Abstract
An indispensable step in protein biosynthesis is the 2′(3′) aminoacylation of tRNA by aminoacyl-tRNA synthetases. Here we show that a similar activity exists in a tiny, 5-nt-long RNA enzyme with a 3-nt active center. The small ribozyme initially trans-phenylalanylates a partially complementary 4-nt RNA selectively at its terminal 2′-ribose hydroxyl using PheAMP, the natural form for activated amino acid. The initial 2′ Phe-RNA product can be elaborated into multiple peptidyl-RNAs. Reactions do not require divalent cations, and have limited dependence on monovalent cations. Small size and minimal requirements for regiospecific translational activity strongly support the hypothesis that minuscule RNA enzymes participated in early forms of translation.
Keywords: aminoacyl-RNA, enzyme, evolution, peptidyl-RNA, RNA
Amino acids enter modern translation via attachment to a 2′(3′) tRNA terminus, a reaction catalyzed by a protein aminoacyl-tRNA synthetase. Because it is implausible that primitive peptides were synthesized using already-formed protein catalysts, the RNA world hypothesis (1, 2) requires peptide synthetic reactions performed by RNA enzymes (3, 4). Indeed, a number of RNAs have been isolated which accelerate related translational reactions (5).
Several ribozymes capable of catalyzing the same chemical group transfer that is today carried out by aminoacyl-tRNA synthetases (6 –9) have been isolated using Systematic Evolution of Ligands by Exponential Enrichment (10, 11). However, none possess all desirable characteristics. First, an RNA world enzyme should be small, accessible after rudimentary RNA synthesis. In addition, it should act in trans, and should use universal biological, water-soluble substrates. Previously isolated ribozymes employ appropriate substrates [amino acids activated as acyladenylates (6, 12)], but are not true enzymes, as they are self-aminoacylators modified by their own reaction. Other ribozymes aminoacylate RNA with turnover; however, the amino acids must be activated as cyanomethyl, 3,5-dinitrobenzyl, or p-chlorobenzyl esters (8, 13), and hence they do not facilitate the biological reaction.
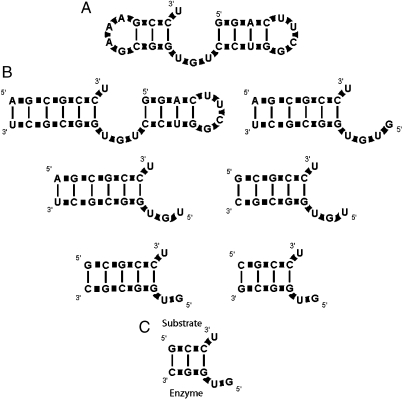
The family of self-aminoacylating ribozymes exemplified by truncate C3 RNA (Fig. 1 A) presented the intriguing possibility of very simple aminoacyl transfer (6). Mutational analyses as well as molecular dynamics and energy minimization of the reactants suggested a tiny active center consisting of only three essential nucleotides—a 3′-terminal U, and a 5′-GU-3′ sequence in a loop apposed to the unpaired 3′-terminal U. Although the C3 RNA family possessed helical elements, adjacent helices appeared nonspecific in sequence, perhaps required only for assembly of the active center (6).
Fig. 1.
Small trans-aminoacylating RNA complexes. (A) C3 RNA. (B) Intermediate trans complexes. (C) Final GUGGC/GCCU complex.
Here we present a radically modified version of C3 ribozyme, unique in three ways: It functions in trans; it has been minimized to a tiny, five-nucleotide ribozyme; and it also supports peptidyl-RNA synthesis. The result is the smallest trans-aminoacylator, and arguably the smallest true ribozyme, ever observed (Fig. S1).
Results
Initially, RNA C3 (6) was separated into “enzyme” and “substrate” modules (Fig. 1 B). Subsequently, a succession of progressively smaller complexes were tested for activity, shortening the helix that linked the ribozymic fragment to the substrate, and also discarding the right-hand hairpin completely (Fig. 1 B). Covalent continuity, half of the substrate helix, and the entire rightward structure, proved dispensable. Each of these modified RNAs made Phe-RNA from PheAMP (Fig. S2). These results are consistent with lack of conservation of the discarded structures in the initially selected RNAs (6). The terminus of this molecular minimization is the 5-nt ribozyme/4-nt substrate complex, GUGGC/GCCU (Fig. 1 C).
