Abstract
The solution structure of the complex containing the isolated kringle 2 domain of human plasminogen (K2Pg) and VEK-30, a 30-amino acid residue internal peptide from a streptococcal M-like plasminogen (Pg) binding protein (PAM), has been determined by multinuclear high-resolution NMR. Complete backbone and side-chain assignments were obtained from triple-resonance experiments, after which structure calculations were performed and ultimately refined by restrained molecular simulation in water. We find that, in contrast with the dimer of complexes observed in the asymmetric unit of the crystal, global correlation times and buoyant molecular weight determinations of the complex and its individual components showed the monomeric nature of all species in solution. The NMR-derived structure of K2Pg in complex with VEK-30 presents a folding pattern typical of other kringle domains, while bound VEK-30 forms an end-to-end α-helix (residues 6–27) in the complex. Most of the VEK-30/K2Pg interactions in solution occur between a single face of the α-helix of VEK-30 and the lysine binding site (LBS) of K2Pg. The canonical LBS of K2Pg, consisting of Asp54, Asp56, Trp60, Arg69, and Trp70 (kringle numbering), interacts with an internal pseudo-lysine of VEK-30, comprising side-chains of Arg17, His18, and Glu20. Site-specific mutagenesis analysis confirmed that the electrostatic field formed by the N-terminal anionic residues of the VEK-30 α-helix, viz., Asp7, and the non-conserved cationic residues of K2Pg, viz., Lys43 and Arg55, play additional important roles in the docking of VEK-30 to K2Pg. Structural analysis and kringle sequence alignments revealed several important features related to exosite binding that provide a structural rationale for the high specificity and affinity of VEK-30 for K2Pg.
Keywords: nmr structure, protein domains, kringle modules, M-protein function, bacterial virulence factors
1. Introduction
Over the past two decades, much evidence supporting a structure of plasminogen (Pg) that comprises a number of independent domains has emerged. Among the modules that constitute this protein are a series of highly homologous kringle domains. Five of these ~80 residue triple-disulfide linked polypeptides, and the intervening linker regions, form the noncatalytic chain of Pg [1]. This polypeptide chain is sequentially followed by an activation peptide region and a latent serine protease domain. Cleavage of one peptide bond in the activation peptide module results in conversion of the zymogen, Pg, to plasmin (Pm), a serine protease that represents the major extracellular protease found in blood.
Kringle domains are responsible for the binding of Pg and Pm to effector molecules, among which include lysine and its analogs [2–5]. While binding of Pg to free lysine is likely of limited significance in vivo, these kringle lysine binding sites (LBS) in Pg, and its activated product, plasmin (Pm), interact with proteins containing COOH-terminal lysine residues, leading to important functional consequences. As one example, binding of kringle domains of Pg/Pm to fibrin clots that develop COOH-terminal Lys/Arg during lysis further enhances the degradation of fibrin by Pm catalysis, and also stimulates activation of Pg by its activators [6,7]. Of additional significance, similar kringle-mediated binding of Pg/Pm to cell surface receptors results in the assembly of Pg/Pm on cells [8]. Such cell-bound proteolytic capabilities are of importance to extracellular-related processes, e.g., extracellular matrix degradation, that can facilitate tumor cell invasion and metastasis [9].
It is now clear that assembly of the Pg/Pm system components on some microorganisms, is vital to their pathogenesis. One notable example includes the receptors for Pg/Pm on some strains of Gram-positive Group A streptococci (GAS) [10], a major cause of skin and mucosal infections in humans [11]. One such Pg/Pm receptor has been identified on subclasses of bacterial M-proteins (emm genes) that contain the Pg-binding group A streptococcal M- or M-like protein (PAM). This protein binds Pg with high affinity, after which Pg is activated to Pm by GAS-secreted streptokinase (SK), thus providing the bacteria with surface protease activity that allows it to invade surrounding tissue [12,13]. A 30-residue internal polypeptide of PAM, VEK-30, represents the major binding determinant of the PAM/Pg interaction. Interestingly, while VEK-30 binding to Pg requires an intact LBS for maximal binding efficiency, VEK-30 does not contain a C-terminal lysine residue, in contrast to many other Pg/Pm receptors. In addition, the binding of VEK-30 to Pg is highly specific for the kringle 2 domain of Pg (K2Pg), despite the fact that K2Pg displays the weakest affinity and specificity for lysine analogs in comparison to the other lysine-binding kringles of Pg.
