Abstract
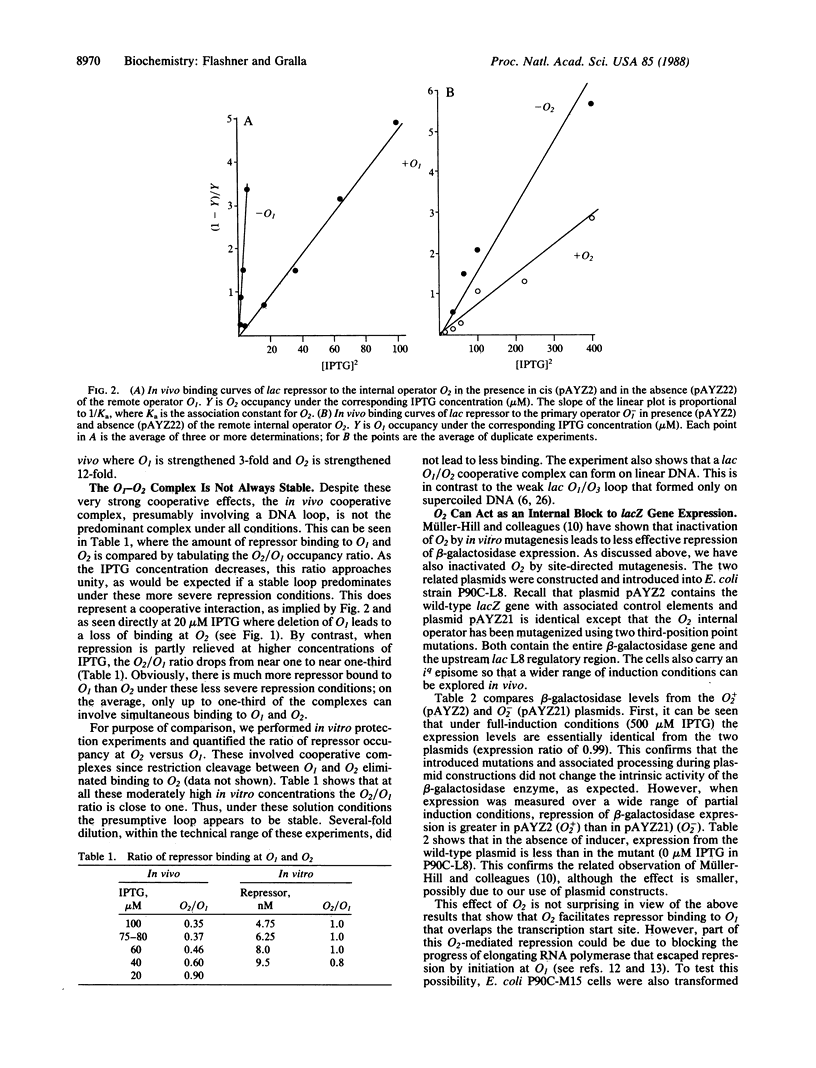
The mechanism by which the internal lacZ gene sequence O2 influences lac repression was investigated by using in vivo footprinting of operon mutants. Quantitative in vivo binding curves show that O2 strengthens by approximately 3-fold repressor binding to O1 that is located 400 base pairs upstream at the transcription start site. The internal O2 sequence also contributes to repression by a second mechanism: repressor bound internally blocks elongation of beta-galactosidase gene expression. This secondary mechanism of repression is facilitated by the remote O1 operator that strengthens binding to O2 12-fold. Thus, lac repression involves two mechanisms, both of which involve cooperation between remote operator elements. During mild repression only the initiation mechanism applies, but more severe repression favors formation of the presumptive O1-O2 repression loop that allows both mechanisms to act simultaneously.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Besse M., von Wilcken-Bergmann B., Müller-Hill B. Synthetic lac operator mediates repression through lac repressor when introduced upstream and downstream from lac promoter. EMBO J. 1986 Jun;5(6):1377–1381. doi: 10.1002/j.1460-2075.1986.tb04370.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betz J. L. Affinities of tight-binding lactose repressors for wild-type and pseudo-operators. J Mol Biol. 1987 Jun 5;195(3):495–504. doi: 10.1016/0022-2836(87)90178-1. [DOI] [PubMed] [Google Scholar]
- Betz J. L., Sasmor H. M., Buck F., Insley M. Y., Caruthers M. H. Base substitution mutants of the lac operator: in vivo and in vitro affinities for lac repressor. Gene. 1986;50(1-3):123–132. doi: 10.1016/0378-1119(86)90317-3. [DOI] [PubMed] [Google Scholar]
- Borowiec J. A., Gralla J. D. High-resolution analysis of lac transcription complexes inside cells. Biochemistry. 1986 Sep 9;25(18):5051–5057. doi: 10.1021/bi00366a012. [DOI] [PubMed] [Google Scholar]
- Borowiec J. A., Zhang L., Sasse-Dwight S., Gralla J. D. DNA supercoiling promotes formation of a bent repression loop in lac DNA. J Mol Biol. 1987 Jul 5;196(1):101–111. doi: 10.1016/0022-2836(87)90513-4. [DOI] [PubMed] [Google Scholar]
- Calos M. P., Johnsrud L., Miller J. H. DNA sequence at the integration sites of the insertion element IS1. Cell. 1978 Mar;13(3):411–418. doi: 10.1016/0092-8674(78)90315-x. [DOI] [PubMed] [Google Scholar]
- Celada F., Zabin I. A dimer--dimer binding region in beta-galactosidase. Biochemistry. 1979 Feb 6;18(3):404–406. doi: 10.1021/bi00570a002. [DOI] [PubMed] [Google Scholar]
- Dandanell G., Hammer K. Two operator sites separated by 599 base pairs are required for deoR repression of the deo operon of Escherichia coli. EMBO J. 1985 Dec 1;4(12):3333–3338. doi: 10.1002/j.1460-2075.1985.tb04085.