Abstract
Isoelectric Focusing (IEF) is the first step for two-dimensional (2D) gel electrophoresis and plays an important role in sample purification for proteomics. However, biases in protein size and pI resolution, as well as limitations in sample volume, gel capacity, sample loss, and experimental time, remain challenges. In order to address some of the limitations of traditional IEF, we present a microfluidic free flow IEF (FF-IEF) device for continuous protein separation into 24 fractions. The device reproducibly establishes a nearly linear pH gradient from 4 to 10. Optimized dynamic coatings of 4% poly (vinyl) alcohol (PVA) minimize peak broadening by transverse electrokinetic flows. Even though the device operates at high electric fields (up to 370V/cm) efficient cooling maintains solution temperature inside the separation channel controllably in the range 2 – 25 °C. Protein samples with a dynamic concentration range between µg/mL to mg/mL can be loaded into the micro device at a flow rate of 1 mL/hr and residence time of ~12 min. By using a protein complex of 9 proteins and 13 isoforms, we demonstrate improved separation with the FF-IEF system over traditional 2D gel electrophoresis. Device to device reproducibility is also illustrated through the efficient depletion of the albumin and hemoglobin assays. Post-device sample concentrations result in a 10 to 20-fold increase, which allow for isolation and detection of low abundance proteins. The separation of specific proteins from a whole cell lysate is demonstrated as an example. The micro device has the further benefits of retaining high molecular weight proteins, providing higher yield of protein that has a broader range in pI, and reducing experimental time compared to conventional IEF IGP gel strip approaches.
Keywords: Microfluidic, Preparative, Free flow, Isoelectric focusing, protein separation, poly(acrylamide) gel, 2D Gel Electrophoresis
INTRODUCTION
Free-flow electrophoresis (FFE), also known as continuous flow electrophoresis, is a popular preparative-scale fractionation and separation techniques. FFE utilizes a thin stream of separation buffer that flows continuously in a laminar fashion between two closed spaced plates and an electric field that is applied perpendicular to the flow. FFE results in a differential deflection of charged sample components as they move toward the collection outlets. This allows the high-throughput separation of samples, such as low-molecular-weight organic compounds, 1 peptides, proteins, protein complexes,2 membranes,3 organelles,4 and whole cells.5 Isoelectric focusing (IEF), one of the modes that FFE supports, is a technique that separates and concentrates proteins based upon their isoelectric point (pI). Proteins migrate until they reach the pH value the gradient at which the charge of the protein becomes zero.
2-D gel electrophoresis is a standard method in which proteins are first separated based on their pI using IEF and then followed with SDS-PAGE to separate the proteins on the basis size. Using an IEF IPG strip, it generally takes 24–36 hrs for the first step of the IEF focusing. Furthermore, a typical 2D gel can take no more than 200 – 500 µg of protein, making it difficult to study low-abundance proteins. Although preparative IEF can be performed on large and thick gels, new instruments for sample fractionation have been developed for preparative IEF in liquid phase, such as Rotofor® cell from BioRad,6 and the ZOOM® IEF fractionator from Invitrogen.7,8 Another fractionation method, the Agilent 3000 OFFGEL fractionator has a frame of 10 wells filled with liquid solution sitting atop of the IPG gel strip. Isoelectric focused proteins inside the gel strip are able to diffuse to the solution for recovery. Weber and his coworkers9,10 developed the first preparative FF-IEF platform, now commercialized as the BD™ Free Flow Electrophoresis System with a 96-well plate interface. In this system, which remains the only commercial FF-IEF instrument available, sample solutions of 1 mg/mL are continuously injected at a flow rate of 1mL/hr to a large rectangular separation chamber (500 × 100 × 0.5 mm L ×W × D) while the separation media sheath flow is loaded into the chamber through multiple inlets at a flow rate of 60 mL/hr. A counterflow is applied at the end of the chamber to facilitate channel flow to the 96 outlets, generating an extra flow of 40 mL/hr. Samples exit the device with a typical residence time of ~25 min. The BD™ FF-IEF system has been widely used as a high-throughput platform for applications of cell organelle purification11,12 and protein isoforms separation.13,14
Whereas analytical IEF is used to separate and identify proteins in complex mixtures, the purpose of preparative IEF is to purify one protein, or a small number of proteins of interest. However, most of the current microfluidic FF-IEF15–21 are on the analytical scale with volumes typically less than 10 µL and the flow rates from sub- µL to µL per minute. Inexpensive and efficient preparative IEF devices have the potential to become a new tool for research involving difficult proteins and protein complexes, reducing laborious sample preparation and increasing assay sensitivity. In addition, in order to perform practical bioseparations, FF-IEF microdevices should preferably fractionate the separated proteins into multiple outlet streams to facilitate downstream processing. In this contribution, we present optimization and validation of a triangular shaped microfluidic preparative FF-IEF device. Dynamic coating to suppress electroosmotic flow (EOF) from the chamber walls is investigated as a means to miniaturize the peak broadening and improve the resolution. Joule heating removal and associated chamber solution temperature control are implemented to reduce fluid thermal distortion and possible sample precipitation. The sample loading flow rate is optimized with respect to fast separation and high throughput. Finally, the microfluidic FF-IEF device is evaluated using multiple protein mixtures and the results compared to those achievable by standard 2D gel procedures.
EXPERIMENTAL SECTION
Reagents and Chemicals
All water used in this work was pre-purified by a Milli-Q Academic (Millipore, Billerica, MA) equipped with an extra 0.22 µm membrane filter. Tris(hydroxymethyl)aminomethane (Tris, 99.9+%), 3-(trimethoxysilyl)propyl methacrylate (98%+), 2,2-dimethoxy-2-phenylacetophenone (DMPA), poly (vinyl alcohol) (PVA, Mw 89,000 ~ 98,000, 99+%), urea (99.5+%), thiourea (99+%), CHAPS (98+%), Methyl Red, Bromothymol Blue, Nonidet P 40 substitute (NP-40), 10X PBS buffer, ethanol (95%), acetone (HPLC grade), Phosphoric acid (85%), acetic acid (≥99.7%), Triton X-100, mineral oil, protein samples of cytochrome C, Carbonic anhydrase I, β-lactoglobulin A were obtained from Sigma Aldrich (St. Louis, MO). Poly(dimethylsiloxane) (PDMS, Sylgard 184, Dow Chemicals, Midland, MI) kit was obtained from Ellsworth Adhesives (Germantown, WI). Acrylamide monomer solutions (PlusOne ReadySol IEF), Immobilines of pK 3.6 and pK 9.3 and pH 3–10 IPG strips were obtained from GE Healthcare, Piscataway, NJ. Cathode electrode buffer (10×) was obtained from Bio-rad Laboratory (Hercules, CA). Zoom® carrier ampholytes 3–10, IEF Markers 3–10 SERVA Liquid Mix, and SimplyBlue™ SafeStain staining reagents were obtained from Invitrogen (Carlsbad, CA). NuSep® mini gels 4–20% T for SDS-PAGE were obtained from Nusep (Lawrenceville, GA).
