Abstract
We used a combination of spectral karyotyping, comparative genomic hybridization and cDNA microarrays to gain insights into the structural and functional changes of the genome in the MCF10A human breast cancer progression model cell lines. SKY data showed several chromosomal aberrations and array comparative genomic hybridization analysis identified numerous genomic gains and losses that might be involved in the progression towards cancer. Analysis of the expression levels of genes located within these genomic regions revealed a lack of correlation between chromosomal gains and losses and corresponding up or down regulation for the majority of the ∼ 1,000 genes analyzed in this study. We conclude that other mechanisms of gene regulation that are not directly related to chromosomal gains and losses play a major role in the progression towards breast cancer.
Introduction
Breast cancer is the most common cancer and accounts for the second highest mortality rate worldwide among cancer-related deaths in women (1). Numerous studies have indicated that initiation and progression towards a breast cancer phenotype is a multi-step process involving accumulation of genomic aberrations (2). In particular, amplifications, deletions and rearrangements have been observed in breast cancer of critical genes involved in cell growth, differentiation and cell death (3). Breast cancers can either be hereditary or sporadic. Germ line mutations in BRCA1 and BRCA2 (4) genes have been linked to the development of familial breast cancer.
Although the genomic and molecular changes in breast cancer have been studied in detail, the initial events leading to tumor formation remain to be elucidated. Cancer progression models have become an invaluable tool in studying the precise genetic aberrations that correlate with a shift from a normal to a disease phenotype. In this investigation we utilized the MCF10 human breast cancer progression model cell lines to study the genetic changes that occur during the transformation of breast epithelial cells into breast cancer. The MCF10 progression model consists of three directly derived cell lines: (1) the spontaneously immortalized cell line MCF10A (5) which do not show any characteristics of invasiveness or tumor formation and hence are considered to be a normal-like breast epithelial cell line (6); (2) MCF10A cells were transformed with c-Ha-Ras to yield the pre-malignant MCF10AT1 cell line (7) (8); (3) MCF10AT1 cells were xenografted on to mice and a third cell line MCF10CA1a was selected that shows all the characteristics of a fully malignant breast cancer cell type (9). Karyotypic analysis of these three cell lines showed a characteristic (3;9) translocation that confirmed a common lineage (9).
Spectral karyotyping (SKY) (10) has been previously employed to study chromosomal aberrations in breast cancer cell lines and patient tumors and led to the identification of numerous chromosomal translocations, deletions and rearrangements (11). Array comparative genomic hybridization (aCGH) is useful in the detection of copy number variations that arise due to the genomic instability in cancer cells or tissues (12) and has led to the detection in breast cancer of gains within chromosome arms 1q, 3p, 4q, 8q, 11q, 17q, 20q and losses in 6q,11q, 8p, 9p, 13q,16q and 17p (13). In most of these studies, the altered regions have been shown to harbor essential gene/s whose deregulation leads to the establishment of tumorogenesis (3). cDNA microarray analysis makes it feasible to study the expression patterns of thousands of genes in cancer (14). This technique has resulted in the identification of numerous changes in gene expression associated with breast cancer (15).
In the present study we attempted to correlate the changes observed in the immortalized normal human breast epithelial cell line MCF10A and its malignant counterpart, MCF10CA1a by combined SKY aCGH and cDNA microarray analysis. We observed numerous changes in gene expression patterns that were indicative of tumor progression. Thus the combination of these three approaches has provided insights into how DNA copy number variations affect the gene expression patterns and for the identification of new candidate genes that might be associated with breast cancer and its progression.
Materials and Methods
Cell Culture
The breast cancer progression model cell lines (MCF10A, MCF10AT1, and MCF10CA1a) used in these studies were obtained from the Barbara Ann Karamanos Center, Detroit, MI. The MCF10A cell line was cultured in DMEM media supplemented with horse serum (5%), insulin (2.5 mg/ml), EGF (50 μg/ml), cholera enterotoxin (150 μg/ml), hydrocortisone (2.5 mg/ml) and HEPES (5 ml) while MCF10AT1 and MCF10CA1a cell lines were cultured in DMEM media supplemented with only horse serum (5%). All three cell lines were grown at 37°C in a 5% carbon dioxide (CO2) substituted incubator. Cells were collected at the same passage and used for each of the analyses (SKY, CGH and gene expression microarray) performed in this study.
Spectral Karyotyping (SKY)
Preparations of metaphase chromosome spreads were subjected to the SKY procedure recommended by Applied Spectral Imaging (ASI, Carlsbad, CA). The images were captured using a combination of rhodamine, Texas-Red, Cy5, FITC and Cy5.5 filter sets mounted on a Nikon fluorescence microscope equipped with a spectral cube and an interferometer module. Karyotypic analysis of the images was carried out using the SKY View software.
aCGH array analysis
DNA printing solutions were prepared from sequence connected RPCI-11 BAC by ligation-mediated PCR as described previously (16). The array contains ∼19,000 BAC clones that were chosen by virtue of their STS content, end-sequence and association with heritable disorders and cancer (17). Each clone is printed in duplicate on amino-silanated glass slides (Schott Nexterion typeA+) using a MicroGrid ll TAS arrayer (Genomic Solutions, Inc.). The BAC DNA products have ∼80 μm diameter spots with 150 μm center to center spacing creating an array of ∼39,000 elements. The printed slides dry overnight and are UV-crosslinked (350 mJ) in a Stratalinker 2400 (Stratagene) immediately before hybridization.