The 5-nt ribozyme/4-nt substrate complex was the smallest that effectively produced aminoacyl-RNA under our standard conditions. A smaller complex with only two base pairs between the enzymic and substrate elements (GUGG/CCU) yielded no observable product(s). The additional GC base pair in GUGGC/GCCU apparently provides marginal stability required for substrate acquisition and/or aminoacyl transfer.
Ribozyme 5′-GUGGC-3′ (GUGGC) catalyzes the addition of phenylalanine to the 3′-terminal uridine of RNA substrate 5′-GCCU-3′ (GCCU), where the underlined ribonucleotides comprise the specific nucleotides of the active site. When GUGGC and 32P-radiolabeled GCCU are incubated with PheAMP, product bands are evident on an acid urea polyacrylamide gel (Fig. 2). These multiple products are shown below to be RNA-Phe, RNA-Phe2, and RNA-Phe3. Thus GUGGC/GCCU complex hosts two of three essential chemical group transfers that occur in biological peptide synthesis: aminoacylation of RNA and subsequent peptide extension.
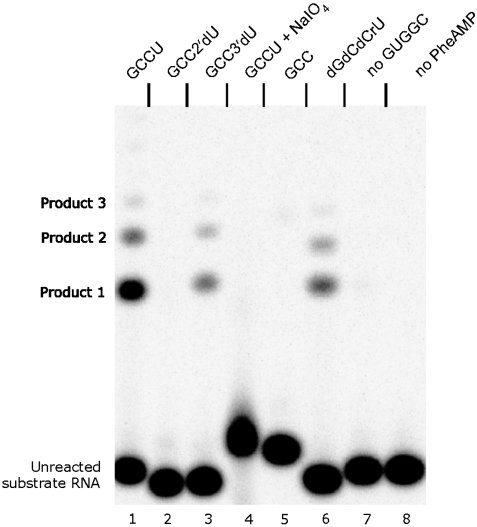
Fig. 2.
Initial reaction occurs only at 2′-OH. Lanes: 1, substrate = GCCU; 2, GCC2′dU; 3, GCC3′dU; 4, GCCU + NaIO4; 5, GCC; 6, dGdCdCrU; 7, GCCU + PheAMP, no GUGGC; 8, GCCU + GUGGC, no PheAMP. Reaction conditions: 10 μM ribozyme GUGGC, 20 μM RNA substrate, 100 mM KCl, 5 mM MgCl2, 100 mM Hepes pH 7.0, 2.0 mM PheAMP; incubation at 4 °C for 30 min.
Shortened substrate lacking unpaired 3′-terminal U (GCC) did not produce detectable product bands when incubated with GUGGC and PheAMP, suggesting the importance of the unpaired 3′-terminal substrate nucleotide in the reaction (Fig. 2, lane 5). Further, no product was formed when GCCU was pretreated with periodate (NaIO4), which selectively oxidizes the 2′(3′)-terminal cis-glycol of ribose, converting the sugar to an open dialdehyde (Fig. 2, lane 4). Gel resolution of the unoxidized GCCU band from the periodate product shows that oxidation was quantitative. These data suggest that phenylalanine transfer is to the ribose hydroxyl(s), as in modern enzymatic aminoacyl-tRNA synthesis.
This transfer was confirmed and defined by use of a synthetic substrate oligomer lacking the terminal 2′-hydroxyl group (GCC2′dU; Fig. 2, lane 2). This molecule yields no visible products under the conditions used here, suggesting that phenylalanine is initially transferred to the 2′-terminal -OH. Initial 2′ aminoacylation was further confirmed by facile Phe-RNA formation when an alternative substrate with a single 2′-terminal hydroxyl, GCC3′dU, was aminoacylated (Fig. 2, lane 3). This reactivity verifies that the 2′-hydroxyl group, but not the 3′, is required. Further, lanes 2, 3, 4, 5, and 6 confirm that internal 2′-OH groups are not the site of aminoacyl transfer: for example, a DNA-RNA hybrid substrate dGdCdCrU retained activity (Fig. 2, lane 6). Accordingly, the terminal ribose 2′-OH is the unique site of initial aminoacyl transfer. These results parallel similar findings for the parental C3 ribozyme (6), and will be relevant below to discussion of multiple products, all now shown to depend on the earliest transfer to terminal 2′-OH.
