Abstract
Background and Aims
DREB proteins are involved mainly in plant responses to abiotic stresses such as cold, drought or high salinity as well as ABA signalling. However, the function of most rice DREB genes and the underlying molecular mechanisms controlling these responses remains elusive. In this study, ARAG1, a rice DREB gene, was functionally analysed.
Methods
Antisense and over-expression constructs of ARAG1 were introduced into rice by an Agrobacterium-mediated method. RT-PCR and western blot were used to detect ARAG1 accumulation in transgenics. PEG and ABA were used to test their response to abiotic stresses.
Key Results
ARAG1 was expressed in inflorescences, roots, immature embryos and germinating seeds, but not in coleoptiles, leaves or mature embryos. Drought stress and ABA treatment increased transcript levels of the gene rapidly. ARAG1 knockdown line was hypersensitive to ABA application during seed germination and seedling growth. However, the line over-expressing ARAG1 behaved similarly to wild type in these circumstances. Knockdown of ARAG1 weakened tolerance of the transgenic seedlings to drought stress, while over-expression of it increased the tolerance slightly. In addition, activity of α-amylases was enhanced in germinating seeds of the knockdown and over-expression lines.
Conclusions
These results indicate that ARAG1 was involved in the ABA signalling and stress responsive pathways.
Keywords: Abscisic acid (ABA), AP2/EREBP, ARAG1, DREB, drought, Oryza sativa
INTRODUCTION
AP2/ethylene-responsive element binding proteins (EREBP), composing a superfamily of plant-specific transcription factors, are characterized by the presence of a highly conserved approx. 70-amino-acid region, termed the AP2 DNA-binding domain (Weigel, 1995; Okamuro et al., 1997). Dehydration-responsive element binding (DREB) proteins, a subgroup of the AP2/EREBP transcription factors, play important roles in plant response and adaptation to abiotic stresses (Sakuma et al., 2002; Gutterson and Reuber, 2004). In arabidopsis, DREB1 and its homologues respond to cold whereas DREB2 and its homologues respond to drought or salt stress, both in an abscisic acid (ABA)-independent manner. Over-expression of DREB1 and DREB2 results in different phenotypes in transgenic plants (Liu et al., 1998). Besides the DREBs of arabidopsis (Stockinger et al.,1997; Gilmour et al., 1998; Liu et al., 1998), characterized members of this subgroup included OsDREB1A, -1B, -1C, -2A and -2B of rice, Pti4, Pti5 and Pti6 of tomato and others from maize, tobacco and cotton (Gu et al., 2002; Niu et al., 2002; Dubouzet et al., 2003).
The DRE/CRT cis-element is identified in the promoter region of stress-responsive genes such as rd29A and kin1 (Yamaguchi-Shinozaki and Shinozaki, 1994, 2005). DREB proteins interact with DRE/CRT by their AP2 DNA-binding domain, thus mediating downstream gene expression in the stress-responsive pathway (Yamaguchi-Shinozaki and Shinozaki, 1994, 2005). In contrast, the ABA-responsive element (ABRE) mainly mediates downstream gene expression in the ABA-signalling pathway. Interestingly, increasing evidence shows that DRE/CRT can act as a coupling element of the ABRE cis-element to regulate downstream gene expression (Narusaka et al., 2003). Thus, there exists a comprehensive connection between stress-responsive and ABA-signalling pathways (Shinozaki and Yamaguchi-Shinozaki, 2000; Shinozaki et al., 2003). As exemplified by ZmABI4, a DREB protein which shows ABA-induced expression, it binds to CE1 and acts as a coupling element of the ABRE in maize (Niu et al., 2002). Consistently, over-expression of DREB1D/CBF4, which is an ABA-responsive gene of arabidopsis, activates expression of drought and cold-related downstream genes that contain the DRE/CRT cis-element (Knight et al., 2004). Another line of evidence comes from microarray analysis, which shows that, among 17 downstream genes of DREB2A, 12 carry both the DRE/CRT and ABRE cis-elements (Maruyama et al., 2004; Sakuma et al., 2006). These results demonstrate that some DREBs are involved in both ABA signalling and stress-responsive pathways.
Recently, Wang et al. (2008) reported that besides responding to exogenous ABA treatment, OsDREB1F was induced by drought or salt stresses in rice, suggesting that OsDREB1F was involved in both the stress-responsive and ABA-signalling pathways. Similarly, over-expression of DBF2, a DREB gene of maize, represses not only the basal promoter activity of its downstream gene, but also the effect of ABA (Kizis and Pagès, 2002). These researches reveal that the plant hormone ABA is intimately linked with DREB protein function and some aspects of their relationship have been addressed. That ABA regulates seed germination and seedling growth has been reported (Finkelstein et al., 2002); however, whether DREB affects theses activities, and through which pathway it regulates them remain to be clarified. In this study, a cDNA sequence of a DREB-like gene, ABA responsive AP2-like gene in rice (ARAG1) was isolated, and its function investigated through transgenic strategies to understand the role it played in ABA-regulated plant activities.
MATERIALS AND METHODS
Plant materials and nuclear acid extraction
Cultivar Zhonghua 10 (Oryza sativa L. ssp. japonica) wild-type (WT)and transgenic rice were grown conventionally in a greenhouse. Plant material was harvested as follows: inflorescences at male meiosis and developing embryo stages; leaves from 2-week-old seedlings; and roots and coleoptiles from seeds germinated for 3 d on paper soaked in sterile water. Germinating embryos were striped from rice seeds after imbibition for 24 h. Total RNA in the endosperm was extracted according to Li's method (Li, 2006). To detect the gene's response to ABA inducement, seedlings 1 week after germination were treated with solutions of different ABA concentrations before RNA was extracted.
Genomic DNA was extracted using cetyltrimethyl ammonium bromide (Murray and Thompson, 1980). Total RNA was extracted using TRIzol reagent (Gibco-BRL) according to the manufacturer's protocol, followed by digestion with RNase-free DNase I (TaKaRa) to remove residual genomic DNA.
