Abstract
Kruppel-like factor 6 (KLF6) is a tumor suppressor gene that is functionally inactivated in human cancer by loss of heterozygosity, somatic mutation, decreased expression, and increased alternative splicing into an oncogenic splice variant, KLF6-SV1. Here we show that increased expression of KLF6-SV1 is associated with decreased survival in patients with lung adenocarcinoma. In addition, KLF6-SV1 is a novel antiapoptotic protein in lung cancer cell lines, and targeted reduction of KLF6-SV1 using siRNA induces apoptosis both alone and in combination with the chemotherapeutic drug cisplatin. Together, these findings highlight a critical role for KLF6-SV1 in lung cancer, and show a potential novel therapeutic strategy for the treatment of lung cancer.
Introduction
The American Cancer Society estimates 162,460 deaths from lung cancer in the United States for 2007, exceeding cancer mortality from colorectal, breast, prostate, and pancreatic cancer combined (1). Adenocarcinoma of the lung is the most common histologic type among all lung cancers diagnosed. In non–small cell lung carcinoma (NSCLC) the tumor-node-metastasis staging system guides treatment and predicts the chance of long-term survival. Patients with stage IA disease are treated with surgical resection resulting in 5-year survival of 70% (2). In stage IB, IIA, and IIB, surgery in combination with chemotherapy increases 5-year survival by 5% to 15% (3, 4). Whereas in stage IV, the 5-year survival is <1%, and chemotherapy and newer targeted molecular agents are used primarily for symptom reduction with a median increase in survival of at most 2 to 4 months (4).
The use of chemotherapy in the treatment of lung cancer is supported by evidence from dozens of randomized controlled trials. There is, however, significant variability in individual response to chemotherapy and the development of resistance after discontinuation of treatment. In fact, data from clinical trials of chemotherapy estimate that >50% of patients derive no benefit from chemotherapy and, thus, can be considered to have chemotherapy-resistant lung cancer (4). In addition to these patients, many others develop chemotherapy resistance during treatment. Therefore, understanding and better defining the molecular mechanisms underlying the development of treatment resistance would represent a major advance in our understanding of disease progression and treatment failure.
KLF6 (Zf9/CPBP; GeneBank accession number AF001461) is a member of the Krüppel-like factor (KLF) family, a group of zinc finger transcription factors that are involved in differentiation and development. Their role in growth-related signal transduction pathways, cell proliferation, apoptosis, and angiogenesis originally drew attention to their potential role in tumor development and progression (5). The KLF6 gene was originally shown to be functionally inactivated by loss of heterozygosity (LOH) and somatic mutation in sporadic prostate adenocarcinomas (6). More recent reports have extended the range of human tumors and the mechanisms by which KLF6 can be inactivated to include deletion of the KLF6 locus and mutation in colorectal cancers (7), hepatocellular and gastric carcinomas (8, 9), LOH in ovarian carcinoma and glioblastoma (10, 11), decreased KLF6 expression in non–small cell lung cancer (12, 13), and hypermethylation of the promoter region in esophageal SCC cell lines and HCC patient samples (14). Most recently, three alternatively spliced KLF6 isoforms have been identified (15), and at least one of them, KLF6-SV1, has been shown to be biologically active, antagonizing the tumor suppressor function of KLF6 and promoting tumor growth (10, 16). Overexpression of KLF6-SV1 in ovarian and prostate cancer cell lines results in increased invasion, tumorigenicity, and dissemination (10, 17).
Although the exact mechanisms underlying the tumor suppressor roles of KLF6 are unknown, a number of highly relevant, overlapping pathways have been described as follows: trans-activation of p21 in a p53-independent manner (6), reduction of cyclin D1/cyclin-dependent kinase 4 complexes via interaction with cyclin D1 (16), inhibition of c-Jun proto-oncoprotein activities (18), decreased vascular endothelial growth factor expression (10), and induction of apoptosis (13). Given the evidence for a role for the KLF6 tumor suppressor gene in lung cancer (12, 13), we sought to define the role of the oncogenic splice variant, KLF6-SV1, in lung cancer development and progression. A role of KLF6 in NSCLC was first suggested by microarray studies (12). COPEB/KLF6 was decreased in malignant compared with benign lung tissue, but high KLF6 expression levels in tumor specimens were associated with advanced disease stages and contributed to a prognostic gene signature of poor survival (19, 20). This apparent paradox suggested to us the possibility that at least two KLF6 splice forms with different or opposing activities could be pertinent to lung cancer biology.