The rate of Phe-RNA formation from 3′-dU substrate (k 2nd order ≅ k cat/K m = 1.2 M-1 min-1) is 18% that of the original RNA GCCU (k 2nd order = 6.5 M-1 min-1) under comparable conditions and where PheAMP instability has been accounted for (Fig. S3). This implies a minor catalytic role for the 3′-hydroxyl group, though it is not required for aminoacylation or peptide formation. This GUGGC/GCCU rate is about an order slower than other small ribozyme self-acylators (12). This minor difference is remarkable in view of the lack of higher-order structure at the active center. In many respects, the small active center appears unchanged from that of parental C3 RNA. The 5′-terminal guanosine of GUGGC is indispensable for the reaction. Both mutation of 5′-G to C, and elimination of 5′-G altogether, completely abolished product formation (Fig. S4). This parallels prior mutational and computational analyses of the active center in C3 RNA, where the analogous G supports critical intermolecular interactions during aminoacyl transfer (6).
GUGGC/GCCU accepts varied aminoacylation substrates, activated as mixed phosphoric anhydrides. Both PheUMP and MetAMP are substrates as monitored by gel electrophoresis (Fig. S5). Both these substrates were also reactive with the C3 ribozyme (6). GUGGC/GCCU therefore tolerates alternative substitution on the phosphate oxygen (A vs. U) and aminoacyl residue (Phe vs. Met), as did C3 RNA. Therefore, both the 2′ regioselectivity and substrate tolerance of the original C3 RNA active center are still demonstrable in GUGGC, even though most of the primary and secondary structure of the initially selected RNA catalyst have been discarded. Indeed, the originally selected RNA molecule (the selected parent of C3 RNA) was a specifically 3′-truncated 69-mer; thus in the present work, 87% of the initial RNA has been deleted.
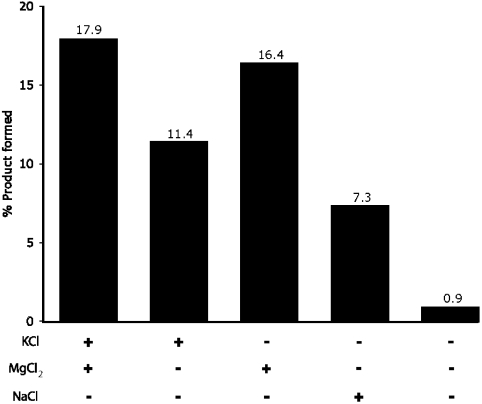
Unlike most ribozymes, GUGGC does not require divalent cations for function. Removal of divalent cations from the reaction reduced activity by only 6.5% (Fig. 3). The implied ionic tolerance is perhaps not surprising because the GUGGC/GCCU complex probably has a relatively simple 3D structure, and thus does not require divalent cations for folding. Magnesium is somewhat stimulatory, however, as are potassium and sodium. We observed slower but significant product formation at 10 mM Na+, but did not test the enzyme’s function in the complete absence of monovalents as some monovalents were introduced during adjustment of the pH of the reaction buffer. Nondiscrimination toward metallocations further supports such enzyme function in primitive environments, where particular divalents and monovalents may have been variably available.
Fig. 3.
Aminoacylation reaction does not require divalent cations. All reactions contained 10 μM GUGGC, 20 μM GCCU, 1.9 mM PheAMP, 100 mM Hepes pH 6.5, 10 mM NaOH. Reactions contained 100 mM KCl, 5 mM MgCl2, or 100 mM NaCl where indicated. Incubation at 4 °C for 60 min.