Reference and test sample genomic DNA (1 μg each) were individually fluorescently labeled using the BioPrime DNA labeling kit (Invitrogen, Carlsbad, CA) for 18 h at 37°C with the appropriate Cy dye (Cy3 or Cy5). After ethanol precipitation, the probes are resuspended in H2O, combined and purified of unincorporated dye by passage over a Qiagen spin column (Valencia, CA). Prior to hybridization, the test and reference probes were resuspended in 110 μl SlideHyb Buffer #3 (Ambion, Foster City, CA) containing 5 μl of 20 μg/μl Cot-1 and 5 μl of 100 μg/μl yeast tRNA (Invitrogen), heated to 95°C for 5 min and placed on ice. Hybridization to the 6k BAC arrays were performed for 16 h at 55°C using a GeneTAC hybridization station (Genomic Solutions Inc, Ann Arbor, MI.) as described (18). The hybridized aCGH slides are scanned using a GenePix 4200A Scanner (Molecular Devices, Sunny Vale, CA) to generate high-resolution (5 μm) images for both Cy3 (test) and Cy5 (control) channels. Image analysis was performed using the ImaGene version 6.1.0 (Bio Discovery, El Segundo, CA). The log2 test/control ratios were normalized using a sub-grid loess correction. Mapping information was added to the resulting log2 test/control values.
cDNA microarray analysis
cDNA microarray analysis was performed on an agilent 44K whole human genome oligo microarray. Total RNA from the cell cultures (MCF10A, MCF10CA1a) were prepared using the RNeasy mini kits (Qiagen Inc, Valencia, CA.) following manufacturer's instructions. After elution, RNA samples were concentrated by ethanol precipitation at −20°C overnight, and resuspended in nuclease-free water. Before labeling, RNA samples were quantified using a Genequant spectrophotometer (GE Healthcare, Niskayuna, NJ) and evaluated for degradation using a 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA).
To screen the samples for gene expression, cRNA was synthesized by in vitro transcription and directly labeled with cyanine 3-CTP or cyanine 5-CTP using the low RNA input linear amplification kit, as per manufacturer's instructions (Agilent Technologies, USA). Initially, first-strand cDNA was synthesized by incubating 0.2 – 2 μg total RNA and spiking controls and T7 oligo(dT) for 10 min at 65 °C. Following the addition of first-strand reaction components, the reaction continued for 2 h at 40°C. cRNA was synthesized by incubating the resuspended double-stranded cDNA with the transcription master mix containing cyanine-CTP, ribonucleotides, and T7 RNA polymerase for 2 h at 40°C. The cyanine-labeled cRNA were recovered by column purification and eluted in 30 μl RNase-free water. The cRNA quality and concentration were assessed using a nanoDrop ND-1000 UV-VIS spectrophotometer (Thermo Scientific, USA). cRNA samples were required to have a yield >750 ng and a specific activity >8.0 pmol Cy3 or Cy5 per μg cRNA, to proceed to the hybridization step.
Prior to hybridization, 0.75 μg of cyanine 5-labeled, linearly amplified cRNA (test) and 0.75 μg of cyanine 3-labeled, linearly amplified cRNA (control) were combined and fragmented by incubating at 60°C for 30 min in fragmentation buffer. The hybridization reaction mixture was prepared by combining 2× hybridization buffer to the fragmented cRNA to a final volume of 210 μl. The hybridization reaction mixture was placed on ice then loaded slowly onto the 44K array, allowing even flow and distribution of hybridization cocktail across the surface. The arrays were secured in a SureHyb chamber cover and then placed in a rotisserie hybridization oven at 65°C for 17 h. After hybridization, the slides were removed from the hybridization oven and washed with Gene Expression Wash Buffer 1 and 2. Following the wash steps, the array images were captured by scanning at 532 nm and 635 nm with an agilent scanner and analyzed with agilent feature extraction software version 8.5.
Data analysis for aCGH and cDNA microarrays
The 19k aCGH array was analyzed using DNAcopy R package which uses the circular binary segmentation algorithm (19). A segment was identified as gain or loss if the estimated copy number ratio is more than 0.2 or less than -0.2 (corresponding to a genomic fold increase of 1.15 and decrease of 1.15) which is roughly eight times the standard error of the mean log ratio on each chip. For agilent gene expression arrays using the dye-swap design, significant changed genes are identified using limma R package (20) which uses a modified t-test with multiple tests in consideration. The gene ontology analysis for the significant changed genes was performed by GOstat software (21). To better plot the aCGH results, an in house ideogram and aCGH data plotting program was developed and used.
Results
Expression analysis of normal and malignant breast cell lines
Microarray expression analysis of immortalized normal MCF10A and malignant MCF10CA1a cell lines was performed to determine the changes in gene expression patterns. A total of 42,000 genes yielded hybridization signals for both samples (Table S1). Analysis of the data yielded a total of about 8000 genes that showed at least two-fold expression level changes (∼3044 genes were up regulated while ∼3829 genes were down regulated) in the MCF10CA1a cell line (Table S2).
A group of genes that were either highly up regulated or greatly repressed in the MCF10CA1a cell line is listed in Table 1. All these genes were chosen based on their functional involvement in breast cancer. Up regulated genes included selenoprotein plasma protein (SEPP1) and decorin (DCN) which have an antimetastatic role in breast cancer (22-24) fibrillin1 (FBN1), aldehyde oxidase 1 (AOX1) and prostagladin E receptor 2 (PTERG2) which are known to promote tumor growth (25-27) and, surprisingly, angiopotein1 (ANGPT1) which when over expressed has antitumor properties (28).
Table 1. Highly up and down regulated genes in MCF10CA1a involved in breast cancer.
| Gene | Fold change (log fold change MCF10CA1a/MCF10A) | Chromosome location |
|---|---|---|
| Selenoprotein plasma protein (SEPP1) | 7.37 | 5p12 |
| Angiopoietin 1 (ANGPT1) | 6.77 | 8q23.1 |
| Decorin (DCN) | 6.70 | 12q21.33 |
| Fibrillin 1 (FBN1) | 5.59 | 15q21.1 |
| Prostaglandin E receptor 2 (PTGER2) | 5.29 | 14q22.1 |
| Aldehyde oxidase 1 (AOX1) | 5.24 | 2q33.1 |
| Aldehyde dehydrogenase 1 family, member A3 (ALDH1A3) | -8.72 | 15q26.3 |
| E-cadherin (epithelial) (CDH1) | -8.65 | 16q22.1 |
| Interleukin 1, beta (IL1B) | -8.11 | 16q22.1 |
| S100 calcium binding protein A14 (S100A14) | -6.15 | 1q21.3 |
| Bradykinin receptor B2 (BDKRB2) | -6.07 | 14q32.2 |
Our results showed a down regulation of E-cadherin (CDH1). Deregulation of this gene was previously observed in breast and other types of cancer (29). The interleukin 1 beta (IL1B) and the S100 calcium binding protein A 14 (S100A14) genes were both greatly repressed which is contrary to the reported high levels of protein expression of these two proteins in breast and other cancers (30). Moreover, the bradykininin receptor B2 (BDKRB2) gene is known to induce mitosis in breast cells and enhance progression of cancer (31). Yet our study shows that this gene was significantly repressed.
