Abstract
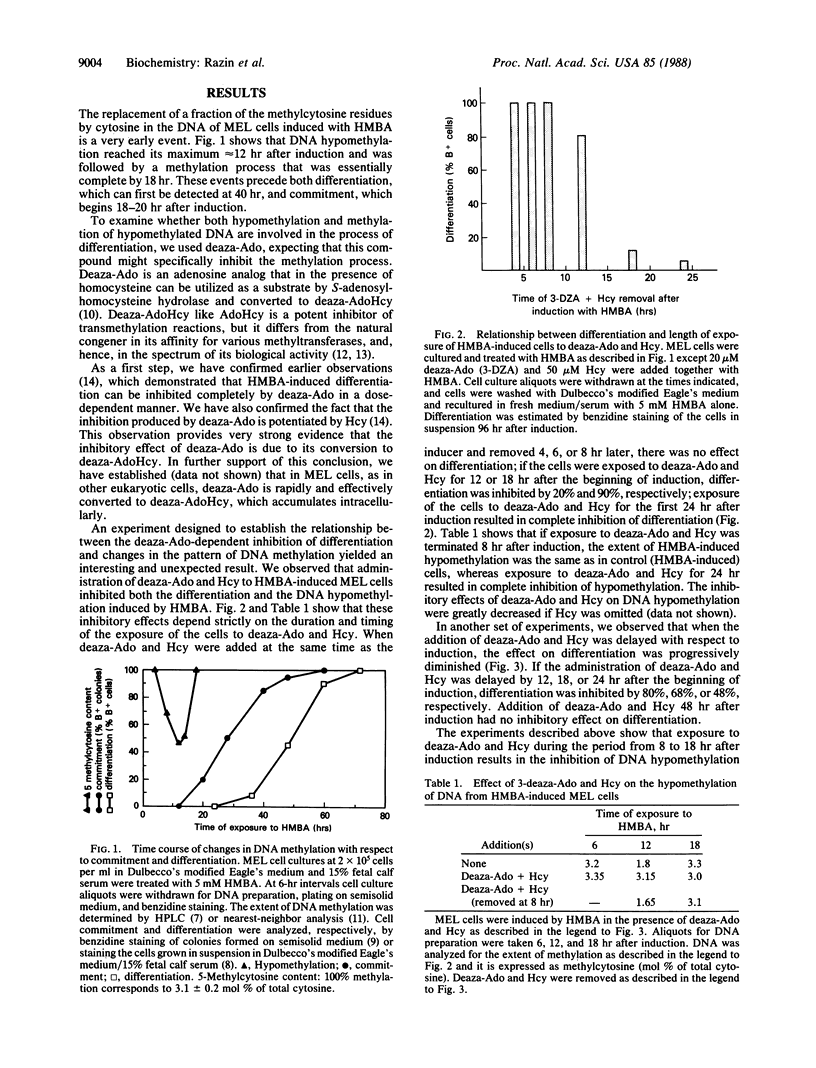
The state of DNA methylation in mouse erythroleukemia (MEL) cells has been analyzed in relation to commitment to differentiation in response to treatment with hexamethylenebisacetamide (HMBA). Previous experiments have shown that induction by HMBA involves transient genome-wide hypomethylation of DNA that is achieved by replacement of 5-methylcytosine with cytosine residues. The experiments described in the present communication revealed that hypomethylation is a very early event in the process of differentiation. Exposure of the cells to 3-deazaadenosine, an adenosine analog, in combination with homocysteine, resulted in the intracellular accumulation of 3-deazaadenosylhomocysteine, which caused an inhibition of HMBA-induced hypomethylation that was correlated with a comparable inhibition of differentiation. While these experiments suggest that hypomethylation is a necessary step in the process of differentiation, other experiments reported here indicate that hypomethylation of DNA may be necessary but not sufficient to trigger the whole program of differentiation in MEL cells. We found, for example that exposure of the cells to cycloheximide during the first 24 hr of induction by HMBA resulted in complete inhibition of differentiation without significant effect on the HMBA-induced hypomethylation. This result also indicates that the enzymatic machinery required for the hypomethylation of DNA is present in uninduced cells.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aksamit R. R., Falk W., Cantoni G. L. Inhibition of chemotaxis by S-3-deazaadenosylhomocysteine in a mouse macrophage cell line. J Biol Chem. 1982 Jan 25;257(2):621–625. [PubMed] [Google Scholar]
- Chiang P. K., Richards H. H., Cantoni G. L. S-Adenosyl-L-homocysteine hydrolase: analogues of S-adenosyl-L-homocysteine as potential inhibitors. Mol Pharmacol. 1977 Sep;13(5):939–947. [PubMed] [Google Scholar]
- Fibach E., Reuben R. C., Rifkind R. A., Marks P. A. Effect of hexamethylene bisacetamide on the commitment to differentiation of murine erythroleukemia cells. Cancer Res. 1977 Feb;37(2):440–444. [PubMed] [Google Scholar]
- Gruenbaum Y., Stein R., Cedar H., Razin A. Methylation of CpG sequences in eukaryotic DNA. FEBS Lett. 1981 Feb 9;124(1):67–71. doi: 10.1016/0014-5793(81)80055-5. [DOI] [PubMed] [Google Scholar]
- Marks P. A., Sheffery M., Rifkind R. A. Induction of transformed cells to terminal differentiation and the modulation of gene expression. Cancer Res. 1987 Feb 1;47(3):659–666. [PubMed] [Google Scholar]
- Nomura S., Yamagoe S., Kamiya T., Oishi M. An intracellular factor that induces erythroid differentiation in mouse erythroleukemia (Friend) cells. Cell. 1986 Feb 28;44(4):663–669. doi: 10.1016/0092-8674(86)90275-8. [DOI] [PubMed] [Google Scholar]
