Abstract
Background
MicroRNAs (miRNAs) are a new class of small, endogenous RNAs that play a regulatory role in the cell by negatively affecting gene expression at the post-transcriptional level. miRNAs have been shown to control numerous genes involved in various biological and metabolic processes. There have been extensive studies on discovering miRNAs and analyzing their functions in model species, such as Arabidopsis and rice. Increasing investigations have been performed on important agricultural crops including soybean, conifers, and Phaselous vulgaris but no studies have been reported on discovering peanut miRNAs using a cloning strategy.
Results
In this study, we employed the next generation high through-put Solexa sequencing technology to clone and identify both conserved and species-specific miRNAs in peanuts. Next generation high through-put Solexa sequencing showed that peanuts have a complex small RNA population and the length of small RNAs varied, 24-nt being the predominant length for a majority of the small RNAs. Combining the deep sequencing and bioinformatics, we discovered 14 novel miRNA families as well as 75 conserved miRNAs in peanuts. All 14 novel peanut miRNAs are considered to be species-specific because no homologs have been found in other plant species except ahy-miRn1, which has a homolog in soybean. qRT-PCR analysis demonstrated that both conserved and peanut-specific miRNAs are expressed in peanuts.
Conclusions
This study led to the discovery of 14 novel and 22 conserved miRNA families from peanut. These results show that regulatory miRNAs exist in agronomically important peanuts and may play an important role in peanut growth, development, and response to environmental stress.
Background
MicroRNAs (miRNAs), initially discovered in C. elegans [1], are a large group of small endogenous RNAs [2-4] that widely exist in animals [5], plants [6], and in some viruses [7]. Increasing evidence demonstrates that miRNAs play an important function in many biological and metabolic processes, including tissue identity, developmental timing, and response to environmental stress [8,9]. However, miRNAs do not directly control plant growth and development. In contrast, miRNAs negatively control gene expression by targeting protein coding gene mRNAs for cleavage or repressing protein translation [2,3].
miRNAs are first transcribed from miRNA genes, located mainly in the intergenic genomic region, by RNA polymerase II [10-12]. There are also a small number of miRNA genes located inside the protein coding genes. For these miRNAs, the transcription orientation is the same as the protein coding gene, indicating that they are transcribed together [2,13]. Following transcription and several post-transcriptional modifications using different enzymes (Dicer, Hen1, and other enzymes), long primary miRNA transcripts (pri-RNAs) are processed to generate miRNA precursors (pre-miRNAs) and eventually mature miRNAs [14]. Although the length of mature miRNA sequences varies from 16 to 29 nucleotides with an average of 22-nt, a majority of mature miRNAs are 21 to 23-nt in length [15]. DCL1 is a key enzyme in miRNA biogenesis and mutating this gene results in globally decreased miRNA levels in plants, ultimately resulting in pleiotropic defects [16,17]. In addition, HEN1 and HYL1 also play important roles in miRNA biogenesis in plants; mutating these two genes results in severe defects during various developmental stages of plant growth, including vegetation maturity and proper formation of reproductive organs [18-20].
miRNAs are involved in plant responses to the environment and several miRNAs are up-regulated or down-regulated by abiotic stress, including high salinity, drought, and low temperatures [21,22]. Furthermore, the targets of several miRNAs are genes that play important roles in stress tolerance, including the gene encoding Cu/Zn SOD [23-25]. miR393 targets auxin receptor genes, such as TIR1, AFB2, and AFB3, which lower auxin signals and inhibit the pathogen P. syringae [26]. miRNAs are also induced by pathogens, which suggests miRNAs are involved in plant-microorganism interactions such as symbiosis events with legumes and rhizobia bacteria [27,28]. Increasing evidence demonstrates that miRNAs might provide a novel platform to better understand plant development and resistance to biotic as well as abiotic stresses.
Currently, 9539 mature miRNAs have been discovered and deposited in the public available miRNA database miRBase (Release 13.0, March 2009; http://microrna.sanger.ac.uk/sequences/index.shtml) [29]. These miRNAs include 1763 miRNAs from 24 plant species. Although numerous miRNAs have been identified in plants, a majority of them were obtained from model species such as Oryza sativa (377), Populus trichocarpa (234), Physcomitrella patens (230), Arabidopsis thaliana (187), and Vitis vinifera (140). This could be attributed to the fact that the entire genomes of these organisms have already been sequenced and are readily available. Even so, few miRNAs have been reported in important agricultural crops. Peanut is widely cultivated and is one of the most important economic and oil crops in China, the USA, and around the world. To date, no miRNA-related research has been performed on peanuts.
There are two major methods used in identifying miRNAs: (1) a direct cloning method, using miRNA-enriched libraries, combined with computational and experimental verification [21,30-32] and (2) computational identification. Several investigations have shown that some miRNAs are highly conserved throughout evolution and can be found in mosses to higher flowering plants [31,33,34] This suggests a powerful strategy for identifying miRNAs using comparative genomics. By performing Blastn searches, using already known miRNAs, against Genbank databases including genome survey sequences (GSS), high through-put genomic sequences (HTGS), expressed sequence tags (ESTs), and non-redundant (NR) nucleotides, hundreds of miRNAs have been identified in plants. Currently, several laboratories have adopted this method in order to identify miRNAs [34-41]. However, this method is limited by the number of nucleotide sequences available in the database. For peanut, the genome has not been completely sequenced and there are only a limited number of peanut ESTs in the database. This does not make computational prediction an effective choice for discovering peanut miRNAs. In this study, we employed the next generation high through-put sequencing technology to sequence and identify peanut miRNAs. Based on our study, we have identified 75 conserved miRNAs as well as 14 novel miRNAs in peanuts. Quantitative real time PCR (qRT-PCR) analysis shows that these miRNAs are expressed in peanuts.
Results and Discussion
Peanut has a complex small RNA population
To date, 92,988 peanut ESTs, including 86,724 ESTs from cultivated peanuts and 6,264 ESTs from wild-type peanuts, have been deposited in the NCBI EST database. These sequences are minor compared with the 2,800-Mb genome of the allotetraploid cultivated peanut or even the genome of the diploid wild-type peanut. Previous studies have demonstrated, using computational approaches and EST analysis, that only three conserved miRNAs exist in peanut [34,38,41]. With the limited amount of peanut ESTs in the EST database, it is not possible to perform a comprehensive study of peanut miRNAs using only a computational analysis. Experimental cloning and subsequent functional analysis, combined with computational prediction, appears to be the most effective method to identify peanut miRNAs.
Next generation high through-put sequencing, including 454 and Solexa technologies, provides a powerful tool for miRNA cloning. By using the high through-put Solexa sequencing technology, a total of 6,009,541 sequences were obtained from a small RNA library, which was constructed from the cultivated peanut variety Fenghua-1. After removing the low quality sequences and adapter sequences, 4,994,631 sequences were obtained with 3-30 nt in length, among which 4,598,005 sequences ranged from 18-30 nt in length. After further removing tRNAs (437,117), rRNAs (133,410), snRNAs (1,282), and snoRNAs (240), a total of 4,025,956 small RNA sequences were obtained. Although some small RNAs were very high in abundance and present thousands of times in our dataset, the majority of small RNAs were sequenced only a few times. For example, 2,232,910 out of 4,598,005 small RNAs were sequenced only one time in our experiment. This result suggests that (1) the expression of different small RNAs in peanut varies drastically and (2) the small RNA survey in peanut is far from saturated. This also suggests that peanut contains a large and diverse small RNA population.