In the present report, we address the question whether KLF6-SV1 expression is dysregulated in NSCLC and whether it contributes to tumor development and progression. We focused our studies specifically on adenocarcinoma, the most common NSCLC histology. We found that KLF6-SV1 was markedly overexpressed in primary human lung adenocarcinoma and was associated with poor clinical outcomes. Targeted down-regulation of KLF6-SV1 using RNA interference (RNAi) in several lung adenocarcinoma cell lines induced marked apoptosis associated with induction of known proapoptotic and inhibition of antiapoptotic regulators. In addition, overexpression of KLF6-SV1 abrogated the proapoptotic effects of chemotherapy on lung cancer cell lines. Finally, combination therapy using siRNA to KLF6-SV1 and cisplatin induced a marked increase in apoptosis compared with either agent alone. Combined, these data suggest an important role for KLF6-SV1 in lung cancer development and points to a novel role for KLF6-SV1 as an antiapoptotic regulator of cell death both alone and in combination with chemotherapy.
Materials and Methods
Cell culture and cell line generation
All cell lines were obtained from the American Tissue Culture Collection. Retroviral infection with KLF6-SV1 was performed as previously described (10).5,6 Transient transfection of nontargeting control and SV1 siRNA was performed with Lipofectamine 2000 (Invitrogen) in the A549, H838, and H1944 lung cancer cell lines. The siRNAs used for these studies have been produced using the siSTABLE v2 modification system patented by Dharmacon, which increases serum stability to over 120 h compared with 12 h for traditional siRNAs. This proprietary modification, which includes a silyl ethers used to protect the 5′-hydroxyl in combination with an acid-labile orthoester protecting group on the 2′-hydroxyl, and sense strand inactivation, which minimizes off-target silencing and enhances target specificity, were used for all culture and xenograft experiments. Additional data and information concerning the siSTABLE v2 system is available at Dharmacon.com.7 For transfection experiments, 100,000 cells per 12-well dish were plated for each cell line. Cells were then transfected with equal amount of the siRNA and harvested at 72 h and 96 h for RNA, protein, and fluorescence-activated cell sorting (FACS) analysis. For chemotherapy experiments, cisplatin (Sicor Laboratories) was added to a final concentration of either 10 or 20 µmol/L 24 h after either plating of the pBABE/SV1 stable cell lines or transfection with si-NTC or si-SV1 in all lung cancer cell lines studied.
Patient samples
After receiving patient consent and project approval by the Institutional Review Board, tumor tissues and adjacent non-neoplastic lung tissues were collected immediately after resection between May 1991 and July 2000 (by General Thoracic Surgery at the University of Michigan Hospital), and a portion of each tumor and/or lung tissue was frozen in liquid nitrogen and then stored at −80°C. Patient medical records were reviewed to obtain tumor staging, pathology, and survival information, and all identifiers were removed to protect confidentiality. Tumor cellularity criteria for inclusion of each tissue sample into this study were reported previously (5). A total of 70 lung tumors and 4 uninvolved lung samples were analyzed. RNA was isolated using Trisol reagent (Life Technologies) and purified using RNeasy columns (Qiagen, Inc.).
Western blot analysis
Cell extracts for Western blotting were harvested in radioimmunoprecipitation assay buffer (standard protocols; Santa Cruz Biotechnology). Tumor tissue extracts were harvested and prepared in the T-PER reagent (Pierce). Equal amounts of protein (50 µg) as determined by the Bio-Rad DC Protein quantification assay were loaded and separated by PAGE and transferred to nitrocellulose membranes. Western blotting was done using a goat polyclonal antibody to Actin; rabbit polyclonal antibodies to NOXA, BCL2, Caspase 3 and 8 (Cell Signaling Technology); and monoclonal antibodies to KLF6 2A2 (Zymed).
Analysis of proliferation
Proliferation was determined by estimating [3H]thymidine incorporation. A549 stable cell lines expressing pBABE and pBABE-SV1 were plated at a density of 50,000 cells per well in 12-well dishes. Forty-eight hours after plating, 1 µCi/mL [3H]thymidine (Amersham) were added. After 2 h, cells were washed four times with ice-cold PBS and fixed in methanol for 30 min at 4°C. After methanol removal and cell drying cells were solubilized in 0.25% sodium hydroxide/0.25% SDS. After neutralization with hydrochloric acid, dpm were estimated by liquid scintillation counting.
RNA and quantitative real-time PCR analysis
A panel of normal tissues was obtained from Clontech; lung cancer patient samples were collected and extracted as described above. Cell line and tumor RNA was extracted using the Rneasy Mini and Midi kit (Qiagen). All RNA was treated with DNase (Qiagen). A total of 1 µg of RNA was reverse transcribed per reaction using first-strand complementary DNA synthesis with random primers (Promega). Quantitative real-time PCR (qRT-PCR) was performed using the PCR primers previously described (10) on an ABI PRISM 7900HT Sequence Detection System (Applied Biosystems). All experiments were done in triplicate and independently validated thrice. All values were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) levels. All primer sequences and methods for quantifying KLF6 alternative splicing were performed as previously described (10, 15).5,6
FACS analysis
Cells were harvested from 12-well dishes and fixed in absolute ethanol (Sigma) for 24 h. On the day of analysis, cells were pelleted and the absolute ethanol was removed. Cells were then stained with a 1-mL solution containing propidium iodide, RNase A, and PBS. FACS analysis was performed on the FACSCalibur instrument (BD Biosciences).