Further analyses of the microarray data were performed to establish the relationships between expression levels and gene function (Tables 2 & 3). Several keratin, connexin and claudin genes involved in the formation of the extracellular matrix and cell-cell communication were down regulated. Genes involved in signal transduction pathways such as erythroblastic leukemia viral oncogene homolog 2 (ERBB2), Harvey rat sarcoma viral oncogene homolog (HRAS), v-raf murine sarcoma viral oncogene homolog B1 (BRAF), vascular endothelial growth factor (VEGF and epidermal growth factor (EGFR) were also down regulated in the cancer cell line. Oncogenes and tumor suppressors including myelocytomatosis viral oncogene (MYC), phosphatase and tensin (PTEN), breast cancer 2 (BRCA2) were down regulated while retinoblastoma 1 (RB1), cyclin-dependent kinase inhibitor (CDKNB1), and cyclin D3 (CCND3) were over-expressed.
Table 2. Selected down regulated genes in MCF10CA1a coding for functionally relevant proteins.
| Genes | Expression (log fold change MCF10CA1a Vs MCF10A) | Chromosome location |
|---|---|---|
| Extracellular matrix: | ||
| Keratin 6 (KRT6) | -6.36 | 12q13.13 |
| Keratin 8 (KRT8) | -1.26 | 12q13.13 |
| Keratin 13 (KRT13) | -5.47 | 17q21.2 |
| Keratin 14 (KRT14) | -3.82 | 17q21.2 |
| Keratin 15 (KRT15) | -5.14 | 17q21.2 |
| Keratin 16 (KRT16) | -3.99 | 17q21.2 |
| Keratin 17 (KRT17) | -3.79 | 17q21.2 |
| Keratin 18 (KRT18) | -2.22 | 12q13.13 |
| Keratin 19 (KRT19) | -2.31 | 17q21.2 |
| Keratin 23 (KRT23) | -5.70 | 17q21.2 |
| Collagen XIII, alpha 1 (COL13A1) | -6.84 | 10q22.1 |
| Cell communication : | ||
| Connexin 26 (GJB2) | -4.81 | 13q12.11 |
| Connexin 43 (GJA1) | -6.05 | 6q22.31 |
| Claudin 1 (CLDN1) | -5.05 | 3q28 |
| Claudin 4 (CLDN4) | -4.23 | 7q11.23 |
| Claudin 7 (CLDN7) | -3.20 | 17p13.1 |
| Claudin 12 (CLDN12) | -1.13 | 7q21.13 |
| Signal Transduction: | ||
| Vascular endothelial growth factor | -1.20 | 6p21.1 |
| V-raf murine sarcoma viral oncogene homolog B1 (BRAF) | -1.24 | 7q34 |
| Erythroblastic leukemia viral oncogene homolog 2 (ERBB2) | -1.57 | 17q12 |
| Epidermal growth factor (EGFR) | -1.61 | 7p11.2 |
| Harvey rat sarcoma viral oncogene homolog (HRAS) | -2.64 | 11p15.5 |
| Oncogenes and Tumor suppressors: | ||
| Breast cancer 2 (BRCA2) | - 1.42 | 13q13.1 |
| Myelocytomatosis viral oncogene (MYC) | -1.55 | 8q24.21 |
| Phosphatase and tensin (PTEN) | -4.28 | 10q23.31 |
| Cytokines: | ||
| Interleukin A (ILA) | -6.67 | 2q13 |
| Interleukin B (ILB) | -8.06 | 2q13 |
| Interleukin 6 (IL6) | -4.20 | 7p15.3 |
| Interleukin 8 (IL8) | -4.71 | 4q13.3 |
| Interleukin 11 (IL11) | -2.50 | 19q13.42 |
Table 3. Selected up regulated genes in MCF10CA1a coding for functionally relevant proteins.
| Genes | Expression (log fold change MCF10CA1a Vs MCF10A) | Chromosome location |
|---|---|---|
| Extracellular matrix: | ||
| Collagen type VI, alpha 1 (COL6A1) | 1.54 | 12q13.13 |
| Fibrillin1 (FBN1) | 2.07 | 15q21.1 |
| Mucin1 (MUC1) | 3.35 | 1q21.3 |
| Matrix Metalloproteinase 2 (MMP2) | 3.62 | 16q12.2 |
| Fibronectin 1 (FN1) | 3.96 | 2q35 |
| Keratin 7 (KRT7) | 5.59 | 21q22.3 |
| Oncogenes and Tumor suppressors: | ||
| Retinoblastoma 1 (RB1) | 1.14 | 13q14.2 |
| Cyclin-dependent kinase inhibitor (CDKNB1) | 1.18 | 12p13.1 |
| Cyclin D3 (CCND3) | 1.36 | 6p21.1 |
| Cytokines: | ||
| Interleukin 7 (IL7) | 1.13 | 8q21.12 |
| Interleukin 18 (IL18) | 1.67 | 11q23.1 |
Several interleukin genes such as Interleukin 6 (IL6), interleukin 8 (IL8), interleukin A (ILA) and interleukin B (ILB) were repressed in MCF10CA1a cell line while interleukin 7 (IL7) and interleukin 18(IL18) genes were expressed at higher levels. Finally, many genes that are involved in the formation of the extracellular matrix such as collagen type VI, alpha 1 (COL6A1), keratin 7 (KRT7), fibrillin1 (FBN1), matrix metalloproteinase 2 (MMP2), mucin1 (MUC1) and fibronectin 1 (FN1) showed higher levels of expression in MCF10CA1a (Table 3).
