Abstract
Nuclei migrate during many events, including fertilization, establishment of polarity, differentiation, and cell division. The C. elegans KASH protein UNC-83 localizes to the outer nuclear membrane where it recruits kinesin-1 to provide the major motor activity required for nuclear migration in embryonic hyp7 cells. Here we show that UNC-83 also recruits two dynein-regulating complexes to the cytoplasmic face of the nucleus that play a regulatory role. One consists of the NudE homolog NUD-2 and the NudF/Lis1/Pac1 homolog LIS-1; the other includes dynein light chain DLC-1, the BicaudalD homolog BICD-1, and the egalitarian homologue EGAL-1. Genetic disruption of any member of these two complexes caused nuclear migration defects that were enhanced in some double mutant animals, suggesting that BICD-1 and EGAL-1 function in parallel to NUD-2. Dynein heavy chain mutant animals also had a nuclear migration defect, suggesting these complexes function through dynein. Deletion analysis indicated that independent domains of UNC-83 interact with kinesin and dynein. These data suggest a model where UNC-83 acts as the cargo-specific adaptor between the outer nuclear membrane and the microtubule motors kinesin-1 and dynein. Kinesin-1 functions as the major force generator during nuclear migration, while dynein is involved in regulation of bidirectional transport of the nucleus.
Keywords: nuclear migration, KASH proteins, nuclear envelope, kinesin, dynein, C. elegans
Introduction
A wide variety of cell and developmental processes, including establishment of polarity, fertilization, cell division, and cell migration, depend on actively positioning the nucleus to a specific location within the cell (Burke and Roux, 2009; Starr, 2007; Starr, 2009). A failure in proper nuclear positioning leads to a wide variety of developmental defects and diseases. For example, the nuclear anchorage genes Syne-1, 2 (Nesprin-1, 2) have been linked to the pathogenesis of an autosomal recessive cerebellar ataxia, Emry-Dreifuss muscular dystrophy, and the premature aging disease Hutchinson-Gilford progeria (Gros-Louis et al., 2007; Kandert et al., 2007; Zhang et al., 2007). In addition, a nuclear migration defect in the developing brain causes lissencephaly, a severe mental retardation disease (Morris et al., 1998). Despite the importance of nuclear migration in development and disease, a detailed mechanistic understanding of nuclear migration remains unclear in most cases.
The connection between the cytoskeleton and the nuclear envelope is essential for moving the nucleus. Microtubules, actin filaments, and intermediate filaments have all been implicated in nuclear positioning (Starr, 2009). Microtubules are required for nuclei to migrate and remain evenly spaced in Aspergillus during hyphal growth (Xiang and Fischer, 2004). Dynein and many of its associated regulatory proteins, including Lis1 and NudE, were discovered to play an important role in this nuclear migration (Efimov and Morris, 2000; Xiang et al., 1995), although it is still unknown how and where dynein is acting to move the nucleus. Dynein is also required for nuclear migration in radially migrating neurons (Tsai et al., 2007; Zhang et al., 2009). During migration, the centrosome moves at a constant rate towards the leading edge of the cell while the nucleus moves in a salutatory manner behind it. Dynein attached to the nuclear envelope may be responsible for providing a pulling force on centrosomal microtubules to help move the nucleus (Tsai et al., 2007; Zhang et al., 2009). Thus, in different nuclear migration events, dynein plays different roles in multiple cellular locations and the exact role of dynein in most examples remains unknown.
The cytoskeleton is linked to the nuclear envelope by the SUN (Sad1 and UNC-84) and KASH (Klarsicht, ANC-1, and Syne Homology) families of proteins (Starr, 2009; Wilhelmsen et al., 2006). SUN proteins are targeted to the inner nuclear membrane and recruit KASH proteins to the outer nuclear membrane through a direct interaction between the SUN and KASH domains in the perinuclear space (Crisp et al., 2006; McGee et al., 2006; Padmakumar et al., 2005). The cytoplasmic domains of KASH proteins are then free to interact with the cytoskeleton and perform a variety of functions, including nuclear positioning (Starr, 2009; Wilhelmsen et al., 2006). In the nuclear-envelope bridging model of how these proteins function, SUN and KASH proteins span both membranes of the nuclear envelope and transfer forces from the cytoskeleton to the nuclear lamina (Starr, 2009). For example, the C. elegans KASH protein ZYG-12 and SUN protein SUN-1 function during pronuclear migration by coupling the centrosome to the nuclear envelope. An interaction between ZGY-12 and dynein mediates attachment of centrosomes to the nuclear envelope, which is essential during pronuclear migration and nuclear positioning in the gonad (Malone et al., 2003; Minn et al., 2009; Zhou et al., 2009).
In C. elegans, hyp7 embryonic hypodermal precursor cells provide an excellent model system for studying the mechanism of nuclear migration. During embryogenesis, left and right groups of dorsal epithelial cells intercalate, and their nuclei migrate contralaterally across the length of the hyp7 cell (Figure 1A). These cells subsequently fuse, forming the dorsal hypodermal syncytium and the nuclei are positioned laterally (Sulston et al., 1983; Williams-Masson et al., 1998). Hyp7 nuclei are easily scored and used to assay the severity of nuclear migration defects. Mutations in unc-84 or unc-83 disrupt hyp7 cell nuclear migration, resulting in nuclei that are mispositioned to the dorsal cord of L1 larvae (Figure 1) (Horvitz and Sulston, 1980; Malone et al., 1999; McGee et al., 2006; Starr et al., 2001). UNC-83 is a KASH protein that localizes to the outer nuclear membrane and interacts in the perinuclear space with UNC-84, a SUN protein of the inner nuclear membrane (McGee et al., 2006). Together UNC-83 and UNC-84 bridge the nuclear envelope to transfer forces for nuclear migration from the cytoskeleton to the nuclear lamina. However, the molecular mechanisms of how UNC-83 interacts with the cytoskeleton to generate and coordinate forces during nuclear migration are poorly understood.
Figure 1.
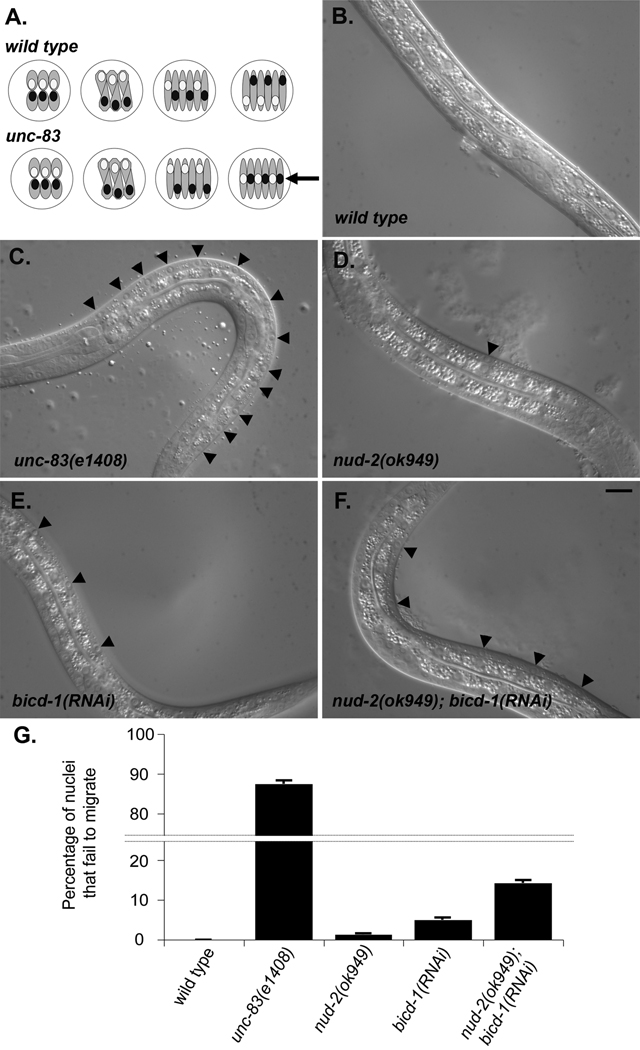
nud-2 and bicd-1 function in hyp7 nuclear migration. (A) A dorsal view of a pre-comma stage embryo illustrating intercalation and nuclear migration of hyp7 precursors in wild-type and unc-83 embryos. The embryo is white, hyp7 precursors are gray, nuclei that migrate from right to left are white, and nuclei that migrate from left to right are black. Anterior is to the left. Arrow indicates the dorsal cord. (B–F) Lateral view of L1 hermaphrodites. Dorsal is upwards. (B) Wild-type N2 animal showing no nuclei in the dorsal cord. (C) unc-83(e1408). (D) nud-2(ok949). (E) bicd-1(RNAi). (F) nud-2(ok949); bicd-1(RNAi). Black arrowheads mark hyp7 nuclei in the dorsal cord. Scale bar is 10 µm. (G) Quantification of hyp7 nuclear migration phenotypes. Error bars are standard error. See Table 2 for details and statistics.
To elucidate a mechanism of UNC-83-mediated nuclear migration, we sought to identify binding partners of the large cytoplasmic domain of UNC-83 using a yeast two-hybrid screen. From the screen we identified several microtubule motor regulating proteins. We recently demonstrated that UNC-83 functions to recruit kinesin-1 to the nuclear envelope (Meyerzon et al., 2009). Mutations in the kinesin-1 light chain klc-2 cause a nuclear migration phenotype similar to unc-83 mutant alleles, suggesting that kinesin-1 provides the major forces to move nuclei (Meyerzon et al., 2009). Here we focus on the interaction between UNC-83 and dynein. Mutations in dynein components lead to a weaker nuclear migration defect, suggesting that dynein plays a regulatory role during hyp7 nuclear migration.
Materials and methods
C. elegans Strains, Genetics, and Phenotypic Analysis
C. elegans were cultured using standard conditions (Brenner, 1974). The Bristol N2 strain was used for wild-type and to generate all other strains. The null allele unc-83(e1408) was described previously (Horvitz and Sulston, 1980; Starr et al., 2001). The sterile allele dhc-1(js121) was from the GFP-balanced stock NM2040 (Koushika et al., 2004), which allowed us to quantify the dhc-1(js121) nuclear migration phenotype in a double blind study. The nud-2(ok949) and bicd-1(ok2731) alleles were generated by the International C. elegans Gene Knockout Consortium (www.celeganskoconsortium.omrf.org). The bicd-1(tm3421) allele was supplied by Shohei Mitani (National Bioresource Project at the Tokyo Women’s Medical University). bicd-1(tm3421) was backcrossed four times to him-8(e1489) males. The deletion was followed by PCR as described (Ahringer, 2006). Since bicd-1(tm3421) was lethal when homozygous, it was balanced over nT1[qIs51](IV;V) from strain RB1276 to make strain UD268. Some nematode strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the NIH National Center for Research Resources (NCRR). Transgenic lines were created by standard DNA microinjection techniques using odr-1::rfp as a transformation marker (Mello et al., 1991; Sagasti et al., 2001). Nuclear migration in embryonic hyp7 precursor cells was scored by counting hyp7 nuclei in the dorsal cord of L1 larvae using DIC optics as in (Starr et al., 2001).
Molecular cloning of plasmids
To clone the yeast two-hybrid bait construct, codons representing residues 137–692 encoded by the unc-83c cDNA yk230e1 were amplified by PCR. The PCR product was fused in-frame to the Gal4 DNA-binding domain (BD) of the pDEST32 vector by Gateway cloning (Invitrogen) to create the bait construct pSL320.