Isolation of the cDNA sequence of ARAG1
According to the predicted sequence of ARAG1 (AJ307662; Genbank), a gene-specific primer pair, P1 (5′-GAGCTCTCTTTCCACGTCGCGAGAG-3′) and P2 (5′-TCTAGACGGGTTGTACATGCAGGCT-3′), were designed. About 5 µg of root total RNA was reversely transcribed into first-strand cDNA by use of the ThermoScript kit (Invitrogen). PCR was performed with the cDNA as template. Purified PCR products were cloned into the pGEM-T vector (Promega) and sequenced.
Semiquantitative RT-PCR analysis of ARAG1 mRNA expression
A total of 5 µg of RNA isolated from different organs or from different treatments were reverse-transcribed into first-strand cDNA. RT-PCR was performed for 30 cycles in a 50-μL mixture that included 1 µL of the first-strand cDNA, 10 pmol each of the gene-specific primers P3 (5′-ATCCATGGACGACTCGTCGTTC-3′) and P4 (5′-CGACTAGTGTAGTACTCCCACAGAAGTG-3′), which were designed to amplify the coding region of ARAG1, 200 µm dNTPs, 1 × PCR buffer and 2·5 U DNA polymerase (5 U µL–1; Takara). The PCR products were separated in 1 % agarose gels and photographed. Tubulin A (Tub A, accession X91806) mRNA was amplified in parallel as a constitutive control as described by Tao et al. (2007).
Preparation of antisense and over-expression constructs of ARAG1
To make an antisense expression construct of ARAG1 (35S–ARAG1 AS), a 963-bp fragment containing the full coding region of ARAG1 was amplified with primer P1 (SacI site added) and P2 (XbaI site added). After digestion, the fragment was inserted into XbaI–SacI doubly digested pBI121 vector in reverse orientation. The resulting construct was digested with HindIII–EcoRI and the smaller fragment harbouring the CaMV 35s promoter, ARAG1 antisense fragment and Nos terminator was sub-cloned into the HindIII–EcoRI polylinker site of the pCAMBIA1301 vector, with beta-glucuronidase (GUS) as reporter gene.
To obtain the ARAG1 over-expression construct (35S–ARAG1 OE), the full coding region of ARAG1 was amplified with primers P3 and P4 (NcoI and SpeI sites added, respectively) and fused into the open reading frame of the GFP (green fluorescence protein) gene in the pCAMBIA1302 vector. The two constructs were introduced separately into rice embryonic calli by Agrobacterium tumefaciens-meditated methods (Hiei et al., 1994).
Subcellular localization of ARAG1 protein
ARAG1 over-expression construct was introduced into onion epidermis cells by Agrobacterium tumefaciens as described by Hu et al. (2008). GFP fluorescence was observed with a microscopic in bright field or through a FITC filter (Zeiss). The transformants were incubated on MS medium at 25 °C in the light for 2·5 d before observation.
Identification of transgenic plants
Hygromycin-resistant transgenic plants were first identified by PCR amplification with primers for the hygromycin phosphotransferase gene: P5 (5′-TGCTGCTCCATACAAGCCAACC-3′) and P6 (5′-AGACCTGCCTGAAACCGAACT-3′). They were further identified by GUS staining in leaves of AS (antisense) lines according to the methods described by Jefferson et al. (1987) as well as by Southern blot in OE (over-expressing) and AS lines. The primer pair P3 and P7 (5′-ATTCTTTTGGTCATGCGTGGAA-3′) were used to amplify transcripts of endogenous ARAG1, and P3 and P4 were used to amplify total ARAG1 transcripts. To ARAG1 over-expression plants, GFP gene-specific primer pairs P8 (5′-GGTCTAGAATGACTAAAGGAGAAG-3′) and P9 (5′-ATGAGCTCGGGCAGATTGTGTGGACA-3′) were used to amplify the expression of ARAG1 from the over-expression construct, and P3 and P4 were used to examine the total level of ARAG1.
Germination, seedling growth and ABA treatment
T1 seeds were germinated on half-strength MS medium contain 50 mg L–1 hygromycin for screening of positive progeny. Offspring from confirmed T1 lines were used for further analysis.
To analyse germination, 30 seeds of the T2 generation from each different transgenic line or WT control were soaked in tap water at 25 °C for 24 h and then allowed to germinate on sterile-water-saturated filter paper at 25 °C with 0 µm, 2 µm, 4 µm, 6 µm or 8 µm ABA. Seeds were regarded as having germinated when the primary root was longer than 2 mm. Germination rates were scored on the 4th, 7th and 10th days in triplicate.
For the seedling growth treatment, transgenic seeds of the T2 generation and WT were allowed to germinate on water-saturated filter paper for 4 d, with a 16 h light/8 h dark regime at 25 °C. After this, they were grown on filter paper supplemented with 0 µm, 2 µm, 4 µm or 8 µm ABA for an additional 10 d. The length of the primary roots and height of the shoots were measured and number of adventitious roots counted on the 10th day. Each datum was the mean value of at least 30 seeds in triplicate.
ARAG1 antibody preparation and western blot analysis
A non-conserved sequence of ARAG1 (encoding amino acids 136 to 225, amplified with primers 5′-ACTGAATTCGGCCTCCTCCGCCAATGC-3′ and 5′-CGACTCGAGTTGTAGTACTCCCACAGAAG-3′) was fused into the glutathione S-transferase (GST) gene in the vector pGEX-4T-3 (Amersham) for protein expression. ARAG1 polyclonal antibody was prepared as described by Hanly et al. (1995) and Worrall (1996).
Total proteins from different organs of transgenic or WT rice were extracted according to the method of Salekdeh et al. (2002). Forty micrograms of the extracted proteins were loaded in each lane. Western blot was performed as described previously (Zhang et al., 2006).
PEG 6000 treatment of the seedlings
To evaluate drought tolerance of ARAG1 transgenic rice, WT and ARAG1 transgenic seedlings were grown hydroponically (Kumar et al., 2003) for 5 weeks in a greenhouse, followed by 2 weeks in which 15 % PEG 6000 was added to the nutrient solution to mimic drought conditions. Afterwards, shoots and roots of the seedlings were harvested for water concentration determination. Their biomasses (dry weight) were measured before and after desiccation in a forced air oven at 70 °C for 3 d.
