Abstract
Background
Studies on the epidemiology of African Animal Trypanosomiasis (AAT) rarely consider the spatial dimension of disease prevalence. This problem is confounded by use of parasitological diagnostic methods of low sensitivity in field surveys. Here we report a study combining highly sensitive and species specific molecular diagnostic methods, and Geographical information system (GIS) for spatial analysis of trypanosome infection patterns, to better understand its epidemiology. Blood samples from 44 and 59 animals randomly selected from Teso and Suba districts respectively were screened for trypanosomes using PCR diagnostic assays. Spatial distribution of the positive cases was mapped and average nearest neighbour analysis used to determine the spatial pattern of trypanosome cases detected.
Findings
Trypanosome prevalence of 41% and 29% in Suba and Teso districts respectively was observed. T. vivax infections were most prevalent in both areas. Higher proportions of T. brucei infections (12%) were observed in Suba, a known sleeping sickness foci compared with 2% in Teso. Average nearest neighbour analysis showed the pattern of trypanosome infections as random. An overlay with tsetse maps showed cases lying outside the tsetse infested areas, mostly being cases of T. vivax which is known to be transmitted both biologically by tsetse and mechanically by biting flies.
Conclusion
These findings suggest a need to design control strategies that target not just the biological vector tsetse, but also the parasite in cattle in order to clear the possibly mechanically transmitted T. vivax infections. There is need to also review the accuracy of available tsetse maps.
Findings
Trypanosomiasis, a disease of humans and animals caused by several species of trypanosomes and spread by tsetse flies is a major constraint to livestock production in 37 countries within the Sub-Saharan region. An estimated 45-50 million cattle are at risk of infection in the region, with an estimated economic loss of up to US$ 1.3 billion in cattle production [1]. Its public health importance has led to attempts to control the disease nationally and regionally with initiatives as Pan Africa tsetse and trypanosomosis eradication program (PATTEC) [2]. These attempts rely on repeated large-scale epidemiological studies and environmental surveys, guiding the design and implementation of intervention strategies. The accuracy of these surveys is limited by use of parasitological diagnostic techniques as microscopy due to low sensitivity [3], and the difficulty in incorporating climatic and environmental data known to influence tsetse distribution, and as a result disease spread [4,5].
The high costs required to produce tsetse distribution maps through ground-based vector surveys have resulted in few studies looking at the spatial dimension of disease prevalence [6]. However, the use Geographical Information System (GIS) software now makes it cheaper and easier to produce maps which can serve as useful tools for policy discussion, and allow for analysis of factors that would influence the disease patterns [7]. Polymerase Chain Reaction (PCR) diagnostic assays overcome the low sensitivity limitations of parasitological techniques. They are powerful tools for identification and diagnosis of trypanosomes in their hosts and vectors although their high cost and need for elaborate expertise has delayed their adoption [8]. However, PCR assays able to detect all pathogenic trypanosome species in a single reaction have been developed [9,10]. These reduce the costs of screening a sample from an endemic area by up to five times, and have been suggested as suitable for large-scale epidemiological studies [11].
Results obtained from molecular studies, associated with geo-referenced information concerning vector, cattle distribution and relevant environment parameters, combined in a GIS have the potential of providing more informative study results. This new approach of studying complex pathogenic system is argued to lead to a better evaluation of the risk of infection, allows for effective risk communication, and gives scientific outputs in ways that are understandable to non-scientists [12]. Here we report a study combining use of highly sensitive and species specific PCR assays, with GIS for spatial analysis of infection patterns. The aim is to provide information about the prevalence and distribution AAT in two areas in Western Kenya, and suggest improved control strategies based on the findings.
