Summary
Objective
Schizophrenia is a chronic psychiatric disorder that affects about 1% of the population globally. A tremendous amount of effort has been expended in the past decade, including more than 2400 association studies, to identify genes influencing susceptibility to the disorder. However, few genes or markers have been reliably replicated. The wealth of this information calls for an integration of gene association data, evidence based gene ranking, and follow-up replication in large sample. The objective of this study is to develop and evaluate evidence based gene ranking methods and to examine the features of top-ranking candidate genes for schizophrenia.
Methods
We proposed a gene-based approach for selecting and prioritizing candidate genes by combining odds ratios (ORs) of multiple markers in each association study and then combining ORs in multiple studies of a gene. We named it combination-combination OR method (CCOR). CCOR is similar to our recently published method, which first selects the largest OR of the markers in each study and then combines these ORs in multiple studies (i.e., selection-combination OR method, SCOR), but differs in selecting representative OR in each study. Features of top-ranking genes were examined by gene ontology terms and gene expression in tissues.
Results
Our evaluation suggested that the SCOR method overall outperforms the CCOR method. Using the SCOR, a list of 75 top-ranking genes was selected for schizophrenia candidate genes (SZGenes). We found that SZGenes had strong correlation with neuro-related functional terms and were highly expressed in brain-related tissues.
Conclusion
The scientific landscape for schizophrenia genetics and other complex disease studies is expected to change dramatically in the next a few years, thus, the gene-based combined OR method is useful in candidate gene selection for follow-up association studies and in further artificial intelligence in medicine. This method for prioritization of candidate genes can be applied to other complex diseases such as depression, anxiety, nicotine dependence, alcohol dependence, and cardiovascular diseases.
Keywords: Schizophrenia, Candidate genes, Odds ratio, Association studies
1. Introduction
Schizophrenia is a common, chronic and debilitating psychiatric disorder that affects about 1% of the global population [1]. The hypothesis that the etiology of schizophrenia is in part genetic has been strongly supported by family, twin, and adoption studies [2, 3]. Schizophrenia is likely genetically heterogeneous: different susceptibility loci may influence liability in different individuals, and liability is likely linked to more than one susceptibility locus [4, 5]. Therefore, identifying all genes susceptible to schizophrenia and their interactions is a great challenge but also an important task for elucidating the genetic basis of schizophrenia. Decades of traditional linkage, association, and gene expression studies have identified many chromosome regions and hundreds of genes that might be linked to schizophrenia. Unfortunately, the results do not support a Mendelian pattern of transmission and the replications of schizophrenia susceptibility genes or markers have been poor [6]. Many factors have been thought to (partially) contribute to the poor replication rate such as variability of the clinical phenotypes, different ethnic background, sample size, genotyping errors, and sample errors.
We have witnessed a rapid increase of both the number of association studies for schizophrenia and the number of studied genes in the past 15 years [7]. As of August 14, 2008, the SchizophreniaGene database has collected and annotated more than 2400 association studies published in the peer-reviewed journals and these studies included more than 700 genes. Besides, many more negative association studies were not reported because of the favor of positive findings in the research community. This large amount of data, along with the poor replication, calls for data integration and meta-analysis; hence the causal relationship between a marker and schizophrenia can be effectively measured. One major goal in meta-analysis is to synthesize the information from multiple studies to provide a more reliable and less biased aggregate estimate of the marker’s risk and its heritability [8]. Several meta-analyses have been reported in schizophrenia association studies [9–11] and more meta-analyses are expected [12].
In a typical association study, one or multiple markers (e.g., single nucleotide polymorphisms (SNPs)), or haplotypes, would be tested in the cases versus controls or in the family based sample. For a gene, different markers or haplotypes have often been employed in the association tests in different studies. The routine meta-analysis is to combine evidence of one marker or one haplotype from multiple association studies. The information of the related variants (markers or haplotypes) at the same gene locus is thus ignored. Given the great variation of study design such as marker selection, sample size, population background and phenotypes, one gene has often had positive association results of multiple but also different markers among all the published studies. This significantly limits the utility of routine meta-analysis. To better utilize the evidence of multiple markers in the association studies, Neal and Sham [13] proposed a gene-based meta-analysis approach in which all studied markers within a candidate gene are considered jointly. Compared to the single-SNP based or single-haplotype based approach, the gene-based approach can easily resolve the inconsistencies arising from different designs in multiple studies (e.g., population difference). This is particularly important because the number of markers within a gene locus in the association studies is expected to increase dramatically in the near future due to the high throughput technologies, but inconsistency of the significant markers at the same gene locus will be even stronger. Until recently we applied the gene-based meta-analysis in schizophrenia, no gene-based meta-analysis has been reported yet [14]. One main reason is that it is not only time-consuming but also difficult to collect and annotate all the markers and their association information in all published studies in each gene. For example, in schizophrenia, some association studies were published before 1980; their detailed association information may not be easily extracted from the publications. The recently established SchizophreniaGenes database, which collects and annotates all the published genetic association studies for schizophrenia phenotypes [9], has largely eased this problem. In our recent study [7], we proposed a combined marker analysis at the gene level, e.g., all the markers in one gene were considered jointly in evaluating the susceptibility of that gene to schizophrenia. However, there are two alternative ways to combine markers when considering all markers in one gene jointly. Given that multiple markers have been studied in an association study, one may select the marker having the largest odds ratio (OR) value to represent the effect size of that gene in that case-control study; alternatively, one may combine the ORs of all markers to represent the effect size. Thus, one of the objectives in this study is to explore which one has a better performance in combining markers for evaluating their evidence in schizophrenia.