Analysis of α-amylase activity in WT and ARAG1 transgenic seeds
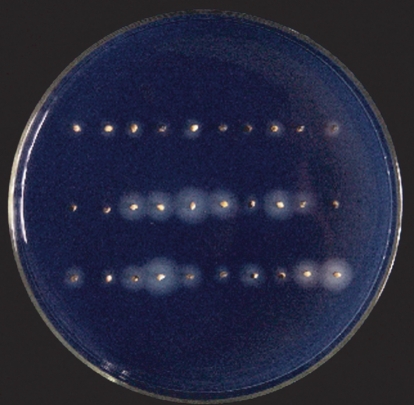
Activity of α-amylases was analysed as described by Ikeda et al. (2001).The embryo halves of WT, OE2 and AS9 seeds were cut away, and the remaining parts were sterilized in 15 % NaClO for 30 min. After rinsing the seed halves with sterilized distilled water five times, they were placed on an agar plate (0·2 % soluble starch,10 mm NaAC3H2O, 2 mm CaCl2, 2 % agar and 1 µm GA3) and incubated at 24 °C for 3 d. Then the plates were exposed to volatilization of iodine gas for 2 min. Activity of α-amylases was indicated by the diameter of the hydrolysis circle on the plates. Each plate contained 30 seeds of the different genotypes in triplicate.
RESULTS
ARAG1 encodes a DREB-like protein
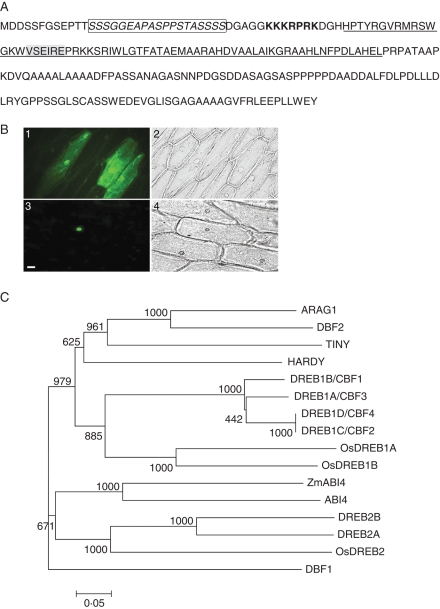
The amplified cDNA sequence of ARAG1 was confirmed by AK108830 on the KOME database (http://cdna01.dna.affrc.go.jp/cDNA/). It includes an open reading frame of 678 bp capable of encoding a 225-amino-acid protein. The N- and C-terminal regions of ARAG1 contain a serine/threonine-rich motif of 21 amino acids (amino acids 13–31) and a highly acidic region of 74 amino acids (134–208; Fig. 1A), respectively. These motifs have been proposed as transcriptional activation domains in other characterized AP2/EREBP members (Jofuku et al., 1994; Elliott et al., 1996). Besides the AP2 DNA binding domain, the protein has a putative nuclear localization signal (NLS) KKKRPRK (Fig. 1A). These features together with the result that ARAG1–GFP fusion targeted to the nucleus (Fig. 1B) suggested that ARAG1 was a potential transcription factor.
Fig. 1.
ARAG1 encodes a DREB-like protein. (A) Amino acid sequence of ARAG1. The AP2/EREBP domain is underlined. The potential nuclear localization signal, the serine/threonine-rich region and VSEIRE motif is marked with bold, italic (in box) and shading, respectively. (B) ARAG1 is a nucleus-localized protein: 1, 2 are transformants with pCAMBIA1302 vector observed in FITC filter and bright field, respectively; 3, 4 are transformants with ARAG1 over-expression construct observed in the FITC filter and bright field, respectively. Scale bar = 30 µm for 1–4. (C) Phylogenetic tree constructed by multiple sequence alignments of the AP2/EREBP domain of ARAG1 and those of other DREB proteins using Clustal W (Higgins et al., 1992) and MEGA3.1 (Kumar et al., 2004). The sequences used are: arabidopsis DREB1A/CBF3, DREB1B/CBF1, DREB1C/CBF2, DREB1D/CBF4, DREB2A, DREB2B, ABI4, HARDY and TINY (Accession nos FJ169301, FJ169278, FJ169318, NM_124578, NM_001036760, NP_187713, NP_181551 NP_181186 and NP_197953, respectively), maize DBF1, DBF2 and ZmABI4 (Accession nos AF493800, AF493799 and AY125490), rice ARAG1, OsDREB1A, OsDREB1B and OsDREB2 (Accession nos BAG98540, AF300970, AF300972, AF300971).
A comparison of amino acid sequences revealed that ARAG1 shared higher sequence similarity with DREB proteins such as maize DBF2 (Kizis and Pages, 2002), arabidopsis TINY (Wilson et al., 1996) and DREB1A–1D/CBF1–4 (Liu et al., 1998; Haake et al., 2002) than other AP2/EREBP subgroup members. Moreover, the similarities were intensively restricted to their AP2/EREBP DNA-binding domain regions in which about 73–92 % amino acids were identical. In addition, ARAG1 contained the 14th valine, 19th glutamic and the quartet amino acids SEIR between the 14th and 19th amino acids (Fig. 1A). These amino acids, which are commonly present in other known DREB proteins, are identified as essential for the recognition and binding of the protein to the target DNA fragment (Sakuma et al., 2002).
Finally, phylogenetic analysis based on multi-sequence alignments of the domains revealed that ARAG1 formed a clade with DBF2 of maize, and it was more close to DREB1 than to DREB2 (Fig. 1C), implying that ARAG1 was a novel DREB-like protein of rice that might be involved in drought response.