Device Fabrication and operation
Devices (Figure 1) were fabricated using standard soft lithography techniques and described in our previous report.22 Briefly, a mold was created from the photopatternable polymer SU-8 2050 (MicroChem, Newton, MA) spin-cast on a featureless 4” silicon wafer with a uniform thickness of 160 µm. The SU-8 was exposed to UV through a transparency mask to transfer the features. Fabrication of the PDMS device (Figure 1) was performed in two steps. A small amount of PDMS was first poured over the mold and cured at 80°C until solid (~20 min). A supporting glass slide was positioned above the first layer to prevent the channel sagging. More was poured and allowed to completely cure. After the PDMS layer was cut and peeled from the master, it was shaped and extra regions were removed by allowing for 15 mm wide on the bottom to the inlet and 2 mm on the top on each side. Access holes (one inlet, 24 outlets) were punched using a blunt-end 20 gauge Luer stub adapter (Becton-Dickinson, Sparks, MD).
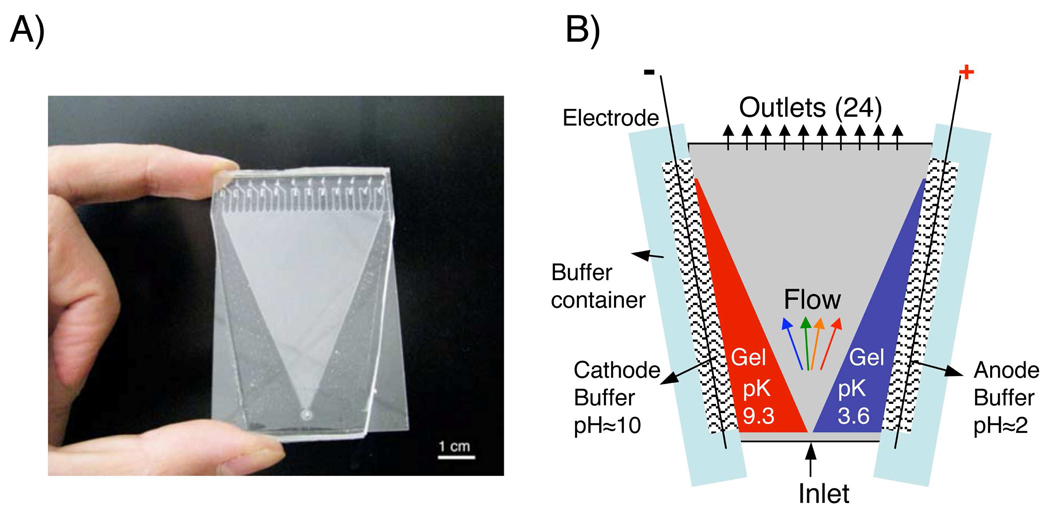
Figure 1.
A) Image of FF-IEF device. B) Schematic of Microfluidic FF-IEF system. Functionalized pH gradient cathode (pK=9.3) and anode (pK=3.6) polyacrylamide gels are polymerized into the gel regions connected to the separation channel. Posts are included in the gel regions for support. The device is 50 mm by 75 mm with the center triangular separation channel of 46 mm as top width and 56 cm in length. All channels and gel sections have a depth of 160 µm.
The PDMS/glass device was sealed by surface oxidation in oxygen plasma (Model PDC-32G, Harrick, Ithaca, NY) on high level. Double-wide microscope slides (75 × 50 mm, Erie Scientific, VWR) were first spin-coated with a thin layer of PDMS at 6000 rpm for 20 sec (Model WS-400B-6NPPL/LITE, Laurell Technologies Cortporation, Wales, PA) and cured at 80°C for 2 hrs. Both PDMS layer and PDMS spin-coated glass were exposed to plasma for 45 sec and then allow to be put together for a permanent bond. In case of situation that PDMS layer was bonded to a plain glass layer, the glass was plasma pretreated for 65 sec before co-treated with PDMS layer. The devices were then filled with 1% 3-(trimethoxysilyl) propyl methacrylate in ethanol (95%) with pH adjusted to ~5 using 6 M acetic acid (~ 1 % v/v) for 1 hr. All the devices were dried, degassed in vacuum overnight at 80°C, and then transferred to an acrylic nitrogen glove box (Air Control, Inc., Henderson, NC) for poly (acrylamide) gel casting. Stock anode and cathode monomer acrylamide gel solution were prepared by mixing 9 mL acrylamide monomer solution containing 40% w/v acrylamide monomer and 3% w/w N,N’-methylenebisacrylamide, 13.5 mL Milli-Q water, and 1.5 mL Immobiline pK 3.6 (stock in water) or Immobiline pK 9.3 (stock in 2-proponal), respectively. Stock gel solutions were stored at 4°C for reuse. Photo initiator was mixed with the acrylamide gel solution right before the polymerization. Polymerization working solution was prepared by adding 6 µL of 10% DMPA (photo initiator) to every 1 mL of the stock monomer solution right before the polymerization step. An extra of 24 µL of 1% v/v Triton X-100 was added to every 1 mL of anode polymerization solution. The monomer solutions were introduced into the edges of the device and pulled through the PDMS/glass device by capillary action before it was held by surface tension at the hydrophobic channel-gel region barriers (a series of posts with 40 µm spacing).22 It is very critical to control the overall hydrophobicity of the polymerization solution (volume of Triton added) so that it will not cross the barriers to the central focusing chamber during the polymerization step. The acrylamide was polymerized by exposure to long-wave UV (354 nm, Spectroline ENF-280C, Spectronics Corporation, Westbury, NY) for 3 min (Figure 1B). The devices were stored in 2% (w/v) PVA solution.