In this paper, we first describe two marker combination methods, both of which are based on combining ORs of the markers in multiple association studies of a gene but differ in selecting representative OR in each study. In these two methods, selection of the marker in each study or each gene is based on the OR, the ratio of allele carriers to non-carriers in cases compared with that in controls for disease. OR is an effective measurement of association evidence. Then we compiled test data sets and evaluated the performance of these two gene-based and evidence-based methods. Based on the evaluation, we chose the better method to rank and prioritize genes for schizophrenia. Finally, we examined functional features of these candidate genes and compared to those of the essential genes or non-disease genes.
2. Methods
2.1. Two gene-based combined OR methods
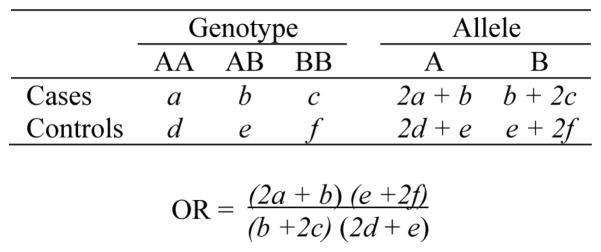
We measure the evidence of positive association by OR. Here, we first describe how to calculate OR in a set of data. Schizophrenia association studies are often based on case-control sample [15], so case-control studies would be used in this work. Cases are those subjects who have been diagnosed with the disease under the study while controls are those who are either known to be unaffected or randomly selected from the population. Frequencies of the alleles, most often the two alleles of a SNP, in cases and controls are counted based on genotyping and then compared. If an allele or a genotype in cases occurs in a significantly higher frequency than controls, its presence in the sample may increase risk for disease. Figure 1 illustrates the calculation of an OR for a SNP marker. After the genotypes have been collected from experiments, frequencies of the two alleles (A and B) can be deducted, and OR can be calculated subsequently using the contingency table. OR is a critical parameter to measure the association between risk markers and disease in case-control studies [2]. When OR is equal to 1, there is no difference between two alleles distributed in the cases and controls. An OR > 1 may suggest that allele A has higher risk for the disease, conversely, OR <1 may suggest that allele B has higher risk [16].
Figure 1.
Genotypes and allele frequencies in cases and controls and calculation of odds ratio.
The marker-based meta-analysis synthesizes the association data of only one marker each time across multiple association studies, thus, it potentially misses the related information of the other markers at the same locus in each analysis. As described in the Introduction, combining of association data of all the studied markers in a gene, each of which has different results in the association studies, may utilize all the available study information more effectively. This gene-based approach has been recently proposed by Neale and Sham [13], but essentially no follow-up application yet. Here, we applied their approach to propose an evidence-based method at the gene level through combining markers’ ORs. All the markers in a gene are considered jointly to evaluate the gene’s susceptibility to schizophrenia. Two alternative ways have been proposed to select the representative OR value of each study. The details are given below.
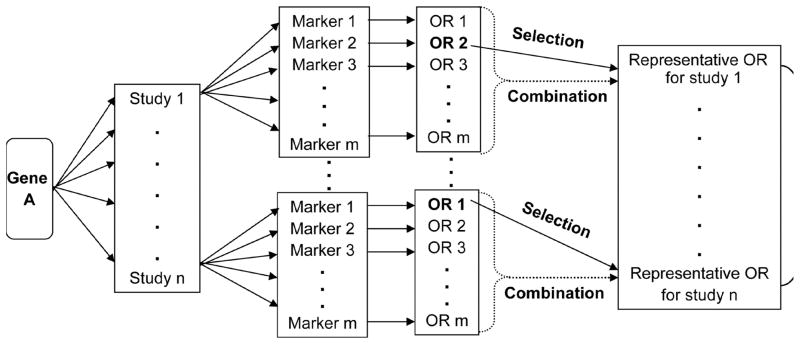
For a gene that has multiple association studies, each of which has one or multiple markers, our goal is to select the markers that have the highest OR value in each study and then to combine them in an effective manner. Figure 2 illustrates a two-step design. In the first step, we calculate OR value for each marker (see Figure 1) and then to select the representative OR for each study. In a simple case, there is only one marker in a study. Its odds ratio is selected to represent that study. Most association studies tested multiple markers. For those studies, two possible ways may be used to select the representative OR value. One is to select the largest OR value among the markers to represent the study, which was described in our recent work and has been applied to gene ranking [7]. Briefly, we first evaluated the risk alleles of all association studies, then calculated the ORs of all markers using their risk alleles (see Figure 1), and finally selected the marker that had the largest OR value in each association study to represent the effect size of that gene. The other is to combine OR values of all markers in the study to calculate a combined OR value to represent the study. A combined OR value is calculated by the “meta” statistical method implemented in the R package “epitools” [17].
Figure 2.
Flowchart of the gene-based combined odds ratio (OR) methods for weighting evidence for schizophrenia candidate gene selection. OR is the ratio of allele carriers to non-carriers in cases compared with that in controls. In the figure, there are two alternative approaches to selecting representative OR of a study: selection (the largest OR value of all the markers being selected, step one in the SCOR method) and combination (the OR values of all markers being combined by the “meta” analysis, step one in the CCOR method).
In the second step, we combine the representative OR values of all studies to generate a final combined OR value. The combined OR value and its statistical significance (p value, 95% confidence interval) is calculated by the R package “meta”, and are used as an aggregate evidence to evaluate the susceptibility of the gene to schizophrenia. Specifically, a Z-test in the R package ‘meta’ is applied to evaluate the significance of the combined OR.
To compare the performance of the two ways of selecting representative OR values in the first step, we named them selection-combination OR method (SCOR) and combination-combination OR method (CCOR), respectively. In both methods, the combined OR across all markers/all studies is computed as a weighted OR with the weights equal to the inverse variance of the OR estimator. Thus, greater weight can be given to larger studies and studies with less random variation than smaller studies.
2.2. Data source
We downloaded the gene information and marker information from the SchizophreniaGene database (http://www.schizophreniaforum.org/res/sczgene/default.asp, accessed: 3 August 2007) [9]. For each gene, we retrieved gene ID, gene symbol, marker ID, alleles of SNP markers, minor allele frequency (MAF) and number of cases and controls.