Results
KLF6-SV1 expression is increased in lung cancer samples and correlates with poor survival
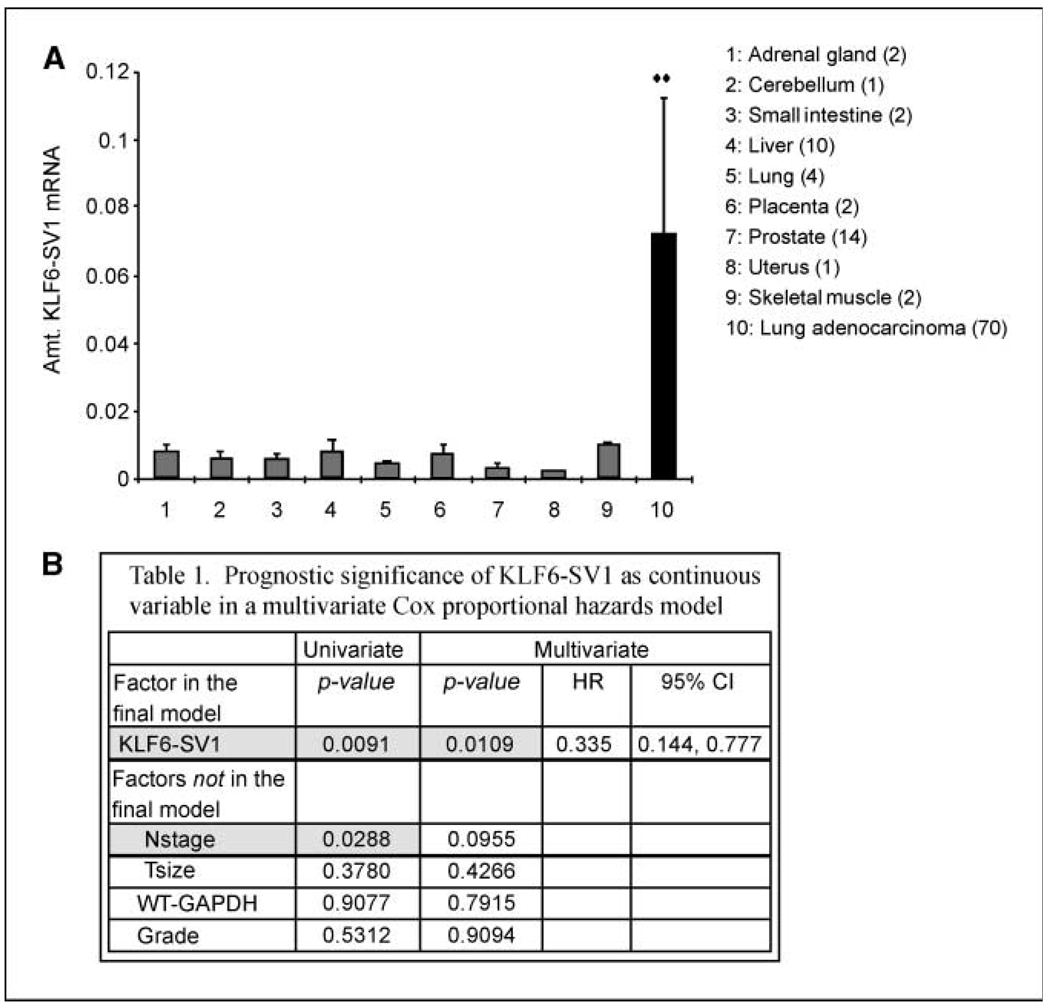
qRT-PCR analysis of a panel of normal tissues and 70 lung adenocarcinoma patient samples revealed a 7-fold increase in KLF6-SV1 expression in lung cancer samples when compared with a panel of normal tissue including four normal lung samples (Fig. 1A; refs. 10, 15, 17). Interestingly, wtKLF6 levels were decreased in lung cancer samples when compared with either all normal tissues or lung tissue specifically (data not shown). We next addressed the possibility that high KLF6-SV1 expression could be an adverse prognostic factor in this same cohort of patients with resected lung adenocarcinomas of mixed stages (IA–IIIB), for whom clinical follow-up for up to 10 years was available. Based on previously published studies in other tumor types (10, 15, 17), we calculated the amount of KLF6-SV1 expression relative to the housekeeping gene GAPDH and treated it as a continuous variable in a Cox multivariate survival analysis, using a model that included tumor size and nodal stage, two clinical prognostic factors that are well-established in NSCLC. In a univariate analysis, KLF6-SV1 expression (P = 0.0091) and nodal stage (P = 0.0288) were associated with survival (Fig. 1B). A post hoc trichotomization of the sample set suggested a 6.5-year difference in median survival between patients in the lowest tertile of KLF-SV1 expression and in the highest. Together these data suggest that KLF6-SV1 is overexpressed in primary lung cancer when compared with normal lung tissue, and that increased KLF6-SV1 expression is associated with poor survival in lung adenocarcinoma patients.