Results
Cross-sectional trypanosomiasis survey
Overall, 29% of the 44 animals and 41% of the 59 animals sampled in Teso and Suba districts respectively had trypanosome infections. Based on the total number of cases detected, T. vivax infections were predominant in both areas (69% in Teso, and 50% in Suba). In Teso, infections with T. congolense savannah and T. brucei showed equal proportions of 8% while T. congolense savannah/T. vivax mixed infections made up 15% of the cases. In Suba, T. brucei made 17% and T. congolense savannah 8% of the cases. Mixed infections between T. congolense savannah and T. vivax made up for 12%, between T. congolense savannah and T. brucei infections 8%, and between T. vivax and T. brucei infections 4%. The prevalence of each of the trypanosome species in the two districts are shown in Table 1. Suba district showed a higher prevalence for T. brucei at 7% compared with Teso at 3%. The mixed infections observed in the two study areas are presented in Table 1. None of the T. brucei tested positive for the Serum Resistance Associated (SRA) gene, present only in the human-infective T. brucei rhodesiense.
Table 1.
The prevalence of different species of trypanosomes in Teso and Suba districts of Western Kenya
| Prevalence in % | ||||
|---|---|---|---|---|
| Trypanosome species | Suba | 95% CI | Teso | 95% CI |
| TCS | 3 | (0 to 13) | 2 | (0 to 13) |
| T.vivax | 20 | (11 to 33) | 20 | (10 to 35) |
| T.brucei | 7 | (2 to 17) | 2 | (0 to 13) |
| TCS/T.vivax | 5 | (1 to 15) | 5 | (0 to 17) |
| TCS/T.brucei | 3 | (0 to 13) | 0 | - |
| T.vivax/T.brucei | 2 | (0 to 10) | 0 | - |
| Total | 41 | (28 to 54) | 29 | (17 to 45) |
TCS - T. congolense savannah
Spatial pattern of the disease
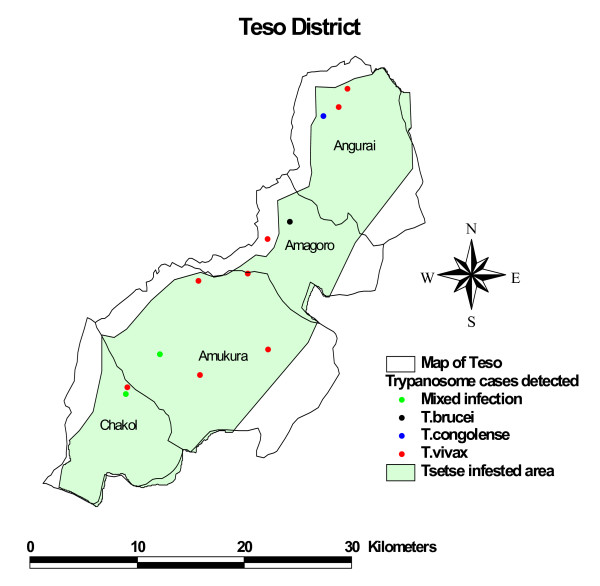
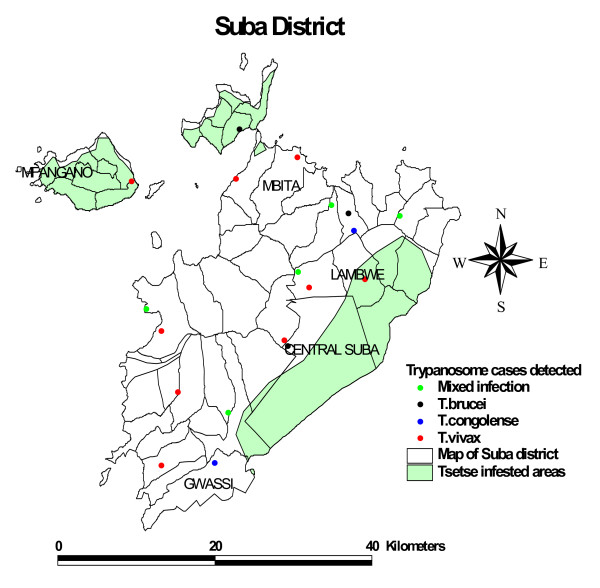
Figures 1 and 2 are maps showing the various trypanosome species detected and the overlay with the tsetse distribution maps for Teso and Suba districts respectively. The results show infection cases falling in the tsetse free areas. Most of these are cases of T. vivax which is known to also be transmitted mechanically through biting flies besides by tsetse. The ratio of the observed mean distance versus expected mean difference was 1.1 and 0.85, and the Z score 0.9 and -1 standard deviations, in Suba and Teso districts respectively. Z-score value is considered significant for dispersion at the confidence level of 0.01 if it is 2.58 or higher. Infection patterns in both Teso and Suba were random.