Constructs used to test interactions by directed yeast-two hybrid were created with Gateway technology (Invitrogen). The sequences encoding full-length DLC-1, full-length NUD-2, NUD-2(1–260), NUD-2(239–293), UNC-83c(137–692), UNC-83c(137–362), and UNC-83c(362–692) were amplified by PCR with attB1 and attB2 overhangs and cloned into pDONR221 with BP clonase (Invitrogen). These sequences were then cloned in frame to both Gal4-AD in pDEST22 (prey) and Gal4-DB in pDEST32 (bait) vectors with LR clonase (Invitrogen). The BICD-1(253–737) construct used was the prey plasmid pulled out of the UNC-83 yeast two-hybrid screen.
GFP transcriptional fusions were cloned as in (Boulin, 2006). Specifically, 3.5kb genomic DNA upstream of the predicted nud-2 ATG and 5.9kb upstream of the bicd-1 predicted ATG, was amplified by PCR with overhanging restriction sites and cloned into the HindIII and BamHI sites or PstI and BamHI sites of pPD96.04, respectively (from the Fire lab vector kit, Addgene, Cambridge, MA).
Constructs encoding fusion proteins for in vitro interaction assays were cloned as follows. For GST-DLC-1, full-length dlc-1 cDNA was amplified by PCR with the appropriate overhanging restriction sites and cloned into the BamHI and EcoRI sites of pGEX-2T (GE Healthcare, Piscataway, NJ), to create pSL292. A portion of cDNA encoding BICD-1(227–737) was amplified by PCR with the appropriate restriction sites and cloned into the BamHI site of pGEX-2T to create a GST-BICD-1 fusion protein (pSL294). Full-length nud-2 cDNA was amplified by PCR with the appropriate overhanging restriction sites and cloned into the PvuI and SacI sites of pGEX-2T to create a GST-NUD-2 fusion protein (pSL246). cDNA encoding NUD-2(239–293) was amplified by PCR with MfeI and BamHI overhanging restriction sites and cloned into the EcoRI and BamHI sites of pGEX-2T to create a GST-NUD-2(239–293) fusion protein (pSL424). Full-length egal-1 cDNA was amplified by PCR with the appropriate overhanging restriction sites and cloned into the BamHI and HindIII sites of pMAL-c2 (New England Biolabs, Ipswich, MA) to create an MBP-EGAL-1 fusion protein (pSL396). cDNA enoding residues 1–698 of UNC-83c were used in the MBP-UNC-83 fusion construct (pDS31; (Starr et al., 2001).
Fusion proteins were made for raising antibodies against BICD-1 and NUD-2. The sequence encoding BICD-1(301–546) was amplified from the ORFeome cDNA (Reboul et al., 2003) and cloned into the BamHI site of pGEX-2T or the BamHI and SalI sites of pMAL-c2. Full-length NUD-2 was amplified by PCR from yk1417c08 (gift of Y. Kohara, National Institute of Genetics, Japan) with the appropriate overhanging restriction sites and cloned into the EcoRI and BamHI sites of pGEX-2T or pMAL-c2.
Clones were made to express C. elegans proteins in mammalian tissue culture cells. cDNAs encoding UNC-83cΔKASH(1–698) and UNC-83c full length were cloned into the XhoI site of pCS2+MT (Rupp et al., 1994) to create pSL446 and 451, respectively. Full-length cDNA encoding NUD-2 was cloned into the SalI and BamHI sites of pEGFP-c3 (BD Biosciences) to create pSL461.
The UNC-83 cytoplasmic domain deletions were constructed in the minimal unc-83 rescuing construct pDS22, a 10.5-kb EagI-to-HpaI genomic fragment of cosmid W01A11 in pBS (Starr et al., 2001) using the DNA SOEing PCR technique (Springer Protocols).
Yeast Two-Hybrid
A yeast two-hybrid screen with the cytoplasmic domain of UNC-83 as bait was performed using the ProQuest system (Invitrogen). The bait construct pSL320 (coding for residues 137–692 of UNC-83c) was transformed into yeast strain AH109 (MATa, trp-901, leu2-3 112, ura3-52, his3-200, gal4Δ, gal80Δ, LYS2::GAL1UAS-GAL1TATA-HIS3, GAL2UAS-GAL2TATA-ADE2, URA3::MEL1UAS-MEL1TATA-lacZ, MEL1) (Clontech). As a control, we showed that when the bait pSL320 was co-transformed with the pEXP-AD control vector, it did not self-activate the reporters. Larger portions of the UNC-83 cytoplasmic domain in the bait construct self activated the reporters (data not shown). The ProQuest C. elegans mixed-stage partial cDNA library made with an oligo dT primer and directionally cloned to the C-terminus of the yeast Gal4 transcriptional activation domain (AD) (Invitrogen) with 5×106 to 1×107 clones was transformed into the AH109 plus pSL320 bait strain. Protein–protein interactions between bait and prey were identified by activation of the HIS3 reporter gene by growth on SD-trp-leu-his dropout plates supplemented with 50 mM 3-amino-1,2,4-triazole. Approximately 1,200 positives were identified from ~1.4×107 clones screened. All of the positives were re-streaked and tested for activation of the lacZ reporter gene by X-gal colony-lift filter assays (Clontech). In this strain, the lacZ reporter is relatively weak; most assays were done to test activation of the HIS3 reporter. Thus, in subsequent experiments, interactions were determined by activation of the HIS3 reporter. Prey plasmids were isolated from 126 colonies (Ausubel, 1987) that activated both reporter genes. After sequencing, the 126 colonies represented 45 different genes (Table 1).
Table 1.
Potential UNC-83 interacting proteins identified by a yeast two-hybrid screen.
| Gene ° | Discription | times isolated † |
|---|---|---|
| Microtubule motor regulators | ||
| klc-2 | *Kinesin light chain | 20 |
| dlc-1 | *Dynein light chain | 4 |
| nud-2 | NudE | 4 |
| bicd-1 | BicaudalD | 3 |
| Cytoskeleton-associated proteins | ||
| gei-16 | *GEX-3 interacting protein | 12 |
| gei-12 | *GEX-3 interacting protein | 1 |
| C36C9.1 | GEI-12 paralog | 2 |
| unc-15 | Paramyosin | 5 |
| ifd-1 | Intermediate filament | 2 |
| ifa-1 | *Intermediate filament | 2 |
| atn-1 | *Alpha actinin | 1 |
| sma-1 | Spectrin β-chain | 1 |
| syd-1 | Synapse defective, RhoGAP | 1 |
| Protein modifiers | ||
| mel-26 | *Ubiquitin ligase subunit | 6 |
| mel-11 | *Phosphatase regulator | 5 |
| gck-4 | *Ste-20 like kinase | 1 |
| mig-5 | *Dishevelled homologue | 1 |
| Unknown function | ||
| C27H5.2 | Novel, coiled-coil | 10 |
| F10E9.3 | Nematode-specific novel protein | 5 |
| math-41 | Novel with meprin-associated Traf domin | 3 |
| bath-44 | BTB/POZ and MATH domains | 3 |
| ess-2 | Nuclear protein ES2 | 3 |
| F54D5.5 | *Nematode-specific novel protein | 3 |
| Y38C9A.1 | Novel | 2 |
| Y37A1A.4 | Nematode-specific novel protein | 2 |
| ril-1 | *Nematode-specific novel protein | 1 |
| H02I12.5 | *Nematode-specific novel protein | 1 |
| Y71H2AM.15 | *Novel | 1 |
| K01A2.10 | Novel, coiled-coil | 1 |
| Y59A8A.3 | Novel | 1 |
| C49H3.6 | Novel, coiled-coil | 1 |
| F25F8.1 | Nematode-specific novel protein | 1 |
| brp-1 | Nematode-specific novel protein | 1 |
| paqr-1 | Progestin and AdipoQ Receptor family | 1 |
| Unlikely candidates | ||
| mep-1 | *Differentiation transcription factor | 2 |
| egl-45 | *Translation initiation factor | 2 |
| col-161 | Collagen | 1 |
| col-94 | *Collagen | 1 |
| ztf-8 | Zinc finger transcription factor | 1 |
| pept-3 | Oligopeptide symporter | 1 |
| C06A5.3 | Transcription coactivator | 1 |
| athp-1 | PHD Zn-finger transcription factor | 1 |
| ZK1098.1 | *Splicing factor | 1 |
| T22D1.3 | IMP dehydrogenase | 1 |
| unc-43 | Calmodulin-dependent kinase | 1 |
Essential for embryonic viability
When a gene has not been given an official three of lour-letter name, the sequence name is shown following C. elegans standard nomenclature (www.wormbase.org)
Since the two-hybrid cDNA library was not normalized, a low number of times isolated could represent a rare transcript.
Many directed two hybrid assays were performed with the various baits and prey constructs cloned as above. All bait constructs failed to activate reporters when transfected with a control prey vector. AH109 yeast were transfected with both prey and bait and interactions were tested for as above.
RNAi experiments
We tested, using RNAi, whether any of the non-essential two-hybrid positives might play a role in hyp7 nuclear migration (Table 1, (Kamath et al., 2003; Sonnichsen et al., 2005). dsRNA was injected into the gonad of adult hermaphrodites (Fire et al., 1998). Injected worms were moved to fresh plates 24 hours after injection. L1 progeny on the new plates from at least five different injected worms were scored for a failure in hyp7 nuclear migration by counting the number of hyp7 nuclei found abnormally in the dorsal cord (Starr et al., 2001). Of the 23 non-essential genes tested, only bicd-1(RNAi) and nud-2(RNAi) caused a nuclear migration phenotype (Table 2 and data not shown). Double-stranded RNA (dsRNA) was synthesized in vitro from PCR products with T7 promoter overhangs using T7 RNA polymerase (Promega) following established protocols (Ahringer, 2006). The source of the template for the dsRNA synthesis varied (Supplemental Table 1).
Table 2.
Nuclear migration phenotypes in dynein regulator mutations.
| Genotype | Percentage of hyp7 nuclei that fail to migrate * |
n ¶ | p-value ° |
|---|---|---|---|
| wild-type | 0 | 40 | |
| unc-83(e1408) | 87.4 ± 0.9 | 33 | <0.0001 ‡ |
| bicd-1(RNAi) | 5.0 ± 0.4 | 151 | <0.0001 ‡ |
| bicd-1(ok2731) | 0.6 ± 0.3 | 50 | 0.07 ‡ |
| bicd-1(tm3421) | 5.7 ± 2.1 | 10 † | <0.0001 ‡ |
| nud-2(RNAi) | 1.0 ± 0.4 | 92 | 0.02 ‡ |
| nud-2(ok949) | 1.3 ± 0.3 | 97 | 0.01 ‡ |
| nud-2(ok949); nud-2(RNAi) | 1.4 ± 0.4 | 88 | 0.006 ‡ |
| egal-1(RNAi) | 3.0 ± 0.5 | 106 | <0.0001 ‡ |
| bicd-1(RNAi); nud-2(ok949) | 14.2 ± 0.6 | 171 | <0.0001 § |
| egal-1 (RNAi); nud-2(ok949) | 9.8 ± 0.7 | 90 | <0.0001 § |
| egal-1(RNAi); bicd-1(RNAi) | 4.6 ± 0.4 | 128 | 0.55 § |
| egal-1(RNAi); bicd-1(RNAi); nud-2(ok949) | 13.2 ± 0.9 | 111 | 0.33 ‖ |
| dhc-1(js121) | 4.6 ± 0.5 | 20 | 0.001 ‡ |
The migrations of 14 hyp7 nuclei were scored per animal. Nuclei abnormally in the dorsal cord were scored as a failed nuclear migration. In unc-83(e1408) mutant animals, 87% of the hyp7 nuclei failed to migrate properly.
n is the number of animals counted
p-value is from a Mann-Whitney test
The bicd-1(tm3421) strain has a high level of embryonic lethality. Only L1 animals healthy enough to count hyp7 cells were scored.
p-value as compared to wild-type
p-value as compared to the stronger of the single mutant lines
p-value as compared to the strongest of the double mutant lines
To determine the role of genes essential for embryonic development in nuclear migration, dsRNA was introduced by the feeding method (Timmons and Fire, 1998). For the dlc-1(RNAi), lis-1(RNAi), dhc-1(RNAi), and dnc-1(RNAi) experiments, N2 animals were fed for 12 hours bacteria expressing dsRNA from the Ahringer RNAi feeding library (Supplemental Table 1; Kamath et al., 2003).