To setup a simple separation system, two strips of 2”-wide Scotch tape were used to stick the device on both edges from the back with two custom buffer containers made from PDMS. The tapes effectively prevented a potential short circuit between the electrode buffers through the device underneath. 5 min@ Epoxy glue (Danvers, MA) were then applied to all the contact between the device and the containers for a complete seal and prevent electrode buffer leak. Two platinum electrodes were inserted through PDMS containers in contact with the cathode buffer (20 mM Arginine, 20 mM Lysine) and anode electrode buffer (0.1 M H3PO4). The device was placed atop a thermoelectric cold plate with a temperature controller (CP-036 and TC-24-10, TE Technology, Inc., Traverse City, MI) to efficiently remove the heat generated during the electrophoresis and to maintain the devices at 2°C. Mineral oil was applied between the cooling plate and device for improved heat transfer and for the prevention of a possible short circuit. Samples were delivered into the device via a syringe pump (Cole-Parmer, Vernon Hills, IL), and the device was powered by a high-voltage power supply (EPS 3501, GE Healthcare, Piscataway, NJ). Maximums of 2 mA in current and 1800 V in voltage were set from the power supply to ensure the device to run under stable conditions, whichever arrives first. Twenty-four 200-µL gel loading capillary pipette tips (VWR, West Chester, PA) were inserted into the PDMS to collect the outlet fractions. The fine plastic capillary of the pipette tip fit snugly into the 20-gauge hole, and were held in place without leaks for the duration of the experiment.
Sample Preparation and Analysis
Urea-based denaturing sample buffer was prepared as 8 M urea, 2 M thiourea, 4% CHAPS, 4% PVA, 0.01% Methyl Red and 2% ZOOM® carrier ampholytes. Protein samples were prepared in a variety of concentrations with the urea denaturing buffer. Protein mixture (cytochrome C, Carbonic anhydrase I, β-lactoglobulin A) stock solution for surface coating optimization experiments was prepared as 4 mg/mL in total concentration (protein ratio of 1:1:1) in urea sample buffer and diluted to 0.2 mg/mL before applied to FF-IEF at a flow rate of 0.6 mL/hr. Stock IEF protein marker 3–10 solution (10 mg/mL) was diluted to 1 mg/mL in urea sample buffer before applied to FF-IEF at a flow rate of 1 mL/hr. Stock HELA whole cell lysate (9.2 mg/mL) was prepared by lysing four 15 cm plates of 80% confluent asynchronious HeLa H3 cells with 8M urea, 2M thiourea, and 4%CHAPS. Lysates were passed though a 30 gauge syringe needle 5 times to shear DNA. Extracts were centrifuged for 20 minutes at 14,000 × g and the supernatant diluted to 0.5 mg/mL in urea sample buffer before loaded to the device at a flow rate of 0.6 mL/hr. All fractionated protein samples were then transferred to the tubes and further analyzed. Fractionated protein samples were then transferred to the tubes and analyzed by SDS-PAGE. Samples for SDS-PAGE experiment were prepared as 10 µL fractions mixed 10 µL 2× SDS sample buffer (containing 2% 2-mercaptoethanol). To each of fractions 17–24, 0.5 µL of 1 M NaOH was added to bring the pH value to neutral range for SDS-PAGE experiment. All samples were heated to 95°C for 5 min and then allowed to cool down to room temperature before 5 µL of each was loaded to the gels. SDS-PAGE was conducted on NuSep® mini gels 4–20% gradient of acrylamide. Proteins were visualized by staining with SimplyBlue™ following the manufacturer’s guidelines. SDS-PAGE gels were then scanned and processed with ImageJ (http://rsb.info.nih.gov/ij/) with brightness and contrast adjusted to approximately the same level for all gels. Samples for western blotting were resolved on 4M urea/10% polyacrylamide gels, transferred to PVDF membrane, and probed using a 14-3-3sigma-specific monoclonal antibody (CS112) as described.23
2D Gel Electrophoresis
IEF 3–10 markers (Invitrogen®) were diluted into sample buffer (8 M urea, 2M thiourea, 4% CHAPS, 2% IPG buffer 3–10, DTT 15 mg/ml) to 1 mg/mL. Ten microgram of sample were cup loaded onto 7 cm, pH 3.10 IPG strips and resolved at 50 mA/strip for 100V for 30 min, 200V for 30 min, 400V for 30 min, 1000V for 60 min, 3500V for 5–6 hr, and 500V to a total of 20,000V·hours using a IPGphor unit (Pharmacia biotech). Strips were then washed in wash solution (50 mM Tris-HCl [pH 8.8], 6 M Urea, 30% Glycerol, 2% SDS) supplemented with 20 mg/ml DTT for 10 min at RT and followed by a wash in wash buffer supplemented with 25 mg/ml iodoacetamide for 10 min. The strips were then loaded onto a standard SDS-PAGE and separated by standard electrophoresis. Gels were fixed in a water/10% acetic acid/10% methanol solution for 1 hr and then stained with Coomassie blue.