We previously solved the X-ray crystal structures of the complexes, VEK-30/K2Pg and VEK-30/angiostatin(K1Pg-K2Pg-K3Pg), and both revealed that an internal “pseudo-lysine”, formed from positive (Arg17 and His18) and negative (Glu20) side-chains, inserts into the LBS of K2Pg [14,15]. However, this unusual binding modality alone cannot explain the high affinity of the binding, nor the specificity of VEK-30 for K2Pg. Indeed, while the structures illuminated numerous exosite interactions which would appear to contribute to the specificity of VEK-30 for K2Pg, their importance to the binding energy is unknown. Furthermore, examination of the crystal packing in both VEK-30/K2Pg and VEK-30/angiostatin revealed that the asymmetric units of both complexes exist in dimeric form. To further identify the potential roles of the exosite interactions in the docking of VEK-30 to K2Pg, and to obtain information on the dynamic aspects of this interaction, we have employed NMR-based methodologies. This communication presents a report of our findings.
2. Experimental
2.1. Protein preparation
Human K2Pg(C4G/E56D/L72Y), a triple variant of K2Pg that displays enhanced affinity for lysine analogs and VEK-30 compared to wild-type-K2Pg, was expressed in Pichia pastoris GS115 cells as described in detail previously [5]. For 15N labeling of the peptide, the recombinant yeast cultures were grown in medium containing (15NH4)2SO4 (99%, Cambridge Isotope Laboratories, Andover, MA). For 15N and 13C labeling, the medium for 15N feeding was used, and glucose and methanol were replaced by [13C]-glucose (99%; Isotec, Champaign, IL) and [13C]-methanol (99%, Isotec). Overall recoveries for the unlabeled and labeled products were 100 mg/l and 80 mg/l, respectively.
To prepare the functional internal peptide, VEK-30, a (His)6-B1 immunoglobulin-binding domain of streptococcal protein G (GB1)-tagged fusion expression system was employed. Briefly, the DNA fragment containing the VEK-30 cDNA was inserted into the bacterial expression vector pET-15b (Novagen, Gibbstown, NJ)/GB1 [16], and expressed in E. coli BL21 (DE3). The final construct contained, sequentially, an ATG initiation codon, a purification (His)6 tag, the GB1 fusion partner for increased solubility, a 9-residue linker, and a thrombin cleavage site, LVPR’GS (‘ representing the site of thrombin-catalyzed cleavage), all sequentially inserted into the parent plasmid [16]. A synthetic gene encoding VEK-30 and a stop codon were inserted immediately downstream of the thrombin cleavage site.
After induction with 0.2 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) for 3 hr at 37 °C, the fusion protein was purified using a Ni2+-Sepharose affinity chromatography column (HisTrap HP; GE Healthcare, Piscataway, NJ). The target fusion protein fractions were pooled and dialyzed against 100 mM NH4HCO3 at 4 °C, and lyophilized. The purified fusion protein (~20 mg), viz., GB1-VEK30, was then cleaved with 100 U thrombin (Enzyme Research Laboratories, South Bend, IN). The cleaved fragments were further separated using a 1 ml HiTrap HP affinity column (GE Healthcare). The flow-through fractions contained VEK-30 (with a GS sequence at the amino-terminus that remained after thrombin cleavage), and were pooled and dialyzed (molecular weight < 2,000 cut-off dialysis membrane), against 50 mM Tris-HCl, pH 7.4/500 mM NaCl overnight at 4 °C. After dialysis, the mixture was applied to a HiTrap-benzamidine FF column (GE Healthcare) to remove the thrombin. The flow-through fraction containing purified VEK-30 was collected and pooled. Finally, the pooled fractions were dialyzed against 100 mM NH4HCO3, 4–6x at 4 °C, and lyophilized. Isotopic labeling was achieved in M9 mineral medium containing 15NH4Cl (99%, Cambridge Isotope Laboratories, Andover, MA) as the sole nitrogen source and/or 13C glucose (99%, Isotec, Champaign, IL) as the sole carbon source. The VEK-30 variants were obtained by site-directed mutagenesis based on pET-15b/GB1/VEK-30, and were over-expressed and purified using the same method as above.