Figure 1.
Expression of KLF6-SV1 in normal tissues and lung cancer patient samples. A, qRT-pCR of a panel of normal tissues and 70 lung adenocarcinoma patient samples using KLF6-SV1 specific real-time primers (10). KLF6-SV1 expression is increased 7-fold in lung cancer specimens when compared with all normal tissue and normal lung specifically; **, P < 0.01. B, the relative amount of KLF6-SV1 to GAPDH expression was determined using qRT-PCR (10, 11) and treated it as a continuous variable in a Cox multivariate survival analysis. In a univariate analysis, KLF6-SV1 expression (P = 0.0091) and nodal stage (P = 0.0288) were associated with survival. WT, wild-type; HR, hazard ratio; 96% CI, 95% confidence interval.
Targeted reduction of KLF6-SV1 expression results in increased apoptosis in lung cancer cell lines
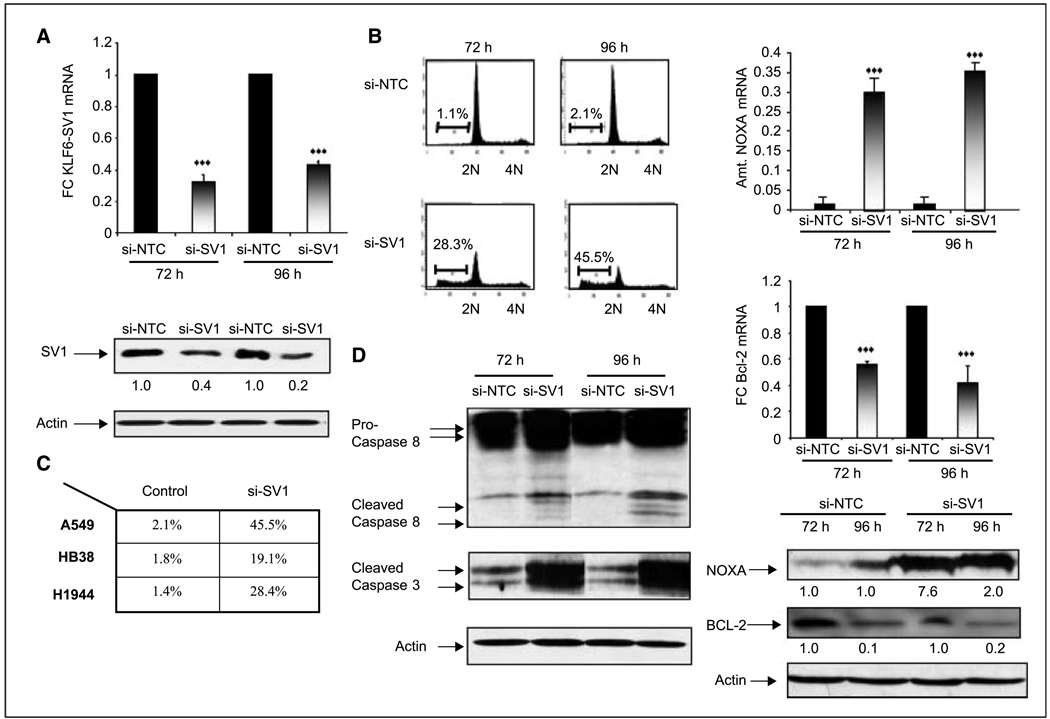
To determine the biological and functional relevance of these findings, we next sought to directly target KLF6-SV1 using RNAi. Using chemically modified siRNA, we explored the biological effect of targeted–reduction of KLF6-SV1 on the behavior of three independent lung cancer cell lines. Transient transfection of KLF6-SV1 specific siRNA resulted in efficient and equal silencing of SV1 expression at both the RNA and protein level at either 72 or 96 h posttransfection in all three lung cancer cell lines tested with no effect on the expression of other KLF6 isoforms (Fig. 2A; data not shown). Consistent with our previous findings in prostate and ovarian cancer cell lines1,2, targeted reduction of KLF6-SV1 resulted in a marked increase in spontaneous apoptosis as shown (Fig. 2B). The induction of apoptosis by targeted reduction of KLF6-SV1 was shown in all three lung cancer cell lines tested (Fig. 2C).
Figure 2.
