is a fusion protein wherein the C-terminal membrane trafficking region of Ras1 is replaced with the C-terminal region of mammalian Rit, a nonCAAX GTPase that is targeted directly to the PM without first trafficking through the endomembrane. Thus, Ras1-C215S is restricted to the endomembrane, and Ras1-Rit is stringently restricted to the PM. Remarkably, Ras1-C215S, whether expressed from the authentic ras1 promoter or overexpressed from a strong promoter in a multicopy plasmid, restored cell morphology of ras1 cells but did not rescue sterility, while the converse was observed with Ras1-Rit (Fig. 2A). Since protein binding studies showed that Ras1-C215S and Ras1-Rit can bind both effectors as efficiently as wild type Ras1, the alternative possibility that the mutations made to the Ras1 C-terminus have somehow altered their ability to interact with the effectors is ruled out, leaving the difference in localization to explain the different activities of the two mutants.
Figure 2.
Spatial segregation of S. pombe Ras1 pathways. The Ras1-Byr2 (MAP kinase) pathway signals from the PM to mediate cross-membrane signaling during mating. The Ras1-Scd1 (Cdc42) pathway regulates uncharacterized intracellular signals to control protein trafficking. Each Ras1 pathway is specifically activated by a GEF. The Ras1-Byr2 pathway is regulated by Ste6, while Efc25 selectively regulates the Ras1-Scd1 pathway. The farnesyl modification is indicated with a branched, unsaturated polyisoprene chain while palmitoylation is indicated with an acyl chain.
Like growth factors in mammalian cells, yeast mating pheromones signal across the PM, so that it seems appropriate for Ras1 to localize to the PM to regulate the MAP kinase pathway. Conversely, the Ras1-Cdc42 pathway has been shown to control, in addition to the cytoskeleton, protein trafficking.20–22 Therefore the endomembrane is a logical platform from which Ras1 might be expected to regulate this function. These results have provided the clearest demonstration thus far that compartmentalized Ras signaling is an evolutionarily conserved mechanism allowing a single Ras protein to control different functions by regulating spatially distinct pathways.
What additional challenges do we face in elucidating compartmentalized Ras signaling? Palmitoylation is the rate-limiting step in determining whether H, N, and K-Ras-4A proteins are trafficked to the PM. Is this step regulated? Magee et al.23 found that the half-life of N-Ras was significantly longer than that of its palmitate group, establishing a depalmitoylation pathway. Recently, Goodwin et al.24 and Rocks et al25 discovered, using GFP-tagged N-Ras and H-Ras, a retrograde pathway of Ras trafficking from the PM to the Golgi that was dependent on a cycle of palmitoylation/depalmitoylation (Fig. 1B). Although these results provide compelling evidence for a palmitoylation/depalmitoylation cycle for N-Ras and H-Ras in mammalian cells, how this cycle is regulated and how it impacts on signaling remain unclear.
Compartmentalized Ras signaling from various subcellular compartments does not fully explain isoform differences in Ras signaling. For example, despite the fact that all Ras isoforms are found at the PM, neither N-Ras nor H-Ras can compensate for the absence of K-Ras in embryonic development. Clearly what is unique about K-Ras is not fully explained by its association with the PM. One extended view of the compartmentalization model suggests that various Ras isoforms may function in different membrane microdomains. Indeed, whereas N-Ras and H-Ras are enriched in liquid-ordered, cholestrol-rich microdomains known as lipid rafts, K-Ras4B is enriched in distinct microdomains that are disordered.26 Much needs to be done before we understand how differential localization to the lipid rafts can lead to distinct signaling output. Nevertheless, the study of Ras and lipid rafts has illuminated the possibility that the PM has a complex geography, which may profoundly impact signaling.
If Ras signaling is compartmentalized, are there upstream and/or downstream signaling elements that are similarly spatially segregated? There is evidence that GEFs and effectors can cosegregate with their cognate Ras proteins. In mammalian cells a Ras GEF called RasGRP1 localizes to the Golgi to activate N- and H-Ras that concentrate on that organelle.27 RasGRP1 is activated by Ca2+ and diacylglycerol, both of which are rich in the endomembrane and thus may explain why Ras1GRP1 has relatively high affinity for Golgi membranes. By contrast, while recruitment of Sos to the PM by the receptor tyrosine kinase-Grb2 complex has long been predicted by protein-protein interaction experiments,28 so far full-length Sos is mainly detectable in the cytosol and its translocation to the PM has not been reported. Thus, it remains possible that many GEFs may cosegregate with Ras only transiently. The S. pombe Ras pathways may again serve as a valuable model system to analyze the roles of GEFs since the two Ras pathways are specifically regulated by two distinct GEFs (Fig. 2A).29 In mammalian cells, activated Ras recruits Raf to the PM30 and in S. pombe Byr2 translocates to the PM upon the onset of sexual differentiation.31 In mammalian cells, Cdc42 localizes to the endomembrane upon release from Rho-GDI,32 and in S. pombe Cdc42 localization is indistinguishable from that of endomembrane-localized Ras1.20 It remains unclear how many GEFs and effectors are intrinsically localized to a particular compartment and how many are somehow recruited to these compartments in response to signals.