The integrity of all protein and peptides were determined by MALDI-TOF mass spectrometry on an Autoflex III (Bruker Daltonics, Bremen, Germany). For all uniformly labelled 15N and 13C/15N species, single mass peaks were obtained of the correct molecular weights, indicating nearly complete incorporation of the heavy isotopes.
2.2. Surface plasmon resonance (SPR)
Binding of recombinant VEK-30 variants to K2Pg was performed using a BIAcore-X SPR system (BIAcore, Uppsala, Sweden) at 25 °C. A 1:1 mixture of 0.05 M N-hydroxysuccinimide (NHS) and 0.2 M N-ethyl-N-dimethylaminopropylcarbodimide (EDC) (provided in the amine coupling kit; BIAcore AB), were added to the sensor chip (CM-5). Subsequently, K2Pg, diluted to a final concentration of 100 μg/ml and redissolved in 0.1 M NaOAc buffer, pH 4.0, was injected to the flow cell 2 (FC2) for immobilization on the sensor surface. After coupling, 1 M ethanolamine was injected to deactivate the sensor chip surface. All binding experiments were conducted by injecting various concentrations (0.2 μM to 5 μM) of VEK-30 and its variants in HBS-EP buffer (10 mM Hepes/150 mM NaCl/3 mM EDTA/0.005% polysorbate 20, pH 7.4) over the K2Pg-coupled chip surface. At the end of each cycle, regeneration of the surfaces was achieved by injecting 20 μl of washing buffer (0.1 M glycine, pH 2.5), which, as shown by control experiments, did not change the binding properties of K2Pg bound to the CM-5 chip.
Sensorgrams were analyzed using BIAevaluation software, version 3.0. The binding data were subtracted from those obtained using a reference flow cell prepared by the same method, with the exception that K2Pg was not immobilized on the chip. The apparent equilibrium dissociation constants (KD) were calculated from the ratio of the dissociation (kd) and association rates (ka).
2.3. Circular dichroism (CD) measurements
CD spectra of the VEK-30 peptides were measured (200–250 nm) with a Jasco J-815 spectrometer using a quartz cell of 1 mm light path at room temperature. The concentrations of VEK-30 were adjusted to 30 μM in 100 mM sodium phosphate, pH 7.4. Protein and peptide concentrations were determined by UV absorption at 280 nm. The α-helical content was determined from mean residue ellipticities at 222 nm using the empirical relationship, fα =(−[θ]222 − 2340)/30,300) [17].
2.4. Analytical ultracentrifugation
Sedimentation equilibrium experiments were performed as described previously [15] in a Beckman XL-I analytical centrifuge operated in absorbance mode with monitoring at 280 nm. VEK-30 and K2Pg samples were individually rotated at speeds of 42,000 and 48,000 rpm for VEK-30 and 32,000 and 40,000 rpm for K2Pg. The VEK-30/K2Pg complex was analyzed at a molar ratio of VEK-30:K2Pg of 2:1 at the speeds employed for K2Pg. The partial specific volumes of VEK-30 and K2Pg were calculated from their primary sequence-based amino acid composition, and were determined to be 0.727 and 0.708 ml/g, respectively.
2.5. NMR Sample preparation
NMR samples were prepared as 1:1 complexes (1 mM of each component), and consisted of [15N]K2Pg or [13C/15N]K2Pg, in the absence and presence of unlabeled VEK-30. Reverse-labeled samples, i.e., [15N]VEK-30 or [13C/15N]VEK-30, bound to 1 eq of unlabeled K2Pg, were also prepared. The sample buffer contained 50 mM Na-Hepes, pH 7.0/1 mM EDTA/200 mM NaCl/3 mM NaN3/10% 2H2O/0.2 mM 2,2-dimethyl-2-silapentane-5-sulfonic acid (DSS). The 1:1 stoichiometry of the K2Pg-VEK-30 interaction [14] was confirmed by the SPR experiments.
2.6. Data collection
All NMR spectra were acquired on a Bruker AVANCE II 800 MHz spectrometer equipped with a 5-mm triple resonance (TCI, 1H,13C,15N) cryoprobe at 298 K. Experimental data were zero-filled to double the original data points and apodized with a 90° to 60° shift square sine bell window function. Proton chemical shifts were referenced to internal DSS. 15N- and 13C-chemical shifts were referenced indirectly to DSS [18]. NMR data were processed with the NMRpipe system [19] and analyzed with NMRview [20]. For 1H/2H exchange experiments, the samples were lyophilized and quickly redissolved in 99.9% 2H2O. The exchange was then monitored by measuring fast 1H-15N HSQC spectra.