- Razin A., Feldmesser E., Kafri T., Szyf M. Cell specific DNA methylation patterns; formation and a nucleosome locking model for their function. Prog Clin Biol Res. 1985;198:239–253. [PubMed] [Google Scholar]
- Razin A., Szyf M. DNA methylation patterns. Formation and function. Biochim Biophys Acta. 1984 Sep 10;782(4):331–342. doi: 10.1016/0167-4781(84)90043-5. [DOI] [PubMed] [Google Scholar]
- Razin A., Szyf M., Kafri T., Roll M., Giloh H., Scarpa S., Carotti D., Cantoni G. L. Replacement of 5-methylcytosine by cytosine: a possible mechanism for transient DNA demethylation during differentiation. Proc Natl Acad Sci U S A. 1986 May;83(9):2827–2831. doi: 10.1073/pnas.83.9.2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuben R. C., Wife R. L., Breslow R., Rifkind R. A., Marks P. A. A new group of potent inducers of differentiation in murine erythroleukemia cells. Proc Natl Acad Sci U S A. 1976 Mar;73(3):862–866. doi: 10.1073/pnas.73.3.862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salditt-Georgieff M., Sheffery M., Krauter K., Darnell J. E., Jr, Rifkind R., Marks P. A. Induced transcription of the mouse beta-globin transcription unit in erythroleukemia cells. Time-course of induction and of changes in chromatin structure. J Mol Biol. 1984 Feb 5;172(4):437–450. doi: 10.1016/s0022-2836(84)80016-9. [DOI] [PubMed] [Google Scholar]
- Scarpa S., Strom R., Bozzi A., Aksamit R. R., Backlund P. S., Jr, Chen J., Cantoni G. L. Differentiation of myoblast cell lines and biological methylation: 3-deazaadenosine stimulates formation of multinucleated myofibers. Proc Natl Acad Sci U S A. 1984 May;81(10):3064–3068. doi: 10.1073/pnas.81.10.3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheffery M., Marks P. A., Rifkind R. A. Gene expression in murine erythroleukemia cells. Transcriptional control and chromatin structure of the alpha 1-globin gene. J Mol Biol. 1984 Feb 5;172(4):417–436. doi: 10.1016/s0022-2836(84)80015-7. [DOI] [PubMed] [Google Scholar]
- Sherman M. L., Shafman T. D., Spriggs D. R., Kufe D. W. Inhibition of murine erythroleukemia cell differentiation by 3-deazaadenosine. Cancer Res. 1985 Nov;45(11 Pt 2):5830–5834. [PubMed] [Google Scholar]
- Singer J., Stellwagen R. H., Roberts-Ems J., Riggs A. D. 5-Methylcytosine content of rat hepatoma DNA substituted with bromodeoxyuridine. J Biol Chem. 1977 Aug 10;252(15):5509–5513. [PubMed] [Google Scholar]
- Watanabe T., Oishi M. Dimethyl sulfoxide-inducible cytoplasmic factor involved in erythroid differentiation in mouse erythroleukemia (Friend) cells. Proc Natl Acad Sci U S A. 1987 Sep;84(18):6481–6485. doi: 10.1073/pnas.84.18.6481. [DOI] [PMC free article] [PubMed] [Google Scholar]