Characterization of bicd-1 locus
The bicd-1 open reading frame was determined by completely sequencing the cDNA C43G2.2 from the ORFeome project (Reboul et al., 2003). The 5’ and 3’ ends of the bicd-1 transcript were identified using the FirstChoice RLM-RACE kit (Ambion). The 3’ RACE product had an alternative splice site that removed the last three amino acids of exon 9 as compared to the ORFeome cDNA. The breakpoints of ok2731 transcript and the identification of a cryptic splice site in exon 6 were determined by RT-PCR. RNA was made using the RNeasy kit (QIAGEN) and RT-PCR was performed using primers in exons 3 and 7 of bicd-1 with SuperScriptIII reverse transcriptase and Platinum Taq DNA polymerase (Invitrogen).
In vitro and in vivo interaction assays
pGEX-2T (GE Healthcare) was used to express GST and pMAL-c2 (New England Biolabs) was used to express MBP. Fusion proteins were expressed in E. coli strain BL21 codon plus (Stratagene) and purified on glutathione sepharose 4B beads (GE Healthcare) or amylose resin (New England Biolabs) and washed of any excess protein in PBS. An excess of purified MBP or UNC-83/MBP was added to beads already bound to GST or GST-fusion proteins with crude E. coli BL21 extract (10mg/ml) in PBS + 10% glycerol. Binding reactions were incubated for 3 hours at 4°C. Beads were washed in PBS five times. Bound protein was boiled off the beads in 2X SDS sample buffer and analyzed by SDS-PAGE and Coomassie blue stain.
To express C. elegans UNC-83cΔKASH(1–698) or full length UNC-83c with NUD-2::GFP in mammalian tissue culture cells, HeLa cells were transiently tansfected using 6 µl of lipofectamine (Invitrogen) and 0.5 µg of plasmids pSL461 and either pSL446 or pSL451 per ml of media. Extracts were made and incubated with 2 µl anti-GFP antibodies per ml of extract (Yang et al., 1999); the NUD-2 complex was purified on protein A beads (Thermo Scientific). UNC-83 was detected by western blot with mouse monoclonal 9E10 anti-myc antibody (1/1000) (Developmental Studies Hybridoma Bank, University of Iowa) and NUD-2 with rabbit polyclonal antibody NB600-308 against GFP (1/1000) (Novus Biologicals). Goat anti-mouse HRP and goat anti-rabbit HRP (Jackson ImmunoResearch) diluted 1/10,000 were used as secondary antibodies.
Antibodies and Immunofluorescence
To raise polyclonal BICD-1 antibodies, two rats were injected with GST-BICD-1(301–546) purified fusion protein. To raise polyclonal NUD-2 antibodies, two guinea pigs were injected with MBP-NUD-2 fusion protein. To raise polyclonal antibodies against the cytoplasmic domain of UNC-83 a rat was injected with MBP-UNC-83 fusion protein. All fusion proteins were purified as above. Animals were injected and cared for by Brett Wilkins and staff at the Laboratory of Comparative Pathology at the School of Veterinary Medicine at UC Davis. Crude serum against UNC-83 was tested for specificity by immunofluorescence as in (Starr et al., 2001). Crude sera against NUD-2 and BICD-1 (at a dilution of 1/5000) were tested for specificity by western blot of wild-type and mutant worm extract as in (Meyerzon et al., 2009). Anti-MBP-NUD-2 serum from guinea pig C was affinity purified against GST-NUD-2 and anti-GST-BICD-1(301–546) serum from rat C was affinity purified against MBP-BICD-1(301’546), using Affi-Gel 15 beads according to the protocol supplied by the manufacturer (BioRad).
For immunofluorescence, late embryos were extruded from slightly starved hermaphrodites, permeabilized by the freeze-crack method, fixed for 10 minutes in −20°C methanol, and blocked in PBST (phosphate-buffered saline + 0.1% Triton X-100) +5% dry milk (Miller and Shakes, 1995). The fixed specimens were stained as described (Miller and Shakes, 1995). Primary NUD-2 and BICD-1 antibodies were diluted 1/500 (crude) or 1/10 (purified) in PBS. Crude UNC-83 rat “H” polyclonal antibody was diluted 1/1000 in PBS. Cy3-conjugated donkey anti-rat IgG and Cy3-conjugated donkey anti-guinea pig (Jackson ImmunoResearch, West Grove, PA) diluted 1/200 in PBS were used as secondary antibodies. DNA was visualized by a 10-minute stain in 1µg/ml of 4,6-diamidino-2-phenylindole (DAPI) in PBS.
HeLa cells tranfected as above were fixed with 3.7% paraformaldehyde in PBS and permeabilized with 0.2% PBST. Mouse monoclonal 9E10 anti-myc antibody (Developmental Studies Hybridoma Bank, University of Iowa) was used at a 1/500 dilution. Rabbit polyclonal antibody NB600-308 against GFP was used at a 1/500 dilution (Novus Biologicals). Cy3 goat anti-mouse IgG and Cy2 goat anti-rabbit IgG (Jackson ImmunoResearch) diluted 1/200 were used as secondary antibodies. DNA was visualized with DAPI.
Results
Identification of UNC-83-interacting proteins
UNC-83 is essential for nuclear migration in many developmental events (Horvitz and Sulston, 1980; McGee et al., 2006; Starr et al., 2001), but its molecular mechanism remains unknown. To identify UNC-83-interacting proteins we performed a yeast two-hybrid screen with the cytoplasmic domain of UNC-83. The bait consisted of amino acids 137–692 of UNC-83c (Figure 2), which does not include the trans-membrane or KASH domains (Meyerzon et al., 2009). Expression of the UNC-83c(137–692) bait with an empty prey vector did not activate the HIS3 or LacZ reporters (Figure 2). Larger UNC-83 bait constructs self activated the reporters (data not shown). We identified 45 different potential interacting partners that activated both the HIS3 and lacZ reporter genes (Figure 2; Table 1). 31 of the 45 different hits were only identified one or two times. Since the library was not normalized, these could represent rare transcripts. However, many are likely false-positive interactions (Yu et al., 2008).
Figure 2.

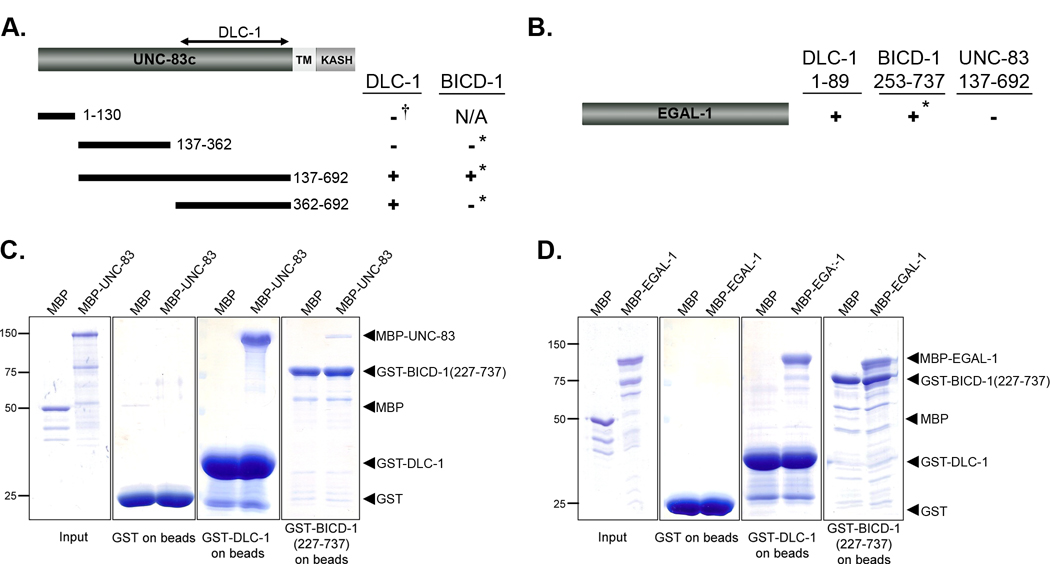
BICD-1, NUD-2, and BICD-1 interact with UNC-83 in a yeast two-hybrid screen. A schematic of full-length UNC-83c and the portion used as bait in the two-hybrid screen (residues 137–692) is shown at top. Bottom left panel shows activity of the HIS3 reporter assayed by yeast growth on minimal media minus Trp, Leu, and His supplemented with 50 mM 3-AT. Successive ten-fold dilutions are shown. All combinations of bait and prey grow on minimal media minus Trp and Leu (middle). Activity of the LacZ reporter is shown in an X-gal assay (right). For all data shown here the UNC-83c(137–692) fused to the DNA-binding domain of Gal4 was used as bait. For prey constructs fused to the activation domain of Gal4, the empty vector pEXP-AD, BICD-1(253–737), or full-length NUD-2 and DLC-1 were used.
The most exciting candidate interacting proteins were the four proteins previously shown to regulate the microtubule motors dynein and kinesin-1 (Table 1). We recently established a role for KLC-2 and the kinesin-1 heavy chain UNC-116 in nuclear migration (Meyerzon et al., 2009). Here we focus on the three dynein regulators identified in our screen, BICD-1, NUD-2, and DLC-1, and their roles in UNC-83-mediated hyp7 nuclear migration. As a control for the UNC-83 yeast two-hybrid interactions with BICD-1, NUD-2, and DLC-1, the bait and prey were switched. The BICD-1 bait self-activated the reporters and was not further examined. Both NUD-2 and DLC-1 bait constructs interacted with UNC-83 prey in the two-hybrid system (data not shown).
Defects in BICD-1 and NUD-2 act synergistically to disrupt nuclear migration
To test the functional significance of the yeast two-hybrid interaction between UNC-83 and the three dynein regulators, we assayed nuclear migration in hyp7 precursor cells from mutant animals (Figure 1, Table 2). In unc-83(e1408) null animals, about 87% of nuclear migrations in hyp7 cells fail and the hyp7 nuclei end up in the dorsal cord of L1 larvae, as compared to 0% in wild-type (Figure 1A–C, G) (Horvitz and Sulston, 1980; Starr et al., 2001). We tested whether the depletion of dynein regulators affected hyp7 nuclear migration. Both bicd-1(RNAi) and nud-2(ok949) animals had a small, but significant number of nuclei in the dorsal cord (Figure 1D–E, G; Table 2).
In animals homozygous for the nud-2(ok949) deletion allele, which starts at the predicted ATG of nud-2 and deletes the entire open reading frame except the last part of the last exon, 1.2% of hyp7 nuclei failed to migrate (Table 2; Figure 1D, G). Although this phenotype was relatively weak as compared to unc-83 or kinesin-1 mutants, animals were observed with two hyp7 nuclei in their dorsal cord (a 14% failure), which was never seen in wild type, suggesting a true nuclear migration defect. It is possible that a small peptide consisting of the C-terminal 33 residues is still expressed in nud-2(ok949) animals and provides some function. We therefore performed RNAi experiments in both wild-type and ok949 backgrounds. Both RNAi treatments resulted in phenotypes similar to nud-2(ok949) alone (Table 2), suggesting that we are observing the null nud-2 phenotype. NUD-2 is the C. elegans homologue of NudE, which was originally identified in Aspergillus and Neurospora from nuclear distribution mutant screens (Efimov and Morris, 2000; Minke et al., 1999) and has subsequently been characterized as a key regulator of dynein function in several systems (Liang et al., 2004; Morris, 2000; Sasaki et al., 2000; Stehman et al., 2007).