2.7. Resonance assignments and structure calculation
Backbone and side-chain 1H, 13C, 15N NMR resonances of the VEK-30/K2Pg complex were assigned by standard methods [21] using 15N- and 13C-edited HSQC spectra and the following triple-resonance experiments, HNCA, HNCO, HNCACB, HBHA(CO)NH, CBCA(CO)NH, (H)CC(CO)NH, and (H)CCH-COSY. These were employed obtain the backbone and side-chain assignments. 15N- and 13C-TOCSY-HSQC spectra, with a spin lock of 60 ms, were used to confirm all intra-residue assignments. Distance restraints were derived from NOE cross-peak volumes observed in 3D 15N- and 13C-NOESY-HSQC and 2D 1H NOESY at mixing times of 120 and 60 ms. Intermolecular NOEs were identified and distinguished from intramolecular NOEs by employing 13C F1-filtered, F3-edited NOESY-HSQC [22]. A 3D 13C-NOESY-HSQC experiment in 2H2O was used to identify aliphatic to aromatic proton NOEs, and 2D NOESY experiments were used to measure aromatic to aromatic proton NOEs. Dihedral angle restraints (φ and ψ) were derived from chemical shift values of 1HN, 15N, 13CO, 13Cα, 13Cβ using TALOS [23]. Slowly exchanging amide protons for K2Pg and VEK-30 were identified from 1H-15N HSQC experiments after redissolving the lyophilized protein in 2H2O.
Structure calculations were carried out using Crystallography & NMR System (CNS) 1.1 software [24]. The normalized NOE intensities were converted into four distance restraint ranges using lower limits of 0 Å and upper limits of 2.8, 3.5, 4.5, and 5.7 Å. At the end of each run, rejected restraints and residual NOE violations were analyzed. The new assignments were checked and introduced, or eliminated, in the subsequent run. This procedure of assignment/refinement was repeated iteratively until complete assignments of NOESY spectra were made. The hydrogen bond restraints for K2Pg and for VEK-30 were added on the basis of well-converged initial structures. In the final iteration, to improve local geometry and electrostatics, the 50 structures with the lowest energy values were further refined in water by restrained molecular dynamics simulation to remove artifacts [25,26]. The 20 lowest energy structures with no distance violations >0.3 Å and no angle violations >5° were selected to form the final ensemble. These were used for statistical analysis and for qualities of the structures with PROCHECK [27] and WHAT-IF [28]. All figures representing structures were generated using PyMOL (http://www.pymol.org/) or MOLMOL [29].
3. Results
3.1. Overall solution structure of the VEK-30/K2Pg complex
The version of K2Pg employed for this work is a previously characterized variant of K2Pg that consists of C4G, E56D, and L72Y replacements [5]. The latter two substitutions were incorporated in order to bring K2Pg into closer homology with the LBS of the other lysine-binding kringles of Pg (i.e., K1, K4 and K5), all of which contain an Asp and Tyr at sequence positions 56 and 72, respectively. These substitutions enhanced the affinity of K2Pg for lysine analogs and VEK-30 by approximately 15-fold compared with the K2Pg parent. Thus, in using this modified kringle, the interpretation of the structural data is simplified, as it eliminates the possibility that the specificity of VEK-30 for K2Pg is based on amino acid differences at the aforementioned positions.
