Abstract
Background and Aims
Interspecific gene flow can occur in many combinations among species within the genus Quercus, but simultaneous hybridization among more than two species has been rarely analysed. The present study addresses the genetic structure and morphological variation in a triple hybrid zone formed by Q. hypoleucoides, Q. scytophylla and Q. sideroxyla in north-western Mexico.
Methods
A total of 247 trees from ten reference and 13 presumed intermediate populations were characterized using leaf shape variation and geometric morphometrics, and seven nuclear microsatellites as genetic markers. Discriminant function analysis was performed for leaf shape variation, and estimates of genetic diversity and structure, and individual Bayesian genetic assignments were obtained.
Key Results
Reference populations formed three completely distinct groups according to discriminant function analysis based on the morphological data, and showed low, but significant, genetic differentiation. Populations from the zone of contact contained individuals morphologically intermediate between pairs of species in different combinations, or even among the three species. The Bayesian admixture analysis found that three main genetic clusters best fitted the data, with good correspondence of reference populations of each species to one of the genetic clusters, but various degrees of admixture evidenced in populations from the contact area.
Conclusions
The three oak species have formed a complex hybrid zone that is geographically structured as a mosaic, and comprising a wide range of genotypes, including hybrids between different species pairs, backcrosses and probable triple hybrids.
Keywords: Altitudinal cline, hybridization, introgression, leaf shape variation, Mexico, nuclear microsatellites, Quercus scytophylla, Quercus sideroxyla, Quercus hypoleucoides, red oak
INTRODUCTION
Hybrid zones are considered an important source of genetic recombination and diversity in plant evolution (Arnold, 1997; Rieseberg, 1997). These zones are most often the product of secondary contact between populations or species that have differentiated previously in allopatry, or may also arise in situ in response to spatially varying selection (Durrett et al., 2000). Nearly all of the hybrid zones that have been so far studied involve two parental taxa and their hybrids. Nevertheless, more complex instances of simultaneous hybridization among three or more parental taxa also occur in nature, but these have been rarely analysed (Arnold, 1993; Kaplan and Fehrer, 2007). Consequently, the genetic structure and the dynamics of gene flow in multispecies hybrid zones are poorly known (Dodd and Afzal-Rafii, 2004; Curtu et al., 2007; Kaplan and Fehrer, 2007; Lepais et al., 2009). These situations require the production of some fertile hybrid genotypes (F1 and/or backcrosses) between at least one species pair. Subsequently, these individuals may disperse throughout the hybrid zone and crosses can occur between hybrids and a third species, or between hybrids from different species combinations (Kaplan and Fehrer, 2007).
According to theoretical models, the evolution of hybrid populations will depend on the relative fitness of the various hybrid and parental genotypes (Barton and Gale, 1993; Arnold, 1997). Nevertheless, the nature and intensity of selective factors are not necessarily homogeneous across space. Both exogenous and endogenous selection have been invoked to explain the structure of hybrid zones (Barton and Hewitt, 1985; Moore and Price, 1993). Exogenous selection implies adaptation to local environments, whereas endogenous selection occurs when hybrids have low fitness due to incompatibilities between the parental genomes (Bronson et al., 2003). Therefore, the selective pressures acting on hybrids may result from the combined effects of environmental factors such as climate, soil and interactions with other organisms (Fitzpatrick and Schaffer, 2004; James and Abbott, 2005; Raudnitschka et al., 2007), and factors independent of environmental variation such as the disruption of co-adapted gene complexes and the action of genes associated with sterility (Barton and Gale, 1993; Rieseberg and Wendell, 1993). If the main selection pressures are exogenous, the likely result is a clinal or mosaic hybrid zone, in which certain genotypes could be spatially segregated according to the distribution of habitats (Endler, 1977). In contrast, if the selection pressures are endogenous, the generation of tension zones is expected, wherein clines are maintained by the equilibrium between the movement of parental individuals and selection against the hybrid genotypes (Barton and Gale, 1993). However, it is not clear which of these models best describe multispecies hybrid zones (Dodd and Afzal-Rafii, 2004).
Although a high frequency of interespecific gene flow in many combinations has been inferred within the genus Quercus from morphological variation, relatively few studies have used genetic markers considering several species simultaneously (Whittemore and Schaal, 1991; Dumolin-Lapègue et al., 1999; Dodd and Afzal-Rafii, 2004; Curtu et al., 2007; Lepais et al., 2009). Of particular interest has been the detection of widespread cytoplasmic introgression even among distantly related species at local scales (Whittemore and Schaal, 1991; Dumolin-Lapègue et al., 1999). Nevertheless, few studies have been conducted using nuclear microsatellites, which allow the identification of different genealogical classes and detailed characterization of the genetic structure of hybrid zones (i.e. Gugerli et al., 2008; Lepais et al., 2009).
Mexico is considered a centre of species diversification for the genus Quercus, with 161 species (32–40 % of the worldwide diversity). In particular, 76 species of red oaks have been reported in Mexico, including 61 endemic species (Valencia, 2004). In recent years, several studies conducted on Mexican red oaks have focused on hybridization between two species (Valencia and Delgado, 2003; González-Rodríguez et al., 2004, 2005; Tovar-Sánchez and Oyama, 2004; Tovar-Sánchez et al., 2008). However, several oak species complexes comprising three or more species that probably hybridize simultaneously have been described in Mexico (Dodd and Kashani, 2003; McCauley et al., 2007). In this context, understanding of the evolutionary dynamics and consequences of hybridization requires the consideration of such multispecies interactions as this is the way the phenomenon occurs in nature. The present study investigated a complex of three red oak species formed by Q. hypoleucoides, Q. scytophylla and Q. sideroxyla. These species are well delimited by diagnostic morphological characters, although some individuals exhibit morphological intermediacy in different combinations in several localities across the Sierra Tarahumara in north-west Mexico where the species coexist. In this region, the species overlap partially in distribution along an altitudinal gradient (1800–2500 m) and also in the timing of their flowering periods (from March to August) (Valencia, 2004).
The purpose of this study was to analyse this multispecies oak hybrid zone by combining morphometric analysis of leaf shape and nuclear microsatellite molecular markers. The particular questions addressed here were: (1) Do hybridization and introgression occur between Q. scytophylla, Q. sideroxyla and Q. hypoleucoides?; (2) What is the geographical structure of morphological and genetic variation among the three species in the contact area; and (3) are there associations between molecular and phenotypic variation with the altitudinal gradient encompassed by the hybrid zone?
