Abstract
Introduction
Human β-defensin-2 (hBD-2) is an antimicrobial peptide, induced by bacterial stimuli and inflammation, that plays a role in mucosal and skin innate immune defense. The nuclear factor-κB (NF-κB) transcription factor family is important in innate and adaptive immune responses to bacteria and proinflammatory cytokines. NF-κB operates via the traditional IKKβ signaling, as well as an alternative pathway utilizing IKKα signaling, which is important in keratinocyte differentiation. Our previous studies showed that pathogenic, but not commensal, bacteria used NF-κB signaling in hBD-2 induction. The objective of this study was to understand which arm of the NF-κB pathway is involved in gingival epithelial cell responses to pathogenic bacteria, including hBD-2 induction.
Methods
Cultured oral epithelial cells were transfected with synthetic small interfering RNAs (siRNAs) specific for various steps in each pathway, namely IKKβ, TRAF6 and MyD88 in the canonical, and IKKα and TRAF3 in the alternative pathway, and subsequently stimulated with various oral bacteria.
Results
The hBD-2 induction level was reduced to 21–61% in cells in which the alternative NF-κB pathway was blocked and subsequently stimulated with pathogenic bacteria, while cells in which the canonical pathway was blocked showed reduction to 78–99%. Cells stimulated with commensals showed little change in hBD-2 induction level regardless of the siRNA used. Microarray analysis showed that oral epithelia differentially regulated numerous innate immune markers in response to pathogens and commensals.
Conclusion
Our data suggest a role for the IKKα/TRAF3 pathway in NF-κB activation by pathogenic bacteria, while commensal bacteria do not utilize either NF-κB pathway, for hBD-2 induction.
Keywords: commensals, gingival epithelium, innate immunity, nuclear factor-κB, pathogens
Epithelial tissues function as the first line of defense between the host and the outside environment. Recent studies have demonstrated that these tissues protect the host by providing not only a physical barrier, but also by innate immune responses in the form of antimicrobial peptides (4, 13). These antimicrobial peptides have broad-spectrum activity against both Gram-negative and Gram-positive bacteria, as well as against yeasts and viruses (25, 34). In humans, these antimicrobial peptides, defined as less than 100 amino acids in size, include β-defensins and a cathelicidin family member LL-37 in skin and oral mucosa and other epithelia (15, 25, 40). The human β-defensins (hBDs) are small, cationic antimicrobial peptides made by epithelial cells and expressed in all human epithelia tested to date, including oral epithelia (5). hBD-1 is expressed constitutively in epithelial tissues, whereas hBD-2 and hBD-3 are expressed when the epithelia are stimulated with bacteria, Candida albicans, interleukin-1 (IL-1), or tumor necrosis factor-α (TNF-α) (17, 22, 30, 32). Although hBD-2 is induced and expressed only in inflamed sites in most tissues, in the oral epithelium it is expressed in normal uninflamed gingival tissue as well, presumably because of the high level of exposure of this tissue to both pathogenic and commensal organisms (6). Although genes for 28 hBDs have been identified (39), only limited information on the regulation of expression of additional defensins is known (34).
Various epithelia of the body are populated with distinct populations of commensal bacteria, and recent conceptual advances suggest that host tissues may promote associations with these commensal bacteria that can be beneficial to the host and may choose not to activate an immune response to eliminate them (35, 36, 42). Commensal and pathogenic bacteria share the same molecular patterns that are recognized by Toll-like receptors (TLR) in innate immune responses, but the commensals do not trigger inflammatory responses. Recent studies have investigated this question in gut and uro-epithelia (12, 28, 35, 37). Nevertheless, mechanisms for the differential response to commensals and pathogens in other tissues are still not clearly understood, and we are attempting to address how the host distinguishes pathogens from commensals in oral and skin epithelia by investigating the molecular mechanisms utilized by epithelia in inducing hBD-2, a marker of innate immunity.
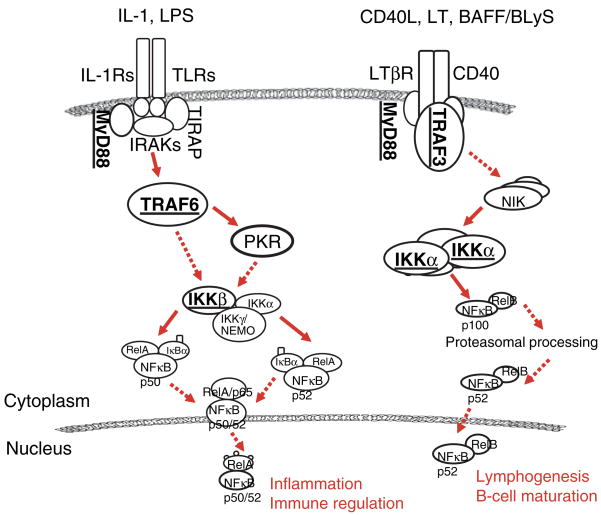
In both oral epithelium and epidermis, multiple pathways are involved in the induction of hBD-2. The hBD-2 is induced in cultured human gingival epithelial cells (GECs) and epidermal keratinocytes (HEKs) from foreskin by proinflammatory cytokines IL-1 and TNF-α, the epithelial cell activator phorbol 12-myrsitate 13-acetate, and by a variety of commensal and pathogenic bacteria (1, 9, 11, 17, 21, 22). Our earlier studies also reported that the upregulation of hBD-2 by the commensal oral bacterium, Fusobacterium nucleatum, was independent of other innate immune responses, such as IL-8 expression, a neutrophil chemoattractant, and that hBD-2 induction by commensal bacteria involved mitogen-activated protein kinase (MAPK) pathways (22). In contrast, responses to pathogenic bacteria involved both MAPK and the NF-κB transcription factors (1, 21, 22). The NF-κB transcription factor pathway is important in the cellular responses to inflammatory stimuli and to the overall response to pathogens in many cell types. TLRs of the innate immune system induce NF-κB upon recognition of bacterial products, resulting in production of proinflammatory cytokines and T-cell stimulation in the adaptive immune response. Traditionally, NF-κB activation has been known to operate through the I-κB kinase (IKKβ) signaling pathway. However, recently a non-canonical pathway for NF-κB activation was described (Fig. 1) (33). This alternative pathway involves the activation of NF-κB through IKKα, which was considered dispensable in the canonical IKKβ pathway. The IKKβ pathway is required for the activation of NF-κB by proinflammatory cytokines, whereas the IKKα pathway is thought to have a role in keratinocyte differentiation, epidermal development, and inflammatory responses (19, 26, 31).