RESULTS AND DISCUSSION
EOF Suppression
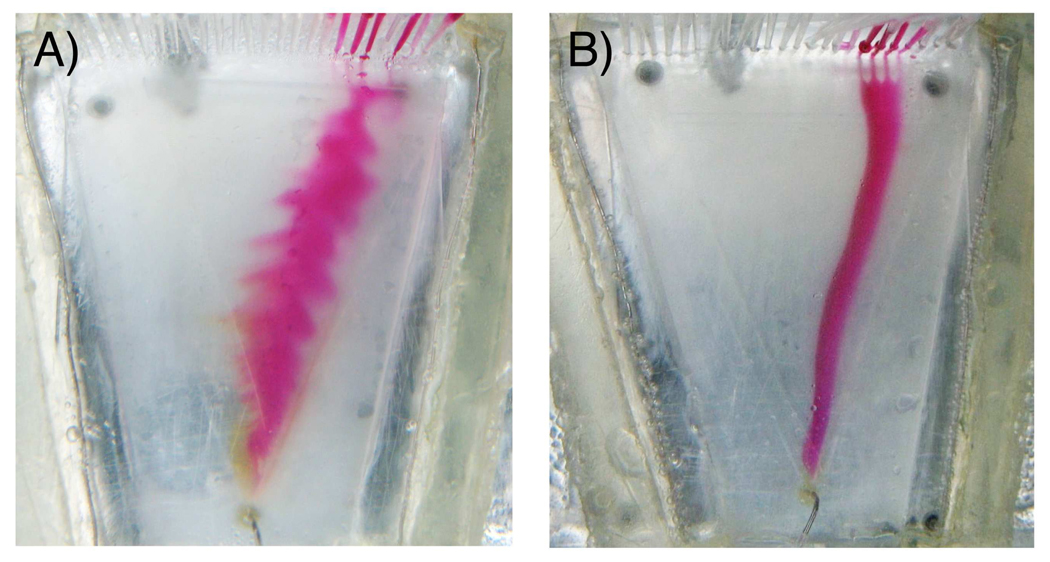
EOF is caused by the charged double layers near the chamber wall, which for the current glass bottom device creates a flow towards the cathode in the electric field. EOF is often eliminated by permanent coatings that are covalently bonded to the channel walls with neutral and hydrophilic polymers, but complex coating procedures and stability of the coatings at alkaline pH have been challenges. 24 Dynamic coating procedures that mix the soluble polymer in the buffer during operation are often more adaptable. A temporary layer of polymer adsorbs on the channel walls and suppresses the EOF as demonstrated for microfluidic glass, 25 PDMS26, 27 or plastic devices.28 Dynamic coating has the additional potential advantage of reducing non-specific adsorption of proteins from the sample.29
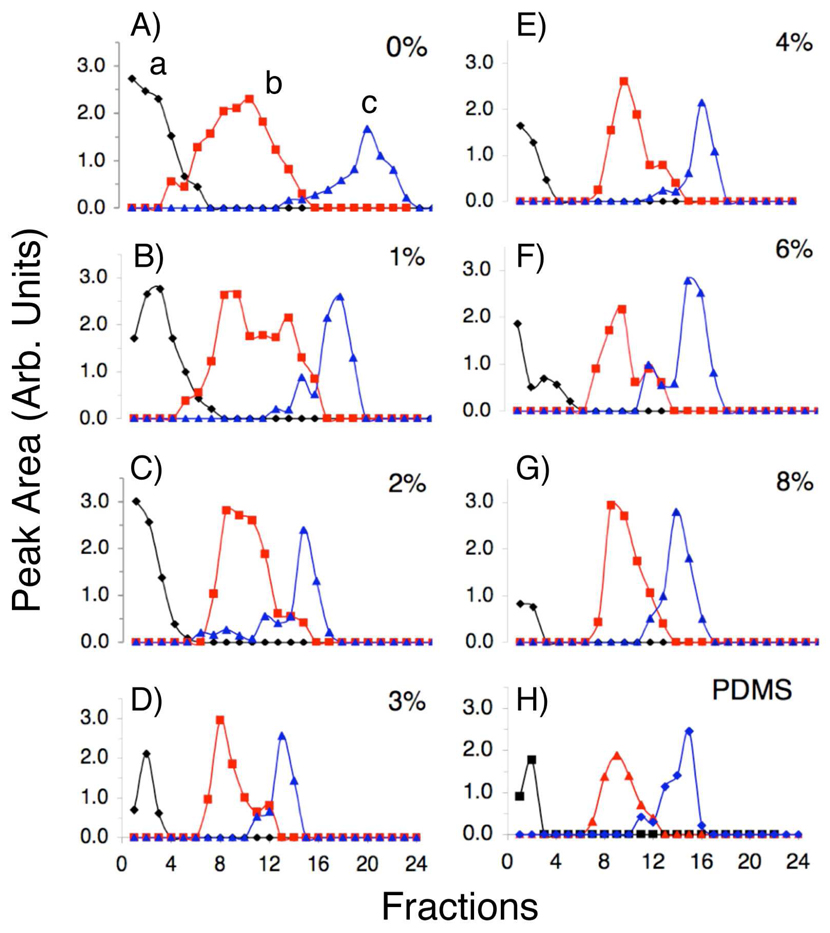
Figure 2 shows the effect of PVA concentration on separation of 3 proteins. The collected fractions were run on standard SDS-PAGE gels. In order to easily compare the separation results from different dynamic coating conditions on different gels, the gels were scanned and ImageJ software was used to read the band intensity of each fraction into a peak format. Peak area was calculated and plotted in Figure 2 versus fraction collected, with the bands normalized for the amount of protein loaded. Even though devices were stored in a 2% PVA water solution, we observed a significant band broadening (Figure 2A) when no PVA was included in the running buffer, presumably caused by the non-uniform EOF.30 Since IEF separation relies on the formation of a linear pH gradient along the separation channel, a transverse EOF greatly affect the separation resolution through the unwanted sample dispersion. In another report, we have shown that the device is able to achieve constant electric fields high as 370 ± 20 V/cm through the entire triangular channel.22 Considering the relatively large size of the device in dimension and high electric field across the separation chamber, the EOF could varies at different locations due to factors such as nonunifom surface coating and the differing zeta potentials of the PDMS and glass surfaces at the top and bottom of the channel, resulting in the ‘wavy’ flow shown in Figure 3A. Similar ‘wavy’ flow as the evidence of the EOF instability has also been reported in microfluidic electrophoresis studies.31 Protein was difficult in focusing and broadened across multiple fractions shown in Figure 2A. The peaks appear to be better resolved because of the narrower pH range of the outlets. EOF would prevent steep pH gradient formation, reducing the overall resolution of the device. Dramatic improvement was observed when the PVA concentration was increased from 2% to 4% in the running buffer. It is also worthwhile to note that the majority of protein b (Carbonic anhydrase I, pI 6.6) and protein c (β-lactoglobulin A, pI 5.1) has shifted from fraction 10 to 8 and18 to 15, respectively upon their complete focusing because of the decreased EOF effect. Both peaks have been much narrower in presence of 4% PVA compared to that in absence of PVA, showing an improved resolution of the device. Increasing the concentration of PVA past 4% produced no significant further improvement (Figure 2 and Figure 3). Thus, 4% PVA included in the running buffer was employed in subsequent experiments.
Figure 2.