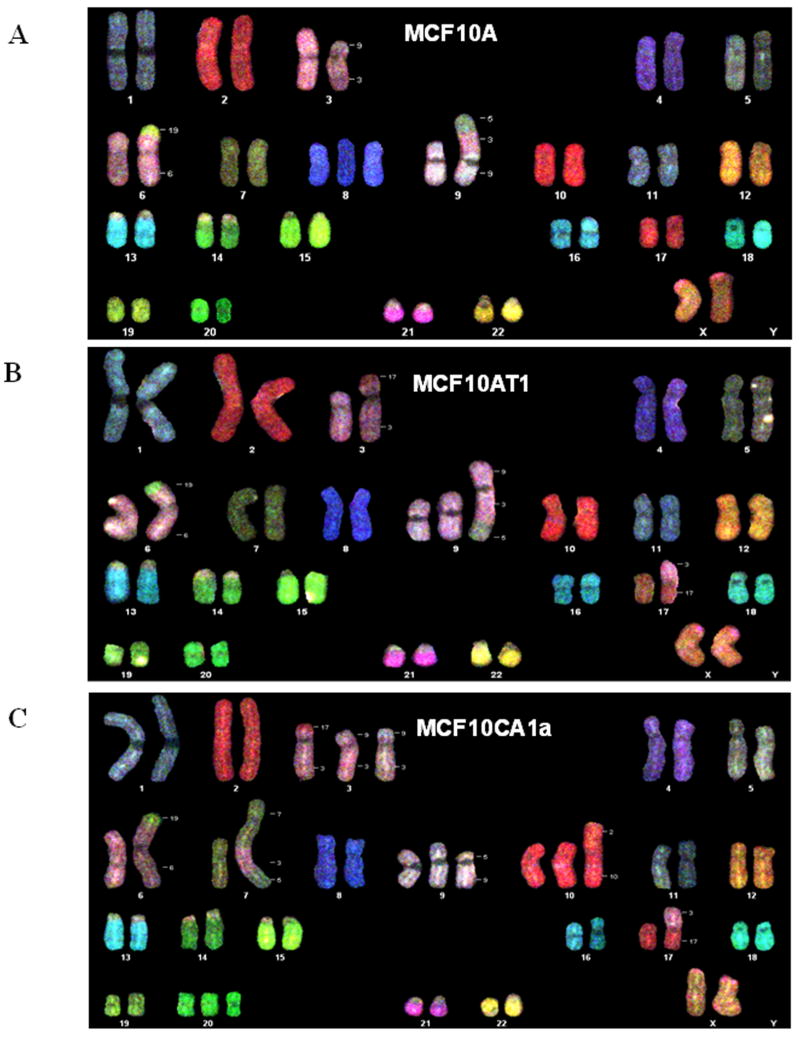
SKY analysis of MCF10 breast cancer progression model cell lines
Spectral karyotyping identified several chromosomal rearrangements in each of the three breast cancer cell lines (Fig 1). A karyotype of 47 chromosomes was found for normal MCF10A and the premalignant MCF10AT1 cell line (Fig 1A, B, Table S3), whereas the malignant MCF10CA1a cell line had 50 chromosomes (Fig 1C, Table S3). Four marker chromosomes were identified in MCF10A, MCF10AT1 and nine in the malignant MCF10CA1a cell line (Table S3). MCF10A demonstrated the characteristic (3; 9) reciprocal translocation that has been described as the single most important event in the immortalization and transformation of the MCF10 breast epithelial cell line (32). Other karyotypic changes including a gain of chromosome 8(+8) and translocations involving chromosomes 5, 3 9 (t (5;3;9)) and chromosomes 6 and 19 (t (6;19)) (Table S3). Karytotypic changes in the MCF10AT1 included a reciprocal translocation involving chromosomes 3 and 17(rept (3;17)) in addition to those found in MCF10A (Table S3). Karyotypic alterations in MCF10CA1a also included a gain of chromosome 20 (+20), translocation between chromosomes 5, 9 (t (5;9)) and chromosomes 2 and 10 (t (2;10)) in addition to the changes that are already present in MCF10A (Table S3).
Figure 1. Spectral karyotyping SKY) analysis of the MCF10 breast cancer progression model cell lines.
(A) MCF10A (B) MCF10AT1 (C) MCF10CA1a
aCGH analyses of chromosomal gains and losses in the breast cancer progression model cell lines
Analysis of aCGH data of the three cell lines revealed chromosomal breakpoints that were previously reported (33) as well as several new chromosomal aberrations. Three independent aCGH experiments were performed in the following manner; MCF10A vs. a normal diploid cell line, MCF10A vs. MCF10AT1, and MCF10A vs. MCF10CA1a. Ideograms of the complete analysis are presented in Supplementary Figure 1 (S1).
The MCF10A cell line showed gains at 5q23.1-35.3, 19q13.11q13.43 and 13q32.1-p32.2 (Table 4 & Fig S1). A gain of 8p23.3-q24.3 was seen in MCF10A as evidenced by the presence of an extra copy of chromosome 8 in this cell line compared to the normal diploid karyotype. Losses were observed at 9p21.3, 3p26.3, 16p11.2, 21p11-q11.2 and 22q11.1 (Table 4 & Fig S1). The premalignant MCF10AT1 cell line showed gains at 3p14.3, 3q13.3, 19q12-q34.3, 10q22.1-q22.2, 16q23.3, and 17p11.2 and losses of 5q12.1, 5q14.3-15, and 15q21.1 (Table 4 & Fig S1). The additional chromosome 8 copy found in the normal MCF10A cell line is deleted in this cell line. The malignant MCF10CA1a cell line showed several more genomic aberrations compared to the normal and premalignant MCF10 cell lines. Gains include 2p25.3-q21.2, 3p14.1-q29, 9p24.3-p11.2, 9p34.13-p34.3, 10q11.1-q26.3, 17p11.2 and 20p13-q13.3 (Table 4 & Fig S1). Moreover, loss of genomic regions 2q21.2-q22.1, 5q12.1, 5q14.3-q15, -8p23.3- q24.3 and 16q23.1 were observed (Table 4 & Fig S1). These cytogenetic gains and losses were concordant with the loss and gain of genes that have been previously reported (33) in these cell lines.