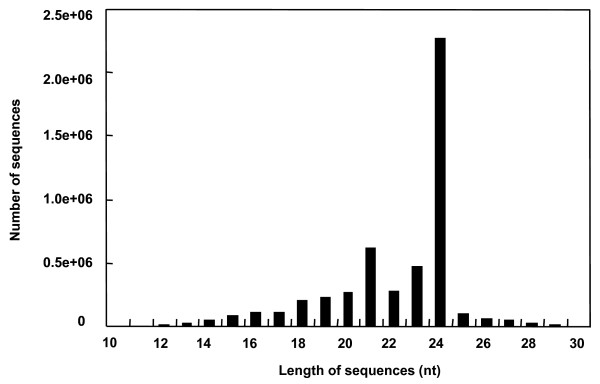
In peanut, the size of the small RNAs was not evenly distributed (Figure 1). Among these sequences, the number of 24-nt sequences was significantly greater than shorter or longer sequences (Figure 1) and accounted for 45% of the total sequence number. This result was consistent with that of Medigcago [42] and rice [43], as well as Arabidopsis 454 sequencing results [44]. In Arabidopsis, the 24-nt small RNAs accounted for about 60% of its small RNA transcriptome [45]. However, the size distribution differs from wheat and conifer sequences obtained through 454 high through-put sequencing [43,46] and Chinese yew sequences obtained through Solexa sequencing [47]. In conifer, the fraction of 24-nt RNAs was very small (2.6%) due to the lack of DCL3, the enzyme that matures 24-nt RNAs in angiosperms [43,48]. In total, 620,060 sequences (13.5%) with 21-nt, which is the typical length of plant mature miRNAs, represented the second highest abundance of sequences in the peanut library.
Figure 1.
Length distribution and abundance of the sequences.
Identification of conserved peanut miRNAs
To identify conserved miRNAs in peanuts, all small RNA sequences were Blastn searched against the currently known miRNAs in the miRNA database miRBase (March 9, 2009). In total, 1,763 known miRNAs from diverse plant species were utilized in order to identify conserved peanut miRNAs from the small RNA dataset.
After Blastn searches and further sequence analysis, a total of 75 conserved miRNAs were identified in peanuts and these miRNAs belong to 22 miRNA families (Table 1). Of the 22 miRNA families, three miRNA families (miR156/157, miR166, and miR167) were predicted [34,38,41] using a comparative genomics-based strategy [38]. The identified miRNA families have been shown to be conserved in a variety of plant species. For example, miR156/157, miR159/319, miR166, miR169, and miR394 have been found in 51, 45, 41, 40, and 40 plant species, respectively [34,38,41]. In this study, we also tried to identify the precursor sequences for the 75 conserved peanut miRNAs. However, due to the fact that the peanut genome has not been fully sequenced, the pre-miRNAs and their secondary structures were only identified for nine miRNAs (Additional file 1).
Table 1.
Conserved miRNAs from peanut
| miRNA family | Name | Sequence(5'-3') | Length (nt) |
Reference miRNA |
Conserved in other plants | Reads | |||
|---|---|---|---|---|---|---|---|---|---|
| ath | ptc | vvi | osa | ||||||
| 156/157 | ahy-MIR156a | ugacagaagagagugagcac | 20 | ath-miR156a | ++ | ++ | ++ | ++ | 17058 |
| ahy-MIR156b | ugacagaagagagugagcaca | 21 | bna-miR156a | + | + | + | + | 255 | |
| ahy-MIR156c | cugacagaagauagagagcac | 21 | smo-miR156b | + | + | + | + | 43 | |
| ahy-MIR156e | ugacagaggagagugagcac | 20 | vvi-miR156e | + | + | ++ | + | 8 | |
| ahy-MIR156 g | cgacagaagagagugagcac | 20 | ath-miR156 g | ++ | + | + | + | 15 | |
| ahy-MIR156 h | ugacagaagaaagagagcac | 20 | ath-miR156 h | ++ | + | + | + | 4 | |
| ahy-MIR156k | ugacagaagagagggagcac | 20 | ptc-miR156k | + | ++ | ++ | + | 69 | |
| ahy-MIR156f | uugacagaagaaagagagcac | 21 | smo-MIR156c | + | + | + | + | 4 | |
| ahy-MIR157a | uugacagaagauagagagcac | 21 | ath-miR157a | ++ | ++ | ++ | + | 95381 | |
| ahy-MIR157d | ugacagaagauagagagcac | 20 | ath-miR157d | ++ | + | ++ | + | 3967 | |
| ahy-MIR157k | ugacagaagagagcgagcac | 20 | zma-miR156k | + | + | + | + | 67 | |
| 159 | ahy-MIR159a | uuuggauugaagggagcucua | 21 | ath-miR159a | ++ | ++ | ++ | + | 66 |
| ahy-MIR159b | uuuggauugaagggagcucuu | 21 | ath-miR159b | ++ | + | + | + | 41 | |
| ahy-MIR319a | uuggacugaagggagcucccu | 21 | ath-miR319a | ++ | + | + | + | 12 | |
| ahy-MIR319b | uuggacugaagggagcuccc | 20 | mtr-miR319 | + | ++ | + | + | 5 | |
| 160 | ahy-MIR160a | ugccuggcucccuguaugcca | 21 | ath-miR160a | ++ | ++ | ++ | ++ | 41 |
| ahy-MIR160b | ugccuggcucccugaaugcca | 21 | osa-miR160f | + | ++ | ++ | ++ | 4 | |
| 162 | ahy-MIR162a | ucgauaaaccucugcauccag | 21 | ath-miR162a | ++ | ++ | ++ | ++ | 94 |
| 164 | ahy-MIR164a | uggagaagcagggcacgugca | 21 | ath-miR164a | ++ | ++ | ++ | ++ | 4116 |
| ahy-MIR164d | uggagaagcagggcacgugcu | 21 | osa-miR164d | + | + | + | ++ | 88 | |
| ahy-MIR164c | uggagaagcagggcacgugcg | 21 | ath-miR164c | ++ | + | + | + | 4 | |
| ahy-MIR164d | uggagaagcaggguacgugca | 21 | osa-miR164c | + | + | + | ++ | 1 | |
| 166 | ahy-MIR165a | ucggaccaggcuucauccccc | 21 | ath-miR165a | ++ | + | + | + | 40 |
| ahy-MIR166a | ucggaccaggcuucauucccc | 21 | ath-miR166a | ++ | ++ | ++ | ++ | 9577 | |