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deuschle U., Gentz R., Bujard H. lac Repressor blocks transcribing RNA polymerase and terminates transcription. Proc Natl Acad Sci U S A. 1986 Jun;83(12):4134–4137. doi: 10.1073/pnas.83.12.4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn T. M., Hahn S., Ogden S., Schleif R. F. An operator at -280 base pairs that is required for repression of araBAD operon promoter: addition of DNA helical turns between the operator and promoter cyclically hinders repression. Proc Natl Acad Sci U S A. 1984 Aug;81(16):5017–5020. doi: 10.1073/pnas.81.16.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eismann E., von Wilcken-Bergmann B., Müller-Hill B. Specific destruction of the second lac operator decreases repression of the lac operon in Escherichia coli fivefold. J Mol Biol. 1987 Jun 20;195(4):949–952. doi: 10.1016/0022-2836(87)90499-2. [DOI] [PubMed] [Google Scholar]
- Fried M., Crothers D. M. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981 Dec 11;9(23):6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert W., Maxam A. The nucleotide sequence of the lac operator. Proc Natl Acad Sci U S A. 1973 Dec;70(12):3581–3584. doi: 10.1073/pnas.70.12.3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gralla J. D. Rapid "footprinting" on supercoiled DNA. Proc Natl Acad Sci U S A. 1985 May;82(10):3078–3081. doi: 10.1073/pnas.82.10.3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith J., Hochschild A., Ptashne M. DNA loops induced by cooperative binding of lambda repressor. Nature. 1986 Aug 21;322(6081):750–752. doi: 10.1038/322750a0. [DOI] [PubMed] [Google Scholar]
- Irani M. H., Orosz L., Adhya S. A control element within a structural gene: the gal operon of Escherichia coli. Cell. 1983 Mar;32(3):783–788. doi: 10.1016/0092-8674(83)90064-8. [DOI] [PubMed] [Google Scholar]
- Krämer H., Niemöller M., Amouyal M., Revet B., von Wilcken-Bergmann B., Müller-Hill B. lac repressor forms loops with linear DNA carrying two suitably spaced lac operators. EMBO J. 1987 May;6(5):1481–1491. doi: 10.1002/j.1460-2075.1987.tb02390.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin K., Huo L., Schleif R. F. The DNA loop model for ara repression: AraC protein occupies the proposed loop sites in vivo and repression-negative mutations lie in these same sites. Proc Natl Acad Sci U S A. 1986 Jun;83(11):3654–3658. doi: 10.1073/pnas.83.11.3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossing M. C., Record M. T., Jr Thermodynamic origins of specificity in the lac repressor-operator interaction. Adaptability in the recognition of mutant operator sites. J Mol Biol. 1985 Nov 20;186(2):295–305. doi: 10.1016/0022-2836(85)90106-8. [DOI] [PubMed] [Google Scholar]
- Mossing M. C., Record M. T., Jr Upstream operators enhance repression of the lac promoter. Science. 1986 Aug 22;233(4766):889–892. doi: 10.1126/science.3090685. [DOI] [PubMed] [Google Scholar]
- Nick H., Gilbert W. Detection in vivo of protein-DNA interactions within the lac operon of Escherichia coli. 1985 Feb 28-Mar 6Nature. 313(6005):795–798. doi: 10.1038/313795a0. [DOI] [PubMed] [Google Scholar]
- Ptashne M. Gene regulation by proteins acting nearby and at a distance. Nature. 1986 Aug 21;322(6081):697–701. doi: 10.1038/322697a0. [DOI] [PubMed] [Google Scholar]