Figure 1.
Map of Teso district showing the areas infested with tsetse flies (grey), and the different species of trypanosomes detected.
Figure 2.
Map of Suba district showing the areas infested with tsetse flies (grey), and the different species of trypanosomes detected.
Discussion
Attempts to control trypanosomiasis are based on large-scale epidemiological studies, active detection and treatment of confirmed cases, combined with tsetse control programmes [2]. The accuracy of these studies depends on the quality of the study design, and on the sensitivity and specificity of diagnostic methods used. Previous studies have for instance reported T. congolense as the predominant species in cattle in various parts of Africa [13,14], whereas this study established T. vivax as the most prevalent species. This finding agrees with other studies conducted in Busia district, Kenya [15], and in Tororo, South East Uganda [16]. T. vivax is pathogenic and sometimes causes a fatal hemorrhagic disease.
Overlays with the tsetse maps showed a large proportion of trypanosome cases falling in tsetse-free areas. Assuming the maps are accurate, we suspect that the transmission in those areas is mechanical since most infections are T. vivax, which is transmitted both by tsetse flies and mechanically by biting flies. A longitudinal study comparing bovine trypanosomiasis in tsetse free and tsetse infested areas of northwest Ethiopia found T. vivax to be responsible for 90.9% of the cattle trypanosome cases in the tsetse free zone support this hypothesis [17]. Animals may move between the tsetse-infested and tsetse-free areas although this is unlikely to adequately explain the high infection numbers in tsetse free areas. Farmers in the study region either tether their animals or graze them communally within limited areas.
The presence of mixed infections emphasizes the need for epidemiologists to focus beyond single species to multiple infections with different parasites which are a norm under field conditions [18]. In addition to trypanosomes, animals used in this study may have been infected with other parasites endemic in the region.
Lower trypanosome prevalence in Teso compared with Suba district could be attributed to the fact that unlike Suba, Teso was part of the FITCA program which focused on tsetse control and animal treatment in selected regions [19]. The small sample sizes used in this study limit its power to adequately detect true differences in the two study districts. A similar study using larger sample sizes is recommended. In Teso, a temporal decline of trypanosome prevalence is reported, with up to 40% prevalence in the late 90's, [20] and only 4% in 2005 [15]. Our study reports a much higher prevalence (29%) possibly reflecting higher sensitivity levels attained while using the PCR assays, or a possible lapse in stringent control measures following cessation of the FITCA campaigns. This apparent resurgence raises a question on sustainability of such control programs after donors pull out. Large differences in prevalence as may be obtained by PCR and parasitological methods could have a significant impact on the control strategies selected.
The tsetse maps used here are dated the year 1998, and land usage could have interfered with the tsetse habitat and hence it's distribution. For instance, in Teso district, the status of tsetse may have changed following the FITCA campaigns. Use of impregnated targets and deltamethrin treated cattle has been shown to reduce tsetse populations by 90% and the incidence of bovine trypanosomiasis from over 30% to below 5% in a 6 month period [21].