Fig. 1.
Canonical (left) and non-canonical (right) pathways for NF-κB activation. The components targeted in this study are underlined.
The goal of this study was to better understand which arm of the NF-κB signaling pathway is involved in the gingival epithelial cell response to commensal and pathogenic bacteria. We used RNA interference (RNAi) to block signaling through the canonical IKKβ and non-canonical IKKα arms of the pathway. We show in this study that pathogenic bacteria preferentially signal via the IKKα/TNF receptor-associated factor 3 (TRAF3) arm of NF-κB activation in the expression of hBD-2, and that commensal bacteria, as expected, do not use either NF-κB pathway in hBD-2 stimulation. Additionally, with a pathway-directed microarray, we show that epithelia differentially utilize the IKKα and IKKβ pathways in response to commensals and pathogens in regulating other innate immune and inflammatory markers.
Materials and methods
Human epithelial cells and bacterial culture conditions
Healthy gingival samples were obtained from patients undergoing third-molar extractions at the Department of Oral Surgery, School of Dentistry, University of Washington. Fresh human neonatal foreskins were collected from the Dermatology Clinic at the University of Washington Medical Center. Tissue was prepared as described earlier, and subsequently isolated GECs and HEKs were grown in keratinocyte basal medium supplemented with keratinocyte growth medium (Cambrex, Walkersville, MD) (1). For most experiments, the cells were grown in media containing 0.15 mM calcium (Ca2+), but for some studies the cells were grown at 0.06 mM or 1.2 mM Ca2+ during small interfering RNA (siRNA) transfection (48 h) and subsequent bacterial stimulation (16 h). Porphyromonas gingivalis (ATCC 33277) cells were cultured in anaerobic conditions (85% N2, 10% H2, 5% CO2) at 37°C in trypticase soy broth (BBL, Sparks, MD) supplemented with 1 g yeast extract, 5 mg hemin and 1 mg menadione per liter. Streptococcus gordonii (Challis DL1) was grown in trypticase soy broth at 37°C under static conditions. F. nucleatum (ATCC 25586) and Aggregatibacter actinomycetemcomitans (ATCC 43718) were grown in Todd–Hewitt broth supplemented with 1 g yeast extract per 100 ml at 37°C in anaerobic conditions. All bacterial species used in this study are from our laboratory stocks. Bacterial numbers were estimated by density in a GENios Multi-detection Reader (Phenix, Hayward, CA).
Transfection of keratinocytes with siRNA
Specific siRNAs were custom-synthesized by Orbigen (San Diego, CA). Cultured GECs and HEKs were grown to 50–60% confluence in a 24-well plate, as described above, and on the day of transfection, cells were incubated in antibiotic-free medium. Fluorescent siRNA oligos were diluted to 30 nM in appropriate buffer and medium according to the manufacturer’s suggestions, and added to each well. Cells transfected with reagents only and with non-silencing siRNA were used as controls. The cells were subsequently stimulated with appropriate bacteria 48 h after transfection, and total RNA and whole cell proteins were extracted after 16 h. Successful knockdown of targeted genes was determined by reverse transcription-polymerase chain reaction (RT-PCR) and Western blot analysis as described below.
Western blot analysis
Whole cell proteins were isolated using a PARIS kit (Ambion, Austin, TX) and the concentration was determined using the Bradford assay. The proteins were resolved on a 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred to nitrocellulose membranes (Schleicher & Schuell, Keene, NH) for 1 h. The membranes were blocked in phosphate-buffered saline (PBS)/0.1% Tween-20/3% non-fat skim milk for 30 min, and incubated overnight at 4°C with a primary antibody specific for each protein (Santa Cruz Biotech, Santa Cruz, CA). The membranes were washed in PBS/0.1% Tween-20, incubated with horseradish peroxidase-conjugated secondary antibody (New England Biolabs, Beverly, MA) for 1 h at room temperature, washed three times, then incubated with enhanced chemiluminescence substrates (Santa Cruz Biotech) for 1 min. The membranes were subsequently exposed to Biomax film (Kodak, Rochester, NY) for various times depending on the intensity of the signals.
Conditions for RT-PCR
Total RNA was extracted from keratinocytes using PARIS kit (Ambion, Austin, TX) according to the manufacturer’s suggestion. Reverse transcription was performed with 1 μg total RNA, 1× reverse transcriptase (RT) buffer, 250 nM oligo-dT primer, 10 mM deoxynucleoside triphosphate (dNTP) mix, 50 U reverse transcriptase, and 13 U RNase inhibitor. Initial denaturation of secondary RNA structure was carried out at 72°C for 2 min, followed by annealing of the primer and template at 42°C for 1 h. The temperature was subsequently raised to 95°C for 10 min to inactivate the reverse transcriptase. Controls without reverse transcriptase were included in each experiment. Amplification of the resulting cDNA was carried out with each 50 μl PCR mixture containing 3 μl cDNA, 1× PCR buffer, 1.5 mM MgCl2, 10 mM dNTP mix, 250 nM each of sense and antisense primers, and 2.5 U Taq DNA polymerase. Ribosomal phosphoprotein (RPO) was used as a housekeeping control gene to determine the total RNA level. The PCR conditions were denaturation at 94°C for 30 s, annealing at 57°C for 30 s, and elongation at 72°C for 2 min for 35 cycles for hBD-2, TRAF3, and TRAF6 and 25 cycles for RPO. The primers for hBD-2 and RPO have been previously described (16, 23). The primer sequences for TRAF3 and TRAF6 are as follow. TRAF3-5′: TGT CAT CAT GCG TGG AGA AT; TRAF3-3′: ACA GTT TGG GCC ACA AAG AC; TRAF6-5′: CTG CAA AGC CTG CAT CAT AA; TRAF6-3′: GGG GAC AAT CCA TAA GAG CA. The final RT-PCR products were visualized with ethidium bromide on an agarose gel, and the semi-quantitative analysis of the mRNA expression levels was performed using FluorChem SP (Alpha Innotech, San Leandro, CA).
Microarray
The effects of silencing IKKα or IKKβ in the expression of other inflammatory markers and cytokines induced by commensals or pathogens were analyzed with the Human Inflammatory Cytokines and Receptors Microarray (Superarray, Frederick, MD). This array contains 128 genes encoding cytokines and interleukins associated with the inflammatory response and their receptors. The cRNA was amplified and biotin-labeled from total RNA using a TrueLabeling AMP kit (Superarray, Frederick, MD) according to the manufacturer’s suggestion. Arrays were hybridized with 4 μg labeled cRNA, and resulting images were analyzed using GEArray Expression Analysis Suite (Superarray, Frederick, MD).