Table 4. Array comparative genomic hybridization analysis of the MCF10 breast cancer progression model cell lines.
| Cell line | aCGH | |
|---|---|---|
| Gains | Losses | |
| MCF10 A | +5q23.1-35.3,+8p23.3-q24.3, +13q32.1-p32.2, +19q13.11q13.43 | -3p26.3, -9p21.3, -16p11.2, -21p11-q11.2,-22q11.1 |
| MCF10AT1 | +3p14.3, +3q13.31, +9p24.3-11.2, +9q12-q34.3, +10q22.1-q22.2, +16q23.3,+17p11.2 | -5q12.1,-5q14.3-15,-8p23.3-q24.3,-15q21.1 |
| MCF10CA1a | +2p25.3-q21.2, +3p14.1-q29, +9p24.3-p11.2, +9q34.3-q34.13, +10q11.1-q26.3, +17p11.2, +20p13-q13.33 | -2q21.2-q22,-5q12.1,-5q14.3-q15, -8p23.3-q24.3, -16q23.1 |
Correlation of chromosomal gains and losses with gene expression
Gene expression changes for 40 genes found in 36 different genomic regions showing gains or losses in MCF10CA1a are shown in Table S4. The ratio of genomic gains range from 1.18 to 1.49, while losses range from 0.71 to 0.77. We also identified seven genes that are important in breast cancer within the regions of genomic loss/gain in MCF10CA1a (Table S4, double asterisks). These genes include carbonic anhydrase VIII (CA8), sulfatase 1 (SULF1), chromosome 8 open reading frame 13 (C8orf13), chromosome 2 open reading frame 32 (C2orf32), tumor-associated calcium signal transducer 1 (TACSTD1), type IX collagen (COL9A3) and tissue plasminogen activator (PLAT). These genes are located within the chromosome bands 8q12.1, 8q13.2, 8p23.1, 2p14, 2p21, 20q13.3 and 8p11.21, respectively. Among the genes found within genomic gain regions, 13 were up-regulated and 18 were down regulated while in the loss regions, 3 genes were down regulated and 6 genes were up regulated (Table S4). This indicated that in many cases there was no direct relationship between gains or losses of chromosomal regions and gene expression in those regions.
We further investigated this question at the global level of gene expression. ∼1000 genes were identified that showed both changes in genomic gains or losses and at least a twofold change in their expression levels (Table S5). To our surprise, we found that all 6 regions showing chromosomal gains had a much higher number of down regulated genes and 3 of 5 regions with genomic losses showed higher numbers of up regulated genes (Table 5). Of the 701 genes identified in the genomic gains regions, 76% were down regulated while 56% of the 301 genes identified in the loss regions were up regulated (Tables 5 and S5).
Table 5. Relationships between genomic gains and losses and gene expression in MCF10CA1a.
| Region | # of up regulated genes | # of down regulated genes |
|---|---|---|
| Gains: | ||
| +2p25.3-q21.2 | 44 | 122 |
| +3p4.1- q29 | 60 | 142 |
| +9p24.3-p11.2 | 11 | 43 |
| +9q34.13-q34.3 | 3 | 24 |
| +10q11.1-q26.3 | 48 | 170 |
| +7p11.2 | 3 | 31 |
| Losses: | ||
| -2q21.2-q22 | 10 | 13 |
| -5q12.1 | 3 | 2 |
| -5q14.3-q15 | 16 | 9 |
| -8p23.3-q24.3 | 150 | 85 |
| -16q23.1 | 4 | 9 |
Discussion
Cancer progression models provide an approach to elucidate the intermediate molecular and genetic changes that ultimately lead to the transformation of a normal cell into a metastatic cell type. In this present study we utilized a combination of SKY and CGH methods to identify regions of chromosomal aberrations in the MCF10 human breast cancer progression model cell lines (9). Gene expression analysis was also performed in the immortalized normal MCF10A and malignant MCF10CA1a cell lines as a step towards identifying novel candidate protein factors that potentially correlate with cancer metastasis. In addition, since cancer is characterized by numerous alterations in the genome compared to normal cells, we have attempted to correlate changes in gene expression with genomic gains or losses of the individual genes in the malignant cell line.
About 8000 genes were identified that showed a two-fold difference in expression level (Table S2). We further classified genes based on their relevance to breast cancer (Table 1). Consistent with previous reports in breast cancer, we detected up-regulation in MCF10CA1a of fibronectin 1 (FN1) and matrix metalloproteinase 2 (MMP2) (34, 35). In contrast, while overexpression of selenoprotein plasma protein (SEPP1) and decorin (DCN) have been shown to have an antimetastatic role in breast cancer (23, 24), these genes are highly expressed in the malignant MCF10CA1a line compared to MCF10A (Table 1). While numerous studies demonstrated that over expression or amplification of the genes ERBB2 and EGFR promotes breast cancer tumorogenesis (36), we find that they are significantly down regulated in the MCF10CA1a cells (Tables 2 &3). Repression of the HRAS gene in MCF10CA1a is consistent with the over-expression of a mutant form of this protein engineered into this cell line (7).
Genes such as myelocytomatosis viral oncogene (MYC), phosphatase and tensin (PTEN), breast cancer 2 (BRCA2) (Table 2) that are involved in cell cycle control and DNA repair are down regulated. Germ line mutations in BRCA2 predisposes humans towards development of inherited breast cancer (4) and studies on PTEN showed that deregulation of this gene is associated with increased cell proliferation and tumorogenicity (37). Cytokines including interleukin A (ILA) and interleukin B (ILB) are highly down regulated in MCF10CA1a (Table 2) despite their reported contribution to the invasiveness and aggressive phenotype of breast cancer (38). In contrast, interleukin 7 (IL7), which has also been implicated in cancer progression (39), is significantly up regulated in MCF10CA1a (Table 3).
Previous SKY demonstrated a variety of chromosomal rearrangements in breast cancer (11). In particular the reciprocal translocation involving chromosomes 3 and 9 (t (3;9) (p14;p21)) identified in our analysis has been described as the single most prominent event in the transformation of this cell type (32, 33). We also observed several other translocations involving chromosomes 2, 3,5,10 and 17. Aberrations in chromosomes 3 and 17 were previously reported in breast cancer (40). Chromosome 17 has several putative breakpoints that might be associated with higher frequency of translocations of this chromosome. A gain of chromosome 8 was observed in MCF10A cells which was lost in the subsequent premalignant (MCF10AT1) and malignant (MCF10CA1a) progression cell lines. We also identified a gain of an entire chromosome 20 in the malignant breast cell line. Consistent with these findings, chromosomal rearrangements and amplification of chromosome 8 (41) and chromosome 20 (42) have been observed frequently in breast cancer.