Markers were filtered out by the following criteria: (1) markers whose sample size was smaller than 100; (2) markers that had significant deviation from the Hardy-Weinberg Equilibrium (HWE, P < 0.001) in controls; and (3) markers whose MAF < 0.01, or counts of minor alleles less than 5. After filtering the markers, we retrieved the numbers of minor and major alleles of each selected marker in cases and controls, placed in a 2 × 2 table, and then systematically calculated the OR and its 95% confidence interval for each marker using the R package “epitools”.
To evaluate the candidate genes selected by the SCOR and CCOR methods, we compiled two other gene lists based on recent meta-analysis or expert review. The first is based on the 22 genes suggested by meta-analyses, which was extracted from the SchizophreniaGene database [9]. The second is based on a comprehensive review of schizophrenia susceptibility genes by Ross et al. [18], who suggested 19 genes being the most important candidate genes based on the evidence in four domains (association with schizophrenia, linkage to gene locus, biological plausibility, and altered expression in schizophrenia). These two lists resulted in a total of 33 non-redundant genes, which were considered schizophrenia susceptibility genes by alternative approaches. We named this gene set ‘SZGenes_33’.
Gene feature analysis
2.3.1. Compilation of essential genes and non-disease genes
To explore the characteristic functional features of schizophrenia candidate genes, we compared them with other genes such as essential genes or non-disease genes. The essential genes may serve as more functional important genes than the schizophrenia genes while the non-disease genes for control purpose. There is no direct experimental approach to test essential genes in humans because they are often lethal. We used ubiquitously-expressed human genes (abbreviated as UEHGs) as an approximation for essential genes, as did in Tu et al. [19]. There were 1789 UEHGs based on experimental gene expression in the original data [19]. We checked those UEHGs using the updated gene information extracted from the file “gene2unigene”. This file was downloaded from the NCBI Gene (ftp://ftp.ncbi.nlm.nih.gov/gene/DATA/, accessed: 17 May 2008) and contains all links between gene IDs and unigenes. We had 1425 genes whose gene IDs could be mapped to unigenes; they were used as essential genes (abbreviated as ESGenes hereafter) in this study.
For non-disease genes, we first downloaded human gene information file from the NCBI Gene (ftp://ftp.ncbi.nlm.nih.gov/gene/DATA/GENE_INFO/Mammalia/, accessed: 20 May 2008) and then extracted the protein-coding genes. There were ~ 26,000 human protein-coding genes in the NCBI Gene database. After excluding all disease genes annotated in the NCBI OMIM database (ftp://ftp.ncbi.nlm.nih.gov/repository/OMIM/, accessed: 20 May 2008) and the essential genes, we had a total of 22,220 genes, which were used as non-disease genes (abbreviated as NDGenes hereafter).
2.3.2. Analysis of functional bias and tissue expression of genes
We retrieved the Gene Ontology (GO) terms of the schizophrenia candidate genes using Gene Set Analysis Toolkit in WebGestalt [20]. A Fisher’s exact test was used to identify enriched GO terms of schizophrenia candidate genes by comparing with essential genes or non-disease genes. We examined tissue expression pattern of the selected genes using human gene expression data from the second version of Gene Expression Atlas [21]. The following three factors were considered whether a gene was expressed in a tissue. (1) A gene was expressed in a tissue when its average difference (AD) value was ≥ 200. (2) Expression probes marked “_x_at” and “_s_at” had low confidence. When there were any high confidence probes available for a gene, the low confidence probes were discarded [22]. (3) A gene expressed in more than 80% of the total 79 tissues was defined to be broadly expressed, in less than 20% of the 79 tissues to be narrowly expressed (tissue-specific), and between 20–80% to be moderately expressed. We found 67 of the 75 SZGenes, 1006 of the 1425 ESGenes, and 8772 of the 22,220 NDGenes had gene expression data in the Gene Expression Atlas.
To further investigate the detailed gene expression pattern in tissues, we used another gene expression dataset from the CGAP-expressed sequence tag (EST) project (http://cgap.nci.nih.gov/Tissues/) [23]. These gene expression data were available for analysis at WebGestalt (http://bioinfo.vanderbilt.edu/webgestalt/, accessed: 28 July 2008) [20]. All the 75 SZGenes, 1277 of the 1425 ESGenes, and 14,304 of the 22,220 NDGenes had gene expression data in WebGestalt dataset. For each gene list, the number of genes expressed in each tissue was summarized. Based on the counts of genes in each tissue and the size of each gene list, we calculated the proportion for each tissue and use Wilcoxon signed-rank test to detect significant difference between SZGenes and ESGenes or between SZGenes and NDGenes.
3. Results and discussion
3.1. Candidate genes selected by the SCOR and CCOR methods
As illustrated in Figure 2, both the SCOR and CCOR methods are based on combining markers via their OR values in multiple studies, but they differ in selecting representative OR in each study. Using all the schizophrenia case-control studies downloaded from the SchizophreniaGene database, the SCOR method generated a list of 75 schizophrenia candidate genes when the cutoff p value in the Z-test from the R package “meta” was set 0.05 [7]. To save the space, we named this set of genes ‘SZGenes_75’. By using the same cutoff p value 0.05 in the Z-test, the CCOR method generated a list of 99 genes. These genes were named ‘SZGenes_99’.
Table 1 lists SZGenes_75 and SZGenes_99. There were 66 genes appeared in both gene lists, accounting for 88.0% of SZGenes_75 and 66.7% of SZGenes_99, respectively. Among those 66 genes, 44 were ranked in the top 50 of SZGenes_75, while 45 were in the top 50 of SZGenes_99. These results indicate that the genes with strong aggregate evidence tended to be ranked top by both the methods.
Table 1.