MATERIALS AND METHODS
Study species
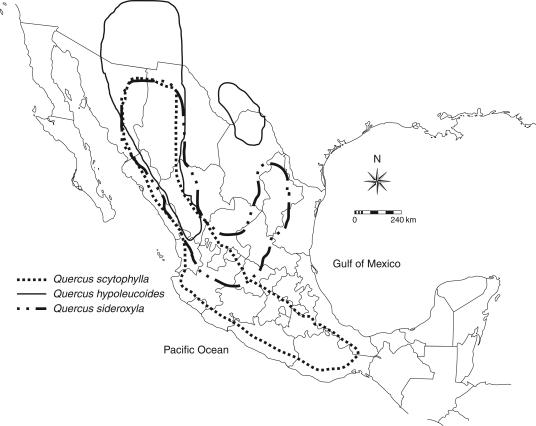
The three species belong to the red oak section Lobatae (Nixon, 1993). Quercus scytophylla Liebm. is a tree about 20 m in height included in the series Scytophyllae (Trelease, 1924). The leaves of this species are elliptical or obovate, 5–17 cm in length and 2·5–8 cm in width, with 1–6 teeth with aristae. The petioles are 9–35 mm in length. Acorns are produced singly or in groups and have a peduncle 3–10 mm in length, with maturation periods of 2 years. It is the most widely distributed of the three species in the complex, present in the Sierra Madre Occidental, the Eje Neovolcánico Transversal and the Sierra Madre del Sur, at altitudes between 1400 and 2500 m (Fig. 1). Quercus sideroxyla Humb & Bonpl is a tree about 10 m in height that belongs to series Sideroxylae (Trelease, 1924), with populations occurring in the Sierra Madre Occidental and the Eje Neovolcánico Transversal, with an altitudinal distribution from 1800 to 2700 m (Fig. 1). It has obovate or oblanceolate leaves 3–6 cm in length and 2·5–3·5 cm in width, with 1–5 distal teeth with aristae, and flattened petioles 3–9 mm long. Fruits are biennial, almost sessile, and solitary or produced in pairs. Finally, Quercus hypoleucoides Camus is a tree 10–20 m in height included in series Hypoleucae (Trelease, 1924), and has a narrower geographical distribution than the other two species, with populations in the north of the Sierra Madre Occidental and in Arizona, New Mexico and Texas, at altitudes ranging from 2000 to 2500 m (Fig. 1). The species has narrow leaves that are lanceolate, elliptical or obovate, 5–11 cm long and 1·5–3 cm wide, with entire margins. Acorns are produced annually or biennially, are sessile or almost sessile and solitary or produced in pairs.
Fig. 1.
Geographical distribution of the three red-oak species studied, Quercus scytophylla, Q. hypoleucoides and Q. sideroxyla.
Sampling procedure
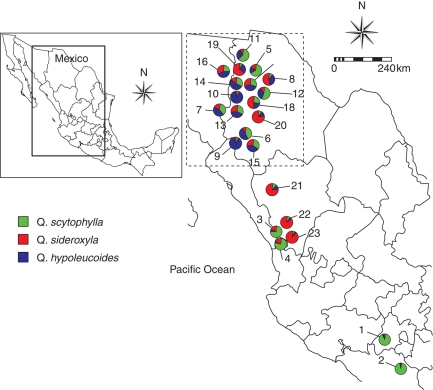
Leaf samples were collected in 13 populations located in the zone of contact (Fig. 2). As reference, two morphologically representative populations situated within the zone of contact were also sampled, and two representative isolated populations situated out of the zone of contact were sampled per species (Fig. 2), with the exception of Q. hypoleucoides, because its geographical distribution within Mexico is restricted to the study zone. Representative populations were chosen based on individuals with typical diagnostic characters of each species. In each locality, 8–12 individuals sampled haphazardly were chosen but at least 50 m apart from each other. Three branches from each individual were collected: one for taxonomic identification, one for morphometric analysis and the third to obtain fresh intact leaves for molecular analysis, which were placed on ice and then at –80 °C until genetic analyses were conducted.
Fig. 2.
Map showing sampling localities representing parental and intermediate populations. Each pie chart represents the proportions in each population of the three genetic groups as assigned by the program STRUCTURE. Green, red and blue represent the genetic groups corresponding to Quercus scytophylla, Q. sideroxyla and Q. hypoleucoides, respectively. Numbers next to each symbol correspond to the population numbers given in Table 1. The area of species contact is delineated with a dashed rectangle.
Leaf shape morphometric analysis
Photographs were taken of the abaxial side of five intact mature light-exposed leaves from each of eight to ten individuals per population. Coordinates ‘x, y’ of 21 unambiguous and repeatable marks (i.e. landmarks and semi-landmarks) were registered along the border of each leaf image using the program TpsDig (Rohlf, 2004). Three marks (apex, lamina base and petiole extreme) corresponded to homologous loci (‘landmarks’ sensu Bookstein, 1991), and the other 18 were semi-landmarks, i.e. morphological points that incorporate information about leaf contour in morphometric analysis, in areas that lack landmarks for all the individuals (Zelditch et al., 2004). Landmarks ‘1’ (lamina base) and ‘12’ (apex) were used to construct a ‘fan’ (radial guidelines with equal angular spacing on images) with 80 radial guidelines covering the whole leaf contour, which was used to digitize the 18 semi-landmarks. The program MakeFan6 within the IMP software package (Integrated Morphometrics Package; http://www.canisius.edu/~sheets/morphsoft.html) was used for this procedure. A Procrustes superimposition analysis was developed with the CoordGen program in IMP. This analysis allows the calculation of leaf shape variation without the effect of the size. The resulting shape variables (Procrustes distances) for all individuals were subjected to a canonical discriminant analysis to determine the variation in leaf shape among individuals of each species and intermediate populations using SPSS 11·0 (Ferran, 1997). Individuals from reference populations of the three species were first analysed to obtain the canonical discriminant functions, and then the discriminant scores calculated with these functions were obtained for individuals from all populations. In this way, five morphological groups were identified, three of which corresponded to pure species individuals and two corresponded to intermediate individuals (see Results).
Microsatellite amplification
Genomic DNA was extracted from 100 mg of leaf material using the method proposed by Lefort and Douglas (1999). Seven nuclear microsatellite loci (OC11, OA01, 1C08, 2M04, OE09, 1H14, 1F07) previously designed for Quercus rubra (Aldrich et al., 2002) were selected based on the quality of preliminary amplification trials. Polymerase chain reactions were carried out in a volume of 25 µL containing 20 ng of template DNA, 2 mm MgCl2, 10 mm Tris-HCl (pH 9), 0·1 mm of each dNTP, 0·5 mg mL−1 bovine serum albumin, 2 µm of each primer and 0·3 units of TaqDNA polymerase (Gibco, Invitrogen, San Diego, CA, USA). The thermal cycling conditions consisted of 40 cycles, each at 94 °C for 1 min, annealing at 50 °C for 1 min and extension at 72 °C for 2 min. A final extension at 72 °C for 10 min was included. Amplified fragments were separated in polyacrylamide gels (6 %) in a semi-automatic sequencer LI-COR 4300 (LI-COR Biosciences, Lincoln, NB, USA). Fragment sizes were calculated using the program ImageJ by comparisons with internal and external standards. All individuals sampled from the 23 populations were genotyped.
Genetic analysis
The mean number of alleles per locus (Na), mean effective number of alleles (Ne), mean observed heterozygosity (HO), mean expected heterozygosity (HE), mean fixation index (F) and their respective standard errors were calculated for each population using FSTAT (Goudet, 1995). Average values were also obtained for each of the five morphological groups defined on the basis of canonical discriminant function analysis. Pairwise genetic differentiation among the five morphological groups was calculated using the method of Weir and Cockerham (1984) with the GENETIX 4 program (Belkhir et al., 2004) with 10 000 permutations for statistical significance. Within each of the morphological groups genetic differentiation among populations was estimated with analyses of molecular variance (AMOVA). The significances of the different variance components were estimated from distributions generated from 10 000 random permutations. These analyses were carried out using ARLEQUIN 3·0 (Excoffier et al., 2005).