Using aCGH, the parental MCF10A breast cell line showed gains in 5q23.1-35.3, 19q13.11q13.43, 13q32.1-p32.2 regions and loss of 9p21.3, 3p26.3, 16p11.2, 21p11-q11.2 and 22q11.1 regions in comparison to a normal diploid cell line (Fig S1 and Table 4). The loss of the 9p21.3 region is consistent with the findings that a deletion in this region was associated with the t (3;9) translocation event that lead to immortalization of this cell line (32). A gain was also detected in the 8p23.3-q24.3 region which is consistent with the gain of an additional copy of chromosome 8 in the SKY analysis. Gain of 3p14.3, 3q13.3, 19q12-q34.3, 10q22.1-q22.2, 16q23.3, 17p11.2 and loss of 5q12.1, 5q14.3-15, and 15q21.1 were observed in the premalignant MCF10AT1 cell line (Table 4 and Fig S1). Cytogenetic changes in chromosome 17 (43) and chromosome 5 (44) have been reported in other studies. In particular, losses in the 5q13-q23.3 region have been associated with breast cancers showing a mutation for the BRCA1 gene (44). This region has also been shown to harbor several putative tumor suppressor genes.
Additional changes in MCF10CA1a include gains of 2p25.3-q21.2, 3p14.1-q29, 9p24.3-p11.2, 9p34.13-p34.3, 10q11.1-q26.3, 20p13-q13.3 and losses of genomic regions 2q21.2-q22.1, 5q12.1, 8p23.3-q24.21, and 16q23.1 (Table 4, Fig S1). The additional chromosome 8 seen in MCF10A was deleted in both the premalignant and malignant cell lines. Deletions of certain genomic regions or the entire chromosome 8 are in agreement with previous studies on chromosome 8 abnormalities in breast cancer (45). The region 8p11-12 harbors about 21 potential oncogenes and is frequently amplified in breast cancer (46).The results of SKY and CGH were in concurrence in nearly all instances thereby substantiating the results from both approaches.
A careful analyses of the aCGH findings revealed that almost all the sub-chromosomal genomic gains and losses were in those chromosomes that were identified as aberrant by the SKY method. Thus combining both these approaches in our cell system has allowed us to identify large-scale genomic aberrations and ploidy changes along with regions of sub-chromosomal abnormalities that might occur in the transformation of a normal cell to a cancerous cell. Overall our SKY and CGH results indicate that a hallmark of progression towards breast cancer involves gradual accumulation of chromosomal aberrations in normal breast cells.
We further correlated genomic alterations with gene expression profiles of normal and malignant MCF10A cell lines. Regions within the MCF10CA1a cell line identified for gains and losses by aCGH were examined for changes in levels of gene expression (Tables 5 and S4). The majority of the ∼1,000 genes investigated showed a lack of direct correlation between the chromosomal aberrations and its expression (Table 5). Previous studies also reported a weak, or in some cases, a complete lack of correlation between genomic gains and losses and gene expression levels in cancer cells (47). Despite this we identified several genes in these regions that are related to breast cancer and show large changes in gene expression in the malignant MCF10CA1a versus the MCF10A breast epithelial cells (Table S4).
Our results, predict that progression towards cancer is a combination of genomic instability as well as gene deregulation. Chromosomal aberrations such as amplifications, deletions and complex chromosomal translocations are a hallmark of solid tumors and are generally believed to occur via telomere dysfunction and the breakage-fusion-bridge mechanism (48). Changes in gene copy number that occur due to the chromosomal aberrations lead to a disruption of the normal expression pattern of the genes therefore causing transformation of a normal cell towards a malignant phenotype.
In conclusion, our combined approach employing SKY, aCGH, and microarray gene expression analysis identified a number of important chromosomal regions and genes whose structural and functional alterations might be involved in the progression and development of breast cancer. Importantly, several of these genomic regions that have been previously shown to be amplified or deleted in breast cancer (49). While the expression levels of many genes agreed with previously published results in breast cancer, there were some significant differences. This could relate to the multiple changes and likely variability in gene expression that occur during cancer progression. Thus a variety of combinations of expression changes in key regulatory factors may lead to the same end of a highly malignant tumor.
We also report a lack of correlation between chromosomal aberrations in cancer and expression patterns of genes within those regions. Other mechanisms of gene regulation are likely involved including epigenetic processes such as methylation (50), histone modification (50) and expression of non coding RNA's (50). Methylation of CpG residues and histone tail modifications are faithfully propagated from one cell generation to another under normal circumstances and any change might lead the cell to become metastatic. In cancer cells, methylation of cytosine in the CpG dinucleotides occurs in the promoter regions which leads to gene inactivation (50). Several genes that are important in cell cycle control, DNA repair, apoptosis have been found to undergo hypermethylation thereby promoting the development of a cancerous phenotype (50). Modification of histone tails such as acetylation, deacteylation and methylation are also known to influence gene expression (50) and in cancer abnormal histone modifications occur that have an effect on gene transcription (50). Recent investigations also demonstrate a role of microRNAs in repression of certain important genes in cancer (50). Our studies, therefore, add to the growing evidence that amplification or deletions of genomic regions are not the sole mechanism for altering gene expression in progression towards cancer.
Supplementary Material
Acknowledgments
This research was supported by a grant from the National Institute of Health (GM-072131) to R Berezney.