Expression of ARAG1 was up-regulated by ABA or drought treatment rapidly and prominently
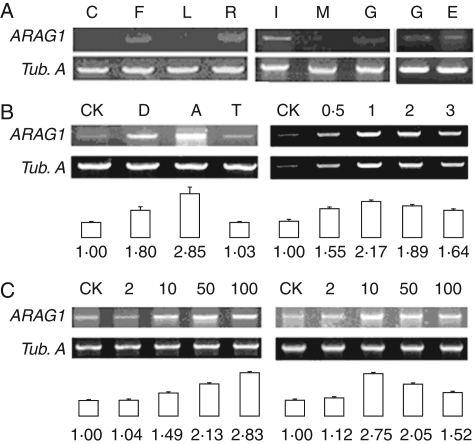
Northern blot failed to detect any signal in different organs of rice even if 30 µg of total RNA was loaded on the agarose gel, suggesting that ARAG1 was expressed at a very low level. Semi-quantitative RT-PCR showed that ARAG1 expressed in roots, inflorescences, endosperms of germinating seeds and developing and germinating embryos but not in coleoptiles, leaves and mature embryos (Fig. 2A).
Fig. 2.
mRNA accumulation pattern of ARAG1 in different tissues or at different development stages examined by semi-quantitative RT-PCR in triplicate. The statistical data presented are the mean ± s.d. Tubulin A (Tub. A) transcripts were used as constitutive controls. The expression intensity was analysed by 2DE Image Master software (2002.01). (A) Expression patterns of ARAG1 transcript in coleoptiles (C), inflorescences with male meiocytes at meiotic stage (F), leaves (L), roots (R), immature embryos (I), germinating embryos (G), endosperms of germinating seeds (E) and mature embryos (M). (B) Accumulation of ARAG1 transcripts in roots subjected to 0·5 h of drought (D), 100 µm ABA (A), and 4 °C low temperature (T), or with 100 µm ABA treatment for 0·5, 1, 2, 3 h, respectively. (C) Accumulation of ARAG1 transcripts in roots after treatment with 2 µm (2), 10 µm (10), 50 µm (50), 100 µm (100) ABA for 30 min (left) or for 60 min (right).
The response of ARAG1 to abiotic stresses and ABA inducement was examined because PLACE (www.dna.affrc.go.jp/PLACE/signalup.html), a database for plant cis-acting element identification (Higo et al., 1999), predicts that three ABRE elements (ACGTG) are presented in its promoter region (–210 to –206, +1604 to +1600 and –1605 to –1601 within 2 kb upstream of the ATG). In arabidopsis, these elements act as cis-acting elements of the ABA-mediated dehydration-responsive expression of rd29B (Yamaguchi-Shinozaki, 1994). As shown in Fig. 2B, compared with the control, expression of ARAG1 was up-regulated by drought and 100 µm ABA treatment after 30 min. However, it seemed insensitive to low temperature (Fig. 2B).
Detailed analysis revealed that the expression of ARAG1 culminated at 1 h of 100 µm ABA treatment, then decreased gradually (Fig. 2B, right). At 30 min of the treatment (Fig. 2C, left), the expression levels of ARAG1 were strengthened with the increase of ABA concentrations from 2 µm to 100 µm. When treatment time extended to 60 min (Fig. 2C, right), the transcripts increased continually by 10 µm ABA treatment. Two micromolar ABA had a similar but less obvious effect. In contrast, 50 µm and 100 µm ABA treatment resulted in decreases of ARAG1 expression, suggesting that self-protective mechanism to against harmful effects of ABA oversupply may exist in this process.
ARAG1 knockdown rice was hypersensitive to ABA inhibition during seed germination and seedling growth
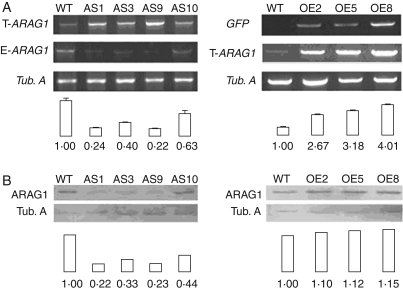
The functions of ARAG1 were investigated using knockdown and over-expression strategies. Three over-expression lines (OE2, OE5 and OE8) and four antisense lines (AS1, AS3, AS9 and AS10) of ARAG1 which had been confirmed by Southern blot or GUS staining (data not shown) were used to analyse expression. ARAG1 transcripts decreased in AS lines and increased in OE lines (Fig. 3A), implying that the constructs worked effectively. Western blot with antibody specific to ARAG1 revealed that the protein level was reduced significantly in AS lines; however, compared with ARAG1 in WT, its increase in OE lines was very limited (<20 %).
Fig. 3.
Expression of ARAG1 in transgenic rice. WT, wild type; AS1, AS9 and AS10, antisense lines 1, 3 and 9, respectively; OE2, OE5 and OE8, over-expression lines 2, 5 and 8, respectively. Left, transgenic AS line; right, transgenic OE line. (A) Semi-quantitative RT-PCR analyses of ARAG1 expression in rice transgenic lines. Tubulin A (Tub. A) transcripts were used as constitutive controls. Quantification of mRNA levels (ratios of normalized data for transgenic lines vs WT) is listed below the figure. The endogenous ARAG1 transcripts (E-ARAG1) were amplified by primers of P3 and P7; the total transcripts of ARAG1 (T-ARAG1) were amplified by primers of P3 and P4; GFP expression level (GFP) represents the over-expression level of ARAG1. (B) Western blot of ARAG1 expression in wild-type and transgenic rice lines.