Bayesian admixture analysis
The genetic ancestry of each individual was inferred with the program STRUCTURE 2·3·1 (Pritchard et al., 2000; Falush et al., 2003; Hubisz et al., 2009). This program is based on a Bayesian model clustering procedure to determine the proportions of ancestry of individuals derived from multiple populations. In this analysis, different probable values of K (number of genetic groups) were assayed, increasing the probable number from K = 1 to 13 (ten times for each value). The program was run using the admixture model with correlated allelic frequencies without prior population information. For all runs, a length of the burn-in period of 503 iterations was used, followed by 106 Markov chain Monte Carlo iterations. Additionally, the most probable value of K was determined using the maximum value of ΔK, following the rules of Evanno et al. (2005), and by the estimated ln probability of data, lnP(D).
Genotype simulation and assignment
To evaluate the ability of the assignment procedure to recognize accurately the different genotypic classes potentially present in our samples (i.e. pure parental, hybrids between the different pairs of species and triple hybrids), the procedure developed by Vähä and Primmer (2006) was followed. Individuals that in the assignment analysis described above had an assignment coefficient (Q) higher than 0·90 (indicating a high probability of belonging to a single genetic cluster) were used to estimate allelic frequencies of the three species. Thirty-nine such individuals were identified for Q. scytophylla, 21 for Q. hypoleucoides and 44 for Q. sideroxyla. Pure species and hybrid genotypes were then simulated with HYBRIDLAB 1·0 (Nielsen et al., 2006) using the calculated allelic frequencies. Four hundred genotypes were simulated for each species, 37 for the different possible F1 hybrids, 113 for backcrosses and 14 for triple hybrids (triple hybrids were considered the product of crossing an F1 hybrid from a pair of species with a pure individual of the third species). The number of simulated hybrid genotypes is similar to the number observed in the real data. The simulated data were then analysed with the program STRUCTURE, with the same number of genetic groups previously inferred (K = 3), and the same settings. Thereafter, the performance of STRUCTURE in hybrid and pure-bred individual identification was evaluated via the parameters of efficiency (number of individuals correctly assigned), accuracy (proportion of an identified group that truly belongs to that category) and performance (efficiency multiplied by accuracy), following Vähä and Primmer (2006). Finally, the optimal threshold values of Q to assign individuals to the different genotypic categories were determined.
Geographical patterns of variation
To determine if variation patterns in genetic composition and morphology of populations could be explained by geographical variables, we conducted stepwise regression analyses of the population average values of canonical discriminant scores and proportions of genetic ancestry in each of the genetic groups inferred against the latitude, longitude and altitude of the populations.
RESULTS
Leaf shape morphometric analysis
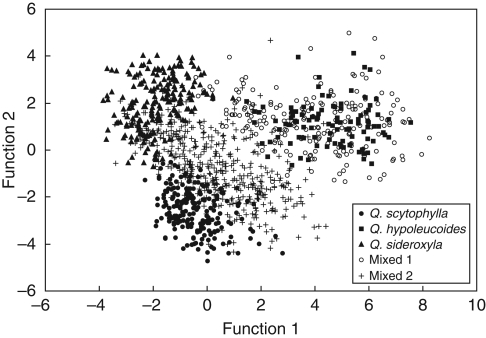
The first two discriminant functions (F1 and F2), derived from the Procrustes analysis of leaf shape, explained all the observed variation (55·1 and 44·9 %, respectively), indicating that discrimination among reference populations of the three species was absolute and highly significant (for F1, Wilks' λ = 0·027, d.f. = 82, P < 0·001; for F2, Wilks' λ = 0·180, d.f. = 40, P < 0·001), supporting the taxonomic delimitation of the three taxa. This is clearly observed in Fig. 3, with the three species forming completely separated clusters. F1 contributed most to the discrimination between Q. hypoleucoides and the other two species, while F2 discriminated between Q. scytophylla and Q. sideroxyla. Individuals from the 13 populations considered to show indications of morphological intermediacy had score values identical to one of the reference clusters or were intermediate either along F1, F2 or both. These populations formed two recognizable groups (Fig. 3). The first group (hereafter called Mixed 1) included individuals from populations Río Chico, Querari and Yécora, most of which were similar to reference individuals of Q. hypoleucoides, and individuals from population Madera that were clearly intermediate between Q. hypoleucoides and Q. sideroxyla. The second group (hereafter Mixed 2) contained individuals similar to Q. scytophylla or intermediate between this species and the other two, and belonged to populations Huracán A, Huracán B, Guadalupe, Km 346, Guachochic, Amarilla B, Poleo, Amarilla A and Babícora.
Fig. 3.
Scatterplot of individual scores from discriminant function analysis of populations in the oak complex based on foliar geometric morphometric data. Closed circles, squares and triangles symbolize representative populations of Quercus scytophylla, Q. hypoleucoides and Q. sideroxyla, respectively, and open circles and crosses are the Mixed 1 and Mixed 2 population groups, respectively. See text for details.
Genetic structure
Seven microsatellite loci revealed high levels of genetic diversity and low but significant genetic differentiation between and within the three species (Tables 1 and 2). The mean number of alleles per locus (Na) within populations varied from 3·71 to 10·42 and the mean of the effective number of alleles (Ne) was between 2·55 and 7·25 (Table 1). High values of HO (ranging from 0·633 to 0·905) and HE (from 0·555 to 0·841) were observed (Table 1). HO, HE, Na and Ne did not differ among the five morphological groups (three reference and two intermediate groups) according to Wilcoxon tests (data not shown). Among 161 inbreeding coefficients (F) calculated (23 populations and seven loci), only four were significant after applying a Bonferroni correction: for locus OA01 in populations Espinazo (F = 0·744, P < 0·00001) and Tecuán (F = 0·226, P < 0·0001), and for locus 2M04 in populations Temosachic (F = −0·337, P < 0·0001) and Tecuán (F = −0·313, P < 0·00001). The program MICROCHECKER (Van Oosterhout et al., 2004) was used to test whether these deviations could be due to the presence of null alleles, stuttering or large allele dropout at those loci. Only population Espinazo showed signs of the presence of a null allele at high frequency (in four of ten individuals) at locus OA01. For further analysis, this locus was coded as missing data in this population.
Table 1.