Targeted reduction of KLF6-SV1 results in a marked increase in spontaneous apoptosis via activation of the intrinsic and extrinsic cell death pathways. A, qtRT-PCR and Western blot analysis of the A549 cell line transfected with a nontargeting control (si-NTC) and a siRNA specific to KLF6-SV1 (si-SV1) show significant down-regulation of KLF6-SV1 at both the mRNA and protein level at both 72 and 96 h; ***, P < 0.001. B and C, FACS analysis of si-NTC– and si-SV1–transfected A549 cells at 72 and 96 h. Targeted reduction of KLF6-SV1 results in a marked increase in spontaneous apoptosis in three independent lung cancer cell lines. Columns, mean of three independent experiments; bars, SD. D, si-SV1 activates both the intrinsic and extrinsic pathways of apoptosis. Western blot analysis shows a marked up-regulation of Caspase 3 and 8 in si-SV1–transfected A549 cell lines. qRT-PCR and Western blot analysis of si-NTC– and si-SV1–transfected cells for NOXA and Bcl-2 shows marked up-regulation of the proapoptotic NOXA (***, P < 0.0001) with concomitant down-regulation of Bcl-2 (***, P < 0.0001).
Based on our previous findings1,2, we analyzed the expression of specific proapoptotic and antiapoptotic genes at both the mRNA and protein level in one of the transfected cell lines with siRNA to SV1. Transfection of si-SV1 in A549 cells resulted in a significant increase in poly(ADP)ribose polymerase cleavage and increased expression of both active Caspase 3 and 8 (Fig. 2D). qRT-PCR for the proapoptotic NOXA and antiapoptotic Bcl-2 was then performed. NOXA mRNA expression was increased 6-fold in cell lines transfected with si-SV1 compared with control, whereas Bcl-2 expression was reduced 80% in this same experiment at both the mRNA and protein level (Fig. 2D). Together, these data suggest that targeted down-regulation of KLF6-SV1 using RNAi results in spontaneous apoptosis through activation of both the intrinsic and extrinsic signaling pathways in lung cancer cell lines.
Overexpression of KLF6-SV1 results in increased cellular proliferation and cell survival
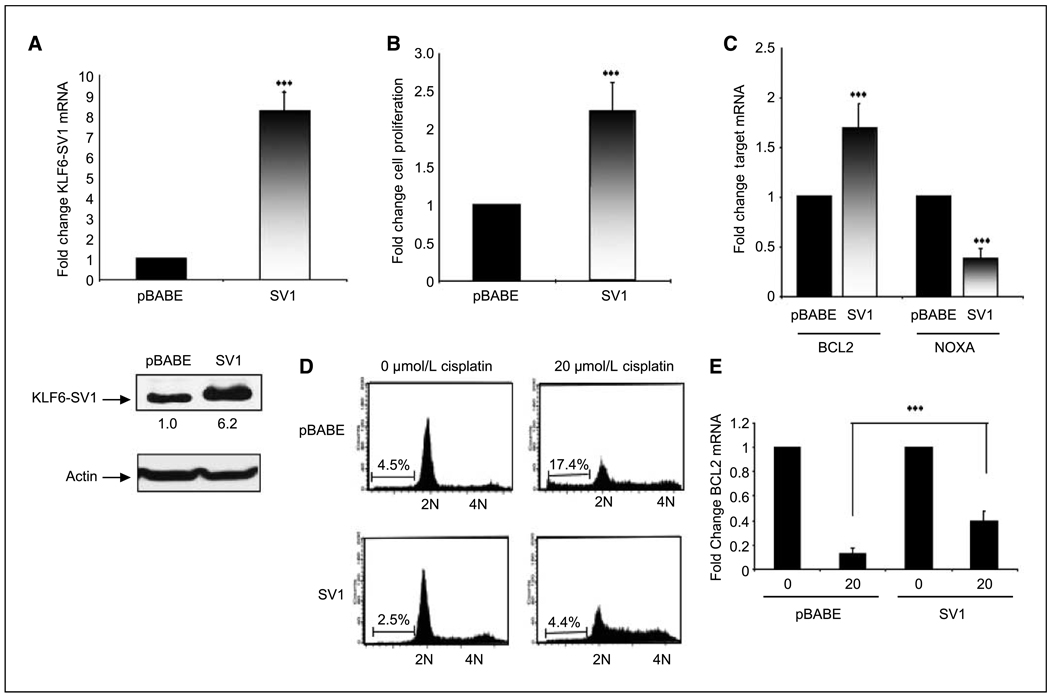
To further characterize the role of KLF6-SV1 and to determine the biological relevance of these findings, we generated stable cell lines overexpressing KLF6-SV1 in the A549 lung cancer cell line. Retroviral infection of this cell line resulted in an 8-fold overexpression of KLF6-SV1 mRNA and protein when compared with pBABE-infected control cell lines (Fig. 3A). Overexpression of SV1 resulted in >2-fold increase in cellular proliferation when compared with control (Fig. 3B). Of significance, increased cellular proliferation was associated with increased expression of Bcl-2 with concomitant down-regulation in NOXA gene expression. Based on our previous findings that targeted reduction of KLF6-SV1 results in a marked increased in apoptosis, we sought to determine if overexpression of SV1 abrogated the ability of the chemotherapeutic agent cisplatin to induce apoptosis. The IC50 for the apoptotic response of the three lung cancer cell lines was determined (data not shown), and cells were treated with that concentration of cisplatin for all subsequent experiments. As predicted, overexpression of SV1 resulted in a significant decrease in apoptosis in response to cisplatin treatment (Fig. 3D). Similar findings were shown in the H388 and H1944 cell line (data not shown). This change was associated with an abrogation of Bcl-2 down-regulation and NOXA up-regulation at the mRNA level (Fig. 3E).