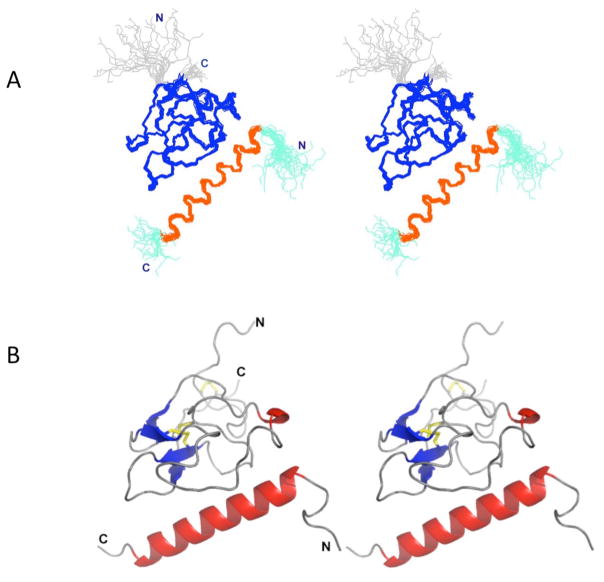
The solution structure of the VEK-30/K2Pg complex was determined using high-resolution multidimensional NMR spectroscopy. A total of 1768 unique distance restraints and 142 dihedral angle restraints for the backbone angles, φ and ψ, were used to calculate the solution structure of the complex (Table 1). A stereoview of 20 superimposed NMR structures and a ribbon representation of the lowest energy structure are shown in Fig. 1A,B, respectively. The overall structure of the 1:1 VEK-30/K2Pg complex displayed overall dimensions of 45 Å (length) x 37 Å (height) x 26 Å (depth). Excluding disordered N-and C-terminal segments, the RMSD, relative to the mean structure, was calculated to be 0.61 Å for backbone atoms and 1.06 Å for all heavy atoms, defined by residues Cys1-Cys78 for K2Pg and residues Ala6-Lys26 for VEK-30. The Ramachandran plot showed 79% of the residues in most favored regions, 19% in additionally allowed regions and 1% of residues in disallowed regions.
Table 1.
NMR and refinement statistics for the VEK-30/K2Pg complex structure
| VEK-30/K2Pg complex | |
|---|---|
| Distance constrains | |
| Total NOEs | 1768 |
| Intra-residue | 801 |
| Inter-residue | |
| Sequential (|i−j|)=1 | 427 |
| Medium-range (|i−j|)≤4 | 202 |
| Long-range (|i−j|)≥5 | 279 |
| Intermolecular | 59 |
| Dihedral angle restraints | |
| φ angle | 71 |
| ψ angle | 71 |
| Hydrogen bonds | 60 |
| PROCHECK-NMR Ramachandran map analysis(%) | |
| Residues in most favored regions | 79.0 |
| Residues in additional allowed regions | 18.3 |
| Residues in generously allowed regions | 1.6 |
| Residues in disallowed regions | 1.1 |
| RMS deviations (Å) among 20 refined structuresa | |
| Backbone | 0.61 ± 0.07 |
| All heavy atoms | 1.07 ± 0.05 |
Residues 1–78 of K2pg, 6–26 of VEK-30 were selected, as the termini were too highly flexible.
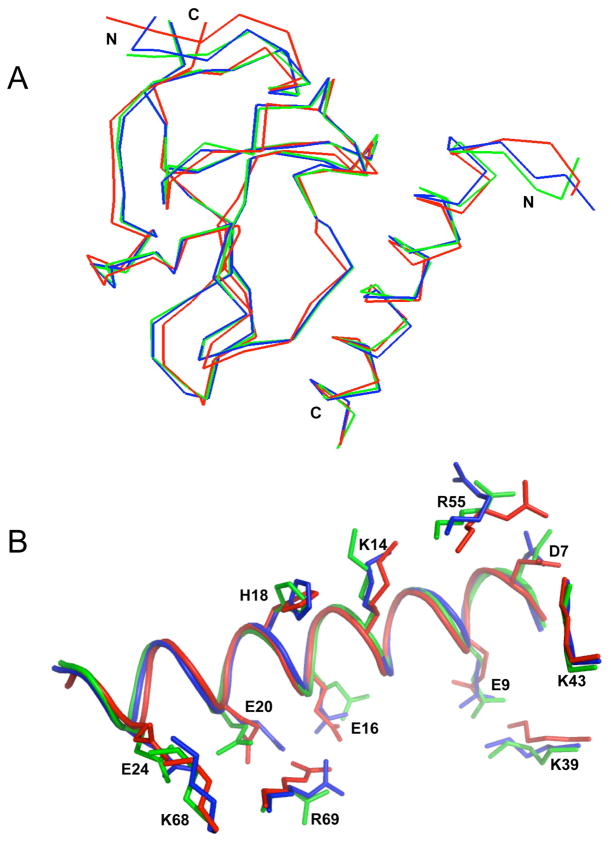
Fig. 1.