Results
Successful knockdown of specific genes determined at both mRNA and protein levels
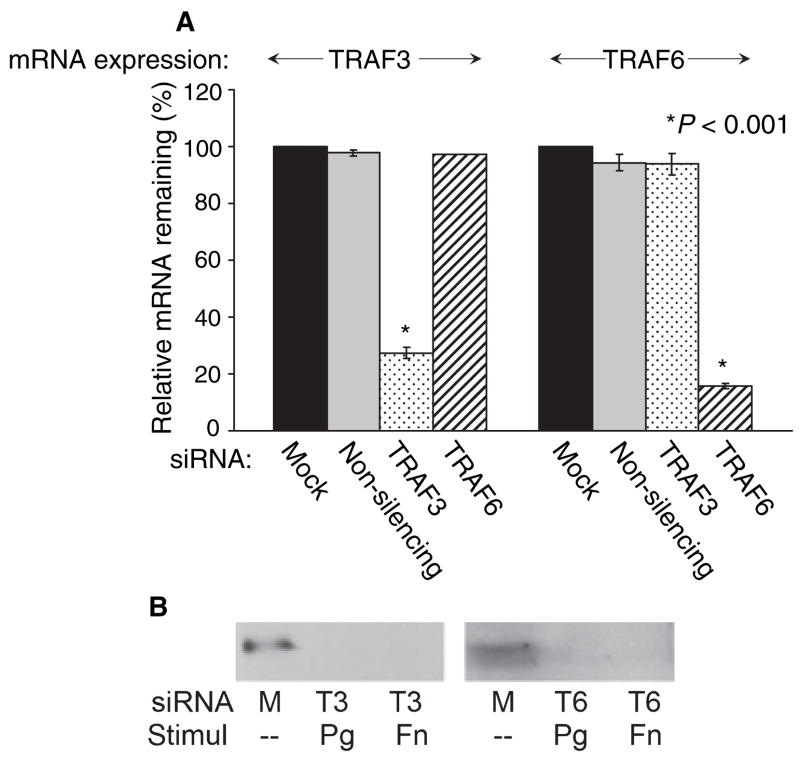
The siRNA-mediated RNAi technique is a highly specific post-transcriptional gene-silencing system. To determine that specific targeted genes were successfully knocked-down, both mRNA and protein expression levels of the targeted genes were determined. Messenger RNA levels for transcripts of interest were determined by RT-PCR. These results are semi-quantitative rather than quantitative because of the presence of fluorescent tag on the siRNA. This tag makes it possible to estimate the efficiency of transfection using fluorescent microscopy, which was between 70% and 80% in these studies. However, the tag prevents the use of quantitative PCR using SYBR Green dye. Therefore, to verify our results, the experiments were performed in duplicate with three separate cell lines, and the results reported here were consistent. GECs transfected with TRAF3 siRNA had 27% TRAF3 mRNA expression remaining, while cells transfected with non-silencing control or TRAF6 siRNA showed TRAF3 expression levels similar to those of the mock-transfection control (Fig. 2A). Likewise, 15% TRAF6 mRNA expression remained in GECs transfected with TRAF6 siRNA, while the TRAF6 expression level remains high in cells with control or TRAF3 siRNA-transfection (Fig. 2A). Knockdown of other specific genes targeted with each siRNA was tested, and the results were as anticipated, with all transfections resulting in less than 27% mRNA expression of the gene of interest remaining (data not shown). Knockdown of each specific targeted gene was also determined at the protein level. When the whole cell protein from GECs transfected with siRNA specific for TRAF3 was incubated with primary antibody for TRAF3, decreased protein expression level was seen, compared to the mock-transfection control, regardless of subsequent bacterial stimulation (Fig. 2B). Similarly, GECs transfected with siRNA specific for TRAF6 showed a decreased TRAF6 protein level (Fig. 2B). A parallel study conducted using proteins from each siRNA-transfection showed similar results (data not shown). Thus, these data confirm that the siRNAs utilized in our study are highly specific and have effectively blocked a gene function at both mRNA and protein levels.
Fig. 2.
Successful gene knockdown after siRNA transfection was determined at the mRNA and protein levels. (A) GECs after control (mock and non-silencing)- or TRAF6 siRNA-transfection show strong TRAF3 mRNA expression, while after TRAF3 siRNA-transfection show TRAF3 mRNA expression level at 27% (left). A parallel RT-PCR was performed to test TRAF6 mRNA expression level after transfection with TRAF3 or TRAF6 siRNA (right). TRAF6 mRNA expression is reduced to 15%. Three different cell lines were tested, and the average values, with standard deviations, are shown. (B) Whole cell protein extract from GECs after mock-transfection show strong TRAF3 protein expression, while after TRAF3 siRNA-transfection (T3) show decreased TRAF3 protein expression (left). The results are the same regardless of subsequent stimulation with pathogenic (Porphyromonas gingivalis: Pg) or commensal (Fusobacterium nucleatum: Fn) bacteria. A parallel study was done to determine TRAF6 protein level after transfection with TRAF6 siRNA (T6) (right).