Figure 3.
Overexpression of KLF6-SV1 in the A549 lung cancer cell line. A, qtRT-PCR analysis of pBABE and SV1 retrovirally infected A549 cells shows an 8-fold overexpression of SV1 in pBABE-SV1–infected cell lines compared with control cells (pBABE; ***, P < 0.001). B, SV1-overexpressing cell lines proliferate significantly more than control cell lines; tritiated thymidine incorporation was determined at 72 h (***, P < 0.001). Columns, mean change in the rate of cellular proliferation from three independent experiments; bars, SD. C, increased cellular proliferation and survival in SV1-overexpressing cell lines is associated with increased expression Bcl-2 and concomitant decrease in NOXA expression as determined by qRT-PCR (***, P < 0.001). D, overexpression of SV1 abrogated the proapoptotic effects of cisplatin; pBABE and pBABE-SV1 cells were plated at equal densities and treated with 20 µmol/L of cisplatin; 72 h after treatment, cells were harvested. FACS analysis of the treated cell lines reveals a marked reduction in the induction of apoptosis in SV1-overexpressing cells treated with cisplatin (17.4% versus 4.4%; **, P < 0.01). This experiment was repeated three independent times; representative FACS data are shown. E, overexpression of SV1 significantly abrogated cisplatin-induced down-regulation of Bcl-2 expression (**, P < 0.001).
Targeted reduction of KLF6-SV1 increases lung cancer cell apoptosis in combination with cisplatin treatment
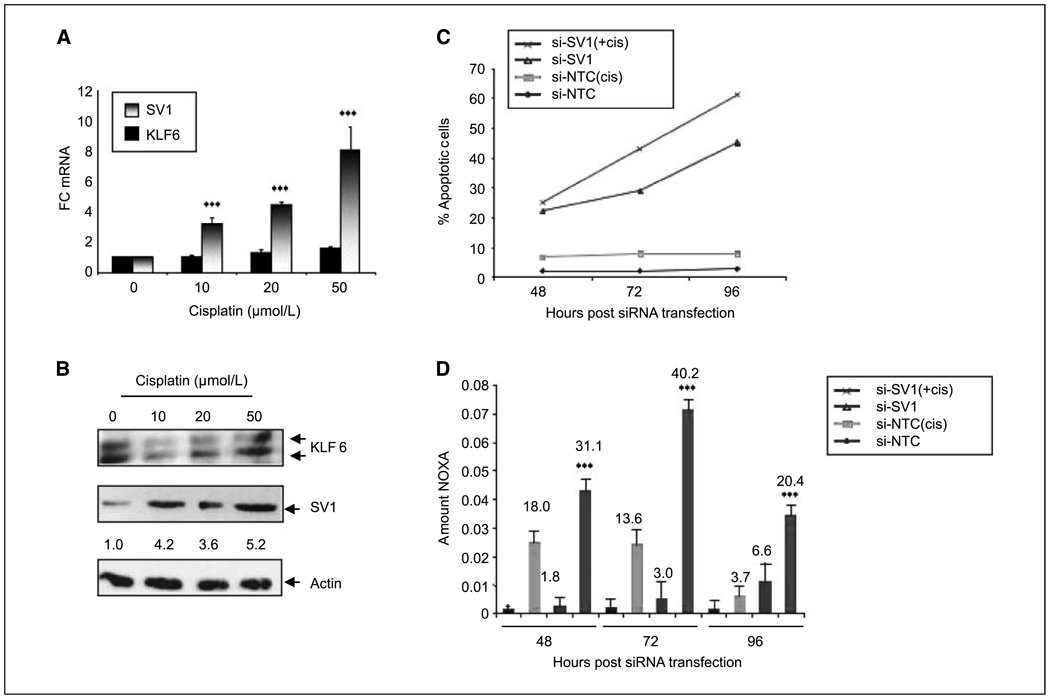
To further characterize the role of KLF6-SV1 in the regulation of cisplatin-mediated DNA damage and apoptosis, we measured the levels of KLF6-SV1 after treatment with cisplatin. Increasing doses of cisplatin resulted in up-regulation of KLF6-SV1 mRNA and protein in the chemotherapy-resistant adherent cells, whereas KLF6 levels remained unchanged (Fig. 4A and B). Based on these findings, we hypothesized that increased expression of KLF6-SV1 may be associated with cisplatin resistance. Combination therapy with siRNA specific to SV1 and cisplatin resulted in the increased activation of the intrinsic pathway of apoptosis (Fig. 4C). The increased apoptosis seen with the combination therapy was associated with a marked increased in NOXA expression at both the mRNA and protein level (Fig. 4D). Combined, these findings highlight a functional role for KLF6-SV1 in chemotherapy resistance and provide a potential biological basis for our finding that increased levels of KLF6-SV1 in lung adenocarcinomas is associated with poor survival.