Effect of dynamic PVA surface coating on protein separation. Sample: a). Cytochrome b, pI 9.5–10.2, b). Carbonic anhydrase I, pI 6.6 and c). β-lactoglobulin A, pI 5.1. Buffer: 8M Urea, 2M Thiourea, 4% CHAPS, 2% Amphyolytes, 0.1% Methyl red. Applied voltage: 1,800V. Current: 1.4 mA. Flow rate: 10 µL/min. [Total protein]: 0.2 mg/mL. Electric field: 370 V/cm. A)-G) increasing PVA concentrations in PMDS devices with glass bottoms. H) All PDMS channel with 4% PVA.
Figure 3.
Flow images visualized by introduction of Methyl Red, (A) without dynamic coating and (B) with 4% PVA. Buffer and separation conditions are the same as in Figure 2.
A spin-coated layer of PDMS (~10 µm in thickness) on the glass prior to device bonding was also investigated since EOF resulting from the glass layer is larger than that of PDMS.32 However, no significant enhancement was observed over those already achieved by using 4% PVA, perhaps because the extraneous peak broadening were a result of EOF. Peak broadening across multiple fractions could also originate in a broad pI range of the protein itself, such as cytochrome C (pI 9.5–10.2) or product purity (multiple isoforms for Carbonic anhydrase II and III). Use of PDMS coated glass substrates did have the additional advantage of improving bonding, but the thickness of the PDMS layer also greatly affected the heat transfer of Joule heating to the cooling plate. PDMS layers had to be coated to thicknesses in the range 5–10 µm (6000 rpm) to avoid boiling of the solution. The devices with PDMS pre-spin coated glass layers were chosen for our further investigations.
Establishment of pH Gradient
IEF is performed through establishment of a pH gradient inside the separation channel. Such a gradient are formed by using a mixture of specially designed acidic and basic amphoteric substances, typically referred to as carrier ampholytes. By including 2% free ampholytes in the solution, the pH gradient is dynamically established and maintained as the sample solution is continuously loaded. Figure 4 shows the pH measurement of each outlet fraction by a micro electrode (Thermo Orion 9802BN, Waltham, MA) averaged from 10 devices. A reproducible and nearly linear pH gradient from 10 to 4 is established along the 24 outlets. The pH drop is observed to be a little steeper between the alkaline and the neutral region (pH 10-5.5, 0.3 pH value internal) relative to that of the acidic region (pH 5.5-4, 0.1 pH value internal). A modestly higher flow variation was found in the central fractions (± 0.4 pH) relative to the end fractions (± 0.1 pH). It is also worthwhile to point out that the pH gradient could be adjusted further by including different immobilines during the device fabrication and carrier ampholytes in the running buffer for different pH ranges, such as 10-8, 8-6, or 6-4.
Figure 4.
Establishment of the pH gradient across the 24 outlet fractions. Buffer and separation conditions are the same as in Figure 2. Data averaged from experiments with 10 different chips.
Temperature Measurements
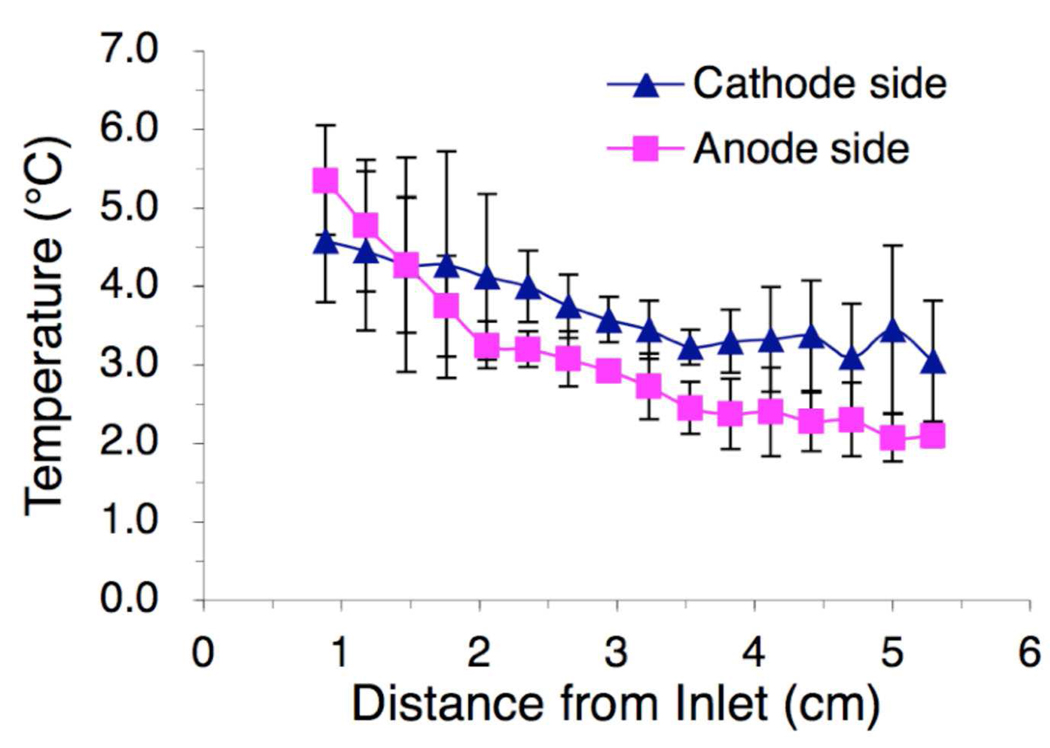
Temperature difference in the separation channel solution could to lead thermally driven convection distorting the separation. To evaluate the effect of Joule heating and efficiency of heat removal, direct temperature measurements in the separation channel were conducted using a thermocouple. An array of 16 points along each side of the separation channel (the central area was blocked by the glass cured inside the PDMS layer) were penetrated by a needle. The thermocouple was inserted into the solution and the local temperature was measured. Sample solutions delivered at room temperature (25°C) were quickly cooled to ~5°C after traveling ~1 cm (~30 s) from the inlet (Figure 5) as the device has been constantly cooled to 2°C. Higher temperature observed close to the inlet than the outlets may also due to the migration of the ampholytes to establish the pH gradient, which could consume more energy and generate more heat, especially at low pH near the anode, as the high electromobility of H+ will increase the local current density and Joule heating. The overall measured solution temperature varied between 2 and 4°C, suggesting sufficient cooling efficiency and the elimination of any local fluid circulation caused by temperature differences. The solution temperature can be easily adjusted by setting the cooling plate, including maintaining the device running at room temperature.