| ahy-MIR166d | ucggaccaggcuucauuccccu | 22 | vvi-miR166d | + | + | ++ | + | 9 | |
| ahy-MIR166 g | ucggaccaggcuucauuccuc | 21 | osa-miR166 g | + | + | ++ | ++ | 3647 | |
| ahy-MIR166 h | ucggaccaggcuucauuccc | 20 | zma-miR166 h | + | + | + | + | 8585 | |
| ahy-MIR166j | ucggaucaggcuucauuccuc | 21 | osa-miR166j | + | + | + | ++ | 8 | |
| ahy-MIR166 m | ucggaccaggcuucauucccu | 21 | osa-miR166 m | + | + | + | ++ | 35 | |
| ahy-MIR166n | ucggaccaggcuucauuccuu | 21 | ptc-miR166n | + | ++ | + | + | 13 | |
| ahy-MIR166e | ucgaaccaggcuucauucccc | 21 | osa-MIR166e | + | + | + | ++ | 3 | |
| ahy-MIR166k | ucggaccaggcuucaaucccu | 21 | osa-miR166k | + | + | + | ++ | 1 | |
| ahy-MIR166b | ucggaccaggcuucauuccccc | 22 | vvi-miR166c | + | + | ++ | + | 5 | |
| 167 | ahy-MIR167a | ugaagcugccagcaugaucua | 21 | ath-miR167a | ++ | ++ | ++ | ++ | 2572 |
| ahy-MIR167b | ugaagcugccagcaugaucuaa | 22 | bna-miR167a | + | + | + | + | 34 | |
| ahy-MIR167c | ugaagcugccagcaugaucuc | 21 | vvi-miR167c | + | + | ++ | + | 15 | |
| ahy-MIR167d | ugaagcugccagcaugaucugg | 22 | ath-miR167d | + | + | + | + | 224 | |
| ahy-MIR167e | ugaagcugccagcaugaucug | 21 | osa-miR167d | + | ++ | ++ | + | 34 | |
| ahy-MIR167f | ugaagcugccagcaugaucuu | 21 | ptc-miR167f | + | ++ | + | + | 8767 | |
| 168 | ahy-MIR168a | ucgcuuggugcaggucgggaa | 21 | ath-miR168a | ++ | ++ | ++ | + | 19898 |
| ahy-MIR168b | ucgcuuggugcagaucgggac | 21 | osa-miR168a | + | + | + | ++ | 86 | |
| 169 | ahy-MIR169b | cagccaaggaugacuugccgg | 21 | ath-miR169b | ++ | ++ | ++ | ++ | 66 |
| ahy-MIR169e | uagccaaggaugacuugccgg | 21 | osa-miR169e | + | + | + | ++ | 1 | |
| ahy-MIR169a | cagccaaggaugacuugccga | 21 | ath-miR169a | ++ | ++ | ++ | ++ | 1 | |
| ahy-MIR169 m | gagccaaggaugacuugccgg | 21 | vvi-miR169 m | + | + | ++ | + | 1 | |
| 171 | ahy-MIR171b | ugauugagccgugccaauauc | 21 | osa-miR171b | + | ++ | + | ++ | 26 |
| ahy-MIR171c | agauugagccgcgccaauauc | 21 | ptc-miR171c | + | ++ | + | + | 1 | |
| ahy-MIR171d | ugauugagccgcgucaauauc | 21 | vvi-miR171b | + | + | ++ | + | 5 | |
| ahy-MIR171f | uugagccgcgccaauaucacu | 21 | vvi-miR171f | + | + | ++ | + | 3 | |
| ahy-MIR171e | uugagccgugccaauaucac | 20 | zma-miR171b | + | + | + | + | 1 | |
| ahy-MIR171a | uugagccgugccaauaucaca | 21 | zma-miR171f | + | + | + | + | 4 | |
| 172 | ahy-MIR172a | agaaucuugaugaugcugcau | 21 | ath-miR172a | ++ | ++ | ++ | ++ | 2176 |
| ahy-MIR172b | agaaucuugaugaugcugca | 20 | zma-miR172a | + | + | + | + | 81 | |
| ahy-MIR172c | agaaucuugaugaugcugcag | 21 | ath-miR172c | ++ | + | + | + | 58 | |
| ahy-MIR172e | ggaaucuugaugaugcugcau | 21 | ath-miR172e | ++ | ++ | + | ++ | 2 | |
| 390 | ahy-MIR390a | aagcucaggagggauagcgcc | 21 | ath-miR390a | ++ | ++ | ++ | ++ | 149 |
| 393 | ahy-MIR393a | uccaaagggaucgcauugaucc | 22 | ath-miR393a | ++ | + | ++ | + | 2 |
| ahy-MIR393b | uccaaagggaucgcauugauc | 21 | osa-miR393 | + | ++ | + | ++ | 6 | |
| ahy-MIR393c | uccaaagggaucgcauugaucu | 22 | osa-miR393b | + | + | + | ++ | 1 | |
| 394 | ahy-MIR394a | uuggcauucuguccaccucc | 20 | ath-miR394a | ++ | ++ | ++ | ++ | 8 |
| 396 | ahy-MIR396a | uuccacagcuuucuugaacug | 21 | ath-miR396a | ++ | ++ | ++ | ++ | 221 |
| ahy-MIR396b | uuccacagcuuucuugaacuu | 21 | ath-miR396b | ++ | ++ | + | ++ | 35 | |
| ahy-MIR396d | uccacaggcuuucuugaacug | 21 | osa-miR396d | + | + | + | ++ | 1 | |
| ahy-MIR396c | uuccacagcuuucuugaacua | 21 | vvi-miR396a | + | + | ++ | + | 5 | |
| ahy-MIR396e | uuccacagcuuucuugaacu | 20 | vvi-miR396b | + | + | ++ | + | 2 | |
| 397 | ahy-MIR397a | ucauugagugcagcguugaug | 21 | ath-miR397a | ++ | ++ | ++ | ++ | 344 |
| ahy-MIR397c | ucauugagugcagcguugaugu | 22 | bna-miR397a | + | + | + | + | 5 | |
| ahy-MIR397b | uuauugagugcagcguugaug | 21 | osa-miR397b | + | + | + | ++ | 1 | |
| 398 | ahy-MIR398b | uguguucucaggucgccccug | 21 | osa-miR398b | + | ++ | ++ | ++ | 12 |
| 399 | ahy-MIR399e | ugccaaaggagauuugcccag | 21 | osa-miR399e | + | + | + | ++ | 1 |
| 408 | ahy-MIR408a | augcacugccucuucccuggc | 21 | ath-miR408 | ++ | ++ | ++ | + | 105 |
| ahy-MIR408b | ugcacugccucuucccuggcu | 21 | ppt-miR408b | + | + | + | + | 5 | |
| 528 | ahy-MIR528 | uggaaggggcaugcagaggag | 21 | osa-miR528 | ++ | 3 | |||
| 535 | ahy-MIR535 | ugacaacgagagagagcacgc | 21 | ppt-miR535a | + | + | 1 | ||
| 894 | ahy-MIR894 | cguuucacgucggguucacc | 20 | ppt-miR894 | 2 | ||||
The abbreviations represent: ath, Arabidopsis thaliana; ptc, Populus trichocarpa; vvi, Vitis vinifera; osa, Oryza sativa. The plus symbols indicate: ++, miRNA sequences of peanut were exactly identical to those in other species; +, miRNA sequences of peanut were conserved in other species but have variations in some nucleotide positions.