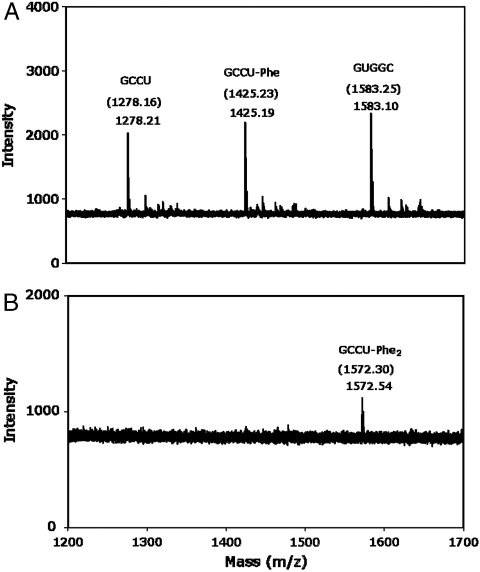
MALDI mass spectrometric analysis of Product 1 confirmed the presence of a compound with the expected mass of quadruply protonated GCCU-Phe (Fig. 4 A and Fig. S6). To summarize, the carboxyl-activated substrate PheAMP transfers the atoms of phenylalanine to ribose 2′-hydroxyl, paralleling the parental self-aminoacylating RNA. Therefore, the first product is the ester Phe-RNA. To further show that the first product formed is an ester, Product 1 was purified and subjected to mild base hydrolysis (pH 8.5, 37 °C). Product 1 hydrolyzed to GCCU (as monitored by gel electrophoresis) at a rate of (0.036 min-1). This rate is close to the previously reported rate of tRNA-Phe hydrolysis under similar conditions (0.042 min-1), confirming that the initial product is RNA-Phe ester (14).
Fig. 4.
MALDI-TOF mass spectrometric analysis of RNA and Products 1 and 2. (A) Product 1 and RNA. (B) Product 2. Numbers in parentheses indicate theoretical mass of protonated products; outside parentheses, observed mass.
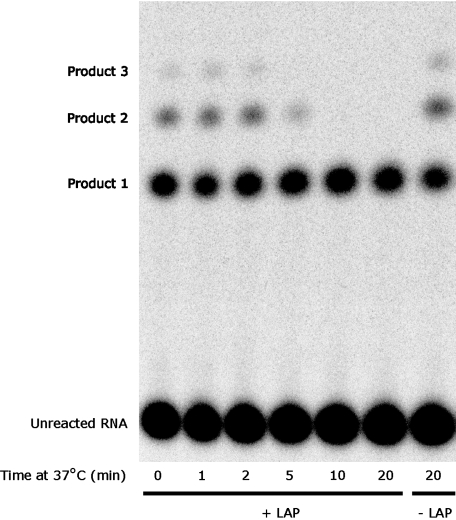
Reaction of GUGGC with GCCU and PheAMP under these conditions yields multiple slower-migrating products, which are RNA peptides. MALDI analysis of Product 2 confirmed a compound with the same mass as quadruply protonated GCCU-Phe2 peptidyl-RNA (Fig. 4 B and Fig. S6). Products 2 and 3 are also susceptible to digestion by exopeptidase, which confirms that they are peptidyl-RNAs (Fig. 5). Treatment of standard reaction products with leucine aminopeptidase, which hydrolyzes N-terminal peptidyl amino acids but not ester bonds, resulted in the degradation of all products to Product 1 (GCCU-Phe ester) (Fig. 5). Thus, the observed products (Figs. 2 and 5) are GCCU-Phe through GCCU-Phe3 peptidyl-RNA. The absence of any products in reactions employing GCC and GCC2′dU instead of GCCU (Fig. 2) is consistent with higher products formed exclusively from initially produced 2′-aminoacyl-RNA.
Fig. 5.
Higher products are RNA peptides. Aminoacylation reaction conditions: 10 μM GUGGC, 20 μM GCCU, 100 mM KCl, 5 mM MgCl2, 100 mM Hepes pH 7.0, 1 mM PheAMP; incubation at 4 °C for 30 min. LAP, leucine aminopeptidase, 23 ng/μL.