Genes ranked by the SCOR and CCOR methods
| Gene | Ranking |
Gene | Ranking |
Gene | Ranking |
|||
|---|---|---|---|---|---|---|---|---|
| SCOR | CCOR | SCOR | CCOR | SCOR | CCOR | |||
| DTNBP1 | 1 | 9 | IL4 | 37 | 66 | GABRG2 | 73 | 22 |
| CHGB | 2 | 39 | NUMBL | 38 | 67 | APOE | 74 | 17 |
| DRD3 | 3 | 4 | MICB | 39 | 21 | ERBB3 | 75 | 99 |
| DAOA | 4 | 5 | PDLIM5 | 40 | 36 | ADCYAP1 | NA | 93 |
| CSF2RA | 5 | 11 | GRM3 | 41 | 55 | ADRA2A | NA | 88 |
| COMT | 6 | 7 | MAGI3 | 42 | 31 | C3 | NA | 78 |
| PIP5K2A | 7 | 53 | GABRA6 | 43 | 15 | CNP | NA | 54 |
| DAO | 8 | 10 | GRIK3 | 44 | 32 | CPLX2 | NA | 56 |
| HTR2A | 9 | 6 | GSK3B | 45 | 48 | CYP2D6 | NA | 80 |
| RGS4 | 10 | 23 | TP53 | 46 | 30 | DBH | NA | 91 |
| MTHFR | 11 | 12 | SOX10 | 47 | NA | DDC | NA | 94 |
| PTPRZ1 | 12 | 3 | FOXP2 | 48 | 28 | DRD1 | NA | 77 |
| DRD4 | 13 | 14 | BDNF | 49 | 38 | DRD1IP | NA | 97 |
| SYN2 | 14 | NA | IL1B | 50 | 47 | FEZ1 | NA | 84 |
| DGCR2 | 15 | NA | ERBB4 | 51 | 59 | FXYD6 | NA | 8 |
| NRG1 | 16 | 2 | NOS1 | 52 | NA | GRIK4 | NA | 45 |
| CHGA | 17 | 42 | PPP3CC | 53 | NA | GRM8 | NA | 27 |
| DRD2 | 18 | 1 | CHL1 | 54 | 37 | HMBS | NA | 70 |
| CLINT1 | 19 | NA | SLC18A1 | 55 | 90 | HP | NA | 52 |
| GSTM1 | 20 | 40 | GABBR1 | 56 | 64 | HTR1B | NA | 87 |
| HTR1A | 21 | 33 | CCKAR | 57 | 34 | HTR4 | NA | 60 |
| MAGI2 | 22 | NA | NOTCH4 | 58 | 29 | HTR6 | NA | 72 |
| DISC1 | 23 | 18 | GABRB2 | 59 | 13 | IL1RN | NA | 79 |
| IL10 | 24 | 49 | HRH1 | 60 | 86 | KMO | NA | 85 |
| CHAT | 25 | NA | CYP1A2 | 61 | 92 | MLC1 | NA | 35 |
| ST8SIA2 | 26 | 83 | TNF | 62 | 25 | NOS1AP | NA | 75 |
| SLC6A4 | 27 | 20 | SYNGR1 | 63 | 24 | OLIG2 | NA | 96 |
| PLXNA2 | 28 | 51 | HTR3A | 64 | NA | OPRS1 | NA | 71 |
| SRR | 29 | 43 | FXYD2 | 65 | 68 | PENK | NA | 61 |
| PRODH | 30 | 82 | CHRFAM7A | 66 | 89 | PLA2G4A | NA | 76 |
| GRIN2B | 31 | 26 | NR4A2 | 67 | 63 | PPP1R1B | NA | 57 |
| MAGI1 | 32 | 69 | GABRA1 | 68 | 74 | RTN4 | NA | 81 |
| GRIN1 | 33 | 44 | TAAR6 | 69 | 73 | SLC6A3 | NA | 65 |
| TPH1 | 34 | 19 | SNAP29 | 70 | 98 | SYN3 | NA | 95 |
| SLC1A2 | 35 | 46 | GAD1 | 71 | 58 | TF | NA | 62 |
| AKT1 | 36 | 41 | GABRP | 72 | 16 | XBP1 | NA | 50 |
SCOR denotes selection-combination OR method and CCOR denotes combination-combination OR method (see Methods). Genes with underline overlap with both the top gene list from the SchizophreniaGene database and the gene list from Ross et al. (2006).
We found 9 genes that were unique in SZGenes_75, with an average rank 34.7. Six of them had rank < 50: SYN2 (rank: 14th), DGCR2 (15th), CLINT1 (19th), MAGI2 (22nd), CHAT (25th) and SOX10 (47th). There were 33 genes appeared in the SZGenes_99 list only, with an average rank 70.0. Most of these uniquely appeared genes had low ranks; only 3 genes had ranks < 50 (FXYD6 (8th), GRM8 (27th) and MLC1 (35th)).
The above comparison of the ranking, which was based on the p value in the meta-analysis, suggests that the genes with strong positive association evidence can be reliably selected by both methods while the CCOR method could select additional genes that had only moderate or weak positive association evidence.
3.2. Evaluation of SCOR and CCOR with SZGenes_33
To evaluate the performance of the SCOR and CCOR methods, we compared SZGenes_75 and SZGenes_99 with SZGenes_33. As described in the Methods, SZGenes_33 contains 33 genes that have shown other positive evidence (meta-analysis and expert review) in the susceptibility to schizophrenia. Among the 33 genes in SZGenes_33, we found 24 in SZGenes_75 and 27 in SZGenes_99 (Table 1). A further check indicated that 23 genes appeared in both the SZGenes_75 and SZGenes_99 lists and their average ranks were nearly the same: 29.4 in SZGenes_75 and 28.2 in SZGenes_99. Statistical test of these genes had insignificant rank order in the two gene lists (Mann-Whitney’s p > 0.05). The genes in the SZGenes_33 that overlapped with only SZGenes_75 or only SZGenes_99 but not the both tended to rank low: PPP3CC (53rd) in SZGenes_75 and DRD1 (77th), FEZ1 (84th), GRIK4 (45th) and HP (52nd) in SZGenes_99. The comparison in subsection 3.1 and evaluation in this subsection indicate that the additional genes generated by the CCOR method had low ranks in the gene list.