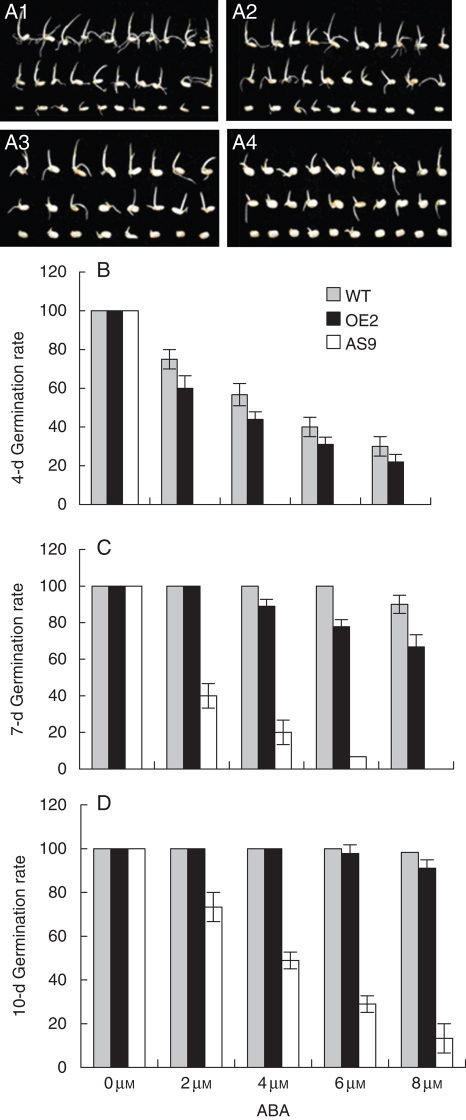
The progenies of positive AS9 and OE2 that did not show genotypic segregation (identified as in the T0 generation) were selected for phenotypic assessment. As ARAG1 was expressed during seed germination (Fig. 2A), ARAG1 transgenic seeds were examined for their ability to germinate, particularly in the presence of ABA application. All concentrations of ABA caused WT seeds to germinate more quickly than the transgenics, and their mean germination rate decreased proportionally with increased ABA concentration (Fig. 4A). At 4 d after imbibition (Fig. 4B), the germination rate of all the seeds reached 100 % in the absence of ABA application. However, when treated with ABA, it decreased obviously in WT and OE lines. No seed germinated in AS lines (Fig. 4B).
Fig. 4.
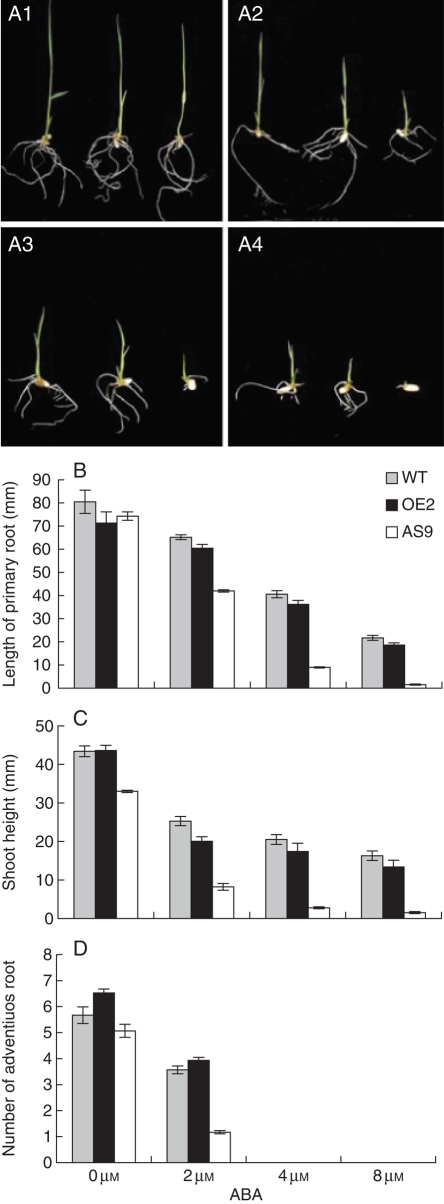
Germination of wild-type and ARAG1 transgenic rice seeds treated with different concentrations of ABA treatments for varying times. A1, A2, A3, A4 are photographs of transgenic and wild-type seeds germinated for 5d with 2, 4, 6 or 8 µm ABA, respectively. In each panel, the upper, middle and lower rows are WT, OE2 and AS9 seeds, respectively. (B–D) show the germination percentage of wild-type and ARAG1 transgenic rice seeds under 0, 2, 4, 6 or 8 µm ABA for 4, 7 or 10 d, respectively. Error bars indicate the s.d. (n = 30).
At 7 d after imbibition, germination rate of WT seeds reached 85 % even if 8 µm ABA was applied. More than half OE seeds germinated under all concentrations of ABA treatment. In addition, all AS seeds, except for those being treated with 8 µm ABA, began to germinate (Fig. 4C). At 10 d after imbibition, most of the WT and OE seeds had germinated under the different ABA concentrations and the AS seeds had began to germinate even when treated with 8 µm ABA (Fig. 4D).
Similar to the observation of seed germination, the seedling growth of WT and transgenic rice showed a similar tendency of inhibition in response to ABA treatments (Fig. 5A). Although the length of the primary root and the height of shoots of these seedlings were not obviously different in normal growth conditions, there was an significant discrepancy when different concentrations of ABA treatments were used – the higher the ABA concentration the more obvious the inhibition (Fig. 5B and C). It is worth noticing that the inhibition was more serious in the AS seedlings compared with their WT and OE counterparts. When treated with 4 or 8 µm ABA, very few adventitious roots were observed in any of the seedlings (Fig. 5D). Taken together, these facts indicate that knockdown of ARAG1 conferred hypersensitivity to ABA inhibition during seed germination and seedling growth of transgenics. In contrast, over-expression of the gene showed only a slight effect on these activities.
Fig. 5.
Response of ARAG1 transgenic and wild-type seedlings to ABA treatment. (A) Wild-type and ARAG1 transgenic rice seedlings after 3 d germination were treated with 0 µm ABA (A1), 2 µm ABA (A2), 4 µm ABA (A3) or 8 µm ABA (A4) for 10 d. In each panel, from left to right, the seedlings are wild type, OE line 2 or AS line 9, respectively. (B–D) Morphological parameters of wild-type and ARAG1 transgenic rice seedlings after treatment for 10 d. (B) Length of primary root (mm); (C) height of shoot (mm); (D) number of adventitious roots. Error bars indicate s.d. (n = 30).