Name, geographical coordinates, altitude, mean observed heterozygosity (HO), mean expected heterozygosity (HE), mean number of alleles (Na) and mean number of effective alleles per locus (Ne) for 23 populations of Quercus scytophylla, Q. hypoleucoides and Q. sideroxyla
| Genetic diversity |
|||||||
|---|---|---|---|---|---|---|---|
| Number and name of locality | State | Coordinates | Sample size | HO | HE | Na | Ne |
| Q. scytophylla | |||||||
| 1. Tejupilco | México | 19°02′/100°03′ | 9 | 0·889 (0·12) | 0·772 (0·13) | 6·85 (2·96) | 5·46 (0·84) |
| 2. Pozo Largo | Guerrero | 17°36′/99°05′ | 10 | 0·742 (0·19) | 0·810 (0·05) | 7·14 (2·05) | 5·61 (0·55) |
| 3. Espinazo | Sinaloa | 23°34′/105°05′ | 10 | 0·814 (0·29) | 0·805 (0·08) | 8·14 (2·26) | 5·83 (0·77) |
| 4. Loberas | Durango | 23°29′/105°51′ | 10 | 0·857 (0·12) | 0·832 (0·03) | 8·42 (0·61) | 6·25 (0·49) |
| Mixed 1 | |||||||
| 5. Río Chico | Chihuahua | 29°36/108°10′ | 12 | 0·657 (0·35) | 0·729 (0·11) | 6·71 (3·03) | 4·52 (0·94) |
| 6. Querari | Chihuahua | 27°11′/107°32′ | 11 | 0·831 (0·24) | 0·819 (0·08) | 9·42 (2·50) | 6·41 (0·85) |
| 7. Yécora | Sonora | 28°22′/109°02′ | 12 | 0·839 (0·13) | 0·831 (0·08) | 9·85 (3·48) | 7·07 (1·03) |
| 8. Madera | Chihuahua | 29°32′/108°09′ | 11 | 0·889 (0·09) | 0·801 (0·05) | 7·86 (0·59) | 5·35 (0·53) |
| Q. hypoleucoides | |||||||
| 9. Batopilas | Chihuahua | 27°08′/107°34′ | 10 | 0·885 (0·16) | 0·782 (0·08) | 7·00 (2·58) | 5·09 (0·58) |
| 10. Temosachic | Chihuahua | 28°59′/108°13′ | 11 | 0·766 (0·22) | 0·728 (0·16) | 7·00 (2·76) | 4·74 (0·85) |
| Mixed 2 | |||||||
| 11. Huracán A | Chihuahua | 29°40′/108°15′ | 12 | 0·853 (0·09) | 0·824 (0·05) | 8·42 (1·51) | 6·13 (0·62) |
| 12. Guadalupe | Chihuahua | 29°11′/107°57′ | 10 | 0·846 (0·17) | 0·765 (0·14) | 7·14 (2·03) | 5·19 (0·68) |
| 13. Km 346 | Chihuahua | 28°26′/108°32′ | 11 | 0·633 (0·47) | 0·555 (0·38) | 3·71 (2·21) | 2·55 (0·67) |
| 14. Amarilla B | Chihuahua | 29°12′/108°14′ | 12 | 0·657 (0·26) | 0·681 (0·14) | 6·42 (3·86) | 4·09 (0·97) |
| 15. Guachochic | Chihuahua | 26°56′/107°08′ | 8 | 0·905 (0·12) | 0·799 (0·03) | 7·57 (1·81) | 5·11 (0·32) |
| 16. Poleo | Sonora | 29°37′/108°98′ | 12 | 0·809 (0·16) | 0·829 (0·09) | 10·42 (3·40) | 7·25 (1·21) |
| 17. Amarilla A | Chihuahua | 29°11′/108°14′ | 10 | 0·778 (0·09) | 0·796 (0·08) | 8·14 (2·11) | 5·53 (0·68) |
| 18. Babícora | Chihuahua | 29°13′/107°49′ | 10 | 0·724 (0·22) | 0·778 (0·13) | 7·57 (2·99) | 5·53 (0·85) |
| 19. Huracán B | Chihuahua | 29°40′/108°15′ | 12 | 0·734 (0·22) | 0·764 (0·08) | 7·71 (2·21) | 4·69 (0·56) |
| Q. sideroxyla | |||||||
| 20. Maycoba | Chihuahua | 28°17′/108°06′ | 10 | 0·857 (0·11) | 0·783 (0·13) | 7·85 (3·02) | 5·75 (0·91) |
| 21. Cuevecillas | Durango | 25°02′/106°16′ | 11 | 0·844 (0·15) | 0·841 (0·07) | 9·71 (2·75) | 7·17 (0·88) |
| 22. Buenos Aires | Durango | 23°42′/105°43′ | 11 | 0·831 (0·11) | 0·841 (0·04) | 8·71 (2·13) | 6·71 (0·62) |
| 23. Tecuan | Durango | 23°55′/105°01′ | 12 | 0·821 (0·04) | 0·833 (0·06) | 10·28 (1·89) | 6·64 (0·79) |
Standard errors are given in parentheses. The five morphological groups (three parental species and two mixed groups) were defined on the basis of geometric morphometrics.
Table 2.
Pairwise values of genetic differentiation (FST) among the five morphological groups estimated with the method of Weir and Cockerham (1984)
| Q. scytophylla | Q. hypoleucoides | Q. sideroxyla | Mixed 1 | Mixed 2 | |
|---|---|---|---|---|---|
| Q. scytophylla | – | 0·022 | 0·071 | 0·031 | 0·039 |
| Q. hypoleucoides | – | 0·036 | 0·022 | 0·035 | |
| Q. sideroxyla | – | 0·029 | 0·054 | ||
| Mixed 1 | – | 0·014 | |||
| Mixed 2 | – |
Numbers in bold indicate statistically significant values (P < 0·05) based on 10 000 random permutations.
According to Weir and Cockerham's θ, genetic differentiation was significant in all pairwise comparisons among the five morphological groups, except between the Mixed 1 and Mixed 2 groups (Table 2). Differentiation among populations was also significant within the three species, but not within the Mixed 1 and Mixed 2 groups (Table 3).
Table 3.
Genetic structure within the five morphological groups estimated with ΦST obtained from AMOVA
| Source of variation |
d.f. | SS | Variance components | Percentage of variation | Fixation index | |
|---|---|---|---|---|---|---|
| FST | ||||||
| Q. scytophylla | ||||||
| Among populations | 3 | 18·1 | 0·157 | 5·01 | ΦST = 0·05*** | |
| Within populations | 74 | 220·06 | 2·973 | 94·99 | ||
| Q. hypoleucoides | ||||||
| Among populations | 1 | 8·89 | 0·292 | 9·54 | ΦST = 0·09*** | |
| Within populations | 40 | 110·89 | 2·772 | 90·46 | ||
| Q. sideroxyla | ||||||
| Among populations | 3 | 17·96 | 0·135 | 4·26 | ΦST = 0·04*** | |
| Within populations | 84 | 254·39 | 3·028 | 95·74 | ||
| Mixed 1 | ||||||
| Among populations | 3 | 7·18 | 0·008 | 0·36 | ΦST = 0·004 n.s. | |
| Within populations | 88 | 194·5 | 2·21 | 99·64 | ||
| Mixed 2 | ||||||
| Among populations | 7 | −3·43 | −0·118 | −6·44 | ΦST = −0·06 n.s. | |
| Within populations | 158 | 307·83 | 1·95 | 106·44 | ||
Asterisks indicate statistically significant values (P < 0·05) and n.s. indicates non-significant values. Tests were based on 10 000 random permutations.