Catalysis on GUGGC/GCCU is seen against a lower background of spontaneous reaction with PheAMP. Under certain conditions (increased pH and/or PheAMP concentration), a dim band of RNA-Phe can be visualized even in reactions that do not contain ribozyme GUGGC. Quantitation of such reactions suggests that GUGGC accelerates formation of RNA-Phe by 25-fold at pH 7.5.
Discussion
These data strongly and collectively support the active site model first suggested for the C3 ribozyme (6): RNA 2′(3′) aminoacylation requires only an unusually simple center composed of three nucleotides.
Simple metal-catalyzed RNA activity is already known. Every ribophosphodiester is subject to hydrolysis catalyzed by divalent cations, and this can be enhanced by small RNA structures that bind metals. The small RNA GAAACp/UUU is cleaved between G and A in the presence of Mn2+ or Cd2+ (15). Further, newly selected RNAs routinely contain nucleotides that can be deleted without abolishing function (16).
Nonetheless, GUGGC/GCCU is unique: It binds a complex heteroatomic substrate and facilitates group transfer from a multipart biochemical. This implies a more complex catalytic interface than in metal-catalyzed hydrolysis employing already poised 2′-hydroxyl and ribonucleotide phosphate. In addition, reactions presented here do not depend on ions infrequent in biological systems, and, in fact, show little ion dependence. Thus, it is more surprising that aminoacyl transfer is accelerated by two normal Us and a G nucleotide, poised at a helix terminus.
Further, these particular reactions are central to metabolism, resembling the substrate and product of biological aminoacyl-tRNA synthesis. Transaminoacylation in this work is performed more simply than elsewhere (9, 12, 17). In addition, observation of RNA-peptide products provides the simplest polypeptide synthesis from aminoacyl adenylate (12, 18, 19), or from any other substrate, in the absence of protein catalysts. Essential intermediates in protein biosynthesis therefore arise surprisingly easily in the presence of very short RNAs.
The ultimate importance of these observations may lie partly in the unknown number of other reactions that can be accelerated by comparably small RNAs. This is because for each such minuscule RNA reaction, there is a prima facie case that it would become accessible even after the most primitive ribonucleotide polymerization.
To see this, consider that, to pick every possible RNA pentamer sequence from arbitrary pentamers (with probability 0.9975), one needs only accumulate 4.1 × 10-18 gm of RNA. To possess every tetramer (with probability 0.9975) from a pool of arbitrary tetramers, one would need 3.4 × 10-18 gm RNA. In a real polymerization, one would have a distribution of lengths; nonetheless, with only attograms of total RNA of distributed short lengths from some geochemical source, one would have not only our ribozyme, but every activity of comparable size.
As an illustration, the ribozymic complexes characterized here demonstrate that aminoacyl-RNA and peptidyl-RNAs could have appeared in the presence of ≥9 nucleotides of polymeric RNA, with six of these free to vary to other base pairs. We have previously estimated that a population containing about 1 ng of arbitrary-sequence RNA would be required before useful ribozymes and other active RNA structures would probably occur among this population (20). This follows the so-called axiom of origin (21), which estimates that the RNA world would begin when the amount of RNA exceeds the threshold for occurrence of ribozymes. The finding of nine-nucleotide active centers reduces the threshold for ribozyme activity about 7 orders of magnitude, to a level much more easily breached by undirected geochemical syntheses, or by RNA-catalyzed RNA synthesis itself (22 –24).
The most intriguing possibility raised by these results is that an RNA reaction center for phosphoester transfer may exist somewhere near this size. This would make the polymerase/replicase needed to initiate Darwinian evolution of RNAs, the founding event of the RNA world, much more likely. On one hand, with this few ribonucleotides to dispose in space, there may not be other similar nucleotide structures that are both stable and capable of catalysis. On the other hand, for obvious reasons, it will be extraordinarily important to look for other tiny RNA active centers, now knowing they can exist.
Methods
RNA and Aminoacyl Substrate Synthesis.