IKKα is preferentially utilized in response to oral pathogenic bacteria in hBD-2 induction
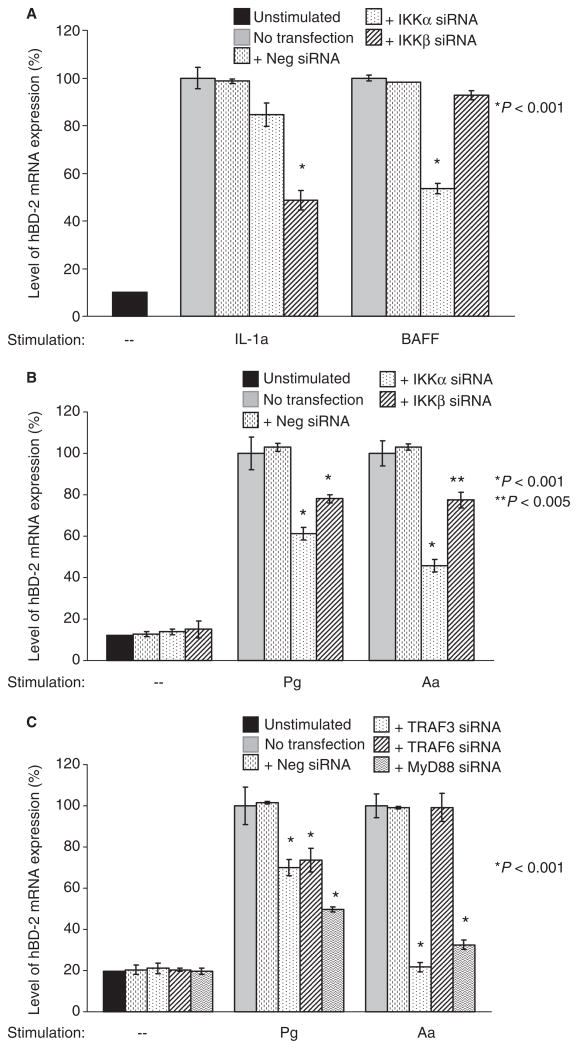
IL-1, a ligand for the canonical arm of the NF-κB signaling pathway, is a known stimulant of hBD-2 (30). To investigate if the alternative arm of the NF-κB signaling pathway is used in the hBD-2 induction, we stimulated GECs with BAFF/BLyS, a ligand for the alternative pathway. After 16 h of stimulation, hBD-2 expression was confirmed by RT-PCR, using three different cell lines (Fig. 3A). As expected, there was significant reduction in the hBD-2 induction level (down to 48%) of GECs after IKKβ siRNA transfection and subsequent stimulation with IL-1a, while hBD-2 induction level was reduced to 53% after IKKα siRNA transfection and subsequent stimulation with BAFF (Fig. 3A). Next, we investigated whether one or both of the two arms of the NF-κB signaling pathway was utilized in hBD-2 induction by pathogenic bacteria, using synthetic siRNAs specific for IKKα and IKKβ of the NF-κB signaling pathway. P. gingivalis is a Gram-negative bacterium strongly implicated as an etiological agent of severe adult periodontitis. A. actinomycetemcomitans is a Gram-negative bacterium associated with localized aggressive periodontitis. When GECs were transfected with siRNA for IKKα, IKKβ, or non-silencing control alone without subsequent bacterial stimulation, no increase in hBD-2 induction was observed compared to the unstimulated control (Fig. 3B). When the IKKβ signaling pathway was blocked and GECs were stimulated with the oral pathogenic bacteria P. gingivalis or A. actinomycetemcomitans, the level of hBD-2 induction was reduced to 78% compared to the controls with mock-transfection or control siRNA transfection (Fig. 3B). In contrast, when cultured GECs were transfected with siRNA for IKKα and cells were subsequently stimulated with P. gingivalis or A. actinomycetemcomitans, hBD-2 mRNA expression levels were reduced to 61% and 45%, respectively, compared to the controls with mock-transfection or non-silencing siRNA transfection (Fig. 3B). Each experiment was repeated using three different cell lines. Our data suggest that the IKKα pathway of NF-κB is the preferred pathway for the epithelial cell response to oral pathogenic bacteria in activating NF-κB transcription factors for hBD-2 induction.
Fig. 3.
RT-PCR analysis of the effect of gene silencing in hBD-2 induction by oral pathogens. Cells were transfected with a specific siRNA for 48 h, then subsequently stimulated with bacteria for 16 h. (A) GECs transfected with IKKα or IKKβ siRNA, and stimulated with IL-1a or BAFF, are used as controls; (B) GECs transfected with IKKα or IKKβ siRNA, and stimulated with Porphyromonas gingivalis (Pg) or Aggregatibacter actinomycetemcomitans (Aa); (C) GECs transfected with TRAF3, TRAF6, or MyD88 siRNA. Unstimulated cells and cells transfected with non-silencing negative control (neg) siRNA were used as controls in each experiment. Data from duplicates with three separate cell lines were normalized to the housekeeping gene control ribosomal phosphoprotein, and the average with standard deviation is shown. Note the preferential utilization of IKKα/TRAF3 signaling pathway by epithelia in response to P. gingivalis and A. actinomycetemcomitans.
TRAF3 is preferentially utilized in response to oral pathogenic bacteria in hBD-2 induction
TRAF3 and TRAF6 are located upstream of IKKα and IKKβ, respectively, in the NF-κB signaling pathway, while MyD88 lies upstream in both pathways (Fig. 1). We next transfected GECs with siRNAs specific for blocking TRAF3, TRAF6, or MyD88 before stimulation with oral pathogenic bacteria. GECs transfected with TRAF3 or MyD88 siRNA and subsequently stimulated with A. actinomycetemcomitans showed 21% and 32% hBD-2 expression levels, respectively, compared to TRAF6 siRNA- or control-transfection (Fig. 3C). This is consistent with the data in which GECs transfected with siRNA for IKKα showed a much more reduced level of hBD-2 induction than when transfected with the siRNA for IKKβ (Fig. 3B), and further suggests that the TRAF3-IKKα arm of NF-κB activation pathway is preferentially used by this pathogen in GECs for hBD-2 upregulation. Similar studies with GECs stimulated with the oral pathogen P. gingivalis showed a decreased level of hBD-2 mRNA expression with both TRAF3 (70%) and TRAF6 (73%) siRNA compared to control-transfection cells (Fig. 3C), and showed an hBD2 mRNA level reduced to 49% when MyD88 was blocked. Thus, our evidence suggests that P. gingivalis does use the TRAF3-IKKα pathway, but may also utilize a TRAF6-containing pathway. As a control, we added 1% human serum to the GECs growth medium at the time of bacterial stimulation to ensure sufficient lipopolysaccharide (LPS)-binding protein and CD14 for utilization of the IKKβ/TRAF6 signaling pathway by pathogens. No difference in hBD-2 induction level was observed after 1% serum was added to the growth medium of control and transfected GECs (data not shown). The variation seen in the response of GECs to the two pathogens suggests that the GEC responses may be tailored to the individual bacteria.