The study of Ras signaling selectivity has more clearly than ever illustrated the importance of the complexity of the architecture of the cell and the dynamic interactions between cell compartments. Thus, to better define the roles of Ras proteins in cancer, it seems insufficient to simply know whether they are overexpressed or carry oncogenic mutations in a particular type of tumor. Rather, one must also decipher where in the cell they concentrate. This information could conceivably facilitate cancer diagnosis and the design of targeted interventions.
ACKNOWLEDGEMENTS
The authors thank Aaron Secrest for technical help and Gary Chamness for reading the manuscript. ECC is supported by grants from the NIH (CA90464 and CA107187), DOD (BC021935), and Susan Komen Foundation (PDF0402733), and MP is supported by grants from the NIH (GM55279, CA116034, and CA118495) and the New York State Breast Cancer Research and Education Program.
References
- 1.Varmus HE. Nobel lecture. Retroviruses and oncogenes. I. Biosci Rep. 1990;10:413–430. doi: 10.1007/BF01152288. [DOI] [PubMed] [Google Scholar]
- 2.Barbacid M. ras oncogenes: Their role in neoplasia. Eur J Clin Invest. 1990;20:225–235. doi: 10.1111/j.1365-2362.1990.tb01848.x. [DOI] [PubMed] [Google Scholar]
- 3.McCormick F. ras GTPase activating protein: Signal transmitter and signal terminator. Cell. 1989;56:5–8. doi: 10.1016/0092-8674(89)90976-8. [DOI] [PubMed] [Google Scholar]
- 4.Vetter IR, Wittinghofer A. The guanine nucleotide-binding switch in three dimensions. Science. 2001;294:1299–1304. doi: 10.1126/science.1062023. [DOI] [PubMed] [Google Scholar]
- 5.Hingorani SR, Tuveson DA. Ras redux: Rethinking how and where Ras acts. Curr Opin Genet Dev. 2003;13:6–13. doi: 10.1016/s0959-437x(02)00017-5. [DOI] [PubMed] [Google Scholar]
- 6.Koera K, Nakamura K, Nakao K, Miyoshi J, Toyoshima K, Hatta T, Otani H, Aiba A, Katsuki M. K-ras is essential for the development of the mouse embryo. Oncogene. 1997;15:1151–1159. doi: 10.1038/sj.onc.1201284. [DOI] [PubMed] [Google Scholar]
- 7.Johnson L, Greenbaum D, Cichowski K, Mercer K, Murphy E, Schmitt E, Bronson RT, Umanoff H, Edelmann W, Kucherlapati R, Jacks T. K-ras is an essential gene in the mouse with partial functional overlap with N-ras. Genes Dev. 1997;11:2468–2481. doi: 10.1101/gad.11.19.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bos JL. ras oncogenes in human cancer: A review. Cancer Res. 1989;49:4682–4689. [PubMed] [Google Scholar]
- 9.Choy E, Chiu VK, Silletti J, Feoktistov M, Morimoto T, Michaelson D, Ivanov IE, Philips MR. Endomembrane trafficking of ras: The CAAX motif targets proteins to the ER and Golgi. Cell. 1999;98:69–80. doi: 10.1016/S0092-8674(00)80607-8. [DOI] [PubMed] [Google Scholar]
- 10.Chiu VK, Bivona T, Hach A, Sajous JB, Silletti J, Wiener H, Johnson RL, IInd, Cox AD, Philips MR. Ras signalling on the endoplasmic reticulum and the Golgi. Nat Cell Biol. 2002;4:343–350. doi: 10.1038/ncb783. [DOI] [PubMed] [Google Scholar]
- 11.Kataoka T, Powers S, McGill C, Fasano O, Strathern J, Broach J, Wigler M. Genetic analysis of yeast RAS1 and RAS2 genes. Cell. 1984;37:437–445. doi: 10.1016/0092-8674(84)90374-x. [DOI] [PubMed] [Google Scholar]
- 12.Toda T, Uno I, Ishikawa T, Powers S, Kataoka T, Broek D, Cameron S, Broach J, Matsumoto K, Wigler M. In yeast, RAS proteins are controlling elements of adenylate cyclase. Cell. 1985;40:27–36. doi: 10.1016/0092-8674(85)90305-8. [DOI] [PubMed] [Google Scholar]
- 13.Sobering AK, Watanabe R, Romeo MJ, Yan BC, Specht CA, Orlean P, Riezman H, Levin DE. Yeast Ras regulates the complex that catalyzes the first step in GPI-anchor biosynthesis at the ER. Cell. 2004;117:637–648. doi: 10.1016/j.cell.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 14.Sobering AK, Romeo MJ, Vay HA, Levin DE. A novel Ras inhibitor, Eri1, engages yeast Ras at the endoplasmic reticulum. Mol Cell Biol. 2003;23:4983–4990. doi: 10.1128/MCB.23.14.4983-4990.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fukui Y, Kozasa T, Kaziro Y, Takeda T, Yamamoto M. Role of a ras homolog in the life cycle of Schizosaccharomyces pombe. Cell. 1986;44:329–336. doi: 10.1016/0092-8674(86)90767-1. [DOI] [PubMed] [Google Scholar]