3.3. Correlation between gene ranking and number of markers/studies
In this study, we evaluated the amount of positive evidence of a gene for association with schizophrenia using all the published case-control studies and their markers. Here, we performed the Pearson’s product-moment correlation tests to examine the effect of number of markers or number of studies on gene ranking. We found a strong correlation between the gene ranking and number of studies (SZGenes_75: r = −0.510, p = 1.82×10−7; SZGenes_99: r = −0.471, p = 8.56×10−7). The correlation between the gene ranking and number of markers was much weaker in SZGenes_99 (r = −0.190, p = 0.02) and even insignificant in SZGenes_75 (r = 0.001, p > 0.05). This suggests that the number of markers on gene ranking in the SCOR method is much weaker than that of the CCOR method.
In summary, we compared candidate genes generated by the SCOR and CCOR methods, evaluated both gene lists using susceptibility genes suggested by other approaches, and examined the effects of number of markers and studies on gene ranking. The comparisons consistently support that the SCOR method is more efficient to generate candidate genes with strong published positive evidence. Thus, the genes in SZGenes_75 generated by SCOR were used for follow-up gene feature analysis. These genes were named SZGenes hereafter to compare the ESGenes (essential genes) and NDGenes (non-disease genes).
3.4. Strong functional bias in schizophrenia candidate genes
We first performed GO-term enrichment test in SZGenes using NDGenes as the reference. Because of the large number of genes and a long list of GO terms, we restricted the GO terms that (1) had been annotated to have at least five genes in a list, (2) located at the fourth hierarchical level or higher in the GO tree and, (3) the p value in the Fisher’s exact tests was < 0.0001. According to these criteria, we found 36 GO terms that were significantly enriched in the SZGenes compared to the NDGenes. The details are shown in Table 2.
Table 2.
GO terms significantly enriched in the schizophrenia candidate genes compared to non-disease genes
| GO code | GO term description | Number of genes | P value1 |
|---|---|---|---|
| Biological process: 11 | |||
| GO:0007268 | Synaptic transmission | 18 | 2.44 × 10−17 |
| GO:0019226 | Transmission of nerve impulse | 18 | 8.40 × 10−17 |
| GO:0007267 | Cell-cell signaling | 21 | 1.99 × 10−13 |
| GO:0007214 | Gamma-aminobutyric acid signaling pathway | 5 | 6.89 × 10−9 |
| GO:0001505 | Regulation of neurotransmitter levels | 6 | 1.03 × 10−8 |
| GO:0050877 | Neurophysiological process | 20 | 1.81 × 10−8 |
| GO:0007166 | Cell surface receptor linked signal transduction | 24 | 4.29 × 10−6 |
| GO:0006807 | Nitrogen compound metabolic process | 9 | 1.08 × 10−5 |
| GO:0007186 | G-protein coupled receptor protein signaling pathway | 16 | 5.09 × 10−4 |
| GO:0006519 | Amino acid and derivative metabolic process | 6 | 5.74 × 10−4 |
| GO:0006811 | Ion transport | 11 | 6.65 × 10−4 |
| Molecular function: 16 | |||
| GO:0005230 | Extracellular ligand-gated ion channel activity | 10 | 1.49 × 10−12 |
| GO:0030594 | Neurotransmitter receptor activity | 11 | 5.49 × 10−12 |
| GO:0015276 | Ligand-gated ion channel activity | 10 | 5.83 × 10−11 |
| GO:0008227 | Amine receptor activity | 7 | 1.40 × 10−9 |
| GO:0016917 | GABA receptor activity | 6 | 6.65 × 10−9 |
| GO:0004890 | GABA-A receptor activity | 5 | 2.03 × 10−7 |
| GO:0004888 | Transmemebrane receptor activity | 23 | 2.13 × 10−7 |
| GO:0005216 | Ion channel activity | 11 | 1.94 × 10−6 |
| GO:0015267 | Alpha-type channel activity | 11 | 3.39 × 10−6 |
| GO:0043168 | Anion binding | 5 | 5.13 × 10−6 |
| GO:0005254 | Chloride channel activity | 5 | 5.13 × 10−6 |
| GO:0031404 | Chloride ion binding | 5 | 5.13 × 10−6 |
| GO:0008066 | Glutamate receptor activity | 5 | 6.93 × 10−6 |
| GO:0005253 | Anion channel activity | 5 | 8.00 × 10−6 |
| GO:0015075 | Ion transmembrane transporter activity | 13 | 1.44 × 10−5 |
| GO:0008509 | Anion transporter activity | 6 | 2.11 × 10−5 |
| Cellular component: 9 | |||
| GO:0045211 | Postsynaptic membrane | 11 | 2.59 × 10−13 |
| GO:0005887 | Integral to plasma membrane | 25 | 5.19 × 10−10 |
| GO:0031226 | Intrinsic to plasma membrane | 25 | 6.51 × 10−10 |
| GO:0044459 | Plasma membrane part | 27 | 8.59 × 10−10 |
| GO:0005886 | Plasma membrane | 29 | 2.13 × 10−9 |
| GO:0044425 | Membrane part | 40 | 4.97 × 10−5 |
| GO:0016020 | Membrane | 44 | 1.00 × 10−4 |
| GO:0016021 | Integral to membrane | 37 | 2.23 × 10−4 |
| GO:0031224 | Intrinsic to membrane | 37 | 2.39 × 10−4 |
P value was calculated by Fisher’s exact test between schizophrenia candidate genes and non-disease genes. Because of the large number of genes and a long list of GO terms, we restricted the GO terms that (1) had been annotated to have at least five genes in a list, (2) located at the fourth hierarchical level or higher in the GO tree and, (3) the p value in the Fisher’s exact tests was < 0.0001.