IKKα/TRAF3 or IKKβ/TRAF6 are not utilized in response to oral commensal bacteria in hBD-2 induction
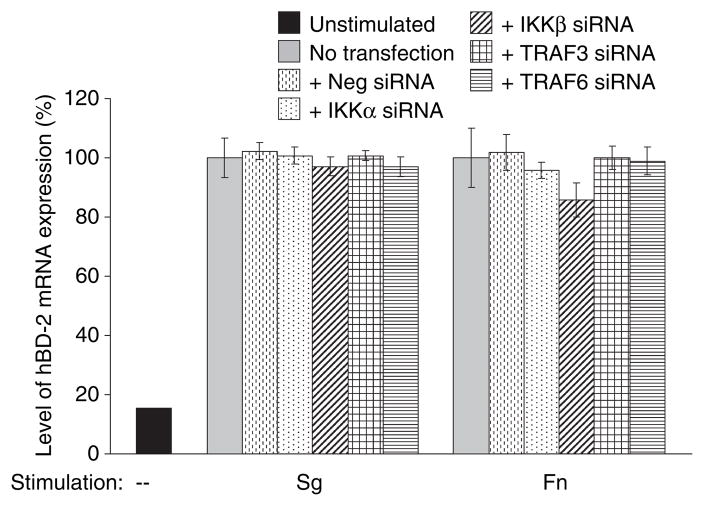
We have reported previously that commensal bacteria did not utilize the NF-κB signaling pathway in hBD-2 induction in either GECs or HEKs in studies using specific inhibitors of NF-κB (1, 21). In this study, we used the more powerful siRNA technique to confirm our previous results. As expected, when either the IKKα or IKKβ signaling pathway was blocked and GECs were stimulated with commensal bacteria S. gordonii or F. nucleatum, the expression levels of hBD-2 between control- and IKKα-transfected cells were similar (100% vs. 95%), while a slight reduction was observed when IKKβ was blocked (100% vs. 86%) (Fig. 4). Likewise, the parallel studies conducted using siRNAs specific for TRAF3 and TRAF6 gave similar results. When TRAF3 or TRAF6 was blocked and the cells were subsequently stimulated with commensal bacteria, the hBD-2 mRNA expression level remained at 97–100% (Fig. 4). Our data suggest that the NF-κB transcription factors are not utilized during hBD-2 induction by commensal bacteria in GECs.
Fig. 4.
RT-PCR analysis of the effect of gene silencing in hBD-2 induction by oral commensals. Cells were transfected with siRNAs specific for IKKα, IKKβ, TRAF3, or TRAF6 for 48 h, then subsequently stimulated with Streptococcus gordonii (Sg) or Fusobacterium nucleatum (Fn) for 16 h. Unstimulated cells and cells transfected with reagents only (mock) or non-silencing negative control (neg) siRNA were used as controls (as presented in Fig. 3). Data from duplicates with three separate cell lines were normalized to the housekeeping gene control ribosomal phosphoprotein, and the average with standard deviation is shown. Regardless of the siRNAs used, no significant reduction in hBD-2 expression was seen after stimulation with commensal bacteria.
The effects of silencing IKKα or IKKβ in the expression of other inflammatory markers and cytokines
To investigate the effects of silencing IKKα or IKKβ on the epithelial expression of other innate immune and inflammatory markers, a microarray containing genes encoding human cytokines and their receptors was utilized. The average density of four replicates was normalized using an interquartile method, and the fold increase in the expression of various innate immune markers as compared to unstimulated controls is listed in Table 1 (P < 0.05). Several different gene expression patterns were noted in response to commensals vs. pathogens. Expression levels of a number of these genes, including CCL20, CCL4 and CXCL14, decreased when either IKKα expression or IKKβ expression was blocked and cells were subsequently stimulated with P. gingivalis. The expression levels of CCL4 and CXCL14 also decreased when IKKα was blocked and cells were subsequently stimulated with A. actinomycetemcomitans. In contrast, with the commensal F. nucleatum, expression levels generally remained similar regardless of the siRNAs used. Of special interest was chemokine ligand CCL20. We have observed that CCL20 follows the expression pattern of hBD-2 (unpublished data). Changes in CCL20 mRNA expression were verified by RT-PCR, and the RT-PCR data were consistent with the array data (data not shown). These preliminary array data support the finding that oral epithelia differentially utilize IKKα and IKKβ in the regulation of innate immune and inflammatory markers in response to commensals and pathogens. Differential regulation of these genes is also seen in the epithelial responses to each bacteria. For example, comparing CCL20 and CCL4 expressions in response to P. gingivalis with those to A. actinomycetemcomitans suggests individualized cell response to each bacteria.
Table 1.
The effects of silencing IKKα or IKKβ in the expression of innate immune and inflammatory markers
| Fold changes with siRNA transfection |
|||||
|---|---|---|---|---|---|
| Stimulated |
|||||
| Genes | Bacterial stimulation | Unstimulated control | Mock | IKKα | IKKβ |
| CCL20 | P. gingivalis | 1.5 | 110 | 28 | 15 |
| A. actinomycetemcomitans | 1.9 | 7.7 | 9.6 | 19 | |
| F. nucleatum | 1.5 | 10 | 13 | 15 | |
| CXCL5 | P. gingivalis | 5 | 6.8 | 5.9 | 4.9 |
| A. actinomycetemcomitans | 4.8 | 5.1 | 1.9 | 3.4 | |
| F. nucleatum | 5 | 3.3 | 8.2 | 20.8 | |
| CCL4 | P. gingivalis | 0.3 | 3.4 | 0.1 | 0.2 |
| A. actinomycetemcomitans | 0.1 | 30 | 6.6 | 60 | |
| F. nucleatum | 0.3 | 0.3 | 0.3 | 01 | |
| CXCL14 | P. gingivalis | 4.2 | 12.2 | 2.5 | 2.5 |
| A. actinomycetemcomitans | 2.4 | 7.2 | 5.1 | 13 | |
| F. nucleatum | 4.2 | 1.6 | 1.3 | 1 | |
| IL-6 | P. gingivalis | 0.7 | 3.5 | 0.8 | 0.7 |
| A. actinomycetemcomitans | 3.3 | 38 | 53 | 130 | |
| F. nucleatum | 01 | 0.4 | 0.4 | 0.9 | |
| IL-17 | P. gingivalis | 10.4 | 10 | 2.7 | 1.2 |
| A. actinomycetemcomitans | 1 | 37 | 22 | 50 | |
| F. nucleatum | 4 | 1 | 01 | 1 | |
| IL-1R | P. gingivalis | 7 | 36 | 5 | 01 |
| A. actinomycetemcomitans | 11 | 2 | 0.1 | 3 | |
| F. nucleatum | 01 | 3 | 2 | 01 | |
| IL-10R | P. gingivalis | 11 | 23 | 16 | 11 |
| A. actinomycetemcomitans | 4.7 | 2.7 | 1.1 | 4.1 | |
| F. nucleatum | 1.2 | 1.7 | 1.1 | 0.9 | |
| hBD-22 | P. gingivalis | 1 | 11.3 | 4.5 | 7.3 |
| A. actinomycetemcomitans | 1 | 5.4 | 1.7 | 5.4 | |
| F. nucleatum | 1 | 10 | 9.1 | 7.8 | |
No spot density detected.