The C. elegans gene bicd-1 (previously named C43G2.2) has not been described previously, but is homologous to Drosophila BicaudalD (BicD). We characterized the bicd-1 cDNA by sequencing the ORFeome RT-PCR product (Reboul et al., 2003) and performing 5’ and 3’ RACE (Supplemental Figure 1). Drosophila BicD functions with dynein to position nuclei in the oocyte and photoreceptor cells (Houalla et al., 2005; Swan et al., 1999).
In C. elegans bicd-1(RNAi) animals, 5% of the hyp7 nuclei failed to migrate properly, significantly more than in wild-type (p<0.0001; Table 2, Figure 1E, G). Up to three nuclei (21.4 %) were seen in the dorsal cord of individual animals. To confirm the bicd-1(RNAi) phenotype, we analyzed two deletion alleles of bicd-1. RT-PCR of bicd-1(ok2731) showed that exon 4 was spliced in frame to a cryptic splice site in the middle of exon 6 and encodes a protein with a 68 residue deletion (Supplemental Figures 1–2). Not surprisingly, bicd-1(ok2731) did not cause a significant hyp7 nuclear migration phenotype (Table 2). bicd-1(tm3421) deletes exon 7, causing a frame shift and a premature stop codon 11 residues into exon 8 (Supplemental Figure 1), suggesting that bicd-1(tm3421) is a likely null. bicd-1(tm341) caused embryonic lethality. Since there was no lethality seen in our RNAi studies and because neurons are known to be resistant to RNAi (Tavernarakis et al., 2000), the lethal phenotype of bicd-1(tm3421) is likely the result of a neuronal defect. bicd-1(tm3421) was balanced and the rare homozygous bicd-1(tm34231) animals that hatched had a 5.7% defect in nuclear migration (n=10), indicating that the RNAi phenotype represents a strong loss-of-function effect on hyp7 nuclear migration (Table 2).
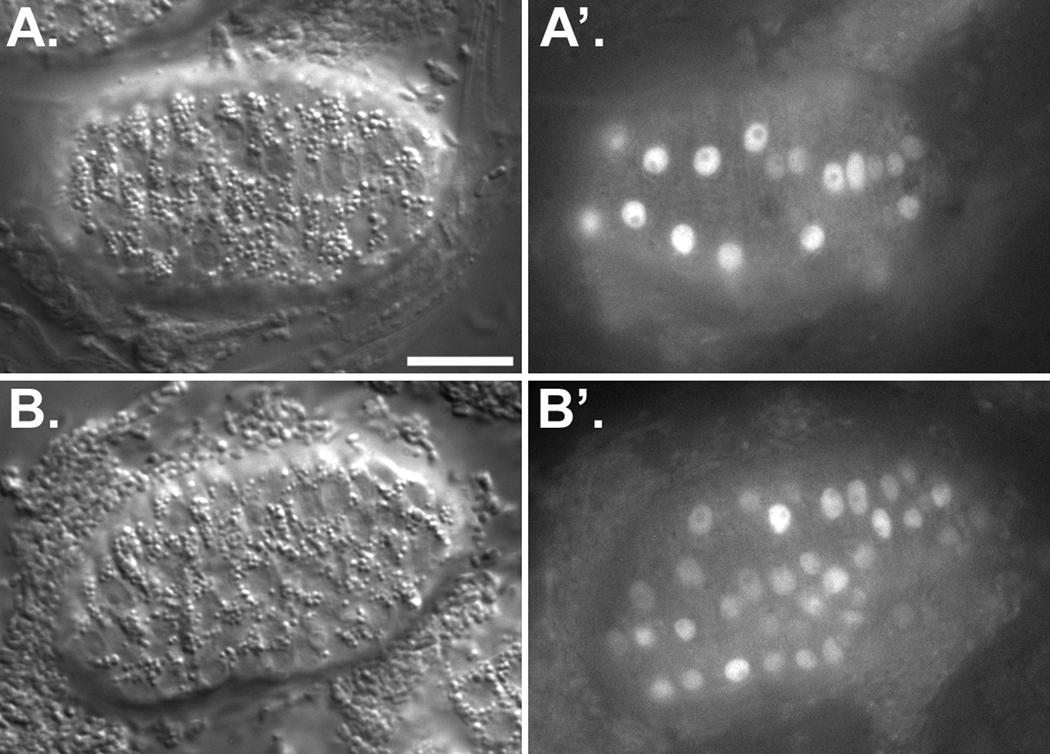
To confirm that BICD-1 and NUD-2 were expressed at the time of hyp7 nuclear migration, GFP reporter constructs of the bicd-1 and nud-2 promoters were engineered and expressed in transgenic animals. Both reporter constructs were expressed in a subset of cells including the hyp7 precursors during the time of nuclear migration (Figure 3A, B), consistent with them functioning through UNC-83 during hyp7 nuclear migration. We were unable to confirm the expression patterns by raising antibodies against NUD-2 and BICD-1 (Supplemental Figure 2). Dynein regulators are likely to bind to multiple cargos throughout the cytoplasm. Furthermore, the volume of hyp7 cells at the time of nuclear migration is very small, compounding the difficulty localizing a ubiquitous protein to the nuclear envelope.
Figure 3.
NUD-2 and BICD-1 are expressed in hyp7 precursors during nuclear migration. Pre-comma stage embryos expressing GFP with a nuclear localization signal from the bicd-1 promoter (A) or the nud-2 promoter (B). (A, B) DIC image showing intercalating hyp7 precursors during nuclear migration. (A’, B’) Fluorescent image showing GFP localized to nuclei. Scale bar is 10 µm.
To test whether bicd-1 and nud-2 were partially redundant, we performed double mutant analysis. The weak bicd-1(RNAi) and nud-2(ok949) phenotypes were significantly enhanced; an average of 14.2% of hyp7 nuclei failed to migrate (n=171) and a maximum of 42.9% of nuclei failed to migrate in one bicd-1(RNAi); nud-2(ok949) animal (Table 2; Figure 1F, G). These data, along with the data below, suggest that NUD-2 and BICD-1 are functioning in separate pathways or complexes during hyp7 nuclear migration.
Disruption of dynein leads to a nuclear migration defect
BICD-1 and NUD-2 homologs have been shown in other systems to function by regulating dynein (Liang et al., 2004; Morris, 2000; Sasaki et al., 2000; Stehman et al., 2007). We therefore examined whether genetic disruptions of dynein heavy chain (dhc-1) cause a hyp7 nuclear migration phenotype similar to those observed in bicd-1 and nud-2 mutant animals. In C. elegans, dhc-1 is an essential gene for pronuclear migration and early embryonic development (Gonczy et al., 2000). Therefore, it is not possible to observe a dynein null phenotype. However, homozygous dhc-1(js121) animals are sterile and 4.6% of hyp7 nuclei fail to migrate (n=20; Table 2 and Figure 4C, E). In support of this, dhc-1 RNAi experiments also produced hyp7 nuclear migration defects. dsRNA was fed to mothers and progeny laid after 12 hours of exposure to the dsRNA were analyzed. This allowed observation of a small window of viable progeny with only a partial loss of dhc-1 gene product. Some of these dhc-1(RNAi) animals had multiple hyp7 nuclei in the dorsal cord (a 14–28% failure of migration), suggesting that dhc-1 also functions during hyp7 nuclear migration. These data are consistent with the hypothesis that BICD-1 and NUD-2 function through dynein to regulate hyp7 nuclear migration.
Figure 4.
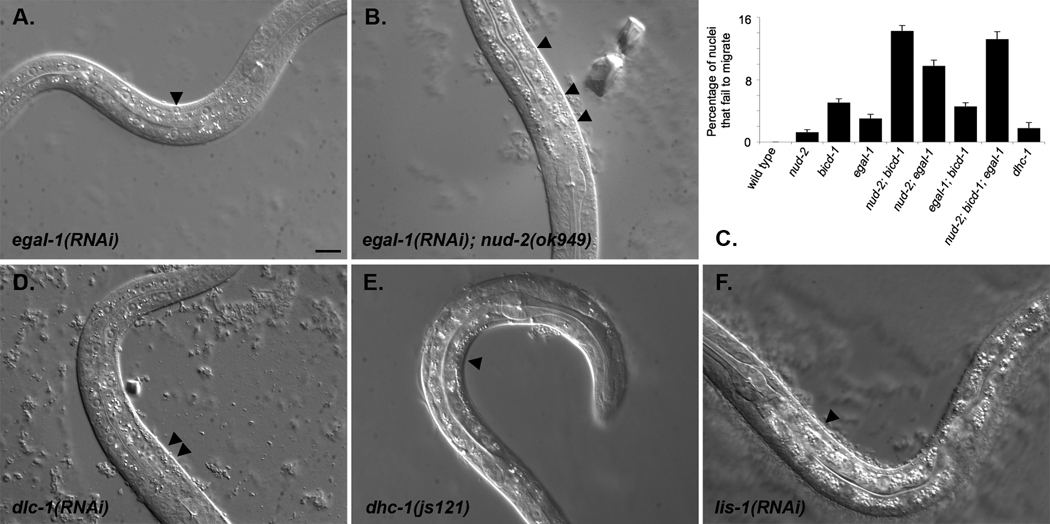
Dynein functions in hyp7 nuclear migration. (A–B, D–F) Lateral view of L1 hermaphrodites. Dorsal is upwards or right. (A) egal-1(RNAi). (B) egal-1(RNAi); nud-2(ok949). (C) Quantification of hyp7 nuclear migration phenotypes. Error bars are standard error. See Table 2 for details and statistics. (D) dlc-1(RNAi). (E) dhc-1(js121). (F) lis-1(RNAi). Black arrowheads mark hyp7 nuclei in the dorsal cord. Scale bar is 10 µm.
UNC-83 interacts with a complex containing BICD-1, DLC-1, and EGAL-1
In Drosophila, Egalitarian binds BicD and dynein light chain and together they regulate dynein (Mach and Lehmann, 1997); (Navarro et al., 2004). We therefore inquired whether the C. elegans homologue of Egalitarian (C10G6.1, which we named egal-1) functions in nuclear migration. egal-1(RNAi) animals had a small but significant nuclear migration defect where an average of 3%, and up to 21% in a single animal, of nuclei failed to migrate (Table 2; Figure 4A, C). Double and triple mutant animals were examined to place egal-1 in either the bicd-1 or nud-2 pathway. The egal-1(RNAi) phenotype was enhanced to an average of 9.8% failed nuclear migration in egal-1(RNAi); nud-2(ok949) animals (Table 2, Figure 4B, C). egal-1(RNAi) did not show a synthetic relationship with bicd-1(RNAi) (Table 2, Figure 4C), consistent with the data below showing EGAL-1 and BICD-1 interact and that double mutants of bicd-1 and egal-1 homologs in Drosophila are not stronger than single mutants (Mach and Lehmann, 1997); (Navarro et al., 2004). Alternatively, the double RNAi phenotype observed could have possibly been reduced due to interference from multiple dsRNA treatments done simultaneously (Piano et al., 2000). Furthermore, triple bicd-1(RNAi); egal-1(RNAi); nud-2(ok949) animals were no more severe than the strongest double mutants (Table 2, Figure 4C). Thus, genetic analyses suggest that bicd-1 and egal-1 function in a pathway together and are partially redundant to a pathway with nud-2.
The C. elegans homolog of the LC8 dynein light chain (dlc-1) was also identified in our yeast two-hybrid screen (Table 1) and has been implicated in BicD and Egalitarian function (Navarro et al., 2004). In C. elegans, dlc-1, like dhc-1, is an essential gene for pronuclear migration and early embryonic development (Gonczy et al., 2000). A small portion of the L1 hatched from mothers fed dlc-1 dsRNA had nuclear migration phenotypes similar to those seen in bicd-1 or egal-1 mutants (Figure 4D).