Stereo view of the solution structure of the VEK-30/K2Pg complex. (A). Backbone trace of the ensemble of 20 superimposed lowest energy structures. Color coding: Cys1-Cys78 of K2Pg and Ala6-Lys27 of VEK-30 are shown in blue and orange, respectively. The N- and C-termini of each chain are shown in gray (K2Pg) and cyan (VEK-30), respectively. (B). Ribbon representation of the lowest-energy structure. Helices are shown in red, β strands are shown in blue, coil and turn regions are shown in gray.
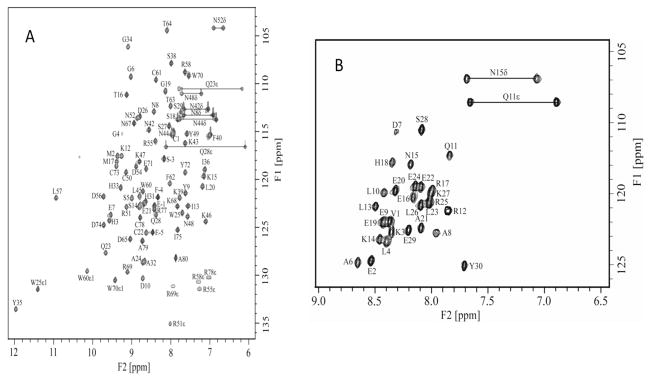
Except for Tyr7 of K2Pg and Ser1, Gly2, and Ser5 of VEK-30, all of the backbone 1H and 15N resonances were assigned, as well as 93% of the side-chain resonances (Fig. 2). The chemical shift assignments and structural constraints have been deposited into the Biological Magnetic Resonance Data Bank (accession number 16311). The structural coordinates for the VEK-30/K2Pg complex have been deposited in the Protein Data Bank (accession code 2KJ4). The asymmetric units of the VEK-30/K2Pg and VEK-20/K1–3Pg crystals have been shown to exist as asymmetric dimers [14,15]. In contrast, our NMR spectra show only one set of resonances for both K2Pg and VEK-30 in the 1:1 complex state (Fig. 2), suggesting that VEK-30/K2Pg exists as a symmetric dimer, or as a monomer, unlike with asymmetric dimer in the crystal. To confirm the monomeric nature of the complex in solution, global correlation times (τm) and apparent molecular weights of free K2Pg and VEK-30/K2Pg were performed. The τm values of the free and bound K2Pg were obtained following common procedures for the determination of rotational diffusion tensors as derived from backbone relaxation (R1 and R2) rates. The diffusion tensors were best defined by isotropic models of rotational tumbling, from which τm estimates of 6.72 ± 0.02 and 7.80 ± 0.02 ns were obtained for apo-K2Pg and VEK-30/K2Pg, respectively. The τm for free K2Pg is in good agreement with the calculated value (6.70 ns) for isotropic tumbling of the molecule, while the experimentally-derived τm for the complex is slightly smaller than the calculated value of 8.5 ns. Sedimentation equilibrium analysis were performed and confirmed the monomeric nature of the complex and each of the individual components. The individually determined apparent molecular weights of VEK-30 and K2Pg were 3,210 ± 260 and 9,660 ± 260, respectively, correlating well with their sequence based molecular weights of 3,600 and 9,570. The apparent molecular weight of the VEK-30/K2Pg complex (1:1 molar ratio) was 12,060 ± 240, in excellent agreement with the combined value (12,870) of the apparent molecular weights of the separate species. These data conclusively support the monomeric nature of the VEK-30/K2Pg complex under solution conditions.
Fig. 2.
NMR spectral analysis of the K2Pg/VEK-30 complex. (A). 2D 1H-15N HSQC spectra of 15N-labeled K2Pg bound to unlabeled VEK-30. (B). 15N-labeled VEK-30 bound to unlabeled K2Pg. Sequence-specific assignments are indicated and peaks corresponding to the NH2 groups of the side-chain amides of Gln and Asn residues are connected by lines.