Semi-quantitative analysis of RT-PCR using FluorChem.
Discussion
In this study we report the preferential utilization of the alternative IKKα/TRAF3 arm of the NF-κB signaling pathway in response to pathogenic bacteria in hBD-2 upregulation by epithelial cells that originate from stratified oral epithelium. This is the first study to demonstrate the use of the alternative NF-κB pathway in the epithelial response to bacterial infection. Our earlier studies indicated that hBD-2 upregulation by commensal bacteria involved MAPK and Ca2+-mediated signaling, while upregulation by pathogenic bacteria involved both MAPK and NF-κB signaling pathways (1, 20, 21). In our current study utilizing small interfering dsRNA-mediated RNAi to post-transcriptionally knockdown genes of interest in the NF-κB signaling pathway, we have further evidence that epithelial cells utilize different signaling pathways in hBD-2 upregulation in response to pathogenic and commensal bacteria. In addition, P. gingivalis upregulated hBD-2 via protease-activated receptor 2 (PAR2), and P. gingivalis proteases play a crucial role in this response (2). The use of several pathways in hBD-2 upregulation by pathogens suggests that multiple bacterial components and multiple host receptors are involved in the stimulation of antimicrobial responses. Thus, epithelial cells have evolved multiple ways to recognize and respond to pathogens and distinguish individual bacterial species.
In tracheal epithelia, bacterial LPS upregulates hBD-2 mRNA transcription, and antibodies against CD14, a cell-surface receptor for LPS, inhibit this transcription (7, 8). CD14 interacts with LPS and TLR4 to activate NF-κB (38). However, LPS from P. gingivalis, Escherichia coli, and F. nucleatum was a poor stimulant of hBD-2 in GECs, suggesting the involvement of signaling pathways other than LPS activation of TLR4 signaling (22). These findings indicate that different cell types utilize different pathways for the stimulation of hBD-2 induction in response to bacteria. Utilizing specific siRNAs, our laboratory has observed that TLR2 and TLR4 do not have a major role in hBD-2 induction by oral bacteria, and the data suggest that hBD-2 induction by pathogenic bacteria may utilize something other than the LPS-TLR4-mediated signaling pathway. In addition, we confirmed the mRNA expression of TLR2, TLR4, and MD2 from four different oral epithelial cell lines. Taken together, these results suggest that epithelial cells recognize and utilize components other than LPS-TLR in distinguishing pathogens from commensals during hBD-2 induction or, alternatively, that there is some cross-talk between various components in this complex pathway. Further studies are needed to test the possible involvement of other TLRs and possible cross-talk with PAR2 or as yet unidentified receptors in hBD-2 induction by pathogenic bacteria.
There is widespread acceptance of the concept of pattern recognition receptors (PRR) and pathogen-associated molecular patterns (PAMP) to describe the structural components of microorganisms recognized by innate immune responses. However, this concept does not explain the fact that commensals and pathogens stimulate different epithelial cell responses and that they differ in terms of the innate immune response leading to inflammation. How the host distinguishes pathogens from commensals is still not well understood, but several recent studies have investigated this question in gut epithelia (10, 14, 18, 28, 37, 42). In addition, a recent study by Fischer et al. shows fimbriae expressed by uropathogenic E. coli triggers pathogen-specific TLR4 activation, and the type of fimbriae determines which adaptor molecules are involved in this signaling (12). Beutler suggests that the terms PRR and PAMP are too general and vague and that what is being recognized by the innate immune system is not molecular patterns, but specific individual molecules on pathogens and commensals, or perhaps specific combinations of multiple PAMPs (12). Our data are consistent with this proposal. What the host cells recognize may not be the components distinguishing Gram-positives from Gram-negatives (i.e. lipoteichoic acid vs. LPS), but the components distinguishing pathogens from commensals, or individual bacterial species. This is reflected in the signaling pathways utilized for innate immune responses by epithelial cells and further supported by our microarray data. In addition to hBD-2, a number of other inflammatory and innate immune markers showed similar patterns of up- or down-regulation as a response to post-transcriptional silencing of the two arms of the NF-κB signaling pathway after stimulation with oral pathogens, even showing slight variations between cells stimulated with P. gingivalis and with A. actinomycetemcomitans. In contrast, the pattern differed in the cells stimulated with oral commensal bacteria. Thus, our microarray data, although preliminary, suggest that epithelial innate immune responses differ when cells are exposed to pathogenic bacteria vs. commensal bacteria, and point to individuality of the cell responses to each bacteria.
In our study, the IKKα/TRAF3 arm of the signaling pathway is preferentially utilized in NF-κB activation during hBD-2 induction by pathogens in GECs, and neither of the NF-κB pathways is utilized by commensal bacteria. Parallel studies were conducted in our laboratory using epidermal keratinocytes stimulated with the skin pathogen Streptococcus pyogenes, a Gram-positive coccus and causative agent of impetigo and necrotizing fasciitis. A similar trend was observed in the results. Our data suggest that the epithelial cells from different body sites share a common utilization pattern in the innate immune response to pathogenic bacteria. In mouse embryonic fibroblasts, defective IKKα or IKKβ subunits failed to activate NF-κB after IL-1 or TNF-α stimulation, resulting in a lack of the inflammatory gene response (29, 31, 41). IKKα-dependent, but IKKβ-independent, activation of NF-κB was also observed in Epstein–Barr virus infection of mouse embryonic fibroblast (27). In contrast, inactivation of IKKα was shown to enhance inflammation and bacterial clearance in mice (24), and in human vascular endothelial cells during Rickettsia rickettsii infections (3, 24), possibly via a failure to limit the inflammatory response. Although these contrasting conclusions may be the result of differences in experiments and types of cells used, they all suggest IKKα as a secondary pathway that is necessary for full activation of NF-κB in inflammation or for control of the inflammatory response. The differences in the biological functions between IKKα and IKKβ, which share high sequence identity, may stem from their interactions with different components of the NF-κB signaling pathway. The molecular mechanism by which the TRAF3-IKKα is activated is still not understood, and the diverse nature of NF-κB inducers suggests that there may be different upstream activators of IKK. We are currently investigating the role of other components of this complex pathway in hBD-2 induction and its possible relationship to PAR2 signaling.