RNA was synthesized by Dharmacon. GUGGC = 5′-GUGGC-3′; GCCU = 5′P-GCCU-3′; 5′OH-GCCU = 5′-GCCU-3′; GCCU2′dU = 5′-GCC-2′-dU; GCC = 5′-GCC-3′; dGdCdCrU = 5′-dGdCdCU-3′.
RNA GCC3′dU was prepared by first synthesizing 5′-O-(4,4′-Dimethoxytrityl)3′-deoxyuridine as follows: 3′-deoxyuridine (MP Biomedicals; 991 mg, 0.434 mmol) was dissolved in 5 mL anhydrous pyridine and pyridine was then removed under vacuum while stirring. Solid was then redissolved in 2 mL pyridine. Dimethoxytrityl chloride (170 mg, 0.499 mmol) was dissolved in 12 mL pyridine and slowly added to 3′-deoxyuridine solution. Solution was stirred at room temperature for 4 h. All solutions were sequestered from exposure to air throughout.
Reaction was then quenched by addition of 5 mL methanol, and solvent was removed by rotary evaporation. Remaining solvent evaporated overnight in a vacuum chamber. Product was then dissolved in 1 mL acetonitrile and purified through a silica column (acetonitrile elution). Final product fractions (confirmed through TLC, 1∶1 hexane:acetonitrile) were pooled and rotary evaporated. Yield was 71%.
Dimethoxytrityl-protected 3′dU was then sent to Dharmacon for immobilization of 3′-dU on glass and synthesis of 5′-GCC-3′-dU.
PheAMP, PheUMP, and MetAMP were synthesized by the method of Berg (25) with modifications and purification as described in ref. 6. Yield was as follows: PheAMP 85%, PheUMP 67%, and MetAMP 36%.
Aminoacylation Assays.
For aminoacylation assays, substrate RNA was labeled with 32P using T4 polynucleotide kinase (New England BioLabs). Kinase conditions were 5 μM substrate RNA, 1X T4 polynucleotide kinase forward reaction buffer (Invitrogen), 0.625–1.25 μM (7.5 mCi/mL) γ-32P-ATP, 0.5 units/μL T4 polynucleotide kinase; 37 °C for 20–30 min. Kinase reaction was then passed through a Micro Bio-Spin 6 chromatography column (Bio-Rad) to remove excess γ-32P-ATP or purified using polyacrylamide gel electrophoresis followed by ethanol precipitation.
Standard aminoacylation conditions were as follows: GUGGC 10 μM, GCCU 20 μM, γ-32P-GCCU ≤ 0.3 μM, KCl 100 mM, MgCl2 5 mM, Hepes pH 7.0 100 mM, PheAMP 1–2 mM; incubation at 4 °C for 30 min. RNA, salts, and buffer were first incubated at 4 °C for 10 min. PheAMP (1–10 μmol) was dissolved in cold water, filtered, and added to RNA solution. After incubation, the reaction was stopped by addition of xylene cyanol/bromophenol blue dye containing sodium acetate, and either frozen on dry ice or loaded directly into gel for electrophoresis. Conditions for product isolation experiments for MS used 25 μM GUGGC, 50 μM GCCU, and 2.5 mM PheAMP.
Periodate reaction contained 100 μM 5′OH-GCCU, ≤ 0.3 μM γ-32P-GCCU, 60 mM KCl, 5 mM MgCl2, 30 mM NaOAc pH 5.2, 8.2 mM NaIO4 (total volume 100 μL). Reaction was incubated on ice in the dark for 2 h. Then glucose was added to 14.9 mM to consume excess periodate and the reaction sat on ice for ∼10 min. Reaction was then passed through a Micro Bio-Spin 6 column (Bio-Rad) and lyophilized. Product was redissolved in 15 μL water and 1 μL was mixed with GUGGC, KCl, MgCl2, Hepes, and PheAMP as in standard assay conditions.
Gel electrophoresis was performed using polyacrylamide gel from 12% acrylamide, 6 M urea, 0.1 M sodium acetate pH 5.2. Gels were run from 6–16 h at 4 °C until bromophenol blue had migrated at least 15 cm. Gels were dried, exposed to a phosphorimager screen, and analyzed with a Bio-Rad Molecular Imager FX and QuantityOne software (Bio-Rad).