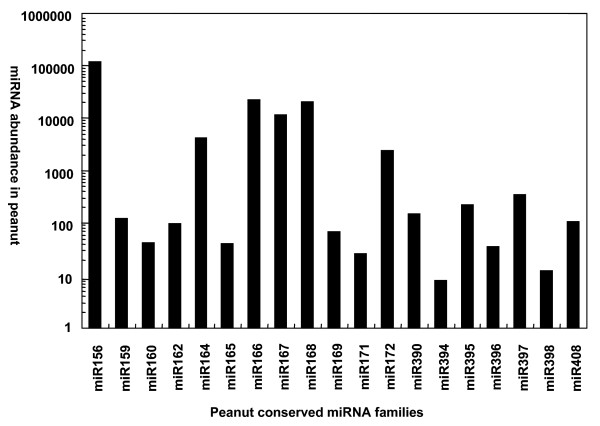
Next generation high through-put sequencing provides an alternative way to estimate expression profiles of protein coding genes and/or miRNA genes [44,46]. Millions of peanut small RNA sequences, generated from Solexa sequencing, allowed us to determine the abundance of various miRNA families and even distinguish between different members of a given family. Interestingly, peanut miRNA families displayed significantly varied abundance from each other. For example, ahy-miR157a, ahy-miR168a, and ahy-miR156a were detected 95,381, 19,898, and 17,058 times respectively (Table 1). In comparison to other plant species, tae-miR169b in wheat and osa-miR169 in rice were the most frequently sequenced miRNAs while miR156 in rice and wheat exhibited low abundance [46]. This suggests a species-specific expression profile for miRNAs. miR156a was also found to be highly expressed in another legume species, Medicago [49]. In Arabidopsis, miR156a, located on chromosome 2 [49], targets 10 mRNAs that code for the squamosa promoter-binding protein (SBP) box, which is involved in leaf morphogenesis [50]. Similar to miR156a, miR157a, which is located on chromosome 1 in Arabidopsis thaliana, was thought to target mRNAs coding for proteins comprising the SBP box [49]. However, the mechanisms, causing the differential expression profile of a same miRNA in different plant species, are unknown. A majority of peanut miRNAs were only sequenced less than 1,000 times, and some rare miRNAs were detected less than 10 times. Compare with the most abundant miRNA ahy-miR157a, their expression level is about 9,500 times lower (Table 1). miRNAs of moderate abundance included ahy-miR157d, ahy-miR164a, ahy-miR166a, ahy-miR166 g, ahy-miR166a, ahy-miR167f, and ahy-miR172a were detected 2,000-10,000 times in the library. The relative abundance of the 22 conserved peanut miRNA families is represented in Figure 2.
Figure 2.
Abundance of peanut-conserved miRNA families.
Next generation high through-put sequencing technology also provides a method for distinguishing and measuring miRNA sequences with only a few nucleotide changes. Based on the results from the Solexa sequencing, different family members displayed drastically different expression levels. For example, the abundance of miR156 family varied from 4 read (ahy-miR156f) to 17,058 reads (ahy-miR156a) in the deep sequencing. This was also the case for some other miRNA families, such as ahy-miR164 (from 1 read to 4,116 reads) and ahy-miR166 (from 1 read to 9577 reads). The existence of a dominant member in a miRNA family may suggest that the regulatory role of this family was performed by the dominant member at the developmental time when the samples were collected for RNA extraction. Abundance comparisons of different members in one miRNA family, during various growth conditions or specific developmental stages, may provide valuable information on the role that miRNAs play in plant growth. Expression levels of two members of the ahy-miR159 family (ahy-miR159a and ahy-miR159b) were similar and were detected 66 and 41 times, respectively (Table 1).
Identification of novel peanut miRNAs
In addition to the identification of conserved miRNAs, 14 novel peanut miRNA families were also identified (Table 2). Only one member was identified in each species-specific miRNA family and the read number for each novel miRNA was much lower than that for the conserved miRNAs. This is consistent with previous conclusions indicating that non-conserved miRNAs are usually expressed at lower levels and with a tissue- or developmental-specific pattern. Therefore, miRNAs identified in this study might represent only a small portion of novel miRNA families found in peanut due to the fact that the small RNA library was constructed from young peanut seedlings grown under normal conditions. Precursors of these novel miRNAs were identified and formed proper secondary hairpin structures, with free energies ranging from -26.91 kcal mol-1 to -132 kcal mol-1 (average of -52.54 kcal mol-1) (Table 2, Additional file 1). More importantly, the identification of an anti-sense miRNA (miRNA*) from five novel miRNA candidates provided more evidence to consider them as novel miRNAs. To investigate the conservation of these 14 novel miRNAs in a wide range of plant species, we used these 14 miRNAs as query sequences to perform Blastn searches against all nucleotide sequences in NCBI databases. No homologs were found in any plant species except miRn1, which has a homolog in the soybean EST CD39249. This suggests that these newly identified miRNAs are all peanut-specific miRNAs except miRn1.
Table 2.
Novel miRNAs identified from peanut
| Name | Count | miRNA sequence | Folding energy |
|---|---|---|---|
| ahy-miRn1 | 656 | UAGAGGGUCCCCAUGUUCUCA | -65.9 |
| ahy-miRn2 | 40 | UCACCGUUAAUACAGAAUCCUU | -70.57 |
| ahy-miRn2* | 3 | AGGAUUCUGUAUUAACGGUGA | -70.57 |
| ahy-miRn3 | 15 | AAUGUAGAAAAUGAACGGUAU | -64.6 |
| ahy-miRn4 | 12 | UGCUGGGUGAUAUUGACAGAAG | -48.72 |
| ahy-miRn5 | 7 | CUGACCACUGUGAUCCCGGAA | -39.5 |
| ahy-miRn6 | 6 | UGACCUUUGGGGAUAUUCGUG | -61.9 |
| ahy-miRn7 | 5 | UCAAUCAAUGACAGCAUUUCA | -39.42 |
| ahy-miRn8 | 4 | UGGUGAUGGUGAAUAUCUUAUC | -38.1 |
| ahy-miRn8* | 1 | AAGGGAGACGUUUGAAUUAUC | -38.1 |
| ahy-miRn9 | 3 | UGGUGAGUCGUAUACAUACUG | -30.91 |
| ahy-miRn10 | 3 | AUACUUGAGAGCCGUUAGAUGA | -52.8 |
| ahy-miRn10* | 1 | AUCUAACGACUCUCAGAUAUAAU | -52.8 |
| ahy-miRn11 | 3 | UUAUACCAUCUUGCGAGACUGA | -49.7 |
| ahy-miRn12 | 4 | UGUUACUAUGGCAUCUGGUAA | -40.2 |
| ahy-miRn12* | 1 | GCCAGGGCCAUGAAUGCAGAU | 40.2 |
| ahy-miRn13 | 3 | CGCAAAUGAUGACAAAUAGA | -26.91 |
| ahy-miRn14 | 11 | UUAAUUUCUGAGUUUGUCAUC | -32.57 |
| ahy-miRn14* | 1 | UUGAUAAGAUAGAAAUUGUAU | -32.57 |
Besides these 14 identified novel candidate miRNAs, we also discovered two small RNAs, with 701 and 159 reads in our small RNA dataset, which correspond to Phaseolus vugaris legume-specific miRS1 and miR2118. These two miRNAs were able to detected in peanut by northern blot analysis [51]. Interestingly, the expression of miR2118 has previously been shown to be induced in Phaseolus vugaris by abiotic stress, especially drought and ABA treatment [51]. We did not include these two sequences in the list of novel peanut miRNAs because we could not find their precursor sequences in the current databases. In addition to miRS1 and miR2118, we also found the third small RNA with 137 reads in our dataset that had only one mismatch with Phaseolus vugaris miR159.2. A fourth 21-nt small RNA with 729 reads was also identified in our dataset, which had 4 mismatches and one nucleotide missing to compare with Phaseolus vugaris miR482*.
Based on the number of detection times and sequences in the small RNA library, novel peanut miRNAs displayed lower expression levels compared to the majority of conserved families. The low abundance of novel miRNAs might suggest a specific role for these miRNAs under various growth conditions, in specific tissues, or during developmental stages. The library enriched only small RNAs that play a role during early seedling stages under normal growth conditions. Whether these low-abundant miRNAs are expressed at higher levels in other tissues and organs, such as flowers, gynophores, pods, or seeds, or whether they are regulated by biotic or abiotic stress, remains to be investigated. Future experiments would provide more insight into the function of these miRNAs.