Whereas this study was not designed to identify the spatial risk factors associated with trypanosomiasis, it shows the importance of simple mapping in helping understand the status of the disease, and possibly guiding future research questions and policy formulation in disease mitigation strategies. For instance, is mechanical transmission of trypanosomiasis significant or is it a case of tsetse habitats having extended to areas captured as tsetse free before? A combination of highly sensitive diagnostic methods which can be extended to study infection in the vector tsetse and GIS provides an opportunity to use evidence based control strategies.
Conclusion
The prevalence maps correlated with tsetse maps show cases lying outside the tsetse areas, mostly being cases of T. vivax which is known to be transmitted both biologically by tsetse flies and mechanically by biting flies. These findings suggest that control of tsetse is not enough and that to clear the possibly mechanically transmitted T. vivax infections, trypanosomes in cattle should also be targeted.
Methods
Study area
The cross-sectional study was carried out between August and September 2006 in Teso and Suba districts of Western Kenya. Both districts fall within the tsetse belt and are endemic for trypanosomiasis. Teso district lying towards north and bordering Uganda had recently been involved in a tsetse control program (Farming in tsetse controlled areas -FITCA), that run from the year 2000 to 2004. Suba district lying towards the south was not part of the FITCA program. Lambwe valley a known sleeping sickness foci, is within Suba district. This region offers a good environment for tsetse flies and wildlife which are the major reservoirs for Human African Trypanosomiasis. This study aims at establishing the prevalence of trypanosomiasis in the two districts, giving a possible indication of the impact of the tsetse control program.
Blood sampling, DNA extraction
A total of 44 and 59 blood samples were obtained from Teso and Suba districts respectively. A multi-stage sampling plan was used. The first was sub-location selection in which all sub-locations in each district were allocated a unique number and a random sample picked. The farms in each sub-location were randomly selected based on a frame supplied by the respective local veterinary office. At the farm level one or two animals were randomly selected depending on availability and total number of animals in each farm. Indigenous cattle of both sexes and all ages except suckling calves were sampled. Genomic DNA for the screening of trypanosomes was extracted from the blood samples using the salt out method [22] with slight modification.
PCR Diagnosis
Samples were screened for pathogenic T. congolense, T. vivax and T. brucei species. Species specific primers [23], and multi-species detecting ITS1 BR and ITS1 CF primers [9], and nested ITS 1, 2, 3 and 4 primers [10] designed to amplify the Internal transcribed spacer of the ribosomal DNA were used to screen each sample. The details of these screenings are well described in a previous paper by the same authors [11]. All T. brucei positives were further tested for the serum resistance associated (SRA) gene, which is known to be present only in the human infective T. brucei rhodesiense [24,25].
Spatial distribution and analysis
Using the geographical coordinates of sampling areas, and results from the PCR screenings, infection distribution maps for each study site were created. Spatial pattern of infections were determined using the average nearest neighbour analysis feature in ESRI ArcGis 9.0. According to the software's manual, this is done by calculating the average distance between each feature and it's nearest neighbouring feature within the dataset. The result is the observed mean distance which is then divided by the expected mean distance to calculate the average nearest neighbour ratio. This value is compared with the average distance value for a hypothetical random distribution containing the same number of features and area. If the average nearest neighbour ratio or index is equal to 1, the pattern is random, a ratio greater than 1 indicates, the pattern is dispersed, while a ratio less than 1 implies the pattern is clustered.
Tsetse distribution
Additional data layers describing the tsetse distribution in the two study areas were obtained from a GIS database available at the International Livestock Research Institute website http://www.ilri.org/GIS. The original map was based on tsetse distribution maps by Ford and Katondo [26], modified using data generated by Kenya Trypanosomiasis Research Institute. These tsetse distribution maps were overlaid on the trypanosome infection maps to determine whether all cases fell within tsetse infested areas.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
SMT drafted the manuscript. SMT, JOJ, FAM and ROM conceived the study. FAM and SMT carried out the molecular and spatial analysis. All authors read and approved the final manuscript.