ARAG1 plays a role in rice drought tolerance
Five-week-old hydroponically grown seedlings of WT and ARAG1 transgenics were prepared to compare their drought tolerance. Seedlings were treated in nutrient solution supplemented with 15 % PEG 6000 to mimic drought stress, with another set of seedlings grown in normal nutrient solution as a control. As shown in Fig. 6A, after 2 weeks of treatment, the AS line had a slightly wilted appearance and the WT line displayed similar but less-severe symptoms. In contrast, the OE seedlings looked normal. To evaluate the effects of the PEG treatment, the water concentration in the shoots of these plants was measured. Under drought stress, as shown in Fig. 6B, compared with the control that was grown in normal conditions, the WT line lost about 4·7 % of water, the OE line lost 4·4 % and the AS line lost 7·0 %. Consistent with the observation made with the seedlings, differences in water concentration between OE and WT seedlings seemed negligible, but they were evident between AS and WT lines. This suggests that ARAG1 was involved in the process associated with tolerance in the seedlings.
Fig. 6.
Comparison of drought tolerance of wild-type and ARAG1 transgenic rice. (A) After being in normal nutrient solution for 5 weeks, WT, OE2 and AS9 seedlings were grown in nutrient solution supplemented with 15 % PEG 6000 for 2 weeks. (B) Mean water concentrations in the shoots of the seedlings in Fig. 6A. Error bars signify standard deviations (n = 8).
α-Amylase activity was markedly enhanced in ARAG1 transgenic rice
As α-amylase activity is an important factor linked with seed germination, it was measured in WT and ARAG1 transgenic seeds. The results showed that, during germination, α-amylase activity increased remarkably in both the AS and the OE seeds compared with WT seeds (Fig. 7). It was contrary to the observation that, in the absence of ABA treatment, ARAG1 transgenic seed had similar germination features to WT seed. This demonstrates that germination and α-amylase activity are separate processes although they are closely linked in seeds. The presence of ARAG1 transcripts in embryos and endosperms of germinating seeds (Fig. 2) suggests that the gene is probably regulated differently in these tissues.
Fig. 7.
Activity of α-amylases in germinating seeds of wild-type and ARAG1 transgenic rice. Activity of α-amylases was indicated by the diameter of the hydrolysis circle on agar plates that contain soluble starch. Upper row, WT; middle row, OE2; lower row, AS9 (n = 30).
DISCUSSION
ABA plays an important role in many plant processes such as formation and dormancy of seeds, inhibition of germination and growth arrest in the early seedling stage under unfavourable environmental conditions (Koornneef and Karssen, 1994; Bewley, 1997; Lopez-Molina et al., 2001; González-Guzmán et al., 2002). Some DREB transcription factors, capable of being activated by ABA, mediate their downstream gene expression to help a plant survive a stressful environment (Kizis and Pagès, 2002; Karaba et al., 2007; Wang et al., 2008). However, during seed germination and seedling growth, little is known about the role of DREB proteins and how they were regulated by ABA. In this study, a DREB-related gene, ARAG1 was cloned and functionally analysed, and the findings may help to understand DREB protein function more comprehensively.
ARAG1 encodes a DREB-like protein containing the characterized AP2 DNA binding domain (Fig. 1). This domain, which was identified in other DREB proteins such as arabidopsis DREB1 and DREB2, rice OsDREBs and maize DBF2, can bind to the DRE/CRT cis-element of the stress-responsive genes, and thus activate the expression of their down-stream genes (Stockinger et al., 1997; Gilmour et al., 1998; Liu et al., 1998; Kizis and Pagès, 2002; Dubouzet et al., 2003). In addition, its relatively hydrophilic and acidic features, combined with the presence of nuclear-targeting motif, suggest that this protein has a potential nucleus-localizing function. The serine- and glutamine-rich motifs, which have been proposed to act as transcriptional activation domains in the maize DBF2 and some other transcriptional factors (Elliott et al., 1996; Kizis and Pagès, 2002) are also presented in ARAG1. These imply that ARAG1 may act as a transcriptional regulator in rice.
Expression of ARAG1 was quickly induced by drought stress, which is similar to maize DBF2 (Kizis and Pagés, 2002). The transgenic seedlings that over-express ARAG1 showed slightly enhanced tolerance, whereas the ARAG1 knockdown lines showed reduced tolerance to drought stress (Fig. 6). Perhaps like HARDY, an arabidopsis DREB gene which significantly increased drought tolerance of transgenics when expressed in rice (Karaba et al., 2007), ARAG1 is potentially useful for improving crop productivity.
α-Amylases are activated to hydrolyse carbohydrates that are stored in endosperm into soluble sugars to meet the needs of seed germination (Paiva and Kriz, 1994). Therefore, when seed is germinated in stressful conditions, α-amylase activity should be enhanced to provide additional energy for survival (Plaxton, 2004, 2006). In accordance with this postulation, the present results showed that activity of α-amylases was increased in ARAG1 transgenic seeds (Fig. 7), implying that when seedlings are grown in a stressful environment, ARAG1 can help them to survive by increasing the α-amylase activity. That the transgenic seedlings had smaller biomass compared with WT plants (data not shown) provided further support for this inference.
Previous findings have shown that transgenics with modified expression of AP2/EREBPs are either hypersensitive or insensitive to ABA treatment (Söderman et al., 2000; Pandey et al., 2005). In the present study, the AS lines were hypersensitive to ABA treatment since their seed germination and seedling growth was obviously repressed compared with that of WT control (Figs 4 and 5). However, little difference in these aspects is found between the ARAG1 OE line and the WT control (Figs 4 and 5). Given the fact that reducing ARAG1 protein conferred hypersensitivity on the AS seedlings to ABA application as compared with the WT, it is probable that ARAG1 acted as a negative regulator in the ABA-dependent pathway.
In drought stress, the OE line showed slightly increased tolerance and the AS line displayed reduced tolerance, implying that ARAG1 took part in this process besides its role in the ABA-dependent pathway. Interestingly, tolerance of drought stress in transgenics seems to be positively related to their ARAG1 level.
Recently, Vinces et al. (2009) reported that promoters containing a sequence of repeats confer higher rates of transcriptional divergence. Analogously, duplicating a copper regulatory element confers efficient copper induction on a heterologous promoter (Thiele and Hamer, 1986). This research indicates that repeating cis-elements are important for regulating the promoted gene's expression. Therefore, the three ABRE elements identified in the promoter region of ARAG1 may help explain the gene's diversified regulation property, which is important for plants to adapt flexibly to the environment.