Validation of peanut miRNAs
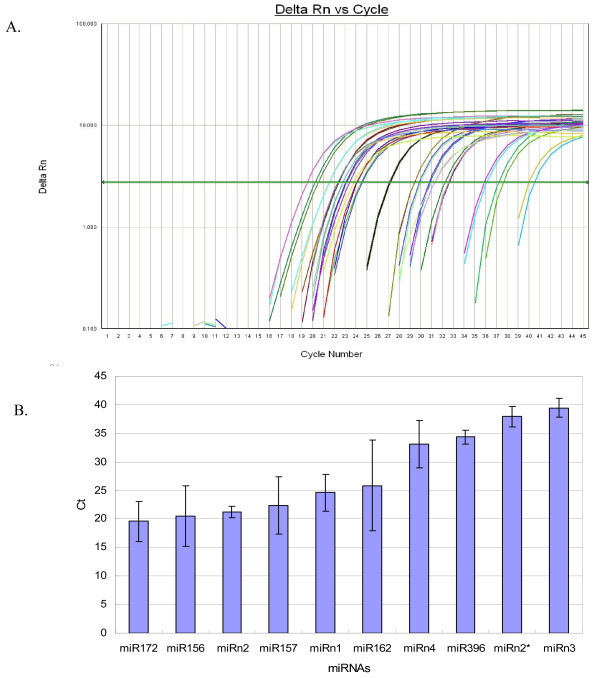
Stem-loop qRT-PCR is a reliable method for detecting and measuring the expression levels of miRNAs. The stem-loop primers increase the sensitivity of the reactions such that this method can significantly distinguish two miRNAs with only one single nucleotide change [52]. In this study, we adopted this technique to validate and measure the expression of 4 novel miRNAs (miRn1, miRn2 and miRn2*, miRn3, and miRn4) as well as 5 conserved miRNAs (miR156, miR157, miR162, miR172, and miR396). All of these miRNAs were identified in peanut by Solexa sequencing. The qRT-PCR results demonstrate that all tested miRNAs, and one miRNA*, are expressed in peanut leaves (Figure 3). However, the expression levels of the different miRNAs varied.
Figure 3.
qRT-PCR validation of the identified peanut miRNAs using high through-put sequencing technology. A. Amplification plot; B. Threshold cycle. Error bars indicate one standard deviation of three different biological replicates (n = 3).
The results of the qRT-PCR reaction show that conserved miRNAs are expressed in peanut. Based on the threshold cycle (CT), miR172 and miR156 were highly expressed with CT values of 19.6 ± 3.5 and 20.5 ± 5.3, respectively. In one of our previous studies, we also found that miR172 is highly expressed in cotton leaves [53]. Other studies have shown that conserved miR172 and miR156 play very important roles in plant growth and development [41]. miR156 is involved in Arabidopsis leaf development by negatively regulating the Squamosa-promoter binding protein (SBP) [38,42]. miR172 controls flower development by regulating the expression of apetala2 (ap2) in Arabidopsis [4,43] and glossy 15 in maize [44]. Aberrant expression of miR156 and miR172 in plants disrupts normal leaf and flower development. Compared with miR156 and miR172, the expression levels of miR157 and miR162 are moderate while the expression of miR396 is low. The expression patterns of these miRNAs appear to be related to their function.
Four novel miRNAs and one miRNA*, all identified by Solexa sequencing, were validated by qRT-PCR. The expression levels of the miRNAs differed from one another in peanut leaves. miRn2 and miRn1 were expressed much higher than other tested peanut-specific miRNAs with a CT value of 21.2 ± 1.0 and 24.6 ± 3.2, respectively. The expression levels are much lower for miRn3 and miRn2* with CT values of 37.9 ± 1.8 and 33.1 ± 4.2. However, more studies need to be performed to elucidate the function that these miRNAS have on the growth and development of peanut.
Target prediction of peanut miRNAs
To better understand the functions of the newly identified species-specific as well as conserved peanut miRNAs, putative targets of these miRNAs were predicted using the described criteria and methods. The target genes of thirteen conserved and seven novel peanut miRNA families were predicted. Transcription factors, including GRAS family transcription factor, nuclear transcription factor Y subunit and NAC1 were predicted to be potential targets of peanut miRNAs. Furthermore, genes directly involved in protein synthesis, e.g., ribosomal protein S12, were targets of peanut miRNAs. A previous study indicates that auxin signaling is regulated by miRNAs [18]; our current result is consistent with this study and the auxin signaling F-box 3 is a potential target of peanut miR393. Resveratrol synthase, NAM (no apical meristem)-like protein, growth regulator factor 5, basic blue copper protein, endonuclease, a protein kinase, transport inhibitor response 1 and a disease resistance response protein were also predicted to be potential targets of identified peanut miRNAs (Additional file 2).
Conclusion
For the first time we discovered, through high through-put Solexa sequencing, 14 novel miRNA families and 75 conserved miRNAs, belonging to 22 families, in peanut. Of these 14 novel peanut miRNAs, 13 are peanut-specific because no homologs have been found in other plant species. qRT-PCR analysis demonstrated that both conserved and peanut-specific miRNAs are expressed in peanuts.
Methods
Plant materials
Peanuts (Arachis hypogaea L. cv. Fenghua-1) were grown in a growth chamber, with a light intensity of 3000 lx, at a relative humidity of 75%, and 26/20°C day/night temperatures. Leaves, stems, and roots from 14-day-old seedlings were collected and immediately stored in liquid nitrogen until total RNA extraction.
RNA extraction and miRNA cloning
Total RNA was isolated from leaves and roots using Trizol agent (TaKaRa, Dalian, China), according to the manufacturer's instructions. Total RNA was isolated from stems using a modified CTAB method with isopropanol instead of lithium chloride for RNA precipitation [54]. Briefly, one gram of stem tissue was ground to a fine powder using liquid nitrogen and mixed thoroughly with 5 ml of pre-warmed (65°C) extraction buffer (2% CTAB, 2% PVP, 0.1 M Tris-HCl, 2.0 M NaCl, 25 mM EDTA, 2% beta-mercaptoethanol, pH 8.0). The mixture was incubated at 65°C for 5 min and shaken three individual times during the incubation period. After a brief cooling of the mixture, 2.5 ml of chloroform and 2.5 ml of isopropanol were added. The mixture was vortexed for 1 min and then centrifuged at 12000 rpm for 15 min at 4°C. After DNase treatment of the extract, RNA was precipitated at room temperature (25°C) for 10 min using an equal volume of isopropanol. The RNA was resuspended in an equal volume of phenol:chloroform:isopropanol (25:24:1), and then resuspended again with an equal volume of chloroform:isopropanol (24:1). A total of 1/10 volume of 3 M NaOAC (pH 5.2) and 2.5 volumes of cold ethanol were added to precipitate the RNA overnight at -20°C.