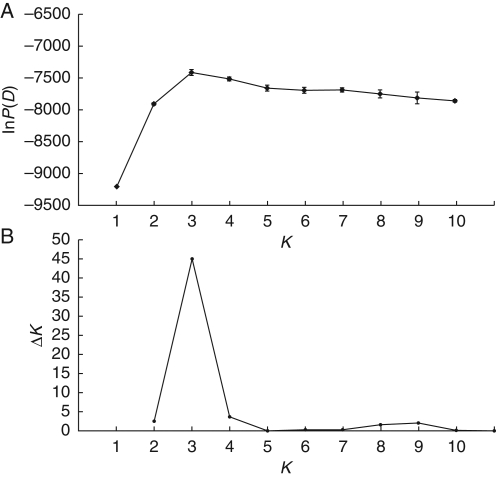
Estimates of admixture
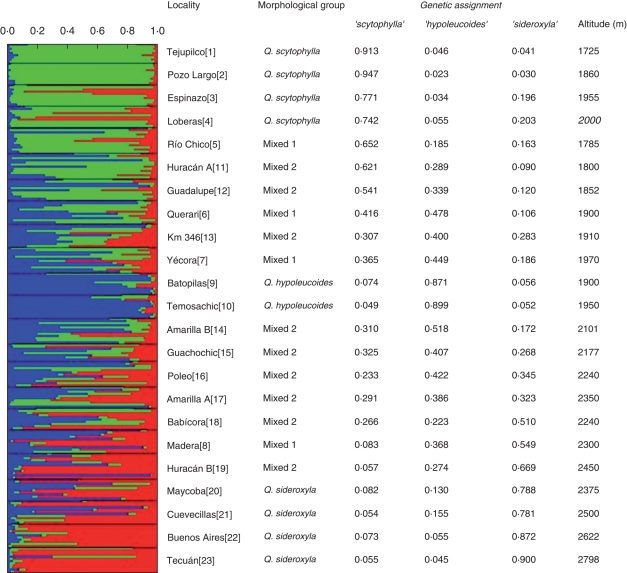
The highest posterior probability was obtained for three genetic clusters according to the values of lnP(D) (Fig. 4A). This result was also confirmed by the ΔK values (Fig. 4B). Using the admixture model in the program STRUCTURE, the proportion of ancestry (Q) of each individual and population in each of the three genetic groups was also estimated. The allopatric reference populations had a high proportion of ancestry from a single genetic group (Figs 2 and 5). For Q. scytophylla, populations Tejupilco and Pozo Largo had genetic ancestries of 0·913 and 0·947 in one genetic cluster and, for Q. sideroxyla, individuals from populations Buenos Aires and Tecuan also were assigned mostly to a single genetic group (Figs 2 and 5). In contrast, morphologically representative populations of the three species from within the contact zone showed some indications of introgression. Populations of Q. hypoleucoides had slight contributions from the other two genetic groups, while populations Espinazo and Loberas, belonging to Q. scytophylla, were influenced by the Q. sideroxyla genetic group. Finally, some introgression of the Q. hypoleucoides genetic cluster was detected in populations Maycoba and Cuevecillas, belonging to Q. sideroxyla (Figs 2 and 5). In the morphologically mixed populations, there was evidence of admixture among the three genetic clusters in different proportions. The Mixed 1 group was characterized by a transition from a higher proportion of the Q. scytophylla genetic cluster at lower altitudes (i.e. populations Río Chico and Huracán A) to more even contributions from the three species at higher altitudes (Km 346 and Yecora). The Mixed 2 group showed a change from even genetic proportions from the three genetic clusters towards a predominance of ancestry from Q. sideroxyla at high altitudes (Figs 2 and 5).
Fig. 4.
(A) Mean and standard deviation of lnP(D) for ten independent runs of STRUCTURE plotted against the number of genetic groups (K) used in the analysis. (B) Values of ΔK plotted against K. In both cases the peak indicates the most probable number of genetic groups given the data.
Fig. 5.
Genetic assignment of individuals and populations according to the Bayesian method implemented in the program STRUCTURE. One sample, considered to explain the data best, of ten iterated runs is shown. Each thin horizontal line represents an individual and the proportion of each colour is the proportion of ancestry derived from each of the three main genetic groups (K = 3) inferred. Populations are separated by black lines. Values of the proportions of ancestry for each population are given in the table.
Performance of STRUCTURE with simulated genotypes
Analysis of the simulated genotypes indicated that a threshold value in the admixture coefficient of Q ≥ 0·90 allowed adequate separation between pure species and hybrid individuals (Table 4). The genetic assignment of pure individuals from the three species showed high levels of performance, with values of 93 % for Q. scytophylla, 96 % for Q. sideroxyla and 95 % for Q. hypoleucoides (Table 4). For hybrid classes, performance values in most cases were moderately lower in comparison with pure-species assignment. Even for triple hybrids there was a reasonable identification rate.
Table 4.
Number of simulated individuals (rows), which were assigned to one of the three species or into a hybrid category (columns)
| Simulated/ assigned | Qscytop | Qhypole | Qsidero | Hyb QscytoQsidero | Hyb QscytoQhypole | Hyb QsideroQhypole | Triple Hybrid | Total |
|---|---|---|---|---|---|---|---|---|
| Qscytop | 391 | – | – | 9 | – | – | – | 400 |
| Qhypole | – | 398 | – | – | 2 | – | – | 400 |
| Qsidero | – | – | 395 | – | – | 5 | – | 400 |
| F1 Qscyto-Qsidero | 2 | – | – | 3 | – | 2 | – | 7 |
| BX Qscyto | 11 | – | – | 24 | – | 2 | – | 37 |
| F1 Qscyto-Qhypole | – | – | – | 2 | 14 | – | – | 16 |
| BX Qhypole | – | 12 | – | – | 32 | – | 44 | |
| F1 Qsidero-Qhypole | – | – | – | – | 1 | 13 | – | 14 |
| BX Qsidero | – | – | 9 | – | 1 | 21 | 1 | 32 |
| Triple Hybrid | – | – | – | 2 | – | 3 | 9 | 14 |
| Total | 404 | 410 | 404 | 40 | 50 | 46 | 10 | 1364 |
| Efficiency | 97·75 | 99·50 | 98·75 | 61·3636 | 76·666 | 73·913 | 64·28 | |
| Accuracy | 96·782 | 97·073 | 97·777 | 67·50 | 92·00 | 73·914 | 90·00 | |
| Performance | 94·60 | 96·58 | 96·55 | 41·42 | 70·53 | 54·63 | 57·85 |
Parameters of efficiency, accuracy and overall performance of the assignment method are given in per cent. Note individuals correctly assigned are in bold type. Hyb, hybrids; F1, first-generation hybrids; BX, backcrosses; Qscyto, Quercus scytophylla; Qhypole, Q. hypoleucoides; Qsidero, Q. sideroxyla.