- 16.Van Aelst L, Barr M, Marcus S, Polverino A, Wigler M. Complex formation between RAS and RAF and other protein kinases. Proc Natl Acad Sci USA. 1993;90:6213–6217. doi: 10.1073/pnas.90.13.6213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang EC, Barr M, Wang Y, Jung V, Xu H, Wigler HM. Cooperative interaction of S. pombe proteins required for mating and morphogenesis. Cell. 1994;79:131–141. doi: 10.1016/0092-8674(94)90406-5. [DOI] [PubMed] [Google Scholar]
- 18.Fukui Y, Yamamoto M. Isolation and characterization of Schizosaccharomyces pombe mutants phenotypically similar to ras1- Mol Gen Genet. 1988;215:26–31. doi: 10.1007/BF00331298. [DOI] [PubMed] [Google Scholar]
- 19.Onken B, Wiener H, Philips RM, Chang EC. Compartmentalized signaling of Ras in fission yeast. Proc Natl Acad Sci USA. 2006;103:9045–9050. doi: 10.1073/pnas.0603318103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murray JM, Johnson DI. Isolation and characterization of Nrf1p, a novel negative regulator of the Cdc42p GTPase in Schizosaccharomyces pombe. Genetics. 2000;154:155–165. doi: 10.1093/genetics/154.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yen HC, Chang EC. INT6: A link between the proteasome and tumorigenesis. Cell Cycle. 2003;2:81–83. [PubMed] [Google Scholar]
- 22.Yen HC, Gordon C, Chang EC. Schizosaccharomyces pombe Int6 and Ras homologs regulate cell division and mitotic fidelity via the proteasome. Cell. 2003;112:207–217. doi: 10.1016/s0092-8674(03)00043-6. [DOI] [PubMed] [Google Scholar]
- 23.Magee AI, Gutierrez L, McKay IA, Marshall CJ, Hall A. Dynamic fatty acylation of p21N-ras. Embo J. 1987;6:3353–3357. doi: 10.1002/j.1460-2075.1987.tb02656.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goodwin JS, Drake KR, Rogers C, Wright L, Lippincott-Schwartz J, Philips MR, Kenworthy AK. Depalmitoylated Ras traffics to and from the Golgi complex via a non-vesicular pathway. J Cell Biol. 2005;170:261–272. doi: 10.1083/jcb.200502063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rocks O, Peyker A, Kahms M, Verveer PJ, Koerner C, Lumbierres M, Kuhlmann J, Waldmann H, Wittinghofer A, Bastiaens PI. An acylation cycle regulates localization and activity of palmitoylated Ras isoforms. Science. 2005;307:1746–1752. doi: 10.1126/science.1105654. [DOI] [PubMed] [Google Scholar]
- 26.Prior IA, Hancock JF. Compartmentalization of Ras proteins. J Cell Sci. 2001;114:1603–1608. doi: 10.1242/jcs.114.9.1603. [DOI] [PubMed] [Google Scholar]
- 27.Bivona TG, Perez De Castro I, Ahearn IM, Grana TM, Chiu VK, Lockyer PJ, Cullen PJ, Pellicer A, Cox AD, Philips MR. Phospholipase Cgamma activates Ras on the Golgi apparatus by means of RasGRP1. Nature. 2003;424:694–698. doi: 10.1038/nature01806. [DOI] [PubMed] [Google Scholar]
- 28.Pawson T. Tyrosine kinase signalling pathways. Princess Takamatsu Symp. 1994;24:303–322. [PubMed] [Google Scholar]
- 29.Papadaki P, Pizon V, Onken B, Chang EC. Two Ras pathways in fission yeast are differentially regulated by two Ras GEFs. Mol Cell Biol. 2002;22:4598–4606. doi: 10.1128/MCB.22.13.4598-4606.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stokoe D, Macdonald SG, Cadwallader K, Symons M, Hancock JF. Activation of Raf as a result of recruitment to the plasma membrane. Science. 1994;264:1463–1467. doi: 10.1126/science.7811320. [DOI] [PubMed] [Google Scholar]
- 31.Ozoe F, Kurokawa R, Kobayashi Y, Jeong HT, Tanaka K, Sen K, Nakagawa T, Matsuda H, Kawamukai M. The 14-3-3 proteins Rad24 and Rad25 negatively regulate Byr2 by affecting its localization in Schizosaccharomyces pombe. Mol Cell Biol. 2002;22:7105–7109. doi: 10.1128/MCB.22.20.7105-7119.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Michaelson D, Silletti J, Murphy G, D’Eustachio P, Rush M, Philips MR. Differential localization of Rho GTPases in live cells: Regulation by hypervariable regions and RhoGDI binding. J Cell Biol. 2001;152:111–126. doi: 10.1083/jcb.152.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]