Purification and Identification of Products 1 and 2.
Removal of salts and buffer from products were necessary for MALDI mass spectrometric analysis. Product 1, GUGGC, and GCCU were purified from a standard aminoacylation reaction using a Waters Sep-Pak C18 cartridge. Sep-Pak (Waters) was equilibrated with 1.8 mL methanol followed by 7 mL water. Completed aminoacylation reaction (100 μL, aqueous) was added to Sep-Pak and washed with 3.5 mL water followed by 200 μL methanol. Then 600 μL methanol was added, collected, and concentrated with a SpeedVac (Savant). Gel electrophoresis and analysis confirmed the presence of radiolabeled GCCU and Product 1 in collected fractions. A parallel Sep-Pak purification with a scaled up (10x volume), cold reaction was performed for mass spectrometric analysis.
A Waters Atlantis 5 μm dC18 column was used to separate Product 2 from other reaction components through HPLC. The gradient was 95% A/5% C for 10 min, ramp to 70% A/30% C for 33 min. The flow was 1 mL/ min. Solvent A = 0.1 M NH4OAc, pH 4.5; solvent C = acetonitrile. Fractions were collected (one per minute) and counted using a scintillation counter to detect elution of products. Product 2 (confirmed by gel electrophoresis) was present in fractions eluted between 32 and 34 min. A parallel HPLC purification was done with a scaled-up cold reaction and fractions 32–34 for both hot and cold reactions were collected, lyophilized, and resuspended in 100 μL water.
A second HPLC purification was necessary to remove ammonium acetate buffer from Product 2. The gradient was 95% B/5% C for 20 min, ramp to 100% C for 10 min. Solvent B = water. Counts were highest in fractions 7–13 for hot reaction; corresponding fractions were collected for scaled-up cold reaction, pooled, and lyophilized.
To measure the rate of ester hydrolysis, Product 1 was first purified through HPLC. Lyophilized product was resuspended in 0.1 M Tris·HCl, pH 8.5, and incubated at 37 °C. Hydrolysis rate was determined from the slope of the natural log of fraction P1 remaining vs. time.
Mass spectrometry.
MS was carried out using a Voyager-DE STR MALDI-TOF mass spectrometer (Applied Biosystems). Dried samples were dissolved in acetonitrile and cospotted with α-cyano-4-hydroxycinnamic acid matrix solution. Analysis was performed in the negative, reflector mode and masses were calibrated using a three-peptide standard. Spectra were averaged over 50 laser shots.
Peptidase reactions.
Leucine aminopeptidase (microsomal from porcine kidney) was purchased from Sigma-Aldrich. Peptidase reactions were carried out in the presence of 0.1 M Hepes pH 7.0.
Supplementary Material
Acknowledgments.
The authors thank Dr. Bruce Eaton and his lab for contribution of equipment and reagents, Dr. Shuji Kato for aiding with mass spectrometry, Dr. Mali Illangasekare for assistance with kinetic experiments, and Dr. Irene Majerfeld and Dr. Teresa Janas for comments on this manuscript. This work was supported by National Institutes of Health research Grant R01 GM 48080 (to M.Y.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0912895107/DCSupplemental.