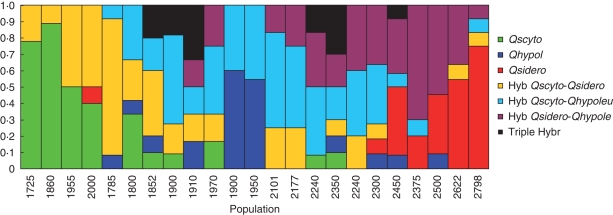
Given these results, individuals from all populations in the real samples were assigned to a genotypic class as follows: individuals with Q ≥ 0·90 were considered to be pure bred, trees with Q < 0·90 from two genetic groups were considered hybrids between two species, and trees with ancestry from the three genetic groups in more or less equal proportions (Q = 0·3–0·4) were considered triple hybrids (Fig. 6). Populations from the contact zone were largely dominated by hybrid individuals (Fig. 6). As previously noted for the admixture coefficient of each population, the proportion of the different genotypes also changed with altitude. At lower altitudes, the predominant genotypes corresponded to crosses between Q. scytophylla and Q. sideroxyla, which were replaced by crosses between Q. scytophylla and Q. hypoleucoides, and finally by crosses between Q. sideroxyla and Q. hypoleucoides with increasing altitude. Triple hybrids were situated in populations at mid altitudes, particularly in populations Km 346 and Amarilla A, where also almost all the other possible genotypes were present.
Fig. 6.
Frequency of the different genotypic classes observed in each population. Individuals were assigned to each category (Q. scytophylla, Q. hypoleucoides, Q. sideroxyla, hybrids between Q. scytophylla and Q. sideroxyla, hybrids among Q. scytophylla × Q. hypoleucoides, hybrids between Q. sideroxyla and Q. hypoleucoides and triple hybrids, as indicated in the key), depending on their individual coefficient of admixture derived from STRUCTURE. Performance of the assignment procedure was previously assessed by analysing simulated genotypes (see text for details).
Associations among leaf shape morphology, genetic ancestry and altitude
The results of the regression analysis indicated that the proportions of genetic ancestry of Q. scytophylla and Q. sideroxyla in the populations were strongly correlated with altitude (R2 = 0·78, P < 0·0001 and R2 = 0·72, P = 0·0002, respectively) but not with latitude or longitude. In turn, the proportion of the Q. hypoleucoides genetic group was weakly correlated with latitude only (R2 = 0·4, P = 0·03). Finally, the canonical scores of morphological variation showed a marginally significant tendency to be correlated with latitude (R2 = 0·3, P = 0·06).
Nevertheless, as reference populations of the three species were situated at the extremes of the altitudinal range, it might be that the significant correlations obtained are mostly due to the different altitudinal requirements of the pure species. To verify this explanation, correlations were calculated again without the reference populations and practically the same results were obtained (data not shown).
DISCUSSION
The combined results of geometric morphometrics of leaf shape and genetic analysis with microsatellites indicate that a high rate of hybridization and introgression have occurred among the three oak species studied, which have resulted in a complex hybrid zone containing a wide array of genotypes. Furthermore, this hybrid zone is geographically structured as a mosaic, with a patchy spatial distribution of pure and mixed populations with different genetic compositions. Altitude was, apparently, the most important geographical variable explaining the distribution of genetic and morphological variation.
The utility of geometric morphometrics to quantify leaf shapes and to assess hybridization in plants is well established (e.g. Jensen et al., 2002). In species of Quercus, foliar shape is particularly informative because other organs (i.e. flowers) show little variation (Kaul, 1985). The morphometric assessment performed in this study, based on Procrustes analysis of landmarks followed by canonical discriminant function analysis, substantiated the complete morphological differentiation of individuals from representative populations of the three oak taxa Q. hypoleucoides, Q. scytophylla and Q. sideroxyla. The morphometric analysis also demonstrated that some individuals were intermediate in various combinations with respect to representative populations, as was initially presumed from field observations. Intermediate individuals were present in a series of populations scattered along a wide area of contact among the three oak species. These populations showed clear differences in their phenotypic composition and could be divided into two groups. The first group included a series of sites (i.e. Río Chico, Querari, Yécora and Madera) in which the predominant morphology was similar to Q. hypoleucoides, Q. sideroxyla or intermediate between them. The second group (i.e. Huracán, Guadalupe, Km 346, Guachochic, Amarilla B, Poleo, Amarilla A and Babícora) consisted of populations showing evidence of morphological intergradation between Q. scytophylla and the other two species.
The degree of genetic differentiation among the three species was significant but very low (Table 2). However, similar comparisons of oak species belonging to several complexes have repeatedly found low interspecific genetic differentiation based on nuclear markers, ranging from 0·02 to 0·17 (Dodd and Kashani, 2003; González-Rodríguez et al., 2005; Muir and Schlötterer, 2005; Craft and Ashely, 2006; Curtu et al., 2007; Gugerli et al., 2007). Although it has been argued that low genetic differentiation among oak species does not necessarily imply introgression, but can be accounted for by the sharing of ancestral polymorphisms (e.g. Muir and Schlötterer, 2005), in most instances interspecific gene flow is a more parsimonious explanation for this observation (Lexer et al., 2006).
In the specific case studied here intermediate populations occurred within a defined area with the characteristics of a mosaic hybrid zone (see below). Furthermore, the Bayesian analysis suggested a clustering into three genetic groups, which agrees with the number of taxa involved in this complex, with isolated representative populations assigned to a single genetic cluster. However, the same analysis also suggested the occurrence of various degrees of admixture among the three genetic groups in populations from within the area of contact. The above evidence strongly suggests that introgression has occurred among the three oak taxa and contributed to shape the patterns of variation observed. As shown, both the morphological and the genetic data indicate that some of the populations in the zone of contact are the result of introgression between a pair of species, but others clearly seem to be an admixture of all three taxa.
At the level of individuals, a high proportion of hybrids resulting from crosses between different species pairs were identified within the contact zone, and 14 trees were assigned as probable triple hybrids. Simulations of genotypes provided reasonable support to the assignment method, providing some confidence on the identification of even these complex hybrids (Vähä and Primmer, 2006). Studies on closely related European white oaks have suggested that five or six microsatellite loci are sufficient to distinguish between pure species and introgressed individuals (Curtu et al., 2007; Gugerli et al., 2007). A recent study involving four oak species suggested that patterns of interspecific crosses may be more complex than those modelled in simulations, possibly leading to the existence of third- or later-generation hybrids as well as to hybridization involving more than two species (Lepais et al., 2009). Triple hybridization first requires the production of fertile hybrid genotypes between at least two species, and then the crossing between hybrids and a third species, or between hybrids from different species combinations (Kaplan and Fehrer, 2007).