Figure 5.
Direct measurements of fluid temperatures in the FF-IEF device.
Separation of Protein pI Standard
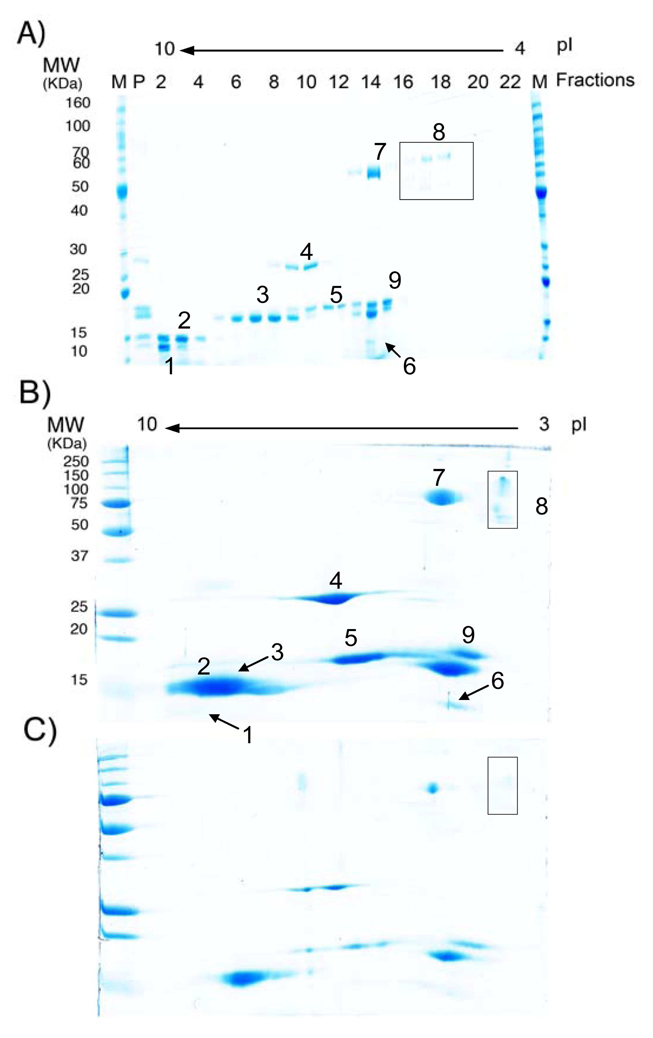
The ability of the preparative device to separate a complex protein mixture was investigated using a standard IEF 3–10 protein marker mixture obtained from Invitrogen®. The protein mixture contained 13 isoforms from 9 proteins of varying molecular size. The sample mixture was separated by on-chip IEF into 24 fractions with the denaturing running/sample buffer at the optimized flow rate of 1 mL/hr and then followed by conventional SDS-PAGE experiments (Figure 6A). To compare the resolution of FF-IEF separation with that achieved by standard 2D gel electrophoresis, the protein complex sample was studied for high gel loading (total protein load of 50 µg; Figure 6B ) and low gel loading (total protein load of 10 µg; Figure 6C). Owing to the limit in the well numbers from mini gels, only fraction collections of 2–22 were validated by SDS-PAGE in Figure 6A. Because of the broad pI range of cytochrome C (pI 9.5–10.7), protein 1 shown in Figure 6A exiting at fraction 2, would extend to fraction 1 at the very basic end. Our results suggest that FF-IEF yields better resolution and yield for alkaline proteins than the conventional 2D gels. One of these alkaline proteins, cytochrome C is difficult to resolve by 2D gel electrophoresis either in Figure 6B or 6C, but is easily resolved by FF-IEF. We also achieve high resolution and separation between Ribonuclease A (protein 2) and isoforms of lectin (protein 3) by the FF-IEF system as compared to conventional 2D gel electrophoresis (Figure 6B or 6C). Both glucose oxidase and amyloglucosidase (protein 8) have been denatured into their monomers presented in the denaturing sample buffer, which shows a single band of ~77 kD at pI 5.5 and dual bands of 89 kD and 70 kD at pI ~5. We are only able to visualize the low abundance proteins of amyloglucosidase and β-Lactoglobulin (protein 6) in the highly loaded 2D gel while sacrificing overall protein resolution as shown in Figure 6B. On the other hand, the preparative FF-IEF approach yields nice resolution by its ability to concentrate protein up to 10–20 fold. Thus, the technique allows for the identification and isolation of low abundance protein for further analysis.
Figure 6.
Comparison of protein separation using A) FF-IEF/SDS-PAGE system, B) 2D gel IEF-electrophoresis, high loading (total protein load of 50 µg), and C) 2D gel IEF-electrophoresis, low loading (total protein load of 10 µg).. Protein samples: 1) Cytochrome C, pI 9.5–10.7, 12.4 kD; 2) Ribonuclease A, pI 9.5, 13.7 kD, 3) Lectin, pI 8.3, 8.0 and 7.8, 17 kD; 4) Carbonic anhydrase, pI 6–6.6, 29 kD; 5) Myoglobin, pI 6.9–7.4, 17.8 kD; 6) β-Lactoglobulin, pI 5.3 and 5.2, 14 kD; 7) Glucose oxidase, pI 5.5, 77 kD; 8) Amyloglucosidase, pI 5, 89 kD and 70 kD. 9) Trypsin inhibitor, pI 4.5, 21.5 kD. Lane markers correspond to marker (M), sample mixture for positive control (P) or device fraction number (high to low pH). Loading [total protein] for FF-IEF system: 1 mg/mL. IEF fractionation flow rate: 1 mL/hr.
Depletion of Albumin and Device Reproducibility
In order to explore the system’s ability to process highly concentrated protein samples along with device reproducibility from batch to batch, we further investigated the IEF isolation using samples with 3 different concentration settings, conducted in 3 different devices. Devices were also fabricated from 3 different batches. Current during device running was held constant once the pH gradient was established. Figure 7A shows the isolation of BSA from cytochrome C at a lower concentration, with 0.25 mg/mL for each protein. Upon focusing, the majority of BSA was found exiting in fraction 12 while the alkaline cytochrome C was located in fractions of 1 and 2. Doubling the concentration from 0.25 mg/mL of each protein to 0.5 mg/mL (Figure 7B) produces enhanced band intensity without further peak broadening or cross fraction contamination.