Among the 36 GO terms, 11 terms belonged to biological process, 16 to molecular function and 9 to cellular component, three GO organization principles. Six (synaptic transmission, transmission of nerve impulse, regulation of neurotransmitter levels, neurophysiological process, neurotransmitter receptor activity, and postsynaptic membrane) were directly related to neurotransmitters and neuroplasticity. It is worth noting that almost all these terms except “neurophysiological process” were listed within the top 5 in each GO organization principle, which strongly supports the neurotransmitters and neuroplasticity theory in schizophrenia [14, 24]. Three GO terms were directly related to GABA neurotransmission: gamma-aminobutyric acid signaling pathway, GABA receptor activity and GABA-A receptor activity. One GO term (glutamate receptor activity) was directly related to glutamate neurotransmission. GABA neurotransmission and glutamate neurotransmission are commonly considered to be involved in the pathophysiology during the development of schizophrenia [25, 26]. Besides, there were 16 GO terms that are biologically related to cell communication: cell-cell signaling, cell surface receptor linked signal transduction, G-protein coupled receptor protein signaling pathway, ion transport, extracellular ligand-gated ion channel activity, ligand-gated ion channel activity, amine receptor activity, transmemebrane receptor activity, ion channel activity, alpha-type channel activity, anion binding, chloride channel activity, chloride ion binding, anion channel activity, ion transmembrane transporter activity, and anion transporter activity. Genes whose proteins involved in cell communication in brain are important in neuronal function and are likely the candidates for nervous system disorders [27]. Of note, we found 8 GO-terms in cellular component are involved in membrane: integral to plasma membrane, intrinsic to plasma membrane, plasma membrane part, plasma membrane, membrane part, membrane, integral to membrane, and intrinsic to membrane. Finally, two metabolic pathways (nitrogen compound metabolic process, amino acid and derivative metabolic process) have been previously thought to serve as bridges or modulators between genes and environment in schizophrenia [28–30]. Overall, these enriched GO terms in SZGenes indicate that (1) schizophrenia candidate genes tend to share very similar functions and (2) development of schizophrenia likely involves several major genetic factors, their interactions, and the interactions with environment. Therefore, more genes and genetic factors need to be considered when the genetic mechanisms of schizophrenia are explored.
We next performed GO-term enrichment test in SZGenes using ESGenes as the reference. We found 42 GO terms that were significantly enriched in the SZGenes compared to ESGenes using the same criteria as to NDGenes (see above). These GO terms included almost all those terms in the comparison with NDGenes (Table 2) with one exception (amino acid and derivative metabolic process). Besides, we identified three development related GO terms: “system development”, “nervous system development” and “growth factor activity”. The results further confirm that schizophrenia is the behavioral outcome of an aberration in neurodevelopmental processes [14, 24].
3.5. Schizophrenia candidate genes highly expressed in the brain-related tissues
The tissue-specific pattern of gene expression may provide information on gene function or phenotype. We used two datasets to examine the tissue expression pattern of SZGenes and compared to ESGenes and NDGenes.
First, we examined how frequently a gene was expressed in the 79 tissues using the second version of Gene Expression Atlas [21]. We define a gene to be broadly expressed when it is expressed in more than 80% of tissues and to be tissue-specific when it is expressed in less than 20% of tissues, otherwise to be moderately expressed [22]. Among the 75 SZGenes, 67 had the expression data. Among these 67 genes, 29 (43.3%) were broadly expressed, 24 (35.8%) were moderately expressed, and 14 (20.9%) were tissue-specific (Table 3). For the broadly expressed genes, SZGenes had slightly higher proportion than NDGenes (41.9%), but both had much lower proportion than ESGenes (90.4%). Conversely, the proportion of tissue-specific genes in SZGenes (20.9%) or NDGenes (20.9%) was substantially higher than ESGenes (3.4%) (Table 3). Overall, no difference in this comparison of gene expression in tissues was observed between schizophrenia genes and non-disease genes, but essential genes, as expected, overwhelmingly expressed in almost all tissues. Furthermore, our gene network analysis revealed that, opposite to cancer or essential genes, schizophrenia candidate genes tend to be non-essential and do not serve as the network super-hubs as shown for cancer genes. Nevertheless, schizophrenia candidate genes have an intermediate level of connectivity in the human protein-protein interaction network and their connectivity is much stronger than non-disease genes (unpublished data). These observations suggest that, rather than a strong casual effect, schizophrenia candidate genes might confer their susceptibility to schizophrenia at a moderate or minor level.
Table 3.
Number and proportion (%) of genes expressed in 79 human tissues
| Expression (% of tissues) | SZGenes (%) | ESGenes (%) | NDGenes (%) |
|---|---|---|---|
| Broadly (≥80) | 29 (43.3) | 909 (90.4) | 3677 (41.9) |
| Moderately (20–80) | 24 (35.8) | 63 (6.3) | 3259 (37.2) |
| Tissue-specific (<20) | 14 (20.9) | 34 (3.4) | 1836 (20.9) |
| Total | 67 | 1006 | 8772 |
SZGenes, ESGenes, NDGenes denote schizophrenia candidate genes, essential genes, and non-disease genes, respectively.