In this study we report that epithelial cells preferentially utilize the IKKα/TRAF3 arm of the NF-κB activation pathway in response to pathogens, but do not use either NF-κB pathway in response to commensals for hBD-2 induction. Our data suggest that epithelial cells distinguish individual bacterial species, or distinguish between commensals and pathogens, for innate immune responses, and utilize the IKKα/TRAF3 arm of the NF-κB pathway, in addition to the MAPK, IKKβ and PAR signaling pathways that have been previously described, in responding to pathogens. Understanding how different signaling pathways are activated in a coordinated way will give us insights into the innate immune responses to bacteria and help find suitable targets for new therapeutics.
Acknowledgments
We thank Carol Belton, Beth Hacker and Teresa Oswald at the University of Washington, Comprehensive Center for Oral Health Research, for providing GECs: Philip Fleckman and Anna Pirrone at the University of Washington, Department of Medicine, Division of Dermatology, for providing HEKs through the Dermatology Keratinocyte Culture Core laboratory: and L. Page Fredericks for her assistance with the diagram. This work was supported by NIDCR grants R01 DE 013573, R21 DE 0159972 and K22 DE 015812.
References
- 1.Chung WO, Dale BA. Innate immune response of oral and foreskin keratinocytes: utilization of different signaling pathways by various bacterial species. Infect Immun. 2004;72:352–358. doi: 10.1128/IAI.72.1.352-358.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chung WO, Hansen SR, Rao D, Dale BA. Protease-activated receptor signaling increases epithelial antimicrobial peptide expression. J Immunol. 2004;173:5165–5170. doi: 10.4049/jimmunol.173.8.5165. [DOI] [PubMed] [Google Scholar]
- 3.Clifton DR, Rydkina E, Freeman RS, Sahni SK. NF-kappaB activation during Rickettsia rickettsii infection of endothelial cells involves the activation of catalytic IkappaB kinases IKKalpha and IKKbeta and phosphorylation-proteolysis of the inhibitor protein IkappaBalpha. Infect Immun. 2005;73:155–165. doi: 10.1128/IAI.73.1.155-165.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dale BA. Periodontal epithelium: a newly recognized role in health and disease. Periodontol 2000. 2002;30:70–78. doi: 10.1034/j.1600-0757.2002.03007.x. [DOI] [PubMed] [Google Scholar]
- 5.Dale BA, Fredericks LP. Antimicrobial peptides in the oral environment: expression and function in health and disease. Curr Issues Mol Biol. 2005;7:119–133. doi: 10.1093/jac/dki103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dale BA, Kimball JR, Krisanaprakornkit S, et al. Localized antimicrobial peptide expression in human gingiva. J Periodontal Res. 2001;36:285–294. doi: 10.1034/j.1600-0765.2001.360503.x. [DOI] [PubMed] [Google Scholar]
- 7.Diamond G, Kaiser V, Rhodes J, Russell JP, Bevins CL. Transcriptional regulation of beta-defensin gene expression in tracheal epithelial cells. Infect Immun. 2000;68:113–119. doi: 10.1128/iai.68.1.113-119.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diamond G, Russell JP, Bevins CL. Inducible expression of an antibiotic peptide gene in lipopolysaccharide-challenged tracheal epithelial cells. Proc Natl Acad Sci USA. 1996;93:5156–5160. doi: 10.1073/pnas.93.10.5156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dinulos JG, Mentele L, Fredericks LP, Dale BA, Darmstadt GL. Keratinocyte expression of human beta defensin 2 following bacterial infection: role in cutaneous host defense. Clin Diagn Lab Immunol. 2003;10:161–166. doi: 10.1128/CDLI.10.1.161-166.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eaves-Pyles T, Murthy K, Liaudet L, et al. Flagellin, a novel mediator of Salmonella-induced epithelial activation and systemic inflammation: I kappa B alpha degradation, induction of nitric oxide synthase, induction of proinflammatory mediators, and cardiovascular dysfunction. J Immunol. 2001;166:1248–1260. doi: 10.4049/jimmunol.166.2.1248. [DOI] [PubMed] [Google Scholar]
- 11.Feucht EC, DeSanti CL, Weinberg A. Selective induction of human beta-defensin mRNAs by Actinobacillus actinomycetemcomitans in primary and immortalized oral epithelial cells. Oral Microbiol Immunol. 2003;18:359–363. doi: 10.1046/j.0902-0055.2002.00097.x. [DOI] [PubMed] [Google Scholar]
- 12.Fischer H, Yamamoto M, Akira S, Beutler B, Svanborg C. Mechanism of pathogen-specific TLR4 activation in the mucosa: fimbriae, recognition receptors and adaptor protein selection. Eur J Immunol. 2006;36:267–277. doi: 10.1002/eji.200535149. [DOI] [PubMed] [Google Scholar]
- 13.Ganz T. Defensins: antimicrobial peptides of vertebrates. C R Biol. 2004;327:539–549. doi: 10.1016/j.crvi.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 14.Gewirtz AT, Navas TA, Lyons S, Godowski PJ, Madara JL. Cutting edge: bacterial flagellin activates basolaterally expressed TLR5 to induce epithelial proinflammatory gene expression. J Immunol. 2001;167:1882–1885. doi: 10.4049/jimmunol.167.4.1882. [DOI] [PubMed] [Google Scholar]
- 15.Hancock RE, Scott MG. The role of antimicrobial peptides in animal defenses. Proc Natl Acad Sci USA. 2000;97:8856–8861. doi: 10.1073/pnas.97.16.8856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harder J, Bartels J, Christophers E, Schroder JM. Isolation and characterization of human beta-defensin-3, a novel human inducible peptide antibiotic. J Biol Chem. 2001;276:5707–5713. doi: 10.1074/jbc.M008557200. [DOI] [PubMed] [Google Scholar]
- 17.Harder J, Meyer-Hoffert U, Teran LM, et al. Mucoid Pseudomonas aeruginosa, TNF-alpha, and IL-1beta, but not IL-6, induce human beta-defensin-2 in respiratory epithelia. Am J Respir Cell Mol Biol. 2000;22:714–721. doi: 10.1165/ajrcmb.22.6.4023. [DOI] [PubMed] [Google Scholar]
- 18.Hayashi F, Smith KD, Ozinsky A, et al. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature. 2001;410:1099–1103. doi: 10.1038/35074106. [DOI] [PubMed] [Google Scholar]