ACKNOWLEDGEMENTS
This work was supported by the Chinese Ministry of Science and Technology (2007AA021402). We thank Linda Abraham who works in Plant Molecular & Cellular Biology Program, University of Florida, for her help in language editing.
LITERATURE CITED
- Bewley JD. Seed germination and dormancy. The Plant Cell. 1997;9:1055–1066. doi: 10.1105/tpc.9.7.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubouzet JG, Sakuma Y, Ito Y, et al. OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. The Plant Journal. 2003;33:751–763. doi: 10.1046/j.1365-313x.2003.01661.x. [DOI] [PubMed] [Google Scholar]
- Elliott RC, Betzner AS, Huttner E, et al. AINTEGUMENTA, an APETALA2-like gene of Arabidopsis with pleiotropic roles in ovule development and floral organ growth. The Plant Cell. 1996;8:155–168. doi: 10.1105/tpc.8.2.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Gampala SS, Rock CD. Abscisic acid signaling in seeds and seedlings. The Plant Cell. 2002;14(Suppl) doi: 10.1105/tpc.010441. S15–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour SJ, Zarka DG, Stockinger EJ, Salazar MP, Houghton JM, Thomashow MF. Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. The Plant Journal. 1998;16:433–442. doi: 10.1046/j.1365-313x.1998.00310.x. [DOI] [PubMed] [Google Scholar]
- González-Guzmán M, Apostolova N, Belles JM, et al. The short-chain alcohol dehydrogenase ABA2 catalyzes the conversion of xanthoxin to abscisic aldehyde. The Plant Cell. 2002;14:1833–1846. doi: 10.1105/tpc.002477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutterson N, Reuber TL. Regulation of disease resistance pathways by AP2/ERF transcription factors. Current Opinion in Plant Biology. 2004;7:465–471. doi: 10.1016/j.pbi.2004.04.007. [DOI] [PubMed] [Google Scholar]
- Gu YQ, Wildermuth MC, Chakravarthy S, et al. Tomato transcription factors pti4, pti5, and pti6 activate defense responses when expressed in Arabidopsis. The Plant Cell. 2002;14:817–831. doi: 10.1105/tpc.000794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haake V, Cook D, Riechmann JL, Pineda O, Thomashow MF, Zhang JZ. Transcription factor CBF4 is a regulator of drought adaptation in Arabidopsis. Plant Physiology. 2002;130:639–648. doi: 10.1104/pp.006478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanly WC, Artwohl JE, Bennett BT. Review of polyclonal antibody production in mammals and poultry. ILAR Journal. 1995;37:93–118. doi: 10.1093/ilar.37.3.93. [DOI] [PubMed] [Google Scholar]
- Hiei Y, Ohta S, Komari T, Kumashiro T. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. The Plant Journal. 1994;6:271–282. doi: 10.1046/j.1365-313x.1994.6020271.x. [DOI] [PubMed] [Google Scholar]
- Higgins DG, Bleasby AJ, Fuchs R. CLUSTAL V: improved software for multiple sequence alignment. Computer Applications in the Biosciences. 1992;8:189–191. doi: 10.1093/bioinformatics/8.2.189. [DOI] [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T. Plant cis-acting regulatory DNA elements (PLACE) database. Nucleic Acids Research. 1999;27:297–300. doi: 10.1093/nar/27.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Zhao L, Chong K, Wang T. Overexpression of OsERF1, a novel rice ERF gene, up-regulates ethylene-responsive genes expression besides affecting growth and development in Arabidopsis. Journal of Plant Physiology. 2008;165:1717–1725. doi: 10.1016/j.jplph.2007.12.006. [DOI] [PubMed] [Google Scholar]
- Ikeda A, Ueguchi-Tanaka M, Sonoda Y, et al. Slender rice, a constitutive gibberellin response mutant, is caused by a null mutation of the SLR1 gene, an ortholog of the height-regulating gene GAI/RGA/RHT/D8. The Plant Cell. 2001;13:999–1010. doi: 10.1105/tpc.13.5.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. The EMBO Journal. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jofuku KD, den Boer BG, Van Montagu M, Okamuro JK. Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. The Plant Cell. 1994;6:1211–1125. doi: 10.1105/tpc.6.9.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaba A, Dixit S, Greco R, et al. Improvement of water use efficiency in rice by expression of HARDY, an Arabidopsis drought and salt tolerance gene. Proceedings of the National Academy of Sciences of the USA. 2007;104:15270–15275. doi: 10.1073/pnas.0707294104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kizis D, Pagès M. Maize DRE-binding proteins DBF1 and DBF2 are involved in rab17 regulation through the drought-responsive element in an ABA-dependent pathway. The Plant Journal. 2002;30:679–689. doi: 10.1046/j.1365-313x.2002.01325.x. [DOI] [PubMed] [Google Scholar]
- Knight H, Zarka DG, Okamoto H, Thomashow MF, Knight MR. Abscisic acid induces CBF gene transcription and subsequent induction of cold-regulated genes via the CRT promoter element. Plant Physiology. 2004;135:1710–1717. doi: 10.1104/pp.104.043562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Karssen CM. Seed dormancy and germination. In: Meyerowitz EM, Somerville CR, editors. Arabidopsis. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1994. pp. 313–334. [Google Scholar]
- Kumar A, Silim SN, Okamoto M, Siddiqi MY, Glass AD. Differential expression of three members of the AMT1 gene family encoding putative high-affinity NH4+ transporters in roots of Oryza sativa subspecies indica. Plant, Cell & Environment. 2003;26:907–914. doi: 10.1046/j.1365-3040.2003.01023.x. [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M. Integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinformatics. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Li H, Zhang P, Zha X, He Z, Xia X. A method for rapid isolation of high-quality total RNA from wheat seeds. Molecular Plant Breeding. 2006;4:877–881. [in Chinese] [Google Scholar]