To identify as many tissue- or developmental-specific miRNAs as possible, we pooled the total RNAs from leaf, stem, and root samples in an equal fraction ratio. miRNA cloning was performed as described previously by Sunkar and Zhu [21]. Briefly, 0.5 M NaCl and 10% PEG8000 were used to precipitate and enrich RNAs with low molecular weight. Next, a 15% polyacrylamide denaturing gel was employed to separate the low-molecular weight RNA. During gel electrophoresis, about 100 μg of total RNA was applied to the gel and two labeled RNA oligonucleotides, approximately 18 and 26 nt in length, were used as size standards. After gel electrophoresis, small RNAs with 18-26 nt were excised from the gel and eluted with 0.4 M NaCl overnight at 4°C. The RNA was dephosphorylated using alkaline phosphatase (New England Biolabs, Beijing China) and recovered by ethanol precipitation. The isolated small RNAs were then sequentially ligated to RNA/DNA chimeric oligonucleotide adapters, reversely transcribed, and amplified by PCR. Finally, Solexa sequencing technology was employed to sequence the small RNAs from pooled peanut samples (BGI, Beijing China).
Identification of conserved and peanut-specific miRNAs
The raw sequences were processed using PHRED and CROSS MATCH programs as previously reported [21,55]. After removing the vector sequences, trimmed sequences longer than 17 nt were used for further analyses. First, rRNA, tRNA, snRNA, and snoRNA, as well as those containing the polyA tail, were removed from the small RNA sequences and the remaining sequences were compared against rice and Arabidopsis ncRNAs deposited in the NCBI Genbank database and Rfam8.0 database. Then, the unique small RNA sequences were used to do a Blastn search against the miRNA database, miRBase 13.0, in order to identify conserved miRNAs in peanuts. Only perfectly matched sequences were considered to be conserved miRNAs. To study potential miRNA precursor sequences, we used the identified peanut mature miRNA sequences to do Blastn searches against peanut ESTs in NCBI. Non-coding ESTs, which met previously described criteria [56], were then considered to be miRNA precursors. Specifically, dominant, mature sequences residing in the stem region of the stem-loop structure and ranging between 20-22 nt with a maximum free-folding energy of -25 kcal mol-1 were considered. A maximum of six unpaired nucleotides between the miRNA and miRNA* was allowed. The distance between the miRNA and miRNA* ranged between 5 and 240-nt. After removing the conserved miRNA sequences, the rest of the small RNA sequences were used to perform Blastn searches against peanut ESTs in order to obtain precursor sequences for novel potential miRNAs. The selected EST sequences were then folded into a secondary structure using an RNA-folding program mFold. If a perfect stem-loop structure was formed, the small RNA sequence was sit at one arm of the stem as well as other criteria were followed, this small RNA was consisted as one novel peanut miRNA.
miRNA validation
Identified peanut miRNAs were validated using quantitative real time PCR (qRT-PCR) using a well-developed strategy. The Applied Biosystems TaqMan® microRNA Assays (Foster City, CA) were employed to detect and compare the expression levels of miRNAs in peanut leaves. TaqMan-based real time quantification of peanut miRNAs includes two important steps: a reverse transcription reaction and a real time quantitative PCR reaction [52]. In this study, 5 conserved miRNAs (miR156, miR157, miR162, miR172, and miR396) and 4 peanut-specific miRNAs (miRn1, miRn2 and miRn2*, miRn3, and miRn4) were validated using qRT-PCR (Table 3). The primer and probe sequences for the 5 conserved miRNAs were purchased from Applied Biosystems and the sequences of the primers for the 4 peanut-specific miRNAs were obtained from Invitrogen. In the reverse transcription reaction, mature miRNAs were reversely transcribed into cDNAs using a miRNA-specific stem-loop RT primer and a reverse transcriptase enzyme. In the qRT-PCR reaction, the expression levels of the 5 conserved and 4 peanut-specific miRNAs were analyzed using miRNA-specific primers (forward and reverse primers) [52].
Table 3.
qRT-PCR-validated miRNAs and their sequences
| miRNA | Sequence |
|---|---|
| miR 156 | UGACAGAAGAGAGUGAGCAC |
| miR 157 | UUGACAGAAGAUAGAGAGCAC |
| miR162 | UCGAUAAACCUCUGCAUCCAG |
| miR172 | AGAAUCUUGAUGAUGCUGCAU |
| miR396 | UUCCACAGCUUUCUUGAACUG |
| miRn1 | UAGAGGGUCCCCAUGUUCUCA |
| miRn2 | UCACCGUUAAUACAGAAUCCUU |
| miRn2* | AGGAUUCUGUAUUAACGGUGA |
| miRn3 | AAUGUAGAAAAUGAACGGUAU |
| miRn4 | UGCUGGGUGAUAUUGACAGAAG |
The RT-PCR and qRT-PCR reactions, for validating and detecting peanut miRNAs, were followed using the same protocols as our previous report [37,53]. Briefly, miRNA reverse transcription reactions were performed in 200 μL PCR tubes, each containing a total of 20 μL of reaction solution. Each reaction solution contained 1000 ng of total leaf RNAs, 3.33 U/μL MultiScribe reverse transcriptase, 1× reverse transcription buffer, 0.25 mM each of dNTPs, and 0.25 U/μL RNase inhibitor; sterilized RNase-free water was used to adjust the total volume of the reverse transcription reaction to 20 μL. The miRNA reverse transcription reactions were incubated in an Eppendorf Mastercycler (Eppendorf North America, Westbury, NY). The RT-PCR temperature program was adjusted to run for 30 min at 16°C, 30 min at 42°C, 5 min at 85°C, and then 4°C until future use. For each miRNA, three biological replicates were performed. After reverse transcription, the products of each reaction were diluted 10 times to avoid potential primer interference in the following qRT-PCR reaction.
Quantitative real time PCR was performed using the TaqMan® microRNA Assay kit (Foster City, CA) on an Applied Biosystems 7300 Sequence Detection System (Foster City, CA). Each reaction consisted of 3 μL of product from the diluted reverse transcription reaction, 2 μL of 20× TaqMan MicroRNA Assay primers (forward and reverse), 12.5 μL of 2× TaqMan Universal PCR Master Mix, and 7.5 μL of nuclease-free water. The reactions were incubated in a 96-well plate at 95°C for 10 min, followed by 45 cycles of 95°C for 15s and 60°C for 60s. After the reactions were completed, the threshold was manually set and the threshold cycle (CT) was automatically recorded. The CT is defined as the fractional cycle number at which the fluorescence signal passes the fixed threshold [52]. All reactions were run in two replicates for each sample.
Target gene prediction
The potential targets of peanut miRNAs were predicted using the psRNATarget program http://bioinfo3.noble.org/psRNATarget/ with default parameters. Newly identified peanut miRNA sequences were used as custom miRNA sequences; Arachis transcript/genomic library (EST, GSS, and nucleotide databases) were used as custom plant databases.
All predicted target genes were evaluated by scoring, and the criteria used were as follows: each G:U wobble pairing was assigned 0.5 points, each indel was assigned 2.0 points, and all other non-canonical Watson-Crick pairings were assigned 1.0 points each. The total score for an alignment was calculated based on 20 nt. When the query was longer than 20 nt, scores for all possible consecutive 20 nt subsequences were computed, and the minimum score was considered the total score for the query-subject alignment. Because targets complementary to the miRNA 5' end appear to be critical, mismatches other than G:U wobbles at positions 2-7 at the 5' end were further penalized by 0.5 points in the final score [57]. Sequences were considered to be miRNA targets if the total score was less than 3.0 points.
Once potential target mRNA sequences were obtained, redundant sequences were removed using the 'contig express' feature of the Vector NTI program. Blastx was performed using the target sequence and the NCBI database to predict functions of potential targets.