- 19.Hu Y, Baud V, Oga T, Kim KI, Yoshida K, Karin M. IKKalpha controls formation of the epidermis independently of NF-kappaB. Nature. 2001;410:710–714. doi: 10.1038/35070605. [DOI] [PubMed] [Google Scholar]
- 20.Krisanaprakornkit S, Jotikasthira D, Dale BA. Intracellular calcium in signaling human beta-defensin-2 expression in oral epithelial cells. J Dent Res. 2003;82:877–882. doi: 10.1177/154405910308201106. [DOI] [PubMed] [Google Scholar]
- 21.Krisanaprakornkit S, Kimball JR, Dale BA. Regulation of human beta-defensin-2 in gingival epithelial cells: the involvement of mitogen-activated protein kinase pathways, but not the NF-kappaB transcription factor family. J Immunol. 2002;168:316–324. doi: 10.4049/jimmunol.168.1.316. [DOI] [PubMed] [Google Scholar]
- 22.Krisanaprakornkit S, Kimball JR, Weinberg A, Darveau RP, Bainbridge BW, Dale BA. Inducible expression of human beta-defensin 2 by Fusobacterium nucleatum in oral epithelial cells: multiple signaling pathways and role of commensal bacteria in innate immunity and the epithelial barrier. Infect Immun. 2000;68:2907–2915. doi: 10.1128/iai.68.5.2907-2915.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krisanaprakornkit S, Weinberg A, Perez CN, Dale BA. Expression of the peptide antibiotic human beta-defensin 1 in cultured gingival epithelial cells and gingival tissue. Infect Immun. 1998;66:4222–4228. doi: 10.1128/iai.66.9.4222-4228.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lawrence T, Bebien M, Liu GY, Nizet V, Karin M. IKKalpha limits macrophage NF-kappaB activation and contributes to the resolution of inflammation. Nature. 2005;434:1138–1143. doi: 10.1038/nature03491. [DOI] [PubMed] [Google Scholar]
- 25.Lehrer RI, Ganz T. Defensins of vertebrate animals. Curr Opin Immunol. 2002;14:96–102. doi: 10.1016/s0952-7915(01)00303-x. [DOI] [PubMed] [Google Scholar]
- 26.Li ZW, Chu W, Hu Y, et al. The IKKbeta subunit of IkappaB kinase (IKK) is essential for nuclear factor kappaB activation and prevention of apoptosis. J Exp Med. 1999;189:1839–1845. doi: 10.1084/jem.189.11.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luftig M, Yasui T, Soni V, et al. Epstein–Barr virus latent infection membrane protein 1 TRAF-binding site induces NIK/IKK alpha-dependent noncanonical NF-kappaB activation. Proc Natl Acad Sci USA. 2004;101:141–146. doi: 10.1073/pnas.2237183100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Macpherson AJ, Uhr T. Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science. 2004;303:1662–1665. doi: 10.1126/science.1091334. [DOI] [PubMed] [Google Scholar]
- 29.Massa PE, Li X, Hanidu A, et al. Gene expression profiling in conjunction with physiological rescues of IKKalpha-null cells with wild type or mutant IKKalpha reveals distinct classes of IKKalpha/NF-kappaB-dependent genes. J Biol Chem. 2005;280:14057–14069. doi: 10.1074/jbc.M414401200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mathews M, Jia HP, Guthmiller JM, et al. Production of beta-defensin antimicrobial peptides by the oral mucosa and salivary glands. Infect Immun. 1999;67:2740–2745. doi: 10.1128/iai.67.6.2740-2745.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mynott TL, Crossett B, Prathalingam SR. Proteolytic inhibition of Salmonella enterica serovar typhimurium-induced activation of the mitogen-activated protein kinases ERK and JNK in cultured human intestinal cells. Infect Immun. 2002;70:86–95. doi: 10.1128/IAI.70.1.86-95.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O’Neil DA, Porter EM, Elewaut D, et al. Expression and regulation of the human beta-defensins hBD-1 and hBD-2 in intestinal epithelium. J Immunol. 1999;163:6718–6724. [PubMed] [Google Scholar]
- 33.Pomerantz JL, Baltimore D. Two pathways for NFkB. Mol Cell. 2002;10:693–701. doi: 10.1016/s1097-2765(02)00697-4. [DOI] [PubMed] [Google Scholar]
- 34.Premratanachai P, Joly S, Johnson GK, McCray PB, Jr, Jia HP, Guthmiller JM. Expression and regulation of novel human beta-defensins in gingival keratinocytes. Oral Microbiol Immunol. 2004;19:111–117. doi: 10.1111/j.0902-0055.2002.00127.x. [DOI] [PubMed] [Google Scholar]
- 35.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 36.Roberts FA, Darveau RP. Beneficial bacteria of the periodontium. Periodontol 2000. 2002;30:40–50. doi: 10.1034/j.1600-0757.2002.03004.x. [DOI] [PubMed] [Google Scholar]
- 37.Sansonetti P. Host–pathogen interactions: the seduction of molecular cross talk. Gut. 2002;50(Suppl 3):III2–III8. doi: 10.1136/gut.50.suppl_3.iii2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schroder NW, Opitz B, Lamping N, et al. Involvement of lipopolysaccharide binding protein, CD14, and Toll-like receptors in the initiation of innate immune responses by Treponema glycolipids. J Immunol. 2000;165:2683–2693. doi: 10.4049/jimmunol.165.5.2683. [DOI] [PubMed] [Google Scholar]
- 39.Schutte BC, Mitros JP, Bartlett JA, et al. Discovery of five conserved beta-defensin gene clusters using a computational search strategy. Proc Natl Acad Sci USA. 2002;99:2129–2133. doi: 10.1073/pnas.042692699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Selsted ME, Ouellette AJ. Mammalian defensins in the antimicrobial immune response. Nat Immunol. 2005;6:551–557. doi: 10.1038/ni1206. [DOI] [PubMed] [Google Scholar]
- 41.Sizemore N, Lerner N, Dombrowski N, Sakurai H, Stark GR. Distinct roles of the Ikappa B kinase alpha and beta subunits in liberating nuclear factor kappa B (NF-kappa B) from Ikappa B and in phosphorylating the p65 subunit of NF-kappa B. J Biol Chem. 2002;277:3863–3869. doi: 10.1074/jbc.M110572200. [DOI] [PubMed] [Google Scholar]
- 42.Wehkamp J, Harder J, Wehkamp K, et al. NF-kappaB- and AP-1-mediated induction of human beta defensin-2 in intestinal epithelial cells by Escherichia coli Nissle 1917: a novel effect of a probiotic bacterium. Infect Immun. 2004;72:5750–5758. doi: 10.1128/IAI.72.10.5750-5758.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]