3.2. The structure of K2Pg in the VEK-30/K2Pg complex
K2Pg in complex with VEK-30 is structurally homogeneous and stably folded as evident from the 15N-HSQC spectrum of 15N-labeled K2Pg (Fig. 2A), which shows the expected backbone amide resonances with uniform intensity. Analogous with the backbone conformation of homologous kringle domains, K2Pg, in complex with VEK-30, exhibited the canonical fold of a kringle domain, including a lack of regular secondary structure (Fig. 1B). In solution, the backbone comprises two short antiparallel β-sheets (paired strands 14–16/21–23 and 60–63/70–72), a short 310-helix (41–43), and several loops and turns. No well-defined α-helix was found. The overall structure of the kringle domain is constrained by six Cys residues with side-chain disulfide bonds at residues 1–78 (C1–C6), 22–61 (C2–C4), and 50–73 (C3–C5). Also, consistent with previous observations, C2–C4 and C3–C5 disulfide bonds are located close and relatively perpendicular to each other [14,30,31]. One notable conserved residue in all of the kringles, Leu45, exhibits an anomalous high-field chemical shift of its two methyl resonances (0.6 ppm and −0.8 ppm) and numerous NOEs to surrounding hydrophobic residues, including Trp25, Tyr35, Pro37, and Pro59. Another strictly conserved kringle residue, Trp25, is buried within the K2Pg fold, and its indole group is positioned centrally in the inner hydrophobic core of the protein, making numerous close contacts with surrounding hydrophobic residues. All of the features mentioned above are recognized to be important in properly folded kringles.
NOE data for six out of seven Pro residues of K2Pg, showed strong Hα(i−1)-ProHδ crosspeaks, consistent with their trans conformation. Only Pro30 showed strong Hα(i−1)-ProHα crosspeaks, indicating adoption of the cis conformation. These Pro conformations were also reflected in their corresponding Cβ and Cγ chemical shifts.
The backbone heavy atom RMSD profile (Fig. 1A) reveals that loop segments in the vicinity of four Cys residues, Cys22, Cys50, Cys61, and Cys73, are well-defined. This suggests that the two inner disulfide bridges, viz., Cys22-Cys61 and Cys50-Cys73, are essential for the kringle domain topology. The two segments in the vicinity of the outer disulfide bridge, Cys1-Cys78, i.e., the Cys1-Gly11 and Asp74-Ala80 loops, near the disordered N-and C-termini of the kringle domain, are the least defined areas in the protein, reflected by the fewer inter-residue NOEs that are observed in these segments.
3.3. The structure of VEK-30 in the VEK-30/K2Pg complex
The 15N-HSQC spectrum of 15N-labeled VEK-30 bound to K2Pg (Fig. 2B) indicates that this peptide exhibits a well-defined secondary structure and stably interacts with K2Pg in the complex. VEK-30 adopts an extended conformation with a distinct α-helix of approximately 6 turns, comprising residues Ala6-Lys27 and running through one side of K2Pg (Fig. 1). The N- and C-terminal regions, Gly2-Thr5 and S28-Tyr30, respectively, are solvent exposed and structurally disordered. On one face of the helix, the hydrophobic and charged residues (Ala6, Asp7, Glu9, Leu10, Leu13, Lys14, Glu16, Arg17, His18 and Glu20 and Ala21, Glu24) interact with K2Pg. Dipolar residues, Gln11, Arg12, Asn15, Glu19, Leu23, Arg25, as well as Ala8 and Leu26, exist on the opposite, solvent-exposed face of the helix.
3.4. Intermolecular interactions in the K2Pg/VEK-30 complex
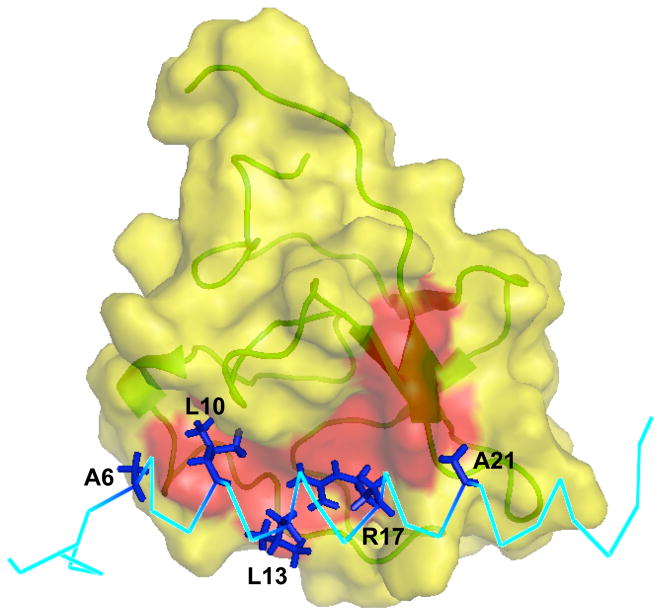
In solution, the K2Pg domain of the complex is a globular structure with an exposed hydrophobic groove consisting of Tyr35, Phe40, Trp60, Phe62, Trp70, and Tyr72 (Fig. 3). Several hydrophobic side-chains of VEK-30, viz., Ala6, Leu10, Leu13, Arg17, and Ala21 (Fig. 3), are all significantly buried within this groove, resulting in a hydrophobic interface between kringle and peptide. In particular, the δ1- and δ2-methyl groups of Leu10 and Leu13 reveal numerous close hydrophobic contacts with the aromatic ring protons of Tyr35 and Phe40 of K2Pg.