References
- 1.Bray F, McCarron P, Parkin DM. The changing global patterns of female breast cancer incidence and mortality. Breast Cancer Res. 2004;6(6):229–39. doi: 10.1186/bcr932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ellsworth RE, Vertrees A, Love B, Hooke JA, Ellsworth DL, Shriver CD. Chromosomal alterations associated with the transition from in situ to invasive breast cancer. Ann Surg Oncol. 2008;15(9):2519–25. doi: 10.1245/s10434-008-0051-7. [DOI] [PubMed] [Google Scholar]
- 3.Reis-Filho JS, Savage K, Lambros MB, et al. Cyclin D1 protein overexpression and CCND1 amplification in breast carcinomas: an immunohistochemical and chromogenic in situ hybridisation analysis. Mod Pathol. 2006;19(7):999–1009. doi: 10.1038/modpathol.3800621. [DOI] [PubMed] [Google Scholar]
- 4.Teng LS, Zheng Y, Wang HH. BRCA1/2 associated hereditary breast cancer. J Zhejiang Univ Sci B. 2008;9(2):85–9. doi: 10.1631/jzus.B0710617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pauley RJ, Soule HD, Tait L, et al. The MCF10 family of spontaneously immortalized human breast epithelial cell lines: models of neoplastic progression. Eur J Cancer Prev. 1993;2 3:67–76. [PubMed] [Google Scholar]
- 6.Soule HD, Maloney TM, Wolman SR, et al. Isolation and characterization of a spontaneously immortalized human breast epithelial cell line, MCF-10. Cancer Res. 1990;50(18):6075–86. [PubMed] [Google Scholar]
- 7.Basolo F, Elliott J, Tait L, et al. Transformation of human breast epithelial cells by c-Ha-ras oncogene. Mol Carcinog. 1991;4(1):25–35. doi: 10.1002/mc.2940040106. [DOI] [PubMed] [Google Scholar]
- 8.Dawson PJ, Wolman SR, Tait L, Heppner GH, Miller FR. MCF10AT: a model for the evolution of cancer from proliferative breast disease. Am J Pathol. 1996;148(1):313–9. [PMC free article] [PubMed] [Google Scholar]
- 9.Santner SJ, Dawson PJ, Tait L, et al. Malignant MCF10CA1 cell lines derived from premalignant human breast epithelial MCF10AT cells. Breast Cancer Res Treat. 2001;65(2):101–10. doi: 10.1023/a:1006461422273. [DOI] [PubMed] [Google Scholar]
- 10.Macville M, Veldman T, Padilla-Nash H, et al. Spectral karyotyping, a 24-colour FISH technique for the identification of chromosomal rearrangements. Histochem Cell Biol. 1997;108(45):299–305. doi: 10.1007/s004180050169. [DOI] [PubMed] [Google Scholar]
- 11.Goodison S, Viars C, Urquidi V. Molecular cytogenetic analysis of a human breast metastasis model: identification of phenotype-specific chromosomal rearrangements. Cancer Genet Cytogenet. 2005;156(1):37–48. doi: 10.1016/j.cancergencyto.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 12.Kallioniemi A, Kallioniemi OP, Sudar D, et al. Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science. 1992;258(5083):818–21. doi: 10.1126/science.1359641. [DOI] [PubMed] [Google Scholar]
- 13.Jonsson G, Staaf J, Olsson E, et al. High-resolution genomic profiles of breast cancer cell lines assessed by tiling BAC array comparative genomic hybridization. Genes Chromosomes Cancer. 2007;46(6):543–58. doi: 10.1002/gcc.20438. [DOI] [PubMed] [Google Scholar]
- 14.DeRisi J, Penland L, Brown PO, et al. Use of a cDNA microarray to analyse gene expression patterns in human cancer. Nat Genet. 1996;14(4):457–60. doi: 10.1038/ng1296-457. [DOI] [PubMed] [Google Scholar]
- 15.Nuyten DS, van de Vijver MJ. Using microarray analysis as a prognostic and predictive tool in oncology: focus on breast cancer and normal tissue toxicity. Semin Radiat Oncol. 2008;18(2):105–14. doi: 10.1016/j.semradonc.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 16.Nowak NJ, Snijders AM, Conroy JM, Albertson DG. The BAC resource: tools for array CGH and FISH. Curr Protoc Hum Genet. 2005 doi: 10.1002/0471142905.hg0413s46. Chapter 4:Unit 4 13. [DOI] [PubMed] [Google Scholar]
- 17.Nowak NJ, Gaile D, Conroy JM, et al. Genome-wide aberrations in pancreatic adenocarcinoma. Cancer Genet Cytogenet. 2005;161(1):36–50. doi: 10.1016/j.cancergencyto.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 18.Cowell JK, Wang YD, Head K, Conroy J, McQuaid D, Nowak NJ. Identification and characterisation of constitutional chromosome abnormalities using arrays of bacterial artificial chromosomes. Br J Cancer. 2004;90(4):860–5. doi: 10.1038/sj.bjc.6601588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olshen AB, Venkatraman ES, Lucito R, Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5(4):557–72. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- 20.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- 21.Beissbarth T, Speed TP. GOstat: find statistically overrepresented Gene Ontologies within a group of genes. Bioinformatics. 2004;20(9):1464–5. doi: 10.1093/bioinformatics/bth088. [DOI] [PubMed] [Google Scholar]
- 22.Squires J, Berry MJ. Selenium, selenoproteins, and cancer. Hawaii Med J. 2006;65(8):239–40. [PubMed] [Google Scholar]
- 23.Baliga MS, Wang H, Zhuo P, Schwartz JL, Diamond AM. Selenium and GPx-1 overexpression protect mammalian cells against UV-induced DNA damage. Biol Trace Elem Res. 2007;115(3):227–42. doi: 10.1007/BF02685998. [DOI] [PubMed] [Google Scholar]
- 24.Goldoni S, Seidler DG, Heath J, et al. An antimetastatic role for decorin in breast cancer. Am J Pathol. 2008;173(3):844–55. doi: 10.2353/ajpath.2008.080275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tseleni-Balafouta S, Gakiopoulou H, Fanourakis G, et al. Fibrillin expression and localization in various types of carcinomas of the thyroid gland. Mod Pathol. 2006;19(5):695–700. doi: 10.1038/modpathol.3800578. [DOI] [PubMed] [Google Scholar]
- 26.Wright RM, McManaman JL, Repine JE. Alcohol-induced breast cancer: a proposed mechanism. Free Radic Biol Med. 1999;26(34):348–54. doi: 10.1016/s0891-5849(98)00204-4. [DOI] [PubMed] [Google Scholar]