Figure 7.
Depletion of Albumin and Hemoglobin in 3 different devices. Buffer and separation conditions are same as in Figure 5. Sample conditions: A) cytochrome C and BSA, 0.25 mg/mL each; B) cytochrome C, BSA, 0.5 mg/mL each; C) BSA and Hemoglobin,0.5 mg/mL each. Flow rate: 1 mL/hr.
Isolation of hemoglobin from albumin serves as a second example. Hemoglobin is a tetramer consisting of two α and two β subunits in the native status and is denatured to monomers under 8M urea denaturing condition. Both subunits have a similar molecular weight (~ 15 KDa) but different theoretical pI (c.f. ExPASy; http://ca.expasy.org). The alpha subunit, consisting of 141 amino acids, has a higher pI value of 8.19 while the beta subunit, consisting of 146 amino acids, has a lower pI value of 7.0. The majority of the denatured hemoglobin focuses to fraction 6 to 11 (8.2 - 6.7) (Figure 7C). Again, majority of BSA was observed well isolated in fraction 12 without cross-fraction broadening. It is worthwhile to note that the IEF procedure not only drives the separation but also the sample concentration. BSA exiting from all of the 3 devices has been concentrated nearly 20-fold whereas cytochrome C (pI 9.5 – 10.7) experienced close to 10-fold increase due to its broad pI range and exited in the first 2 fractions. No protein precipitation upon focusing was observed in any of the devices during the entire separation time, in part because of the denaturing conditions of the 8 M urea and zwitterionic detergent of CHAPS applied to keep the protein soluble. The system was very robust and ran continuously for at least 5 hrs (the longest time we have performed) without intensive supervision.
Whole Cell Lysate Separation
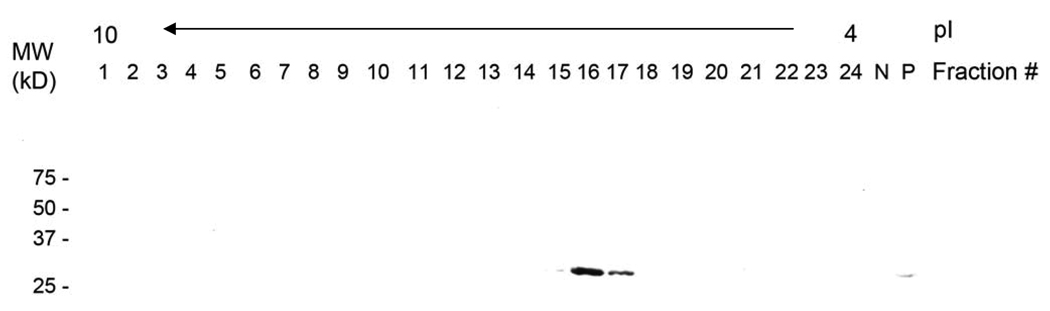
Separation of protein from a crude HELA whole cell lysate was chosen as a demonstration of the ability of this preparative FF-IEF to handle complex samples with potential application to early stage protein screening. A total of 1 mL of diluted whole cell lysate (0.5 mg/mL) was loaded to the device and ~ 40 uL fractions were collected through its 24 outlets. All the 24 fractions were loaded on a Western blot and a target protein, 14-3-3 sigma, was screened. 14-3-3 sigma is an acidic tumor suppressor protein with the pI range 4.8 to 5.2 that is known to be altered or absent in cancer cells. 33, 34 The vast majority of the target protein isolated through the device was observed only fraction 16 and 17 (pI range 5.0–5.3) with a minor focus in fraction 15 (Figure 8). Compared to the original sample concentration shown as the positive control, the isolated target protein has been greatly concentrated with a fold of 10–20. These results demonstrate the potential usefulness of the device crude sample complex separation and isolation with potential relevance to proteomic characterization in human disease.
Figure 8.
Western blot result of 14-3-3 sigma from HELA whole cell lysate separation. FF-IEF separation conditions: Voltage: 1800 V. Current: 1.2 mA. [Cell lysate]: 0.5 mg/mL. Flow rate: 0.6 mL/hr. Lane markers correspond to marker (M), sample mixture for negative control (N), positive control (P) or device fraction number (high to low pH).
CONCLUSIONS
We have characterized and optimized a preparative micro device for continuous protein IEF separation involving milligrams sample in the mass and milliliters in volume. A reproducible, nearly linear pH gradient from pH 10 to 4 was established across the 24 outlet fractions, and the cooling system was ably to effectively remove any Joule heating. Dynamic surface coatings with PVA for EOF suppression were optimized to 4% (w/v) for minimal peak broadening. The performance of this preparative micro device for protein complex isolation was compared to the standard 2D gel IEF-electrophoresis and revealed higher separation resolution, in particular for low abundance proteins. The free flow electrophoresis system also demonstrated advantages in retaining high molecular weight proteins and depletion of high concentration of albumin or hemoglobin with high reproducibility from device to device. Protein precipitation was eliminated with the denaturing buffer investigated. These results and the crude whole cell lysate separation show promise for the use inexpensive preparative FF-IEF with disposable micro devices in early stage protein screening studies and later stage protein purification.
ACKNOWLEDGEMENT
Funding for this work was provided by U.S. Army Research Office under contract W911NF-07-D-0004 (KFJ), and NIH grants GM60594 (MBY) and GM68762 (MBY and KFJ).