Based on the genetic data presented above, we propose that BICD-1, EGAL-1 and DLC-1 function together in a complex that is recruited to the nuclear envelope by an interaction with UNC-83. To test this hypothesis and to map the interaction domains, directed two-hybrid and in vitro pull down assays were performed. Although we were not able to further map the BICD-1 interaction domain of UNC-83 using directed yeast two-hybrid assays, we did determine that amino acids 362–692 of UNC-83c are sufficient for interaction with DLC-1 (Figure 5A). In addition, EGAL-1 interacted with both DLC-1 and BICD-1 but not UNC-83 in the yeast two-hybrid system (Figure 5B). All of these yeast two-hybrid interactions were confirmed by in vitro GST pulldown experiments. GST-DLC-1(full-length) and GST-BICD-1(227–737) expressed in E. coli and purified on glutathione beads were used to pull down MBP-UNC-83 fusion protein. MBP-UNC-83c(1–698) (the full cytoplasmic domain) bound to both GST-DLC-1(full-length) and GST-BICD-1(227–737) (Figure 5C). Furthermore, an MBP-EGAL-1(full-length) fusion protein interacted with the GST-DLC-1(full-length) and GST-BICD-1(227–737) fusion proteins (Figure 5D). We therefore conclude that EGAL-1, BICD-1, DLC-1 and UNC-83 interact in a complex.
Figure 5.
DLC-1 and BICD-1 bind to UNC-83 and EGAL-1. (A–B) Directed yeast two-hybrid assays were performed between DLC-1 or BICD-1 and parts of UNC-83 (A) or full-length EGAL-1 (B). ‘+’ indicates that an interaction was detected by growth on His dropout plates. All bait constructs alone, except UNC-83(1–130) and BICD-1 (253–737) failed to activate the reporters. In cases marked with the asterisk (*), only the UNC-83 constructs were used as baits because BICD-1 self activated as bait. In the DLC-1 to UNC-83(1–130) test (†), only DLC-1 was used as bait. In all other cases, bait and prey constructs were switched, and the indicated interactions occurred in both directions. All the DLC-1 constructs were full length. (C–D) Coomassie blue stained SDS-PAGE gels are shown to demonstrate interactions between GST-DLC-1 and GST-BICD-1(227–737) with both MBP-UNC-83c(1–698) (C) and MBP-EGAL-1 (D). In the first panel, 1% of the input proteins are shown. The second panel shows the proteins pulled down by beads with GST only. The third and fourth panels show MBP and MBP-UNC-83 or MBP and MBP-EGAL-1 fusion proteins pulled down by GST-DLC-1 or GST-BICD-1(227–737) beads.
UNC-83 interacts with a complex containing NUD-2 and LIS-1
Our genetic analysis predicts that the second complex to interact with UNC-83 and to regulate dynein contains NUD-2. lis-1, which is required for dynein-mediated pronuclear migration and early embryonic viability (Cockell et al., 2004), has been shown to interact in vitro with NUD-2 (Locke et al., 2006). LIS-1 homologs are well-characterized dynein regulators that have been shown to function to increase the processivity of dynein motor activity and to participate in nuclear positioning events in many model systems (Swan et al., 1999; Tsujikawa et al., 2007; Wynshaw-Boris, 2007; Xiang and Fischer, 2004). The role of lis-1 in hyp7 nuclear migration was examined using the RNAi feeding method described above. As was the case for dlc-1 and dhc-1, lis-1(RNAi) displayed nuclear migration defects as was evident by the presence of nuclei in the dorsal cord (7–14% defective migration in some animals), suggesting a role for lis-1 in this process (Figure 4F). These data are consistent with the hypothesis that a complex consisting of at least NUD-2 and LIS-1 interacts with UNC-83 during nuclear migration.
Using directed yeast two-hybrid assays we determined that amino acids 362–692 of UNC-83c are sufficient for interaction with NUD-2 (Figure 6A). The two-hybrid interaction was verified by in vitro GST pulldown experiments where GST-NUD-2(full-length) expressed in E. coli and purified on glutathione beads was able to pull down MBP-UNC-83c(1–698) fusion protein (Figure 6B). To further map the UNC-83 interaction domain of NUD-2, smaller pieces of NUD-2 were used in the directed yeast two-hybrid system. NUD-2(1–260), missing only the last 33 amino acids, failed to interact with UNC-83c (Figure 6A). Expression of NUD-2(239–293), which contains just the last 55 amino acids, restored the interaction with both UNC-83c(137–692) and UNC-83c(362–692) (Figure 6A). These data were also verified by in vitro GST pull-down experiments. GST-NUD-2(239–293) expressed in E. coli and purified on glutathione beads was able to pull down MBP-UNC-83c(1–698) fusion protein (Figure 6B). Together, these data show that the very C-terminus of NUD-2 is necessary and sufficient for interacting with UNC-83.
Figure 6.
The C-terminus of NUD-2 is necessary and sufficient for an interaction with UNC-83. (A) Yeast two-hybrid was used to map the UNC-83 and NUD-2 interaction domains as in Figure 5. All bait constructs alone, except UNC-83(1–130), failed to activate the reporters. In cases marked with the asterisk (*), only the NUD-2 construct was used as the bait because UNC-83(1–130) self activated as a bait. In all other cases, bait and prey constructs were switched, and the indicated interactions occurred in both directions. (B) In vitro co-sedimintation assays. Coomassie blue stained SDS-PAGE gels demonstrate interactions between GST-NUD-2 and MBP-UNC-83c(1–698). In the first panel, 1% of the input proteins are shown. The second panel shows protein pulled down by beads with GST only. The third and fourth panels show MBP and MBP-UNC-83 fusion proteins pulled down by GST-NUD-2 or GST-NUD-2(239-293) beads.
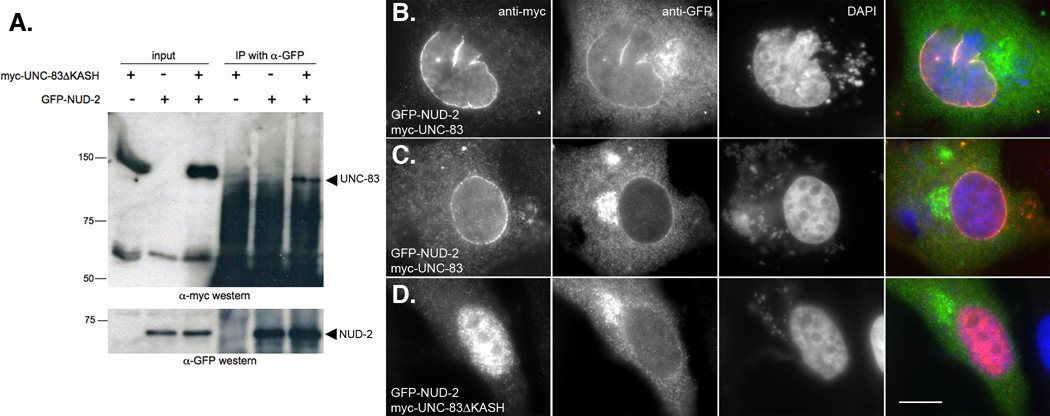
To further confirm our in vitro and yeast two-hybrid interaction results, and because immunolocalization of NUD-2 in C. elegans was uninformative, UNC-83 and NUD-2 were co-expressed in a heterologous mammalian tissue culture system and their localization patterns were examined. HeLa cells were transiently transfected with plasmids encoding a myc-tagged version of the cytoplasmic domain of UNC-83c and GFP-tagged NUD-2. Immunoprecipitation with the anti-GFP antibody pulled down myc-UNC-83ΔKASH (Figure 7A), confirming our results that UNC-83 interacts with NUD-2. These results were also supported by immunofluorescence studies of transfected HeLa cells. It was previously shown that C. elegans UNC-83 localizes to the outer nuclear membrane in transiently transfected mammalian tissue culture cells (McGee et al., 2006). Here, the cytoplasmic domain of UNC-83c, with and without its transmembrane and KASH domains, was co-expressed with NUD-2 in HeLa cells. As expected, UNC-83 localized to the nuclear envelope in a KASH-dependent manner. Full-length UNC-83 recruited NUD-2 to the nuclear envelope (Figure 7B, C), but UNC-83ΔKASH did not (Figure 7D). These data demonstrate that UNC-83 recruits NUD-2 to the nuclear envelope through a direct interaction in vivo.
Figure 7.
C. elegans UNC-83 and NUD-2 interact in a heterologous HeLa cell system. (A) myc-UNC-83ΔKASH was co-immunoprecipitated with GFP-NUD-2. A western blot probed with anti-myc antibodies is shown in the top panel. Lanes 1–3 show transfected HeLa cell extracts used as inputs. Lanes 4–6 show what was immunoprecipitated by anti-GFP antibodies from the same extracts. The lower panel shows the same blot probed with anti-GFP antibodies to recognize GFP-NUD-2. (B–D) HeLa cells transiently transfected with DNA encoding GFPNUD-2 and (B, C) full-length myc-UNC-83 or (D) myc-UNC-83ΔKASH. Immunolocalization of myc-UNC-83 are shown in the left-most column, GFP-NUD-2 in the second column, followed by DAPI staining of chromatin in the third column, and a merge of UNC-83 (red), NUD-2 (green), and DAPI (blue) in the right-most column. Scale bar is 10 µm.
UNC-83 has independent interaction domains for dynein regulators and kinesin light chain
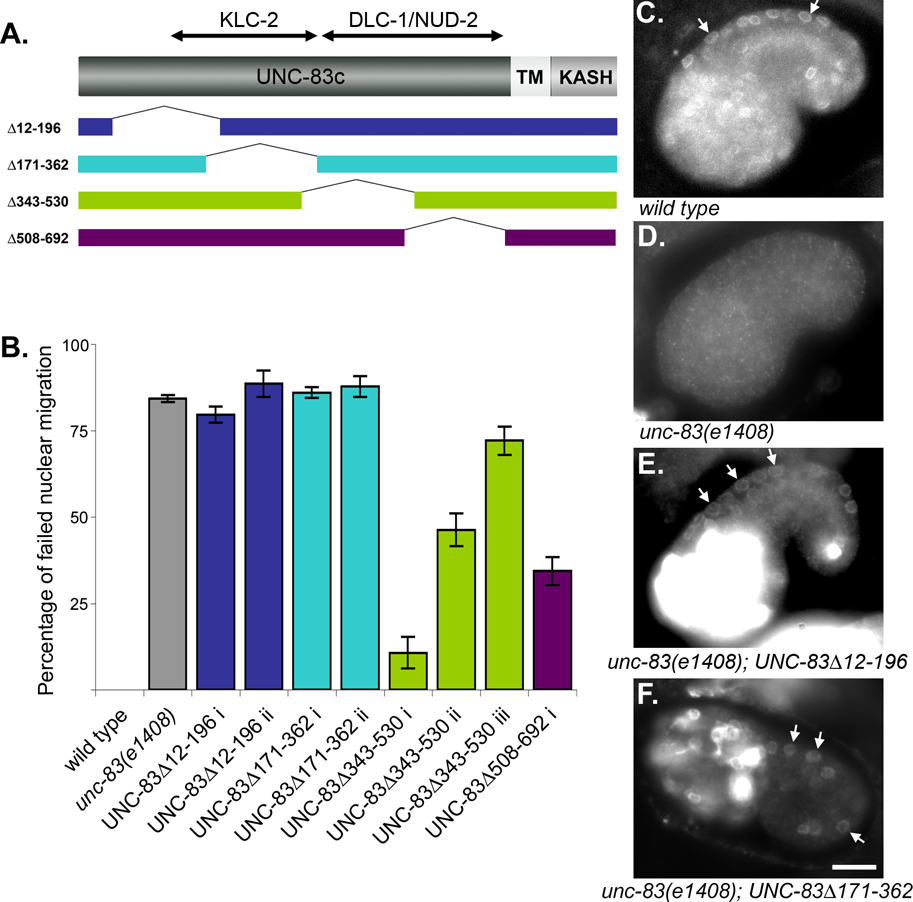
Our two-hybrid and in vitro interaction assays suggested that KLC-2 and the dynein regulators NUD-2 and DLC-1 bind to different domains of UNC-83. KLC-2 bound to residues 137–362 of UNC-83c (Meyerzon et al., 2009) while both NUD-2 and DLC-1 bound to residues 362–692 (Figure 5A, 6A). The phenotypes of kinesin mutants, which were similar to the strong unc-83 defects, suggest that kinesin-1 provides the bulk of the force during hyp7 nuclear migration (Meyerzon et al., 2009). Therefore, we hypothesized that deletion of the KLC-2 binding domain of UNC-83 should lead to a strong nuclear migration defect while deletion of the NUD-2-DLC-1 binding domain should lead to a more minor nuclear migration defect consistent with the nud-2 and bicd-1 phenotypes. To test the hypothesis, an in vivo deletion analysis of the cytoplasmic domain of UNC-83c was conducted.