- Reitzer L. J., Magasanik B. Transcription of glnA in E. coli is stimulated by activator bound to sites far from the promoter. Cell. 1986 Jun 20;45(6):785–792. doi: 10.1016/0092-8674(86)90553-2. [DOI] [PubMed] [Google Scholar]
- Reznikoff W. S., Miller J. H., Scaife J. G., Beckwith J. R. A mechanism for repressor action. J Mol Biol. 1969 Jul 14;43(1):201–213. doi: 10.1016/0022-2836(69)90089-8. [DOI] [PubMed] [Google Scholar]
- Reznikoff W. S., Winter R. B., Hurley C. K. The location of the repressor binding sites in the lac operon. Proc Natl Acad Sci U S A. 1974 Jun;71(6):2314–2318. doi: 10.1073/pnas.71.6.2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadler J. R., Smith T. F. Mapping of the lactose operator. J Mol Biol. 1971 Nov 28;62(1):139–169. doi: 10.1016/0022-2836(71)90136-7. [DOI] [PubMed] [Google Scholar]
- Sasse-Dwight S., Gralla J. D. Probing co-operative DNA-binding in vivo. The lac O1:O3 interaction. J Mol Biol. 1988 Jul 5;202(1):107–119. doi: 10.1016/0022-2836(88)90523-2. [DOI] [PubMed] [Google Scholar]
- Schleif R. Gene regulation: why should DNA loop? Nature. 1987 Jun 4;327(6121):369–370. doi: 10.1038/327369a0. [DOI] [PubMed] [Google Scholar]
- Sellitti M. A., Pavco P. A., Steege D. A. lac repressor blocks in vivo transcription of lac control region DNA. Proc Natl Acad Sci U S A. 1987 May;84(10):3199–3203. doi: 10.1073/pnas.84.10.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefano J. E., Ackerson J. W., Gralla J. D. Alterations in two conserved regions of promoter sequence lead to altered rates of polymerase binding and levels of gene expression. Nucleic Acids Res. 1980 Jun 25;8(12):2709–2723. doi: 10.1093/nar/8.12.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentin-Hansen P., Albrechtsen B., Løve Larsen J. E. DNA-protein recognition: demonstration of three genetically separated operator elements that are required for repression of the Escherichia coli deoCABD promoters by the DeoR repressor. EMBO J. 1986 Aug;5(8):2015–2021. doi: 10.1002/j.1460-2075.1986.tb04458.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitson P. A., Hsieh W. T., Wells R. D., Matthews K. S. Supercoiling facilitates lac operator-repressor-pseudooperator interactions. J Biol Chem. 1987 Apr 15;262(11):4943–4946. [PubMed] [Google Scholar]
- Winter R. B., von Hippel P. H. Diffusion-driven mechanisms of protein translocation on nucleic acids. 2. The Escherichia coli repressor--operator interaction: equilibrium measurements. Biochemistry. 1981 Nov 24;20(24):6948–6960. doi: 10.1021/bi00527a029. [DOI] [PubMed] [Google Scholar]
- Wood W. I., Gitschier J., Lasky L. A., Lawn R. M. Base composition-independent hybridization in tetramethylammonium chloride: a method for oligonucleotide screening of highly complex gene libraries. Proc Natl Acad Sci U S A. 1985 Mar;82(6):1585–1588. doi: 10.1073/pnas.82.6.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoller M. J., Smith M. Oligonucleotide-directed mutagenesis of DNA fragments cloned into M13 vectors. Methods Enzymol. 1983;100:468–500. doi: 10.1016/0076-6879(83)00074-9. [DOI] [PubMed] [Google Scholar]