Mosaic hybrid zones are described as areas where pure species populations and mixed populations are patchily distributed across a zone of overlap (Harrison and Rand, 1989). The morphological variation in the contact area in the Sierra Tarahumara clearly follows this pattern. Populations with typical parental species morphology and those belonging to the Mixed 1 and Mixed 2 morphological groups are geographically scattered. Genotypic variation was also distributed as a mosaic and showed little correlation with latitude and longitude. Strong associations among specific genotypes and certain habitats are also characteristic of mosaic hybrid zones, and exogenous selection is considered important in such structuring (Harrison and Rand, 1989). From the analyses presented, it was evident that the genetic and morphological composition of the oak populations studied is strongly associated with altitude. However, as can be judged from the lack of similarity among geographically proximate populations, dispersal does not seem to be the factor determining these correlations. Alternatively, the influence of environmental variables on the structure of this hybrid zone can be suggested as an explanation for these results. In their study on four Californian red oak species, Dodd and Afzal-Rafii (2004) obtained results implying that environmental gradients rather than pollen dispersal determine the extent of introgression. In other plant species, hybrid zones associated with altitudinal gradients have been reported and the patterns of variation are often coupled to environmental variables that change with elevation (e.g. James and Abbott, 2005; Kimball, 2008). For oaks, there is some direct evidence indicating that the frequency of hybridization, the direction of introgression and the performance of hybrids depend on habitat conditions (Williams and Ehleringer, 2000; Williams et al., 2001; Himrane et al., 2004), but also on species' relative abundance (Lepais et al., 2009). These factors could be contributing to the structure of the hybrid zone in the Sierra Tarahumara. Nevertheless, ecological studies are required to gain more detailed insight into the dynamics of this hybrid zone. In conclusion, the evidence presented supports the existence of a complex hybrid zone that has formed from pairwise and triple hybridization among Q. hypoleucoides, Q. scytophylla and Q. sideroxyla in the mountains of the Sierra Tarahumara in north-western Mexico, which is structured as a mosaic probably in response to environmental variables associated with altitude.
ACKNOWLEDGEMENTS
We thank E. de Luna, A. Guerrero, A. Albarrán-Lara, E. Pascual, L. Herrera, O. Chassin-Noria, P. Cuevas-Reyes, N. Pérez-Nasser, V. Rocha, J. J. Fuentes-Junco and F. Alvarado-Ramos for technical assistance and suggested improvements to the manuscript. We thank Alex Buerkle, Felix Gugerli and an anonymous reviewer for their suggestions that greatly improved the manuscript. J.M.P.-R. received a PhD scholarship from CONACyT. This project was supported by CONACyT [grant 38550-V to K.O.], CONACyT-SEMARNAT [23728 to K.O.] and CONACyT-ECOS NORD [M03-A01] to A. K. and K.O.
LITERATURE CITED
- Aldrich PR, Michler CH, Sun W, Romero-Severson J. Microsatellites markers for northern red oak (Fagaceae: Quercus rubra) Molecular Ecology Notes. 2002;2:472–474. [Google Scholar]
- Arnold ML. Iris nelsonii (Iridaceae): origin and genetic composition of a homoploid hybrid species. American Journal of Botany. 1993;80:577–583. doi: 10.1002/j.1537-2197.1993.tb13843.x. [DOI] [PubMed] [Google Scholar]
- Arnold ML. Natural hybridization and evolution. Oxford: Oxford University Press; 1997. [Google Scholar]
- Barton NH, Gale KS. Genetic analysis of hybrid zones. In: Harrison RG, editor. Hybrid zones and the evolutionary process. Oxford: Oxford University Press; 1993. pp. 13–45. [Google Scholar]
- Barton NH, Hewitt GM. Analysis of hybrid zones. Annual Review of Ecology and Systematics. 1985;16:113–148. [Google Scholar]
- Belkhir K, Borsa P, Chikhi L, Raufaste N, Bonhomme F. GENETIX 4·05, logiciel sous Windows TM pour la génétique des populations. Montpellier: Laboratoire Génome; 1996–2004. Populations, Interactions, CNRS UMR 5171, Université de Montpellier II. [Google Scholar]
- Bookstein FL. Morphometric tools for landmarks data: geometry and Biology. Cambridge: Cambridge University Press; 1991. [Google Scholar]
- Bronson CL, Grubb TC, Jr, Braun MJ. A test of the endogenous and exogenous selection hypotheses for the maintenance of a narrow avian hybrid zone. Evolution. 2003;57:630–637. doi: 10.1111/j.0014-3820.2003.tb01554.x. [DOI] [PubMed] [Google Scholar]
- Curtu AL, Gailing O, Finkeldey R. Evidence for hybridization and introgression within a species-rich oak (Quercus spp.) community. BMC Evolutionary Biology. 2007;7:218–233. doi: 10.1186/1471-2148-7-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craft KJ, Ashley MV. Population differentiation among three species of white oak in northeastern Illinois. Canadian Journal of Forest Research. 2006;36:206–215. [Google Scholar]
- Dodd RS, Afzal-Rafii Z. Selection and dispersal in a multispecies oak hybrid zone. Evolution. 2004;58:261–269. [PubMed] [Google Scholar]
- Dodd RS, Kashani N. Molecular differentiation and diversity among the California red oaks (Fagaceae; Quercus section Lobatae) Theoretical and Applied Genetics. 2003;197:884–892. doi: 10.1007/s00122-003-1290-4. [DOI] [PubMed] [Google Scholar]
- Dumolin-Lapègue S, Kremer A, Petit RJ. Are chloroplast and mitochondrial DNA variation species independent in oaks. Evolution. 1999;53:1406–1413. doi: 10.1111/j.1558-5646.1999.tb05405.x. [DOI] [PubMed] [Google Scholar]
- Durrett R, Buttel L, Harrison R. Spatial models for hybrid zones. Heredity. 2000;84:9–19. doi: 10.1046/j.1365-2540.2000.00566.x. [DOI] [PubMed] [Google Scholar]
- Endler JA. Geographic variation, speciation, and clines. Princeton, NJ: Princeton University Press; 1977. [PubMed] [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular Ecology. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evolutionary Bioinformatics Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferran M. SPSS para WINDOWS. Programación y análisis estadístico. Madrid: McGraw Hill; 1997. [Google Scholar]
- Fitzpatrick BM, Schaffer HB. Environment-dependent admixture dynamics in a tiger salamander hybrid zone. Evolution. 2004;58:1282–1293. doi: 10.1111/j.0014-3820.2004.tb01707.x. [DOI] [PubMed] [Google Scholar]
- González-Rodríguez A, Arias DM, Valencia S, Oyama K. Morphological and RAPD analysis of hybridization between Q. affinis and Q. laurina (Fagaceae), two Mexican red Oaks. American Journal of Botany. 2004;91:401–409. doi: 10.3732/ajb.91.3.401. [DOI] [PubMed] [Google Scholar]
- González-Rodríguez A, Arias DM, Oyama K. Genetic variation and differentiation of populations within the Quercus affinis – Quercus laurina (Fagaceae) complex analyzed with RAPD markers. Canadian Journal of Botany. 2005;83:153–162. [Google Scholar]
- Goudet J. Fstat version 1.2: a computer program to calculate F statistics. Journal of Heredity. 1995;86:485–486. [Google Scholar]