REFERENCES
- 1.Patel PD, Weber G. Journal of Biological Physics and Chemistry. 2003;3:60–73. [Google Scholar]
- 2.Loseva OI, Gavryushkin AV, Osipov VV, Vanyakin EN. Electrophoresis. 1998;19:1127–1134. doi: 10.1002/elps.1150190712. [DOI] [PubMed] [Google Scholar]
- 3.Bardy N, Carrasco A, Galaud JP, Pont-Lezica R, Canut H. Electrophoresis. 1998;19:1145–1153. doi: 10.1002/elps.1150190715. [DOI] [PubMed] [Google Scholar]
- 4.Grab DJ, Webster P, Lonsdale-Eccles JD. Electrophoresis. 1998;19:1162–1170. doi: 10.1002/elps.1150190717. [DOI] [PubMed] [Google Scholar]
- 5.Lu H, Gaudet S, Schmidt MA, Jensen KF. Anal. Chem. 2004;76:5705–5712. doi: 10.1021/ac049794g. [DOI] [PubMed] [Google Scholar]
- 6.Bier M. Electrophoresis. 1998;19:1057–1063. doi: 10.1002/elps.1150190703. [DOI] [PubMed] [Google Scholar]
- 7.Zuo X, Speicher DW. Anal. Biochem. 2000;284:266–278. doi: 10.1006/abio.2000.4714. [DOI] [PubMed] [Google Scholar]
- 8.Zuo X, Echan L, Hembach P, Tang HY, Speicher KD, Santoli D, Speicher DW. Electrophoresis. 2001;22:1603–1615. doi: 10.1002/1522-2683(200105)22:9<1603::AID-ELPS1603>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 9.Burggraf D, Weber G, Lottspeich F. Electrophoresis. 1995;16:1010–1015. doi: 10.1002/elps.11501601169. [DOI] [PubMed] [Google Scholar]
- 10.Weber G, Bocek P. Electrophoresis. 1996;17:1906–1910. doi: 10.1002/elps.1150171216. [DOI] [PubMed] [Google Scholar]
- 11.Zischka H, Lichtmannegger J, Jägemann N, Jennen L, Hamöller D, Huber E, Walch A, Summer KH, Göttlicher M. Methods Mol Biol. 2008;424:333–348. doi: 10.1007/978-1-60327-064-9_26. [DOI] [PubMed] [Google Scholar]
- 12.Mohr H, Voelkl A. Electrophoresis. 2002;23:2130–2137. doi: 10.1002/1522-2683(200207)23:13<2130::AID-ELPS2130>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 13.Islinger M, Weber G. Methods Mol Biol. 2008;432:199–215. doi: 10.1007/978-1-59745-028-7_14. [DOI] [PubMed] [Google Scholar]
- 14.Xie H, Bandhakavi S, Roe MR, Griffin TJ. J Proteome Res. 2007;6:2019–2026. doi: 10.1021/pr060691j. [DOI] [PubMed] [Google Scholar]
- 15.Cabrera CR, Yager P. Electrophoresis. 2001;22:355–362. doi: 10.1002/1522-2683(200101)22:2<355::AID-ELPS355>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 16.Xu Y, Zhang C, Janasek D, Manz A. Lab Chip. 2003;3:224–227. doi: 10.1039/b308476k. [DOI] [PubMed] [Google Scholar]
- 17.Fonslow BR, Bowser MT. Anal. Chem. 2005;77:5706–5710. doi: 10.1021/ac050766n. [DOI] [PubMed] [Google Scholar]
- 18.Kohlheyer D, Besselink GJ, Schlautmann S, Schasfoort RBM. Lab Chip. 2006;6:374–380. doi: 10.1039/b514731j. [DOI] [PubMed] [Google Scholar]
- 19.Kohlheyer D, Eijkel JCT, Schlautmann S, van den Berg A, Schasfoort RBM. Anal. Chem. 2007;79:8190–8198. doi: 10.1021/ac071419b. [DOI] [PubMed] [Google Scholar]
- 20.Albrecht JW, Jensen KF. Electrophoresis. 2006;27:4960–4969. doi: 10.1002/elps.200600436. [DOI] [PubMed] [Google Scholar]
- 21.Albrecht JW, El-Ali J, Jensen KF. Anal. Chem. 2007;79:9364–9371. doi: 10.1021/ac071574q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wen J, Albrecht JW, Jensen KF. Electrophoresis to appear [Google Scholar]
- 23.Wilker EW, van Vugt MA, Artim SA, Huang PH, Petersen CP, Reinhardt HC, Feng Y, Sharp PA, Sonenberg N, White FM, YaffeM B. Nature. 2007;15:329–332. doi: 10.1038/nature05584. [DOI] [PubMed] [Google Scholar]
- 24.Horvath J, Dolnik V. Electrophoresis. 2001;22:644–655. doi: 10.1002/1522-2683(200102)22:4<644::AID-ELPS644>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 25.Mazereeuw M, Best CM, de. Tjaden UR, Irth H, van der Greef J. Anal. Chem. 2000;72:3881–3886. doi: 10.1021/ac991202k. [DOI] [PubMed] [Google Scholar]
- 26.Wu D, Luo Y, Zhou X, Dai Z, Lin B. Electrophoresis. 2005;26:211–218. doi: 10.1002/elps.200406157. [DOI] [PubMed] [Google Scholar]
- 27.Cui H, Horiuchi K, Dutta P, Ivory CF. Anal. Chem. 2005;77:1303–1309. doi: 10.1021/ac048915+. [DOI] [PubMed] [Google Scholar]
- 28.Das C, Fan ZH. Electrophoresis. 2006;27:3619–3626. doi: 10.1002/elps.200600013. [DOI] [PubMed] [Google Scholar]
- 29.Srinivasan K, Pohl C, Avdalovic N. Anal. Chem. 1997;69:2798–2805. [Google Scholar]
- 30.Keely CA, Vandegoor TAAM, Mcmanigill D. Anal. Chem. 1994;66:4236–4242. [Google Scholar]
- 31.Herr AE, Mikkelsen JC, Santiago JG, Kenny TW. Micro-Electro-Mechanical Systems. 2000;2:513–518. [Google Scholar]
- 32.Wang W, Zhao L, Jiang L, Zhang J, Zhu J, Chen H. Electrophoresis. 2006;27:5132–5137. doi: 10.1002/elps.200600110. [DOI] [PubMed] [Google Scholar]
- 33.Wilker EW, Grant RA, Artim SC, Yaffe MB. J Biol Chem. 2005;13:18891–18898. doi: 10.1074/jbc.M500982200. [DOI] [PubMed] [Google Scholar]
- 34.Wilker E, Yaffe MB. J Mol Cell Cardiol. 2004;37:633–642. doi: 10.1016/j.yjmcc.2004.04.015. [DOI] [PubMed] [Google Scholar]