Four overlapping deletions in the minimal unc-83c rescuing construct (McGee et al., 2006) were engineered and assayed for their ability to rescue the unc-83(e1408) hyp7 nuclear migration defect by counting the number of hyp7 nuclei in the dorsal cords of transgenic animals (Figure 8). In unc-83(e1408) mutants, 84.2 % of hyp7 nuclei were observed in the dorsal cord (n=71) as compared to 0 % in wild type (n=30). Our wild-type minimal rescuing construct in the unc-83(e1408) background resulted in a partial rescue of hyp7 nuclear migration where averages of 5.0 to 33.6 % of hyp7 nuclei were observed in the dorsal cords of three independent transgenic lines (McGee et al., 2006). Because of the variability of the partial rescue in independent lines, it was not possible to conclude rescue of the dynein-dependent phenotypes. UNC-83 deletion constructs were injected into unc-83(e1408) hermaphrodites. Both UNC-83cΔ12-196 and UNC-83cΔ171-362, which contain part of the KLC-2 interaction domain, failed to rescue the hyp7 nuclear migration defect (Figure 8B). Therefore, amino acids 12–362 of UNC-83c, the KLC-2 binding domain, were essential to UNC-83’s function in nuclear migration. UNC-83cΔ343-530 and UNC-83cΔ508-692, which contain the DLC-1 and NUD-2 binding domains, were able to partially rescue the unc-83(e1408) nuclear migration phenotype (Figure 8B). To ensure that the transgenic lines expressed the deleted versions of UNC-83 and that it was targeted to the nuclear envelope, the UNC-83cΔ12-196 and UNC-83cΔ171-362 transgenic lines were further examined for localization of UNC-83 by immunofluorescence (Figure 8C–E). The wild-type minimal rescuing construct expressed in unc-83(e1408) embryos localizes normally to the nuclear envelope of hyp7 precursors but is also expressed at extremely high levels in the cytoplasm of a large number of cells in the anterior half of the embryo (McGee et al., 2006). In transgenic lines expressing UNC-83cΔ12-196 (Figure 8D) and UNC-83cΔ171-362 (Figure 8E), antibodies against UNC-83 localize normally to the nuclear envelope of hyp7 cells. We therefore conclude that the portion of UNC-83 encoded by aa343-692 that interacts with dynein regulators is not essential for nuclear migration, but instead plays an important regulatory role in the process, consistent with the phenotypes of dynein-regulatory-complex components.
Figure 8.
UNC-83 has independent interaction domains for dynein regulators and kinesin light chain. (A) Four overlapping deletions were constructed in the cytoplasmic portion of UNC-83. (B) The average percentages of hyp7 nuclei in the dorsal cord, representing failed nuclear migration events are shown. The roman numerals (i, ii, and iii) refer to independent transgenic lines. For UNC-83Δ12-196 ii n = 10; UNC-83Δ343-350 i n = 4; in all other cases, n ≥ 15. SE bars are shown. (C–F) Images of anti-UNC-83 immunofluorescence are shown in (C) wild type, (D) unc-83(e1408) or (E–F) the indicated transgenic line. White arrows indicate examples of clear hyp7 nuclear envelope staining in the wild-type or rescued animals. Anterior is left, dorsal is upwards; scale bar is 10 µm.
Discussion
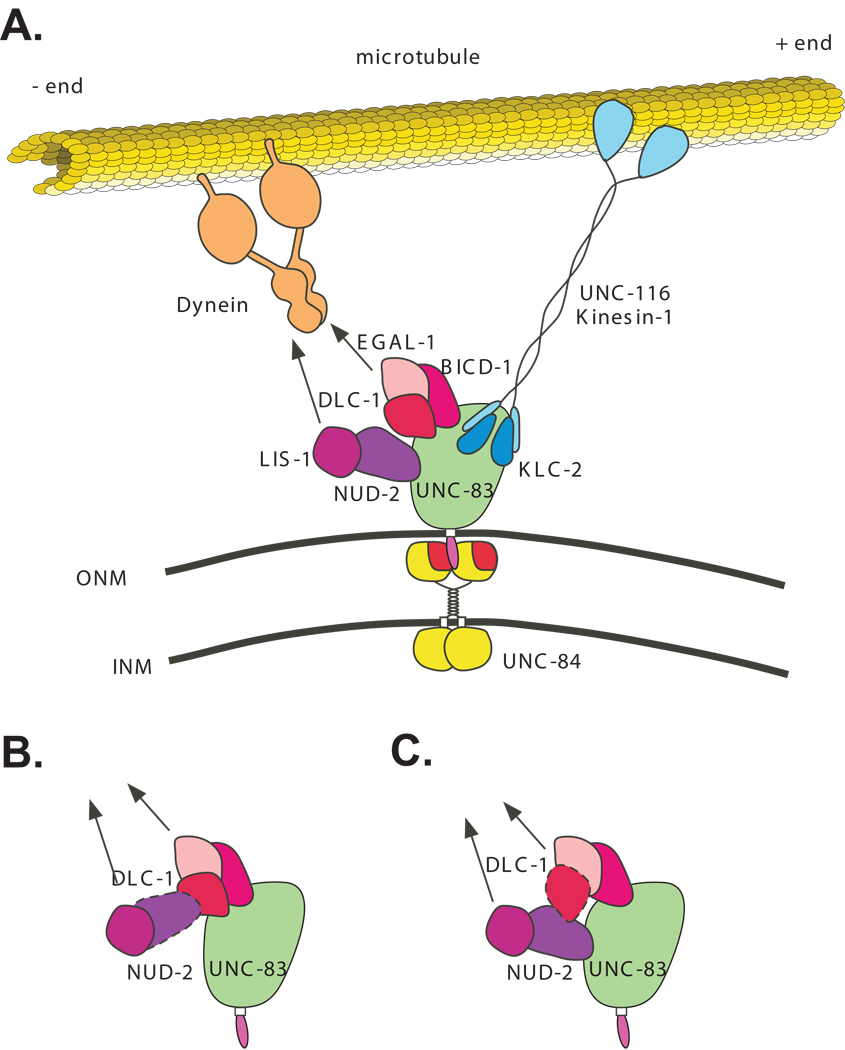
UNC-83 is proposed to extend into the cytoplasm from the outer surface of the nucleus to attach the outer nuclear membrane to force-generating molecules in the cytoplasm during nuclear migration. Forces generated in the cytoplasm are transferred across the nuclear envelope to the structural components of the nuclear lamina through the SUN protein UNC-84 in the inner nuclear membrane (McGee et al., 2006; Starr et al., 2001). However, the mechanisms of how forces are generated in the cytoplasm and transferred to the UNC-83-UNC-84 nuclear envelope bridge were unknown. We recently described how UNC-83 targets kinesin-1 to the outer surface of the nucleus where it provides the major forces to translocate the nucleus along microtubules in hyp7 cells (Meyerzon et al., 2009). Here, we show that UNC-83 interacts with two complexes to recruit dynein to the surface of the nucleus where it regulates nuclear migration. In our model, UNC-83 functions to coordinate the major translocation forces generated by kinesin-1 and the regulatory roles of dynein at the nuclear envelope during migration (Figure 9).
Figure 9.
Model for UNC-83-mediated nuclear migration in C. elegans. (A) UNC-84 (red and yellow) localizes to the inner nuclear membrane (INM) and UNC-83 (green and pink) localizes to the outer nuclear membrane (ONM). The UNC-84 SUN domain (red) interacts with the UNC-83 KASH domain (pink) in the perinuclear space. The cytoplasmic domain of UNC-83 (green) extends into the cytoplasm where it interacts with kinesin-1 through KLC-2 (dark blue). The motor activity of UNC-116 (light blue) moves hyp7 nuclei toward the plus ends of microtubules (yellow). UNC-83 also interacts with two dynein-regulating complexes, one consisting of NUD-2 and LIS-1 (shades of purple) and the other containing DLC-1, EGAL-1 and BICD-1 (shades of pink). These two complexes recruit the minus-end directed microtubule motor dynein to the nuclear envelope. We propose that dynein regulates nuclear migration, perhaps through kinesin-1. (B–C) Alternative models for interactions between the dynein-regulating complexes.
UNC-83 recruits dynein to the nuclear envelope through two complexes
How microtubule motors are targeted to different cargos at different times during the development of a cell is poorly understood (Welte, 2004). Even in different nuclear migration events, relative roles of different microtubule motors vary drastically. For example, contrast the mechanisms of UNC-83-mediated nuclear migration of hyp7 nuclei to the mechanisms of pronuclear migration in the newly fertilized embryo. Dynein is required at the nuclear envelope during pronuclear migration in the newly fertilized embryo where it functions to separate the centrosomes on the male pronucleus and to translocate the female pronucleus along microtubule asters (Gonczy et al., 1999; Reinsch and Gonczy, 1998). No role for kinesin-1 has been demonstrated in pronuclear migration. Here and in Meyerzon et al. (2009), we showed that dynein plays a regulatory role and kinesin-1 provides the major motor activity in hyp7 nuclear migration.
Since dynein plays different roles in pronuclear versus hyp7 nuclear migration, one would expect that different cargo adaptors be used to recruit dynein to the nuclear envelope at different developmental stages. The KASH protein ZYG-12, LIS-1 and dynactin function to target dynein to pronuclei (Cockell et al., 2004; Gonczy et al., 1999; Malone et al., 2003) while BICD-1, NUD-2 and EGAL-1 have no function in the one-cell zygote. However, dynactin and ZYG-12 do not apparently function during hyp7 nuclear migration, because dnc-1(RNAi) animals had no nuclei in the dorsal cord (data not shown) and ZYG-12 is not expressed after early embryogenesis (Malone et al., 2003). Instead, UNC-83 interacted with two dynein-regulating complexes; one consisting of BICD-1, EGAL-1, and DLC-1, and a second that includes NUD-2 and LIS-1. Disruptions in either of these complexes led to weak nuclear migration phenotypes and simultaneous disruption of both led to slightly more severe defects. Although the hyp7 nuclear migration phenotypes reported here were much less severe than those in unc-83, unc-84, or klc-2 mutant animals, they were significant as compared to the wild-type background where zero nuclei were observed in the dorsal cord of L1 animals.