Figure 4.
Targeted down-regulation of KLF6-SV1 increases the induction of apoptosis in combination with cisplatin. A, increasing doses of cisplatin result in increased KLF6-SV1 expression with no changes in wild-type KLF6 levels as measured by qRT-PCR, using wild-type KLF6– and SV1-specific primers (10). RNA and protein were harvested from adherent cells only after cisplatin treatment. B, Western blot of A549 cells treated with increasing doses of cisplatin probed with a KLF6 monoclonal antibody shows increased KLF6-SV1 expression with higher doses of cisplatin. C, targeted reduction of KLF6-SV1 in combination with subtherapeutic doses of cisplatin results in a significant increase in apoptosis in the A549 cell line (**, P < 0.001). Cells were first transfected with the indicated siRNA [nontargeting control (NTC) or SV1 (si-SV1)]; 24 h after transfection, 10 µm of cisplatin or vehicle control was added to the cells. FACS analysis was performed at the indicated time points (48, 72, or 96 h; *, P < 0.01). D, qRT-PCR revealed an additive effect in NOXA up-regulation after treatment of A549 cells with si-SV1 and cisplatin at all time points assayed (48, 72, and 96 h; **, P < 0.001). Numbers above the bars, fold up-regulation in NOXA expression for each condition. All experiments were repeated three independent times. Column, mean of all three experiments; bars, SD.
Discussion
Lung cancer is the leading cause of cancer-related death in the United States. Patients with early stage disease can be treated with surgery and have a 5-year survival exceeding 70%; however, the prognosis of patients with metastatic lung cancer remains dismal (4). A better understanding of the molecular mechanisms underlying lung cancer progression will allow for both markers for risk prognostication to better guide treatment decisions and for the development of targeted therapies. Here, we show that an alternative splice variant of the KLF6 tumor suppressor gene, KLF6-SV1, is specifically up-regulated in lung adenocarcinoma, and that increased KLF6-SV1 expression is associated with poor survival. Targeted reduction of KLF6-SV1 using RNAi resulted in a marked induction of apoptosis in several lung cancer cell lines. In addition, SV1 overexpression abrogated the proapoptotic effects of cisplatin, and combination treatment with si-SV1 and cisplatin resulted in a marked increase in apoptosis. Combined, these data suggest that KLF6-SV1 is an important regulator of lung cancer survival and tumorigenicity, and that targeted reduction of KLF6-SV1 may represent a novel therapeutic strategy for the treatment of lung cancer.
Acknowledgments
We thank Dr. Andreas Beutler for contributions in the statistical analysis of the data and for technical assistance with manuscript preparation.
Footnotes
A. DiFeo, G. Narla, F. Huang, et al. Inhibition of KLF6-1 increases ovarian cancer survival through the regulation of NOXA, submitted.
G. Narla, A. DiFeo, Y. Fernandez, et al. KLF6-1 is a key regulator of metastasis and survival in prostate cancer, submitted.