- Liu Q, Kasuga M, Sakuma Y, et al. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. The Plant Cell. 1998;10:1391–1406. doi: 10.1105/tpc.10.8.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, Chua NH. A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proceedings of the National Academy of Sciences of the USA. 2001;98:4782–4787. doi: 10.1073/pnas.081594298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama K, Sakuma Y, Kasuga M, et al. Identification of cold-inducible downstream genes of the Arabidopsis DREB1A/CBF3 transcriptional factor using two microarray systems. The Plant Journal. 2004;38:982–993. doi: 10.1111/j.1365-313X.2004.02100.x. [DOI] [PubMed] [Google Scholar]
- Murray MG, Thompson WF. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Research. 1980;8:4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narusaka Y, Nakashima K, Shinwari ZK, et al. Interaction between two cis-acting elements, ABRE and DRE, in ABA-dependent expression of Arabidopsis rd29A gene in response to dehydration and high-salinity stresses. The Plant Journal. 2003;34:137–148. doi: 10.1046/j.1365-313x.2003.01708.x. [DOI] [PubMed] [Google Scholar]
- Niu X, Helentjaris T, Bate NJ. Maize ABI4 binds coupling element1 in abscisic acid and sugar response genes. The Plant Cell. 2002;14:2565–2575. doi: 10.1105/tpc.003400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamuro JK, Caster B, Villarroel R, Van Montagu M, Jofuku DK. The AP2 domain of APETALA2 defines a large new family of DNA binding proteins in Arabidopsis. Proceedings of the National Academy of Sciences of the USA. 1997;94:7076–7081. doi: 10.1073/pnas.94.13.7076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paiva R, Kriz AL. Effects of abscisic acid on embryo-specific gene expression during normal and precocious germination in normal and viviparous maize (Zea mays) embryos. Planta. 1994;192:332–339. [Google Scholar]
- Pandey GK, Grant JJ, Cheong YH, Kim BG, Li L, Luan S. ABR1, an APETALA2-domain transcription factor that functions as a repressor of ABA response in Arabidopsis. Plant Physiology. 2005;139:1185–1193. doi: 10.1104/pp.105.066324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaxton WC. Plant response to stress: biochemical adaptations to phosphate deficiency. In: Goodman RM, editor. Encyclopedia of plant and crop science. New York, NY: Marcel Dekker; 2004. pp. 976–980. [Google Scholar]
- Plaxton WC. Metabolic flexibility helps plants to survive stress. In: Taiz L, Zeiger E, editors. Plant Physiology. 4th edn. Heidelberg: Sinauer Associates; 2006. [Google Scholar]
- Sakuma Y, Liu Q, Dubouzet JG, Abe H, Shinozaki K, Yamaguchi-Shinozaki K. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochemical and Biophysical Research Communications. 2002;90:998–1009. doi: 10.1006/bbrc.2001.6299. [DOI] [PubMed] [Google Scholar]
- Sakuma Y, Maruyama K, Osakabe Y, et al. Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. The Plant Cell. 2006;18:1292–1309. doi: 10.1105/tpc.105.035881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salekdeh GH, Sinopongco J, Wade LJ, Ghareyazie B, Bennett J. Proteomic analysis of rice leaves during drought stress and recovery. Proteomics. 2002;2:1131–1145. doi: 10.1002/1615-9861(200209)2:9<1131::AID-PROT1131>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K. Molecular responses to dehydration and low temperature: differences and cross-talk between two stress signaling pathways. Current Opinion in Plant Biology. 2000;3:217–223. [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K, Seki M. Regulatory network of gene expression in the drought and cold stress responses. Current Opinion in Plant Biology. 2003;6:410–417. doi: 10.1016/s1369-5266(03)00092-x. [DOI] [PubMed] [Google Scholar]
- Söderman EM, Brocard IM, Lynch TJ, Finkelstein RR. Regulation and function of the Arabidopsis ABA-insensitive4 gene in seed and abscisic acid response. Plant Physiology. 2000;124:1752–1765. doi: 10.1104/pp.124.4.1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockinger EJ, Gilmour SJ, Thomashow MF. Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proceedings of the National Academy of Sciences of the USA. 1997;94:1035–1040. doi: 10.1073/pnas.94.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao J, Zhang L, Chong K, Wang T. OsRAD21-3, an orthologue of yeast RAD21, is required for pollen development in Oryza sativa. The Plant Journal. 2007;51:919–930. doi: 10.1111/j.1365-313X.2007.03190.x. [DOI] [PubMed] [Google Scholar]
- Thiele DJ, Hamer DH. Tandemly duplicated upstream control sequences mediate copper-induced transcription of the Saccharomyces cerevisiae copper-metallothionein gene. Molecular Cell Biology. 1986;6:1158–1163. doi: 10.1128/mcb.6.4.1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinces MD, Legendre M, Caldara M, Hagihara M, Verstrepen KJ. Unstable tandem repeats in promoters confer transcriptional evolvability. Science. 2009;324:1213–1216. doi: 10.1126/science.1170097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Guan Y, Wu Y, Chen H, Chen F, Chu C. Overexpression of a rice OsDREB1F gene increases salt, drought, and low temperature tolerance in both Arabidopsis and rice. Plant Molecular Biology. 2008;67:589–602. doi: 10.1007/s11103-008-9340-6. [DOI] [PubMed] [Google Scholar]
- Weigel D. The APETALA2 domain is related to a novel type of DNA binding domain. The Plant Cell. 1995;7:388–389. doi: 10.1105/tpc.7.4.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson K, Long D, Swinburne J, Coupland G. A Dissociation insertion causes a semidominant mutation that increases expression of TINY, an Arabidopsis gene related to APETALA2. The Plant Cell. 1996;8:659–671. doi: 10.1105/tpc.8.4.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worrall DM. Extraction of recombinant protein from bacteria. Methods in Molecular Biology. 1996;59:31–37. doi: 10.1385/0-89603-336-8:31. [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. The Plant Cell. 1994;6:251–264. doi: 10.1105/tpc.6.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends in Plant Science. 2005;10:88–94. doi: 10.1016/j.tplants.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Zhang L, Tao J, Wang S, Chong K, Wang T. The rice OsRad21-4, an orthologue of yeast Rec8 protein, is required for efficient meiosis. Plant Molecular Biology. 2006;60:533–554. doi: 10.1007/s11103-005-4922-z. [DOI] [PubMed] [Google Scholar]