Contributor Information
Samuel M Thumbi, Email: sm.thumbi@cgiar.org.
Joseph O Jung'a, Email: owinojj@hotmail.com.
Reuben O Mosi, Email: oyoomosi@uonbi.ac.ke.
Francis A McOdimba, Email: francis.mcodimba@aku.edu.
Acknowledgements
The authors thank ASARECA-AARNet for funding the project and the BecA-ILRI hub for providing the laboratory facilities. They also thank Patrick Kariuki and Shem Kifugo of ILRI for the assistance with spatial analysis.
References
- Kristjanson PM, Swallow BM, Rowlands GJ, Kruska RL, Leeuw PNd. Measuring the costs of African animal trypanosomosis, the potential benefits of control and returns to research. Agricultural Systems. 1999;59:79–98. doi: 10.1016/S0308-521X(98)00086-9. [DOI] [Google Scholar]
- Schofield CJ, Kabayo JP. Trypanosomiasis vector control in Africa and Latin America. Parasit Vectors. 2008;1(1):24. doi: 10.1186/1756-3305-1-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picozzi K, Tilley A, Fevre EM, Coleman PG, Magona JW, Odiit M, Eisler M, Welburn S. The diagnosis of trypanosome infections: applications of novel technology for reducing disease risk. African Journal of Biotechnology. 2002;1(2):39–45. [Google Scholar]
- Hendrickx G, Napala A, Slingenbergh JH, De Deken R, Vercruysse J, Rogers DJ. The spatial pattern of trypanosomosis prevalence predicted with the aid of satellite imagery. Parasitology. 2000;120(Pt 2):121–134. doi: 10.1017/S0031182099005338. [DOI] [PubMed] [Google Scholar]
- Gage KL, Burkot TR, Eisen RJ, Hayes EB. Climate and vectorborne diseases. Am J Prev Med. 2008;35(5):436–450. doi: 10.1016/j.amepre.2008.08.030. [DOI] [PubMed] [Google Scholar]
- Hendrickx G, Napala A, Dao B, Batawui K, Bastiaensen P, De Deken R, Vermeilen A, Vercruysse J, Slingenbergh JH. The area-wide epidemiology of bovine trypanosomosis and its impact on mixed farming in subhumid West Africa; a case study in Togo. Vet Parasitol. 1999;84(1-2):13–31. doi: 10.1016/S0304-4017(99)00086-2. [DOI] [PubMed] [Google Scholar]
- Hay SI, Omumbo JA, Craig MH, Snow RW. Earth observation, geographic information systems and Plasmodium falciparum malaria in sub-Saharan Africa. Adv Parasitol. 2000;47:173–215. doi: 10.1016/s0065-308x(00)47009-0. full_text. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magona JW, Mayende JS, Olaho-Mukani W, Coleman PG, Jonsson NN, Welburn SC, Eisler MC. A comparative study on the clinical, parasitological and molecular diagnosis of bovine trypanosomosis in Uganda. Onderstepoort J Vet Res. 2003;70(3):213–218. [PubMed] [Google Scholar]
- Njiru ZK, Constantine CC, Guya S, Crowther J, Kiragu JM, Thompson RC, Davila AM. The use of ITS1 rDNA PCR in detecting pathogenic African trypanosomes. Parasitol Res. 2005;95(3):186–192. doi: 10.1007/s00436-004-1267-5. [DOI] [PubMed] [Google Scholar]
- Cox A, Tilley A, McOdimba F, Fyfe J, Eisler M, Hide G, Welburn S. A PCR based assay for detection and differentiation of African trypanosome species in blood. Exp Parasitol. 2005;111(1):24–29. doi: 10.1016/j.exppara.2005.03.014. [DOI] [PubMed] [Google Scholar]
- Thumbi SM, McOdimba FA, Mosi RO, Jung'a JO. Comparative evaluation of three PCR base diagnostic assays for the detection of pathogenic trypanosomes in cattle blood. Parasit Vectors. 2008;1(1):46. doi: 10.1186/1756-3305-1-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell BS, Hoskins RE, Pickle LW, Wartenberg D. Current practices in spatial analysis of cancer data: mapping health statistics to inform policymakers and the public. Int J Health Geogr. 2006;5:49. doi: 10.1186/1476-072X-5-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott J, Woitag T, Sidibe I, Bauer B, Diarra B, Ouedraogo D, Kamuanga M, Peregrine A, Eisler M, Zessin KH. Field studies of drug-resistant cattle trypanosomes in Kenedougou Province, Burkina Faso. Acta Trop. 2003;86(1):93–103. doi: 10.1016/S0001-706X(03)00019-6. [DOI] [PubMed] [Google Scholar]
- Leak SGA. Tsetse biology and ecology: their role in the epidemiology and control of trypanosomosis. Elsevier Science Ltd; 1999. [Google Scholar]
- Karanja SM. Epidemiology and importance of trypanosomosis, helminthosis and tick-borne diseases on the performance of cattle in Busia district, Kenya. Freie Universität Berlin. PhD Thesis. 2005.