References
- 1.Gilbert W. The RNA world. Nature. 1986;319:618. [Google Scholar]
- 2.White HB., III Coenzymes as fossils of an earlier metabolic state. J Mol Evol. 1976;7:101–104. doi: 10.1007/BF01732468. [DOI] [PubMed] [Google Scholar]
- 3.Guerrier-Takada C, Gardiner K, Marsh T, Pace N, Altman S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983;35:849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- 4.Kruger K, et al. Self-splicing RNA: Autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell. 1982;31:147–157. doi: 10.1016/0092-8674(82)90414-7. [DOI] [PubMed] [Google Scholar]
- 5.Yarus M. On translation by RNAs alone. Cold Spring Harb Sym. 2001;66:207–215. doi: 10.1101/sqb.2001.66.207. [DOI] [PubMed] [Google Scholar]
- 6.Chumachenko NV, Novikov Y, Yarus M. Rapid and simple ribozymic aminoacylation using three conserved nucleotides. J Am Chem Soc. 2009;131:5257–5263. doi: 10.1021/ja809419f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Illangasekare M, Sanchez G, Nickles T, Yarus M. Aminoacyl-RNA synthesis catalyzed by an RNA. Science. 1995;267:643–647. doi: 10.1126/science.7530860. [DOI] [PubMed] [Google Scholar]
- 8.Lee N, Suga H. A minihelix-loop RNA acts as a trans-aminoacylation catalyst. RNA. 2001;7:1043–1051. doi: 10.1017/s1355838201010457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murakami H, Saito H, Suga H. A versatile tRNA aminoacylation catalyst based on RNA. Chem Biol. 2003;10:655–662. doi: 10.1016/s1074-5521(03)00145-5. [DOI] [PubMed] [Google Scholar]
- 10.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 11.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 12.Illangasekare M, Yarus M. A tiny RNA that catalyzes both aminoacyl-RNA and peptidyl-RNA synthesis. RNA. 1999;5:1482–1489. doi: 10.1017/s1355838299991264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murakami H, Bonzagni NJ, Suga H. Aminoacyl-tRNA synthesis by a resin-immobilized ribozyme. J Am Chem Soc. 2002;124:6834–6835. doi: 10.1021/ja025872a. [DOI] [PubMed] [Google Scholar]
- 14.Chousterman S, Hervé G, Chapeville F. Hydrolyse alcaline de la liaison ester de quelques aminoacyl-tARN normaux et modifiés. Bull Soc Chim Biol. 1966;48:1295–1303. [PubMed] [Google Scholar]
- 15.Kazakov S, Altman S. A trinucleotide can promote metal ion-dependent specific cleavage of RNA. Proc Natl Acad Sci USA. 1992;89:7939–7943. doi: 10.1073/pnas.89.17.7939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anderson PC, Mecozzi S. Unusually short RNA sequences: Design of a 13-mer RNA that selectively binds and recognizes theophylline. J Am Chem Soc. 2005;127:5290–5291. doi: 10.1021/ja0432463. [DOI] [PubMed] [Google Scholar]
- 17.Tamura K, Schimmel P. Chiral-selective aminoacylation of an RNA minihelix. Science. 2004;305:1253. doi: 10.1126/science.1099141. [DOI] [PubMed] [Google Scholar]
- 18.Tamura K, Schimmel P. Peptide synthesis with a template-like RNA guide and aminoacyl phosphate adaptors. Proc Natl Acad Sci USA. 2003;100:8666–8669. doi: 10.1073/pnas.1432909100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang B, Cech TR. Peptide bond formation by in vitro selected ribozymes. Nature. 1997;390:96–100. doi: 10.1038/36375. [DOI] [PubMed] [Google Scholar]
- 20.Knight R, et al. Abundance of correctly folded RNA motifs in sequence space, calculated on computational grids. Nucleic Acids Res. 2005;33:5924–5935. doi: 10.1093/nar/gki886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yarus M, Knight RD. In: The Genetic Code and the Origin of Life. Pouplana LR, editor. Georgetown, TX: Landes Bioscience; 2004. pp. 75–91. [Google Scholar]
- 22.Doudna J. A multisubunit ribozyme that is a catalyst of and template for complimentary strand RNA synthesis. Science. 1991;251:1605–1609. doi: 10.1126/science.1707185. [DOI] [PubMed] [Google Scholar]
- 23.McGinness KE, Wright MC, Joyce GF. Continuous in vitro evolution of a ribozyme that catalyzes three successive nucleotidyl addition reactions. Chem Biol. 2002;9:585–596. doi: 10.1016/s1074-5521(02)00136-9. [DOI] [PubMed] [Google Scholar]
- 24.Zaher HS, Unrau PJ. Selection of an improved RNA polymerase ribozyme with superior extension and fidelity. RNA. 2007;13:1017–1026. doi: 10.1261/rna.548807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berg PJ. The chemical synthesis of amino acyladenylates. J Biol Chem. 1958;233:608–611. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.