- 27.Wang D, Dubois RN. Prostaglandins and cancer. Gut. 2006;55(1):115–22. doi: 10.1136/gut.2004.047100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hayes AJ, Huang WQ, Yu J, et al. Expression and function of angiopoietin-1 in breast cancer. Br J Cancer. 2000;83(9):1154–60. doi: 10.1054/bjoc.2000.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nollet F, Berx G, van Roy F. The role of the E-cadherin/catenin adhesion complex in the development and progression of cancer. Mol Cell Biol Res Commun. 1999;2(2):77–85. doi: 10.1006/mcbr.1999.0155. [DOI] [PubMed] [Google Scholar]
- 30.Pietas A, Schluns K, Marenholz I, Schafer BW, Heizmann CW, Petersen I. Molecular cloning and characterization of the human S100A14 gene encoding a novel member of the S100 family. Genomics. 2002;79(4):513–22. doi: 10.1006/geno.2002.6744. [DOI] [PubMed] [Google Scholar]
- 31.Greco S, Elia MG, Muscella A, Romano S, Storelli C, Marsigliante S. Bradykinin stimulates cell proliferation through an extracellular-regulated kinase 1 and 2-dependent mechanism in breast cancer cells in primary culture. J Endocrinol. 2005;186(2):291–301. doi: 10.1677/joe.1.06052. [DOI] [PubMed] [Google Scholar]
- 32.Cowell JK, LaDuca J, Rossi MR, Burkhardt T, Nowak NJ, Matsui S. Molecular characterization of the t(3;9) associated with immortalization in the MCF10A cell line. Cancer Genet Cytogenet. 2005;163(1):23–9. doi: 10.1016/j.cancergencyto.2005.04.019. [DOI] [PubMed] [Google Scholar]
- 33.Worsham MJ, Pals G, Schouten JP, et al. High-resolution mapping of molecular events associated with immortalization, transformation, and progression to breast cancer in the MCF10 model. Breast Cancer Res Treat. 2006;96(2):177–86. doi: 10.1007/s10549-005-9077-8. [DOI] [PubMed] [Google Scholar]
- 34.Christensen L. The distribution of fibronectin, laminin and tetranectin in human breast cancer with special attention to the extracellular matrix. APMIS Suppl. 1992;26:1–39. [PubMed] [Google Scholar]
- 35.Mendes O, Kim HT, Lungu G, Stoica G. MMP2 role in breast cancer brain metastasis development and its regulation by TIMP2 and ERK1/2. Clin Exp Metastasis. 2007;24(5):341–51. doi: 10.1007/s10585-007-9071-0. [DOI] [PubMed] [Google Scholar]
- 36.Moasser MM. The oncogene HER2: its signaling and transforming functions and its role in human cancer pathogenesis. Oncogene. 2007;26(45):6469–87. doi: 10.1038/sj.onc.1210477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maehama T. PTEN: its deregulation and tumorigenesis. Biol Pharm Bull. 2007;30(9):1624–7. doi: 10.1248/bpb.30.1624. [DOI] [PubMed] [Google Scholar]
- 38.Balasubramanian SP, Azmy IA, Higham SE, et al. Interleukin gene polymorphisms and breast cancer: a case control study and systematic literature review. BMC Cancer. 2006;6:188. doi: 10.1186/1471-2407-6-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Al-Rawi MA, Mansel RE, Jiang WG. Interleukin-7 (IL-7) and IL-7 receptor (IL-7R) signalling complex in human solid tumours. Histol Histopathol. 2003;18(3):911–23. doi: 10.14670/HH-18.911. [DOI] [PubMed] [Google Scholar]
- 40.Anamthawat-Jonsson K, Eyfjord JE, Ogmundsdottir HM, Petursdottir I, Steinarsdottir M. Instability of chromosomes 1, 3, 16, and 17 in primary breast carcinomas inferred by fluorescence in situ hybridization. Cancer Genet Cytogenet. 1996;88(1):1–7. doi: 10.1016/0165-4608(95)00203-0. [DOI] [PubMed] [Google Scholar]
- 41.Mark HF, Taylor W, Afify A, et al. Stage I and stage II infiltrating ductal carcinoma of the breast analyzed for chromosome 8 copy number using fluorescent in situ hybridization. Pathobiology. 1997;65(4):184–9. doi: 10.1159/000164121. [DOI] [PubMed] [Google Scholar]
- 42.Hodgson JG, Chin K, Collins C, Gray JW. Genome amplification of chromosome 20 in breast cancer. Breast Cancer Res Treat. 2003;78(3):337–45. doi: 10.1023/a:1023085825042. [DOI] [PubMed] [Google Scholar]
- 43.Andersen CL, Monni O, Wagner U, et al. High-throughput copy number analysis of 17q23 in 3520 tissue specimens by fluorescence in situ hybridization to tissue microarrays. Am J Pathol. 2002;161(1):73–9. doi: 10.1016/S0002-9440(10)64158-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Johannsdottir HK, Jonsson G, Johannesdottir G, et al. Chromosome 5 imbalance mapping in breast tumors from BRCA1 and BRCA2 mutation carriers and sporadic breast tumors. Int J Cancer. 2006;119(5):1052–60. doi: 10.1002/ijc.21934. [DOI] [PubMed] [Google Scholar]
- 45.Forozan F, Mahlamaki EH, Monni O, et al. Comparative genomic hybridization analysis of 38 breast cancer cell lines: a basis for interpreting complementary DNA microarray data. Cancer Res. 2000;60(16):4519–25. [PubMed] [Google Scholar]
- 46.Yang ZQ, Streicher KL, Ray ME, Abrams J, Ethier SP. Multiple interacting oncogenes on the 8p11-p12 amplicon in human breast cancer. Cancer Res. 2006;66(24):11632–43. doi: 10.1158/0008-5472.CAN-06-2946. [DOI] [PubMed] [Google Scholar]
- 47.Jiang M, Li M, Fu X, et al. Simultaneously detection of genomic and expression alterations in prostate cancer using cDNA microarray. Prostate. 2008;68(14):1496–509. doi: 10.1002/pros.20756. [DOI] [PubMed] [Google Scholar]
- 48.Chin K, de Solorzano CO, Knowles D, et al. In situ analyses of genome instability in breast cancer. Nat Genet. 2004;36(9):984–8. doi: 10.1038/ng1409. [DOI] [PubMed] [Google Scholar]
- 49.Hampton OA, Den Hollander P, Miller CA, et al. A sequence-level map of chromosomal breakpoints in the MCF-7 breast cancer cell line yields insights into the evolution of a cancer genome. Genome Res. 2009 doi: 10.1101/gr.080259.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Clark SJ. Action at a distance: epigenetic silencing of large chromosomal regions in carcinogenesis. Hum Mol Genet. 2007;16:R88–95. doi: 10.1093/hmg/ddm051. Spec No 1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.