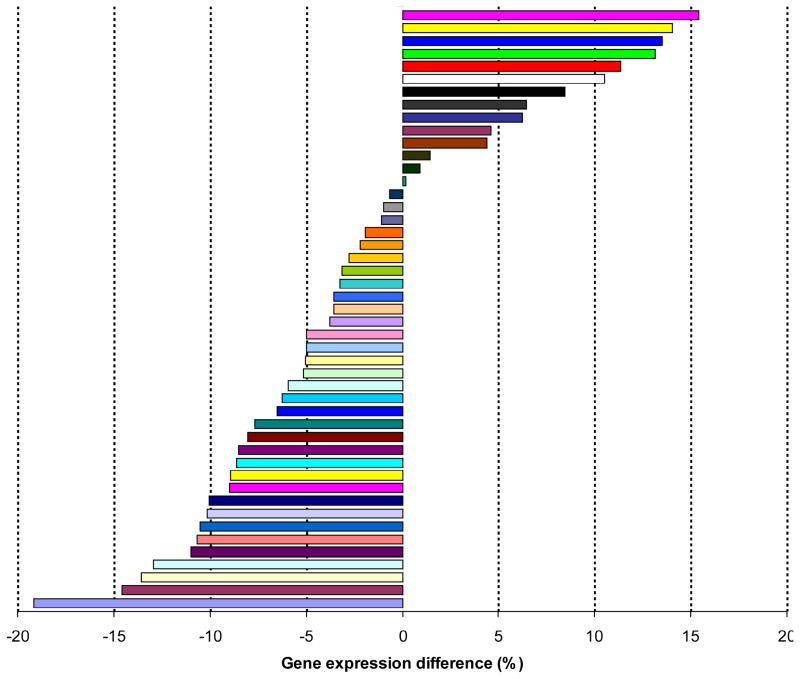
Second, we examined more detailed gene expression patterns using another gene expression dataset from the CGAP-EST project available on the WebGestalt website (see the Methods). When we examined the detailed frequency of genes in each gene set among the total 47 tissues, we found a statistically significant difference in expression pattern among the 47 tissues between SZGenes and NDGenes (Wilcoxon signed-rank test, p = 0.03), but this significance is much weaker than that between SZGenes and ESGenes (Wilcoxon signed-rank test, p = 1.4 × 10−14). We found a higher proportion of SZGenes than that of NDGenes in some brain or nerve tissues, including brain (proportion difference: +13.5%), cerebellum (+11.4%), cerebrum (+10.5%), eye (+13.2%), peripheral nervous system (+15.4%), and nerve (+14.0%) (Figure 3). Conversely, a smaller proportion of SZGenes than NDGenes was found in tissues such as lymph node (−19.0%), bone (−15.0%), cervix (−14.0%), skin (−13.0%), thymus (−11.0%), lymphoreticular (−11.0%), bone narrow (−11.0%), placenta (−10.0%) and genitourinary (−10.0%). Moreover, the proportion of ESGenes was remarkably higher in all tissues than that of SZGenes or NDGenes, which is consistent with the gene expression analysis using the Gene Expression Atlas data (Table 3). In summary, when compared to non-disease genes, SZGenes were more likely expressed in brain or nerve tissues.
Figure 3.
Proportion difference (%) of gene expression in the 47 tissues between schizophrenia candidate genes and non-disease genes. The x-axis represents gene expression difference (%) between schizophrenia candidate genes and non-disease genes. The corresponding tissues are shown in the right panel.
4. Conclusions
In this study, we proposed and compared two gene-based combined odds ratio methods (SCOR and CCOR) for weighting positive association evidence from multiple markers in multiple studies in a gene. The statistical significance in its Z-test was used to select and prioritize schizophrenia candidate genes. Extensive evaluation of these two methods consistently supports that the SCOR method is more appropriate for selecting candidate genes with strong positive evidence based on association studies. The genes selected and ranked by the SCOR method were then used for functional feature analysis. GO-term enrichment tests revealed that schizophrenia genes were strongly involved in neurodevelopment. Examination of tissue expression patterns indicated that schizophrenia genes were more frequently expressed in brain and nerve related tissues than non-disease genes. Our functional feature analyses strongly support the neurotransmitters and neuroplasticity theory in schizophrenia. Importantly, our gene-based meta-analysis method can be applied to candidate gene selection for other complex diseases such as depression, anxiety, drug abuse and cardiovascular diseases.
Acknowledgments
We thank the financial support of this project from NIH grants (AA017437 and LM009598), NARSAD Young Investigator Award and Thomas F. and Kate Miller Jeffress Memorial Trust grant to Z. Zhao. We thank Drs. Po-Hsiu Kuo and Ayman Fanous for their valuable discussion.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Johns LC, van Os J. The continuity of psychotic experiences in the general population. Clin Psychol Rev. 2001;21(8):1125–41. doi: 10.1016/s0272-7358(01)00103-9. [DOI] [PubMed] [Google Scholar]
- 2.Cardno AG, Marshall EJ, Coid B, Macdonald AM, Ribchester TR, Davies NJ, et al. Heritability estimates for psychotic disorders: the Maudsley twin psychosis series. Arch Gen Psychiatry. 1999;56(2):162–8. doi: 10.1001/archpsyc.56.2.162. [DOI] [PubMed] [Google Scholar]
- 3.Riley B, Kendler KS. Molecular genetic studies of schizophrenia. Eur J Hum Genet. 2006;14(6):669–80. doi: 10.1038/sj.ejhg.5201571. [DOI] [PubMed] [Google Scholar]
- 4.Crow TJ. Molecular pathology of schizophrenia: more than one disease process? Br Med J. 1980;280(6207):66–8. doi: 10.1136/bmj.280.6207.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McGrath J. Schizophrenia genesis: the origins of madness. Aust New Zeal J Psychiatr. 1997;31:894–5. [Google Scholar]
- 6.Botstein D, Risch N. Discovering genotypes underlying human phenotypes: past successes for mendelian disease, future approaches for complex disease. Nat Genet. 2003;33 (Suppl):228–37. doi: 10.1038/ng1090. [DOI] [PubMed] [Google Scholar]
- 7.Sun J, Kuo PH, Riley BP, Kendler KS, Zhao Z. Candidate genes for schizophrenia: A survey of association studies and gene ranking. Am J Med Genet B Neuropsychiatr Genet. 2008;147B(7):1173–1181. doi: 10.1002/ajmg.b.30743. [DOI] [PubMed] [Google Scholar]