How the BICD-1/EGAL-1/DLC-1 and NUD-2/LIS-1 complexes interact with one another could have implications in the control of nuclear migration. The C-terminal 50 residues of NUD-2 were necessary and sufficient for interaction with UNC-83. The C-terminus of the mammalian NudE interacts with the dynein light chain (Stehman et al., 2007). Thus, UNC-83 could regulate interactions where either NUD-2 or DLC-1 interacts with UNC-83 (Figure 9B, C). Perhaps both UNC-83 and the BICD-1/EGAL-1/DLC-1 complexes are needed to fully recruit the NUD-2/LIS-1 complex or vice-versa. This would be consistent with the molecular and genetic data that the two complexes are partially redundant. Similar redundant mechanisms function to recruit dynein to mammalian kinetochores. For example, ZW10 functioning through dynactin, and NudE interacting with the dynein light chain, coordinate dynein targeting to the prophase kinetochore (Starr et al., 1998; Stehman et al., 2007).
Dynein and its associated proteins have been shown to function in nuclear positioning in other systems. Nuclear migration is an important step in the development of the Drosophila eye disc. Additionally, in stage 8 of Drosophila oogenesis, the nucleus is repositioned from the posterior to a dorsal anterior position. Lis-1, BicD, Egl, and dynein are required for both of these nuclear positioning events (Swan et al., 1999; Swan and Suter, 1996). Similarly, Pac1, the Lis1 yeast homolog, functions at the cortex of the bud to pull the nucleus from the mother cell into the bud neck (Lee et al., 2003). Instead of functioning at the cell cortex, the interactions described here between UNC-83 and dynein regulators support a model that dynein functions at the cytoplasmic surface of the nucleus during C. elegans hyp7 nuclear migration. These data once again highlight the importance of studying dynein in multiple systems, as it role varies in different nuclear migration events.
We were unable to clearly observe any of the dynein regulators at the surface of the nucleus during hyp7 nuclear migration. This was likely due to the facts that many different cargos utilize the dynein adaptors at any one time and because hyp7 cells are very small and embedded in tissue (Supplemental Figure 2; Meyerzon et al., 2009). Nonetheless, we hypothesize that dynein does localize to the nuclear envelope at the time of nuclear migration. Dynein and LIS-1 are clearly enriched at the nuclear envelope in migrating pronuclei in the 1-cell embryo, which is much better suited for immunofluorescence localization (Cockell et al., 2004; Gonczy et al., 1999). NudE also clearly localizes to the nuclear envelope of Drosophila spermatocytes, a large, easily isolated cell type, although the same group failed to detect NudE at the nuclear envelope in the much smaller larval neuroblasts (Wainman et al., 2009). Finally, we were able to recruit significant amounts of NUD-2 to the nuclear envelope in HeLa cells transfected with UNC-83. Thus, our model proposes that UNC-83 functions to recruit dynein to the outer surface of the nucleus during nuclear migration.
UNC-83 regulates bi-directional transport of nuclei in hyp7 cells
We propose that UNC-83 functions at the surface of the nucleus to act as a regulator controlling the relative outputs of kinesin-1 and dynein. This would allow migrating nuclei in hyp7 cells to switch directions and avoid obstacles, ensuring a more efficient migration. If you knock out this function, by blocking the ability of UNC-83 to interact with dynein, most nuclei would still migrate normally. However, occasionally a nucleus could get stuck at a microtubule intersection or some other block, disrupting nuclear migration. This model fits with the phenotypic analysis of dynein regulators reported here. Furthermore, this model would predict that the kinesin-1 and dynein double mutant phenotype would not be worse than the kinesin-1 mutant phenotype alone. Unfortunately, we were unable to determine the effects of impairing both kinesin-1 and dynein on hyp7 nuclear migration because klc-2(km11); nud-2(ok949) double mutants were embryonic lethal (data not shown).
Most cargos of microtubule motors appear to be moving unidirectionally, but upon closer examination, actually display short runs in both directions (Welte, 2004). Such bidirectional movement could allow the nucleus to switch microtubule tracks, avoid obstacles, or correct errors (Welte, 2004). For example, the larger, flexible step size of dynein could allow the nucleus to move past a microtubule intersection (Ross et al., 2008). Bidirectional movement might not always be essential, but would be used to ensure 100% efficiency of the movement. The coupling of kinesin and dynein activity on a single cargo has been observed during lipid droplet transport in Drosophila embryogenesis (Shubeita et al., 2008). Measured stall forces of lipid droplets in vivo indicate that in kinesin mutants the minus end directed stall force is also reduced; suggesting dynein activity is impaired in these mutants. During axonal transport, inhibition of dynein prevents proper transport of organelles in both directions (Martin et al., 1999), which supports the hypothesis that kinesin and dynein are interdependent on each other during bidirectional transport. These examples suggest that kinesin and dynein function together during bidirectional transport and inhibition of one motor may have unforeseen adverse affects on the other.
Our model of hyp7 nuclear migration in C. elegans is an excellent system to tease apart the relative contributions of dynein and kinesin-1 in a movement of a cargo. During hyp7 nuclear migration, it is thought that microtubules form bundles with their plus ends at the growing end of the cell; nuclei then migrate toward the plus end (Meyerzon et al., 2009; (Williams-Masson et al., 1998). In this model, kinesin would supply the major force to move nuclei while dynein would act as a regulator. UNC-83 is ideally located on the outer surface of the nucleus to act as a cargo adaptor and to coordinate interactions between microtubule motors and the nucleus. As suggested by our functional deletion analysis of UNC-83 (Figure 8), UNC-83 can interact with both dynein and kinesin-1 complexes at the same time. Thus, this is the first report showing that a single cargo adaptor at the surface of the nucleus interacts with both kinesin-1 and dynein. Also, this is the first report demonstrating that both kinein-1 and dynein function together during a nuclear migration event in metazoans.
Supplementary Material
Acknowledgements
We thank many colleagues in the Department of Molecular and Cell Biology and members of the Starr lab for helpful comments. This research was supported by grant 5R01GM073874 from the NIH. H.N.F. was supported by NIH training grant 5T32GM007377.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahringer J. In: Reverse Genetics. T. C. e. r. community, editor. WormBook; 2006. [Google Scholar]
- Ausubel FM. Greene Publishing Associates ; J. Wiley, order fulfillment. Brooklyn, N. Y: Media, Pa; 1987. Current protocols in molecular biology. [Google Scholar]
- Boulin T, Etchberger J, Hobert O. In: Reporter gene fusions. T. C. e. r. community, editor. WormBook; 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke B, Roux KJ. Nuclei take a position: managing nuclear location. Dev Cell. 2009;17:587–597. doi: 10.1016/j.devcel.2009.10.018. [DOI] [PubMed] [Google Scholar]
- Cockell MM, Baumer K, Gonczy P. lis-1 is required for dynein-dependent cell division processes in C. elegans embryos. J Cell Sci. 2004;117:4571–4582. doi: 10.1242/jcs.01344. [DOI] [PubMed] [Google Scholar]
- Crisp M, Liu Q, Roux K, Rattner JB, Shanahan C, Burke B, Stahl PD, Hodzic D. Coupling of the nucleus and cytoplasm: role of the LINC complex. J Cell Biol. 2006;172:41–53. doi: 10.1083/jcb.200509124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efimov VP, Morris NR. The LIS1-related NUDF protein of Aspergillus nidulans interacts with the coiled-coil domain of the NUDE/RO11 protein. J Cell Biol. 2000;150:681–688. doi: 10.1083/jcb.150.3.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Gonczy P, Echeverri C, Oegema K, Coulson A, Jones SJ, Copley RR, Duperon J, Oegema J, Brehm M, Cassin E, Hannak E, Kirkham M, Pichler S, Flohrs K, Goessen A, Leidel S, Alleaume AM, Martin C, Ozlu N, Bork P, Hyman AA. Functional genomic analysis of cell division in C. elegans using RNAi of genes on chromosome III. Nature. 2000;408:331–336. doi: 10.1038/35042526. [DOI] [PubMed] [Google Scholar]
- Gonczy P, Pichler S, Kirkham M, Hyman AA. Cytoplasmic dynein is required for distinct aspects of MTOC positioning, including centrosome separation, in the one cell stage Caenorhabditis elegans embryo. J Cell Biol. 1999;147:135–150. doi: 10.1083/jcb.147.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gros-Louis F, Dupre N, Dion P, Fox MA, Laurent S, Verreault S, Sanes JR, Bouchard JP, Rouleau GA. Mutations in SYNE1 lead to a newly discovered form of autosomal recessive cerebellar ataxia. Nat Genet. 2007;39:80–85. doi: 10.1038/ng1927. [DOI] [PubMed] [Google Scholar]
- Horvitz HR, Sulston JE. Isolation and genetic characterization of cell-lineage mutants of the nematode Caenorhabditis elegans. Genetics. 1980;96:435–454. doi: 10.1093/genetics/96.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houalla T, Hien Vuong D, Ruan W, Suter B, Rao Y. The Ste20-like kinase misshapen functions together with Bicaudal-D and dynein in driving nuclear migration in the developing drosophila eye. Mech Dev. 2005;122:97–108. doi: 10.1016/j.mod.2004.08.005. [DOI] [PubMed] [Google Scholar]
- Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, Welchman DP, Zipperlen P, Ahringer J. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- Kandert S, Luke Y, Kleinhenz T, Neumann S, Lu W, Jaeger VM, Munck M, Wehnert M, Muller CR, Zhou Z, Noegel AA, Dabauvalle MC, Karakesisoglou I. Nesprin-2 giant safeguards nuclear envelope architecture in LMNA S143F progeria cells. Hum Mol Genet. 2007;16:2944–2959. doi: 10.1093/hmg/ddm255. [DOI] [PubMed] [Google Scholar]
- Koushika SP, Schaefer AM, Vincent R, Willis JH, Bowerman B, Nonet ML. Mutations in Caenorhabditis elegans cytoplasmic dynein components reveal specificity of neuronal retrograde cargo. J Neurosci. 2004;24:3907–3916. doi: 10.1523/JNEUROSCI.5039-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WL, Oberle JR, Cooper JA. The role of the lissencephaly protein Pac1 during nuclear migration in budding yeast. J Cell Biol. 2003;160:355–364. doi: 10.1083/jcb.200209022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Yu W, Li Y, Yang Z, Yan X, Huang Q, Zhu X. Nudel functions in membrane traffic mainly through association with Lis1 and cytoplasmic dynein. J Cell Biol. 2004;164:557–566. doi: 10.1083/jcb.200308058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke CJ, Williams SN, Schwarz EM, Caldwell GA, Caldwell KA. Genetic interactions among cortical malformation genes that influence susceptibility to convulsions in C. elegans. Brain Res. 2006;1120:23–34. doi: 10.1016/j.brainres.2006.08.067. [DOI] [PubMed] [Google Scholar]