- McOdimba FA. Epidemiology of Vector borne diseases in cattle from SE Uganda. PhD Thesis. 2006.
- Cherenet T, Sani RA, Speybroeck N, Panandam JM, Nadzr S, Bossche P Van den. A comparative longitudinal study of bovine trypanosomiasis in tsetse-free and tsetse-infested zones of the Amhara Region, northwest Ethiopia. Vet Parasitol. 2006;140(3-4):251–258. doi: 10.1016/j.vetpar.2006.04.004. [DOI] [PubMed] [Google Scholar]
- Cox FE. Concomitant infections, parasites and immune responses. Parasitology. 2001;122(Suppl):S23–38. doi: 10.1017/S003118200001698X. [DOI] [PubMed] [Google Scholar]
- FITCA-K. Farming in Tsetse Controlled Areas; Second Annual Progress Report and Financial Statement of Project Activities. Delegation of the European Commission, Nairobi, Kenya; 2001. [Google Scholar]
- Angus SD. The epidemiology of trypanosomiasis in village livestock in an endemic sleeping sickness area of Western Kenya. PhD Thesis. 1996.
- Bauer B, Amsler-Delafosse S, Kabore I, Kamuanga M. Improvement of cattle productivity through rapid alleviation of African animal trypanosomosis by integrated disease management practices in the agropastoral zone of Yale, Burkina Faso. Trop Anim Health Prod. 1999;31(2):89–102. doi: 10.1023/A:1005115707181. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. 2. Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- Masiga DK, Smyth AJ, Hayes P, Bromidge TJ, Gibson WC. Sensitive detection of trypanosomes in tsetse flies by DNA amplification. Int J Parasitol. 1992;22(7):909–918. doi: 10.1016/0020-7519(92)90047-O. [DOI] [PubMed] [Google Scholar]
- Gibson WC. The SRA gene: the key to understanding the nature of Trypanosoma brucei rhodesiense. Parasitology. 2005;131(Pt 2):143–150. doi: 10.1017/S0031182005007560. [DOI] [PubMed] [Google Scholar]
- Welburn SC, Picozzi K, Fevre EM, Coleman PG, Odiit M, Carrington M, Maudlin I. Identification of human-infective trypanosomes in animal reservoir of sleeping sickness in Uganda by means of serum-resistance-associated (SRA) gene. Lancet. 2001;358(9298):2017–2019. doi: 10.1016/S0140-6736(01)07096-9. [DOI] [PubMed] [Google Scholar]
- Ford J, Katondo KM. Maps of tsetse fly (Glossina) distribution in Africa, 1973, according to sub-generic groups on a scale of 1:5,000,000. Bulletin of Animal Health and Production in Africa 15 188-193 Nairobi. 1977.