- 8.Hettema JM, Neale MC, Kendler KS. A review and meta-analysis of the genetic epidemiology of anxiety disorders. Am J Psychiatry. 2001;158(10):1568–78. doi: 10.1176/appi.ajp.158.10.1568. [DOI] [PubMed] [Google Scholar]
- 9.Allen NC, Bagade S, McQueen MB, Ioannidis JP, Kavvoura FK, Khoury MJ, et al. Systematic meta-analyses and field synopsis of genetic association studies in schizophrenia: the SzGene database. Nat Genet. 2008;40(7):827–34. doi: 10.1038/ng.171. [DOI] [PubMed] [Google Scholar]
- 10.Li D, He L. Meta-analysis shows association between the tryptophan hydroxylase (TPH) gene and schizophrenia. Hum Genet. 2006;120(1):22–30. doi: 10.1007/s00439-006-0190-5. [DOI] [PubMed] [Google Scholar]
- 11.Munafo MR, Thiselton DL, Clark TG, Flint J. Association of the NRG1 gene and schizophrenia: a meta-analysis. Mol Psychiatry. 2006;11(6):539–46. doi: 10.1038/sj.mp.4001817. [DOI] [PubMed] [Google Scholar]
- 12.Levinson DF. Meta-analysis in psychiatric genetics. Curr Psychiatry Rep. 2005;7(2):143–51. doi: 10.1007/s11920-005-0012-9. [DOI] [PubMed] [Google Scholar]
- 13.Neale BM, Sham PC. The future of association studies: gene-based analysis and replication. Am J Hum Genet. 2004;75(3):353–62. doi: 10.1086/423901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lang UE, Puls I, Muller DJ, Strutz-Seebohm N, Gallinat J. Molecular mechanisms of schizophrenia. Cell Physiol Biochem. 2007;20(6):687–702. doi: 10.1159/000110430. [DOI] [PubMed] [Google Scholar]
- 15.Schulz KF, Grimes DA. Case-control studies: research in reverse. Lancet. 2002;359(9304):431–4. doi: 10.1016/S0140-6736(02)07605-5. [DOI] [PubMed] [Google Scholar]
- 16.Levin KA. Study design V. Case-control studies Evid Based Dent. 2003;7:83–4. doi: 10.1038/sj.ebd.6400436. [DOI] [PubMed] [Google Scholar]
- 17.Aragon T. [accessed: 20 March 2007];EpiTools: R package for epidemiologic data and graphics. 2007 Version 0.4–8: http://www.epitool.net.
- 18.Ross CA, Margolis RL, Reading SA, Pletnikov M, Coyle JT. Neurobiology of schizophrenia. Neuron. 2006;52(1):139–53. doi: 10.1016/j.neuron.2006.09.015. [DOI] [PubMed] [Google Scholar]
- 19.Tu Z, Wang L, Xu M, Zhou X, Chen T, Sun F. Further understanding human disease genes by comparing with housekeeping genes and other genes. BMC Genomics. 2006;7:31. doi: 10.1186/1471-2164-7-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang B, Kirov S, Snoddy J. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 2005;33:W741–8. doi: 10.1093/nar/gki475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci USA. 2004;101(16):6062–7. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang C, Han L, Su B, Li WH, Zhao Z. Features and trend of loss of promoter-associated CpG islands in the human and mouse genomes. Mol Biol Evol. 2007;24(9):1991–2000. doi: 10.1093/molbev/msm128. [DOI] [PubMed] [Google Scholar]
- 23.Strausberg RL. The Cancer Genome Anatomy Project: new resources for reading the molecular signatures of cancer. J Pathol. 2001;195(1):31–40. doi: 10.1002/1096-9896(200109)195:1<31::AID-PATH920>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 24.Rapoport JL, Addington AM, Frangou S, Psych MR. The neurodevelopmental model of schizophrenia: update 2005. Mol Psychiatry. 2005;10(5):434–49. doi: 10.1038/sj.mp.4001642. [DOI] [PubMed] [Google Scholar]
- 25.Toda M, Abi-Dargham A. Dopamine hypothesis of schizophrenia: making sense of it all. Curr Psychiatry Rep. 2007;9(4):329–36. doi: 10.1007/s11920-007-0041-7. [DOI] [PubMed] [Google Scholar]
- 26.Lewis DA, Gonzalez-Burgos G. Pathophysiologically based treatment interventions in schizophrenia. Nat Med. 2006;12(9):1016–22. doi: 10.1038/nm1478. [DOI] [PubMed] [Google Scholar]
- 27.Ford JM, Krystal JH, Mathalon DH. Neural synchrony in schizophrenia: from networks to new treatments. Schizophr Bull. 2007;33(4):848–52. doi: 10.1093/schbul/sbm062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Palha JA, Goodman AB. Thyroid hormones and retinoids: a possible link between genes and environment in schizophrenia. Brain Res Rev. 2006;51(1):61–71. doi: 10.1016/j.brainresrev.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 29.Mackay-Sim A, Feron F, Eyles D, Burne T, McGrath J. Schizophrenia, vitamin D, and brain development. Int Rev Neurobiol. 2004;59:351–80. doi: 10.1016/S0074-7742(04)59014-1. [DOI] [PubMed] [Google Scholar]
- 30.Guo AY, Sun J, Riley BP, Thiselton DL, Kendler KS, Zhao Z. The dystrobrevin-binding protein 1 gene: features and networks. Mol Psychiatry. 2008;14(1):18–29. doi: 10.1038/mp.2008.88. [DOI] [PMC free article] [PubMed] [Google Scholar]