References
- 1.Jemal A, Murray T, Ward E, et al. Cancer statistics. CA Cancer J Clin. 2005;55:10–30. doi: 10.3322/canjclin.55.1.10. [DOI] [PubMed] [Google Scholar]
- 2.Strauss GM. Adjuvant chemotherapy of lung cancer: methodologic issues and therapeutic advances. Hematol Oncol Clin North Am. 2005;19:263–281. doi: 10.1016/j.hoc.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 3.Farray D, Mirkovic N, Albain KS. Multimodality therapy for stage III non small-cell lung cancer. J Clin Oncol. 2005;23:3257–3269. doi: 10.1200/JCO.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 4.Shepherd FA. Chemotherapy for advanced non-smallcell lung cancer: modest progress, many choices. J Clin Oncol. 2000;18:35–8S. [PubMed] [Google Scholar]
- 5.Black AR, Black JD, Azizkhan-Clifford J. Sp1 and kruppel-like factor family of transcription factors in cell growth regulation and cancer. J Cell Physiol. 2001;188:143–160. doi: 10.1002/jcp.1111. [DOI] [PubMed] [Google Scholar]
- 6.Narla G, Heath KE, Reeves HL, et al. KLF6, a candidate tumor suppressor gene mutated in prostate cancer. Science. 2001;294:2563–2566. doi: 10.1126/science.1066326. [DOI] [PubMed] [Google Scholar]
- 7.Reeves HL, Narla G, Ogunbiyi O, et al. Kruppel-like factor 6 (KLF6) is a tumor-suppressor gene frequently inactivated in colorectal cancer. Gastroenterology. 2004;126:1090–1103. doi: 10.1053/j.gastro.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 8.Kremer-Tal S, Reeves HL, Narla G, et al. Frequent inactivation of the tumor suppressor kruppel-like factor 6 (KLF6) in hepatocellular carcinoma. Hepatology. 2004;40:1047–1052. doi: 10.1002/hep.20460. [DOI] [PubMed] [Google Scholar]
- 9.Cho YG, Kim CJ, Park CH, et al. Genetic alterations of the KLF6 gene in gastric cancer. Oncogene. 2005;24:4588–4590. doi: 10.1038/sj.onc.1208670. [DOI] [PubMed] [Google Scholar]
- 10.DiFeo A, Narla G, Hirshfeld J, et al. Roles of KLF6 and KLF6-1 in ovarian cancer progression and intraperitoneal dissemination. Clin Cancer Res. 2006;12:3730–3739. doi: 10.1158/1078-0432.CCR-06-0054. [DOI] [PubMed] [Google Scholar]
- 11.Jeng YM, Hsu HC. KLF6, a putative tumor suppressor gene, is mutated in astrocytic gliomas. Int J Cancer. 2003;105:625–629. doi: 10.1002/ijc.11123. [DOI] [PubMed] [Google Scholar]
- 12.Kettunen E, Anttila S, Seppanen JK, et al. Differentially expressed genes in nonsmall cell lung cancer: expression profiling of cancer-related genes in squamous cell lung cancer. Cancer Genet Cytogenet. 2004;149:98–106. doi: 10.1016/S0165-4608(03)00300-5. [DOI] [PubMed] [Google Scholar]
- 13.Ito G, Uchiyama M, Kondo M, et al. Kruppel-like factor 6 is frequently down-regulated and induces apoptosis in non-small cell lung cancer cells. Cancer Res. 2004;64:3838–3843. doi: 10.1158/0008-5472.CAN-04-0185. [DOI] [PubMed] [Google Scholar]
- 14.Yamashita K, Upadhyay S, Osada M, et al. Pharmacologic unmasking of epigenetically silenced tumor suppressor genes in esophageal squamous cell carcinoma. Cancer Cell. 2002;2:485–495. doi: 10.1016/s1535-6108(02)00215-5. [DOI] [PubMed] [Google Scholar]
- 15.Narla G, Difeo A, Reeves HL, et al. A germline DNA polymorphism enhances alternative splicing of the KLF6 tumor suppressor gene and is associated with increased prostate cancer risk. Cancer Res. 2005;65:1213–1222. doi: 10.1158/0008-5472.CAN-04-4249. [DOI] [PubMed] [Google Scholar]
- 16.Benzeno S, Narla G, Allina J, et al. Cyclin-dependent kinase inhibition by the KLF6 tumor suppressor protein through interaction with cyclin D1. Cancer Res. 2004;64:3885–3891. doi: 10.1158/0008-5472.CAN-03-2818. [DOI] [PubMed] [Google Scholar]
- 17.Narla G, DiFeo A, Yao S, et al. Targeted inhibition of the KLF6 splice variant, KLF6 SV1, suppresses prostate cancer cell growth and spread. Cancer Res. 2005;65:5761–5768. doi: 10.1158/0008-5472.CAN-05-0217. [DOI] [PubMed] [Google Scholar]
- 18.Slavin DA, Koritschoner NP, Prieto CC, Lopez-Diaz FJ, Chatton B, Bocco JL. A new role for the kruppel-like transcription factor KLF6 as an inhibitor of c-jun proto-oncoprotein function. Oncogene. 2004;23:8196–8205. doi: 10.1038/sj.onc.1208020. [DOI] [PubMed] [Google Scholar]
- 19.Glinsky GV, Glinskii AB, Stephenson AJ, Hoffman RM, Gerald WL. Gene expression profiling predicts clinical outcome of prostate cancer. J Clin Invest. 2004;113:913–923. doi: 10.1172/JCI20032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Singh D, Febbo PG, Ross K, et al. Gene expression correlates of clinical prostate cancer behavior. Cancer Cell. 2002;1:203–209. doi: 10.1016/s1535-6108(02)00030-2. [DOI] [PubMed] [Google Scholar]