- Gugerli F, Walser JC, Dounavi K, Holderegger R, Finkeldey R. Coincidence of small-scale spatial discontinuities in leaf morphology and nuclear microsatellite variation of Quercus petraea and Q. robur in a mixed forest. Annals of Botany. 2007;99:713–722. doi: 10.1093/aob/mcm006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gugerli F, Brodeck S, Holderegger R. Utility of multilocus genotypes for taxon assignment in stands of closely related European white oaks from Switzerland. Annals of Botany. 2008;102:855–863. doi: 10.1093/aob/mcn164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison RG, Rand DM. Mosaic hybrid zones and the nature of species boundaries. In: Otte D, Endler J, editors. Speciation and its consequences. Sunderland, MA: Sinauer Associates; 1989. pp. 110–133. [Google Scholar]
- Himrane H, Camarero JJ, Gil-Pelegrín E. Morphological and ecophysiological variation of the hybrid oak Quercus subpyrenaica (Q. faginea × Q. pubescens) Trees. 2004;18:566–575. [Google Scholar]
- Hubisz M, Falush D, Stephens M, Pritchard J. Inferring weak population structure with the assistance of sample group information. Molecular Ecology Resources. 2009;9:1322–1332. doi: 10.1111/j.1755-0998.2009.02591.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James JK, Abbott RJ. Recent, allopatric, homoploid hybrid speciation: the origin of Senecio squalidus (Asteraceae) in the British Isles from a hybrid zone on Mount Etna, Sicily. Evolution. 2005;59:2533–2547. [PubMed] [Google Scholar]
- Jensen RJ, Ciofani KM, Miramontes LC. Lines, outlines, and landmarks: morphometric analyses of leaves of Acer rubrum, Acer saccharinum (Aceraceae) and their hybrid. Taxon. 2002;51:475–492. [Google Scholar]
- Kaul RB. Reproductive morphology of Quercus (Fagaceae) American Journal of Botany. 1985;72:1962–1977. [Google Scholar]
- Kaplan Z, Fehrer J. Molecular evidence for a natural primary triple hybrid in plants revealed from direct sequencing. Annals of Botany. 2007;99:1213–1222. doi: 10.1093/aob/mcm072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimball S. Links between floral morphology and floral visitors along an elevational gradient in a Penstemon hybrid zone. Oikos. 2008;117:1064–1074. [Google Scholar]
- Lefort F, Douglas GC. An efficient micro-method of DNA isolation from mature leaves of four hardwood tree species Acer, Fraxinus, Prunus and Quercus. Annals of Forest Science. 1999;56:259–263. [Google Scholar]
- Lepais O, Petit RJ, Guichoux E, et al. Species abundance and direction of introgression in oaks. Molecular Ecology. 2009;18:2228–2242. doi: 10.1111/j.1365-294X.2009.04137.x. [DOI] [PubMed] [Google Scholar]
- Lexer C, Kremer A, Petit RJ. Shared alleles in sympatric oaks: recurrent gene flow is a more parsimonious explanation than ancestral polymorphism. Molecular Ecology. 2006;15:2007–2012. doi: 10.1111/j.1365-294X.2006.02896.x. [DOI] [PubMed] [Google Scholar]
- McCauley RA, Cortés-Palomec AC, Oyama K. Phylogeography and historical gene flow patterns in disjunct Quercus across the Sierra Madre Occidental and southern Cordillera of Mexico. 2007 Botanical Society of America/American Society of Plant Biologists, Chicago, IL. Available at http://www.2007.botanyconference.org/engine/search/index.php . [Google Scholar]
- Moore WS, Price JT. Nature of selection in the northern flicker hybrid zone and its implications for speciation theory. In: Harrison RG, editor. Hybrid zones and the evolutionary process. New York: Oxford University Press; 1993. pp. 196–255. [Google Scholar]
- Muir G, Schlötterer C. Evidence for shared ancestral polymorphism rather than recurrent gene flow at microsatellite loci differentiating two hybridizing oaks (Quercus spp.) Molecular Ecology. 2005;14:549–561. doi: 10.1111/j.1365-294X.2004.02418.x. [DOI] [PubMed] [Google Scholar]
- Nielsen EE, Bach LA, Kotlick P. Hybrilab (version 1.0): a program for generating simulated hybrids from population samples. Molecular Ecology Notes. 2006;6:971–973. [Google Scholar]
- Nixon KC. The genus Quercus in Mexico. In: Ramamoorthy TP, Bye R, Lot A, Fa J, editors. Biological diversity of Mexico: origins and distribution. New York: Oxford University Press; 1993. pp. 447–458. [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raudnitschka D, Hensen I, Oberprieler C. Introgressive hybridization of Senecio hercynus and S. ovatus (Compositeae, Senecioneae) along an altitudinal gradient in Harz national Park (Germany) Systematics and Biodiversity. 2007;5:333–344. [Google Scholar]
- Rieseberg LH. Hybrid origins of plant species. Annual Review of Ecology and Systematics. 1997;28:359–389. [Google Scholar]
- Rieseberg LH, Wendell JF. Introgression and its consequences in plants. In: Harrison RG, editor. Hybrid zones and the evolutionary process. New York: Oxford University Press; 1993. pp. 70–109. [Google Scholar]
- Rohlf FJ. Stony Brook, NY: Department of Ecology and Evolution, State University of New York; 2004. TPSDIG, digitize landmarks and outlines, version 2.0. [Google Scholar]
- Tovar-Sánchez E, Oyama K. Natural hybridization and hybrid zones between Quercus crassifolia and Quercus crassipes (Fagaceae) in Mexico: morphological and molecular evidence. American Journal of Botany. 2004;91:1352–1363. doi: 10.3732/ajb.91.9.1352. [DOI] [PubMed] [Google Scholar]
- Tovar-Sánchez E, Mussali-Galante P, Esteban-Jiménez R, et al. Chloroplast DNA polymorphism reveals geographic structure and introgression in Quercus crassifolia × Q. crassipes complex in México. Botany. 2008;86:228–239. [Google Scholar]
- Trelease W. The American Oaks. Memories of the National. Academy of Sciences. 1924;20:1–255. [Google Scholar]
- Vähä J-P, Primmer CR. Efficiency of model-based Bayesian methods for detecting hybrid individuals under different hybridization scenarios and with different numbers of loci. Molecular Ecology. 2006;15:63–72. doi: 10.1111/j.1365-294X.2005.02773.x. [DOI] [PubMed] [Google Scholar]
- Valencia S. Diversidad del género Quercus (Fagaceae) en México. Boletín de la Sociedad Botánica de México. 2004;75:33–53. [Google Scholar]
- Valencia S, Delgado A. Los tricomas foliares en la caracterización de un grupo de especies del género Quercus, sección Lobatae (Fagaceae) Anales del Instituto de Biología. 2003;74:5–15. [Google Scholar]
- Van Oosterhout C, Hutchinson WF, Wills DPM. Micro-Checker: software for identifying and correcting genotyping errors in microsatellite data. Molecular Ecology Notes. 2004;4:535–538. [Google Scholar]
- Weir BS, Cockerham CC. Estimating F-Statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- Williams DG, Ehleringer JR. Carbon isotope discrimination and water relations of oak hybrid populations in southwestern Utah. Western North American Naturalist. 2000;60:121–129. [Google Scholar]
- Williams JH, Boecklen W J, Howard DJ. Reproductive processes in two oak (Quercus) contact zones with different levels of hybridization. Heredity. 2001;87:680–690. doi: 10.1046/j.1365-2540.2001.00968.x. [DOI] [PubMed] [Google Scholar]
- Whittemore AT, Schaal BA. Interspecific gene flow in sympatric oaks. Proceedings of the National Academy of Sciences USA. 1991;88:2540–2544. doi: 10.1073/pnas.88.6.2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelditch ML, Swiderski DL, Sheets HD, Fink WL. Geometric morphometrics for biologists: a primer. New York: Elsevier; 2004. [Google Scholar]