- Mach JM, Lehmann R. An Egalitarian-BicaudalD complex is essential for oocyte specification and axis determination in Drosophila. Genes Dev. 1997;11:423–435. doi: 10.1101/gad.11.4.423. [DOI] [PubMed] [Google Scholar]
- Malone CJ, Fixsen WD, Horvitz HR, Han M. UNC-84 localizes to the nuclear envelope and is required for nuclear migration and anchoring during C. elegans development. Development. 1999;126:3171–3181. doi: 10.1242/dev.126.14.3171. [DOI] [PubMed] [Google Scholar]
- Malone CJ, Misner L, Le Bot N, Tsai MC, Campbell JM, Ahringer J, White JG. The C. elegans hook protein, ZYG-12, mediates the essential attachment between the centrosome and nucleus. Cell. 2003;115:825–836. doi: 10.1016/s0092-8674(03)00985-1. [DOI] [PubMed] [Google Scholar]
- Martin M, Iyadurai SJ, Gassman A, Gindhart JG, Jr., Hays TS, Saxton WM. Cytoplasmic dynein, the dynactin complex, and kinesin are interdependent and essential for fast axonal transport. Mol Biol Cell. 1999;10:3717–3728. doi: 10.1091/mbc.10.11.3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGee MD, Rillo R, Anderson AS, Starr DA. UNC-83 Is a KASH Protein Required for Nuclear Migration and Is Recruited to the Outer Nuclear Membrane by a Physical Interaction with the SUN Protein UNC-84. Mol Biol Cell. 2006;17:1790–1801. doi: 10.1091/mbc.E05-09-0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. Embo J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DM, Shakes DC. Immunofluorescence microscopy. Methods Cell Biol. 1995;48:365–394. [PubMed] [Google Scholar]
- Minke PF, Lee IH, Tinsley JH, Bruno KS, Plamann M. Neurospora crassa ro-10 and ro-11 genes encode novel proteins required for nuclear distribution. Mol Microbiol. 1999;32:1065–1076. doi: 10.1046/j.1365-2958.1999.01421.x. [DOI] [PubMed] [Google Scholar]
- Minn IL, Rolls MM, Hanna-Rose W, Malone CJ. SUN-1 and ZYG-12, mediators of centrosome-nucleus attachment, are a functional SUN/KASH pair in Caenorhabditis elegans. Mol Biol Cell. 2009;20:4586–4595. doi: 10.1091/mbc.E08-10-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris NR. Nuclear migration. From fungi to the mammalian brain. J Cell Biol. 2000;48:1097–1101. doi: 10.1083/jcb.148.6.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris SM, Albrecht U, Reiner O, Eichele G, Yu-Lee L. The lissencephaly gene product Lis1, a protein involved in neuronal migration, interacts with a nuclear movement protein, NudC. Curr. Biol. 1998;8:603–606. doi: 10.1016/s0960-9822(98)70232-5. [DOI] [PubMed] [Google Scholar]
- Navarro C, Puthalakath H, Adams JM, Strasser A, Lehmann R. Egalitarian binds dynein light chain to establish oocyte polarity and maintain oocyte fate. Nat Cell Biol. 2004;6:427–435. doi: 10.1038/ncb1122. [DOI] [PubMed] [Google Scholar]
- Padmakumar VC, Libotte T, Lu W, Zaim H, Abraham S, Noegel AA, Gotzmann J, Foisner R, Karakesisoglou I. The inner nuclear membrane protein Sun1 mediates the anchorage of Nesprin-2 to the nuclear envelope. J Cell Sci. 2005;118:3419–3430. doi: 10.1242/jcs.02471. [DOI] [PubMed] [Google Scholar]
- Piano F, Schetter AJ, Mangone M, Stein L, Kemphues KJ. RNAi analysis of genes expressed in the ovary of Caenorhabditis elegans. Curr Biol. 2000;10:1619–1622. doi: 10.1016/s0960-9822(00)00869-1. [DOI] [PubMed] [Google Scholar]
- Reboul J, Vaglio P, Rual JF, Lamesch P, Martinez M, Armstrong CM, Li S, Jacotot L, Bertin N, Janky R, Moore T, Hudson JR, Jr., Hartley JL, Brasch MA, Vandenhaute J, Boulton S, Endress GA, Jenna S, Chevet E, Papasotiropoulos V, Tolias PP, Ptacek J, Snyder M, Huang R, Chance MR, Lee H, Doucette-Stamm L, Hill DE, Vidal M. C. elegans ORFeome version 1.1: experimental verification of the genome annotation and resource for proteome-scale protein expression. Nat Genet. 2003;34:35–41. doi: 10.1038/ng1140. [DOI] [PubMed] [Google Scholar]
- Reinsch S, Gonczy P. Mechanisms of nuclear positioning. J Cell Sci. 1998;111:2283–2295. doi: 10.1242/jcs.111.16.2283. [DOI] [PubMed] [Google Scholar]
- Ross JL, Ali MY, Warshaw DM. Cargo transport: molecular motors navigate a complex cytoskeleton. Curr Opin Cell Biol. 2008;20:41–47. doi: 10.1016/j.ceb.2007.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupp RA, Snider L, Weintraub H. Xenopus embryos regulate the nuclear localization of XMyoD. Genes Dev. 1994;8:1311–1323. doi: 10.1101/gad.8.11.1311. [DOI] [PubMed] [Google Scholar]
- Sagasti A, Hisamoto N, Hyodo J, Tanaka-Hino M, Matsumoto K, Bargmann CI. The CaMKII UNC-43 activates the MAPKKK NSY-1 to execute a lateral signaling decision required for asymmetric olfactory neuron fates. Cell. 2001;105:221–232. doi: 10.1016/s0092-8674(01)00313-0. [DOI] [PubMed] [Google Scholar]
- Sasaki S, Shionoya A, Ishida M, Gambello MJ, Yingling J, Wynshaw-Boris A, Hirotsune S. A LIS1/NUDEL/cytoplasmic dynein heavy chain complex in the developing and adult nervous system. Neuron. 2000;28:681–696. doi: 10.1016/s0896-6273(00)00146-x. [DOI] [PubMed] [Google Scholar]
- Shubeita GT, Tran SL, Xu J, Vershinin M, Cermelli S, Cotton SL, Welte MA, Gross SP. Consequences of motor copy number on the intracellular transport of kinesin-1-driven lipid droplets. Cell. 2008;135:1098–1107. doi: 10.1016/j.cell.2008.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnichsen B, Koski LB, Walsh A, Marschall P, Neumann B, Brehm M, Alleaume AM, Artelt J, Bettencourt P, Cassin E, Hewitson M, Holz C, Khan M, Lazik S, Martin C, Nitzsche B, Ruer M, Stamford J, Winzi M, Heinkel R, Roder M, Finell J, Hantsch H, Jones SJ, Jones M, Piano F, Gunsalus KC, Oegema K, Gonczy P, Coulson A, Hyman AA, Echeverri CJ. Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature. 2005;434:462–469. doi: 10.1038/nature03353. [DOI] [PubMed] [Google Scholar]
- Starr DA. Communication between the cytoskeleton and the nuclear envelope to position the nucleus. Mol Biosyst. 2007;3:583–589. doi: 10.1039/b703878j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr DA. A nuclear-envelope bridge positions nuclei and moves chromosomes. J Cell Sci. 2009;122:577–586. doi: 10.1242/jcs.037622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr DA, Hermann GJ, Malone CJ, Fixsen W, Priess JR, Horvitz HR, Han M. unc-83 encodes a novel component of the nuclear envelope and is essential for proper nuclear migration. Development. 2001;128:5039–5050. doi: 10.1242/dev.128.24.5039. [DOI] [PubMed] [Google Scholar]
- Starr DA, Williams BC, Hays TS, Goldberg ML. ZW10 helps recruit dynactin and dynein to the kinetochore. J Cell Biol. 1998;142:763–774. doi: 10.1083/jcb.142.3.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehman SA, Chen Y, McKenney RJ, Vallee RB. NudE and NudEL are required for mitotic progression and are involved in dynein recruitment to kinetochores. J Cell Biol. 2007;178:583–594. doi: 10.1083/jcb.200610112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston JE, Schierenberg E, White JG, Thomson JN. The embryonic cell lineage of the nematode Caenorhabditis elegans. Devel. Biol. 1983;100:64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- Swan A, Nguyen T, Suter B. Drosophila Lissencephaly-1 functions with Bic-D and dynein in oocyte determination and nuclear positioning. Nat Cell Biol. 1999;1:444–449. doi: 10.1038/15680. [DOI] [PubMed] [Google Scholar]
- Swan A, Suter B. Role of Bicaudal-D in patterning the Drosophila egg chamber in midoogenesis. Development. 1996;122:3577–3586. doi: 10.1242/dev.122.11.3577. [DOI] [PubMed] [Google Scholar]
- Tavernarakis N, Wang SL, Dorovkov M, Ryazanov A, Driscoll M. Heritable and inducible genetic interference by double-stranded RNA encoded by transgenes. Nat Genet. 2000;24:180–183. doi: 10.1038/72850. [DOI] [PubMed] [Google Scholar]
- Timmons L, Fire A. Specific interference by ingested dsRNA. Nature. 1998;395:854. doi: 10.1038/27579. [DOI] [PubMed] [Google Scholar]
- Tsai JW, Bremner KH, Vallee RB. Dual subcellular roles for LIS1 and dynein in radial neuronal migration in live brain tissue. Nat Neurosci. 2007;10:970–979. doi: 10.1038/nn1934. [DOI] [PubMed] [Google Scholar]
- Tsujikawa M, Omori Y, Biyanwila J, Malicki J. Mechanism of positioning the cell nucleus in vertebrate photoreceptors. Proc Natl Acad Sci U S A. 2007;104:14819–14824. doi: 10.1073/pnas.0700178104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wainman A, Creque J, Williams B, Williams EV, Bonaccorsi S, Gatti M, Goldberg ML. Roles of the Drosophila NudE protein in kinetochore function and centrosome migration. J Cell Sci. 2009;122:1747–1758. doi: 10.1242/jcs.041798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welte MA. Bidirectional transport along microtubules. Curr Biol. 2004;14:R525–R537. doi: 10.1016/j.cub.2004.06.045. [DOI] [PubMed] [Google Scholar]
- Wilhelmsen K, Ketema M, Truong H, Sonnenberg A. KASH-domain proteins in nuclear migration, anchorage and other processes. J Cell Sci. 2006;119:5021–5029. doi: 10.1242/jcs.03295. [DOI] [PubMed] [Google Scholar]
- Williams-Masson EM, Heid PJ, Lavin CA, Hardin J. The cellular mechanism of epithelial rearrangement during morphogenesis of the Caenorhabditis elegans dorsal hypodermis. Dev Biol. 1998;204:263–276. doi: 10.1006/dbio.1998.9048. [DOI] [PubMed] [Google Scholar]
- Wynshaw-Boris A. Lissencephaly and LIS1: insights into the molecular mechanisms of neuronal migration and development. Clin Genet. 2007;72:296–304. doi: 10.1111/j.1399-0004.2007.00888.x. [DOI] [PubMed] [Google Scholar]
- Xiang X, Fischer R. Nuclear migration and positioning in filamentous fungi. Fungal Genet Biol. 2004;41:411–419. doi: 10.1016/j.fgb.2003.11.010. [DOI] [PubMed] [Google Scholar]
- Xiang X, Osmani AH, Osmani SA, Xin M, Morris NR. NudF, a nuclear migration gene in Aspergillus nidulans, is similar to the human LIS-1 gene required for neuronal migration. Mol Biol Cell. 1995;6:297–310. doi: 10.1091/mbc.6.3.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Hong SH, Privalsky ML. Transcriptional anti-repression. Thyroid hormone receptor beta-2 recruits SMRT corepressor but interferes with subsequent assembly of a functional corepressor complex. J Biol Chem. 1999;274:37131–37138. doi: 10.1074/jbc.274.52.37131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Bethmann C, Worth NF, Davies JD, Wasner C, Feuer A, Ragnauth CD, Yi Q, Mellad JA, Warren DT, Wheeler MA, Ellis JA, Skepper JN, Vorgerd M, Schlotter-Weigel B, Weissberg PL, Roberts RG, Wehnert M, Shanahan CM. Nesprin-1 and -2 are involved in the pathogenesis of Emery Dreifuss muscular dystrophy and are critical for nuclear envelope integrity. Hum Mol Genet. 2007;16:2816–2833. doi: 10.1093/hmg/ddm238. [DOI] [PubMed] [Google Scholar]
- Zhang X, Lei K, Yuan X, Wu X, Zhuang Y, Xu T, Xu R, Han M. SUN1/2 and Syne/Nesprin-1/2 complexes connect centrosome to the nucleus during neurogenesis and neuronal migration in mice. Neuron. 2009;64:173–187. doi: 10.1016/j.neuron.2009.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou K, Rolls MM, Hall DH, Malone CJ, Hanna-Rose W. A ZYG-12-dynein interaction at the nuclear envelope defines cytoskeletal architecture in the C. elegans gonad. J Cell Biol. 2009;186:229–241. doi: 10.1083/jcb.200902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.