Authors' contributions
XW conceived the intellectual design of the project and wrote the manuscript. HX and CZ undertook most of the sequence analysis to identify miRNAs, secondary structures, and prediction of target genes. They also participated in part of the manuscript writing, namely, the method section. TPF and BZ performed the RT-PCR and qRT-PCR experiments and also gave intellectual suggestion for the manuscript writing. YY, YB and AL carried out plant growth, RNA preparation, miRNA library construction. ML and CL completed database searching, data management and processing. All authors read and approved the final version of manuscript.
Supplementary Material
Secondary structures of conserved and novel miRNAs in peanuts.
The putative target genes of identified miRNAs.
Contributor Information
Chuan-Zhi Zhao, Email: huyjh@126.com.
Han Xia, Email: coldxia@126.com.
Taylor Price Frazier, Email: fraziertp@ecu.edu.
Ying-Yin Yao, Email: yingyin@cau.edu.cn.
Yu-Ping Bi, Email: yupingbi@vip.sina.com.
Ai-Qin Li, Email: liaiqin1970@163.com.
Meng-Jun Li, Email: lmj199612@yahoo.com.cn.
Chang-Sheng Li, Email: shzyyxgs@126.com.
Bao-Hong Zhang, Email: zhangb@ecu.edu.
Xing-Jun Wang, Email: xingjunw@hotmail.com.
Acknowledgements
This work was supported by National Natural Science Foundation of China (30871324) and grants 2006BS06024 2006YBS001 and 2007YCX001 to XW. This work is also partially supported by the North Carolina Biotechnology Center grant to BZ.
References
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–854. doi: 10.1016/0092-8674(93)90529-Y. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Carrington JC, Ambros V. Role of microRNAs in plant and animal development. Science. 2003;301(5631):336–338. doi: 10.1126/science.1085242. [DOI] [PubMed] [Google Scholar]
- Ambros V. microRNAs: Tiny regulators with great potential. Cell. 2001;107(7):823–826. doi: 10.1016/S0092-8674(01)00616-X. [DOI] [PubMed] [Google Scholar]
- Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Zhang BH, Pan XP, Cobb GP, Anderson TA. Plant microRNA: A small regulatory molecule with big impact. Developmental Biology. 2006;289(1):3–16. doi: 10.1016/j.ydbio.2005.10.036. [DOI] [PubMed] [Google Scholar]
- Cullen BR. Viruses and microRNAs. Nature Genetics. 2006;38:S25–S30. doi: 10.1038/ng1793. [DOI] [PubMed] [Google Scholar]
- Jones-Rhoades MW, Bartel DP, Bartel B. MicroRNAs and their regulatory roles in plants. Annual Review of Plant Biology. 2006;57:19–53. doi: 10.1146/annurev.arplant.57.032905.105218. [DOI] [PubMed] [Google Scholar]
- Zhang BH, Wang QL, Pan XP. MicroRNAs and their regulatory roles in animals and plants. Journal of Cellular Physiology. 2007;210(2):279–289. doi: 10.1002/jcp.20869. [DOI] [PubMed] [Google Scholar]
- Moss EG, Poethig RS. MicroRNAs: Something new under the Sun. Current Biology. 2002;12(20):R688–R690. doi: 10.1016/S0960-9822(02)01206-X. [DOI] [PubMed] [Google Scholar]
- Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nature Reviews Molecular Cell Biology. 2005;6(5):376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- Chen XM. A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science. 2004;303(5666):2022–2025. doi: 10.1126/science.1088060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Hannon GJ. MicroRNAs: Small RNAs with a big role in gene regulation. Nature Reviews Genetics. 2004;5(7):522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- Schauer SE, Jacobsen SE, Meinke DW, Ray A. DICER-LIKE1: blind men and elephants in Arabidopsis development. Trends Plant Sci. 2002;7(11):487–491. doi: 10.1016/S1360-1385(02)02355-5. [DOI] [PubMed] [Google Scholar]
- Zhang BH, Stellwag EJ, Pan XP. Large-scale genome analysis reveals unique features of microRNAs. Gene. 2009;443(1-2):100–109. doi: 10.1016/j.gene.2009.04.027. [DOI] [PubMed] [Google Scholar]
- Schwartz BW, Yeung EC, Meinke DW. Disruption of morphogenesis and transformation of the suspensor in abnormal suspensor mutants of Arabidopsis. Development. 1994;120(11):3235–3245. doi: 10.1242/dev.120.11.3235. [DOI] [PubMed] [Google Scholar]
- Ray S, Golden T, Ray A. Maternal effects of the short integument mutation on embryo development in Arabidopsis. Developmental Biology. 1996;180(1):365–369. doi: 10.1006/dbio.1996.0309. [DOI] [PubMed] [Google Scholar]
- Vazquez F, Gasciolli V, Crete P, Vaucheret H. The nuclear dsRNA binding protein HYL1 is required for MicroRNA accumulation and plant development, but not posttranscriptional transgene silencing. Current Biology. 2004;14(4):346–351. doi: 10.1016/j.cub.2004.01.035. [DOI] [PubMed] [Google Scholar]
- Park W, Li JJ, Song RT, Messing J, Chen XM. CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Current Biology. 2002;12(17):1484–1495. doi: 10.1016/S0960-9822(02)01017-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Fedoroff N. A mutation in the arabidopsis HYL1 gene encoding a dsRNA binding protein affects responses to abscisic acid, auxin, and cytokinin. Plant Cell. 2000;12(12):2351–2365. doi: 10.1105/tpc.12.12.2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R, Zhu JK. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell. 2004;16(8):2001–2019. doi: 10.1105/tpc.104.022830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HH, Tian X, Li YJ, Wu CA, Zheng CC. Microarray-based analysis of stress-regulated microRNAs in Arabidopsis thaliana. RNA. 2008;14(5):836–843. doi: 10.1261/rna.895308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Rhoades MW, Bartel DP. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Molecular Cell. 2004;14(6):787–799. doi: 10.1016/j.molcel.2004.05.027. [DOI] [PubMed] [Google Scholar]
- Fujii H, Chiou TJ, Lin SI, Aung K, Zhu JK. A miRNA involved in phosphate-starvation response in Arabidopsis. Current Biology. 2005;15(22):2038–2043. doi: 10.1016/j.cub.2005.10.016. [DOI] [PubMed] [Google Scholar]
- Sunkar R, Kapoor A, Zhu JK. Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell. 2006;18(8):2051–2065. doi: 10.1105/tpc.106.041673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro L, Dunoyer P, Jay F, Arnold B, Dharmasiri N, Estelle M, Voinnet O, Jones JDG. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science. 2006;312(5772):436–439. doi: 10.1126/science.1126088. [DOI] [PubMed] [Google Scholar]
- Katiyar-Agarwal S, Gao S, Vivian-Smith A, Jin H. A novel class of bacteria-induced small RNAs in Arabidopsis. Genes & Development. 2007;21(23):3123–3134. doi: 10.1101/gad.1595107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian S, Fu Y, Sunkar R, Barbazuk WB, Zhu JK, Yu O. Novel and nodulation-regulated microRNAs in soybean roots. BMC Genomics. 2008. doi:10.1186/1471-2164-9-160. [DOI] [PMC free article] [PubMed]
- Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Research. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrea C, Erica M, Giorgio G, Massimo P, Claudio M, Mario Enrico P, Andrea S. Cloning and characterization of small non-coding RNAs from grape. The Plant Journal. 2009;59(5):750–763. doi: 10.1111/j.1365-313X.2009.03906.x. [DOI] [PubMed] [Google Scholar]
- Arazi T, Talmor-Neiman M, Stav R, Riese M, Huijser P, Baulcombe DC. Cloning and characterization of micro-RNAs from moss. Plant Journal. 2005;43(6):837–848. doi: 10.1111/j.1365-313X.2005.02499.x. [DOI] [PubMed] [Google Scholar]
- Barakat A, Wall K, Leebens-Mack J, Wang YJ, Carlson JE, dePamphilis CW. Large-scale identification of microRNAs from a basal eudicot (Eschscholzia californica) and conservation in flowering plants. Plant Journal. 2007;51(6):991–1003. doi: 10.1111/j.1365-313X.2007.03197.x. [DOI] [PubMed] [Google Scholar]
- Axtell MJ, Snyder JA, Bartell DP. Common functions for diverse small RNAs of land plants. Plant Cell. 2007;19(6):1750–1769. doi: 10.1105/tpc.107.051706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang BH, Pan XP, Cannon CH, Cobb GP, Anderson TA. Conservation and divergence of plant microRNA genes. Plant Journal. 2006;46(2):243–259. doi: 10.1111/j.1365-313X.2006.02697.x. [DOI] [PubMed] [Google Scholar]
- Pan XP, Zhang BH, SanFrancisco M, Cobb GP. Characterizing viral microRNAs and its application on identifying new microRNAs in viruses. Journal of Cellular Physiology. 2007;211(1):10–18. doi: 10.1002/jcp.20920. [DOI] [PubMed] [Google Scholar]
- Zhang BH, Pan XP, Anderson TA. Identification of 188 conserved maize microRNAs and their targets. FEBS Letters. 2006;580:3753–3762. doi: 10.1016/j.febslet.2006.05.063. [DOI] [PubMed] [Google Scholar]
- Zhang BH, Pan XP, Stellwag EJ. Identification of soybean microRNAs and their targets. Planta. 2008;229(1):161–182. doi: 10.1007/s00425-008-0818-x. [DOI] [PubMed] [Google Scholar]
- Zhang BH, Pan XP, Wang QL, Cobb GP, Anderson TA. Identification and characterization of new plant microRNAs using EST analysis. Cell Research. 2005;15(5):336–360. doi: 10.1038/sj.cr.7290302. [DOI] [PubMed] [Google Scholar]
- Zhang BH, Wang QL, Wang KB, Pan XP, Liu F, Guo TL, Cobb GP, Anderson TA. Identification of cotton microRNAs and their targets. Gene. 2007;397(1-2):26–37. doi: 10.1016/j.gene.2007.03.020. [DOI] [PubMed] [Google Scholar]
- Wang XW, Zhang J, Gu J, He T, Zhang XG, Li YD. MicroRNA identification based on sequence and structure alignment. Bioinformatics. 2005;21(18):3610–3614. doi: 10.1093/bioinformatics/bti562. [DOI] [PubMed] [Google Scholar]
- Sunkar R, Jagadeeswaran G. In silico identification of conserved microRNAs in large number of diverse plant species. BMC Plant Biology. 2008;8:13. doi: 10.1186/1471-2229-8-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szittya G, Moxon S, Santos DM, Jing R, Fevereiro MPS, Moulton V, Dalmay T. High-throughput sequencing of Medicago truncatula short RNAs identifies eight new miRNA families. Bmc Genomics. 2008. doi: 10.1186/1471-2164-9-593. [DOI] [PMC free article] [PubMed]
- Morin RD, Aksay G, Dolgosheina E, Ebhardt HA, Magrini V, Mardis ER, Sahinalp SC, Unrau PJ. Comparative analysis of the small RNA transcriptomes of Pinus contorta and Oryza sativa. Genome Research. 2008;18(4):571–584. doi: 10.1101/gr.6897308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan R, Vaucheret H, Trejo J, Bartel DP. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes & Development. 2006;20(24):3407–3425. doi: 10.1101/gad.1476406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Tej SS, Luo SJ, Haudenschild CD, Meyers BC, Green PJ. Elucidation of the small RNA component of the transcriptome. Science. 2005;309(5740):1567–1569. doi: 10.1126/science.1114112. [DOI] [PubMed] [Google Scholar]
- Yao YY, Guo GG, Ni ZF, Sunkar R, Du JK, Zhu JK, Sun QX. Cloning and characterization of microRNAs from wheat (Triticum aestivum L.) Genome Biology. 2007;8(6) doi: 10.1186/gb-2007-8-6-r96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu D, Pan X, Wilson IW, Ketchum REB, Li F, Liu M, Teng W, Zhang BH. High throughput sequencing technology reveals that the taxoid elicitor methyl jasmonate regulates microRNA expression in Chinese yew (Taxus chinensis) Gene. 2009;436(1-2):37–44. doi: 10.1016/j.gene.2009.01.006. [DOI] [PubMed] [Google Scholar]
- Dolgosheina EV, Morin RD, Aksay G, Sahinalp SC, Magrini V, Mardis ER, Mattsson J, Unrau PJ. Conifers have a unique small RNA silencing signature. RNA. 2008;14(8):1508–1515. doi: 10.1261/rna.1052008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP. MicroRNAs in plants. Genes & Development. 2002;16(13):1616–1626. doi: 10.1101/gad.1004402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP. Prediction of plant microRNA targets. Cell. 2002;110(4):513–520. doi: 10.1016/S0092-8674(02)00863-2. [DOI] [PubMed] [Google Scholar]
- Arenas-Huertero C, Perez B, Rabanal F, Blanco-Melo D, De la Rosa C, Estrada-Navarrete G, Sanchez F, Covarrubias AA, Reyes JL. Conserved and novel miRNAs in the legume Phaseolus vulgaris in response to stress. Plant Molecular Biology. 2009;70(4):385–401. doi: 10.1007/s11103-009-9480-3. [DOI] [PubMed] [Google Scholar]
- Chen CF, Ridzon DA, Broomer AJ, Zhou ZH, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Research. 2005;33(20):e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang BH, Pan XP. Expression of microRNAs in cotton. Molecular Biotechnology. 2009;42(3):269–274. doi: 10.1007/s12033-009-9163-y. [DOI] [PubMed] [Google Scholar]
- Carra A, Gambino G, Schubert A. A cetyltrimethylammonium bromide-based method to extract low-molecular-weight RNA from polysaccharide-rich plant tissues. Analytical Biochemistry. 2007;360(2):318–320. doi: 10.1016/j.ab.2006.09.022. [DOI] [PubMed] [Google Scholar]
- Sunkar R, Girke T, Zhu JK. Identification and characterization of endogenous small interfering RNAs from rice. Nucleic Acids Research. 2005;33(14):4443–4454. doi: 10.1093/nar/gki758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V, Bartel B, Bartel DP, Burge CB, Carrington JC, Chen XM, Dreyfuss G, Eddy SR, Griffiths-Jones S, Marshall M. A uniform system for microRNA annotation. RNA. 2003;9(3):277–279. doi: 10.1261/rna.2183803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YJ. MiRU: an automated plant miRNA target prediction server. Nucleic Acids Research. 2005;33:W701–W704. doi: 10.1093/nar/gki383. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Secondary structures of conserved and novel miRNAs in peanuts.
The putative target genes of identified miRNAs.