Fig. 3.
Surface representation of the binding interface between VEK-30 and K2Pg. The exposed hydrophobic groove (formed by the aromatic residues Tyr35, Phe40, Trp60, Phe62, Trp70 and Tyr72) on the surface of the K2Pg domain is highlighted red. The hydrophobic residues and Arg17 of VEK-30 involved are labeled and depicted as sticks (dark blue).
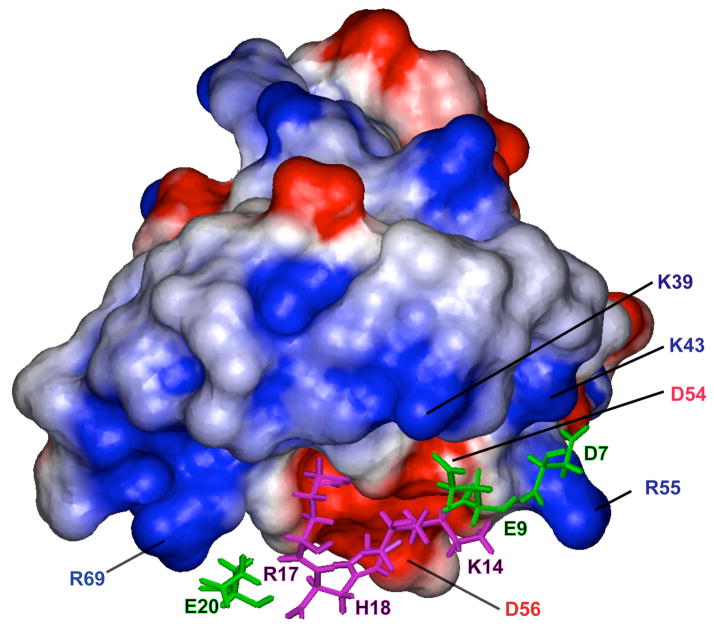
Previous X-ray crystallographic and mutagenesis studies have identified kringle residues Asp54/Asp56 and Arg69 as foci of an electrostatic field that interact with the carboxylate moieties of lysine-type ligands, with additional binding energy deriving from hydrophobic contacts between a hydrophobic kringle cleft and the aliphatic portion of the ligand. The solution structure of the VEK-30/K2Pg complex presented herein supports this general model. Fig. 4 displays a surface representation of K2Pg, color-coded according to the electrostatic potential. This representation shows that both termini of the exposed hydrophobic groove are positively charged due to the presence of Lys39, Lys43, and Arg55 at one end of the cleft and Arg69 at the other. Correspondingly, Asp7 and Glu9 make electrostatic contacts with the cluster of positive charge. Specifically, Lys43 and Arg55 of K2Pg both form salt bridges with Asp7 of VEK-30, resulting in a sandwiched electrostatic field at the N-terminus of the binding interface that stabilizes the orientation of Arg55 and Lys43 side-chains. Additionally, a hydrogen-bonded salt bridge appears to exist between the side-chains of Lys39 of K2Pg and Glu9 of VEK-30. The opposite end of the binding interface is stabilized by ionic contacts between Arg69 of K2Pg and Glu20 of VEK-30. Also, as illustrated in Fig. 4, Asp54 and Asp56 form an anionic locus that interacts with Lys14, Arg17, and His18 of VEK-30, in agreement with previous structural and mutagenesis studies [14,32]. Thus, the anionic and cationic centers of K2Pg, formed by negatively charged residues, Asp54 and Asp56, and positively charged residues, Lys39, Lys43, Arg55, and Arg69, are arranged to produce an electrostatic field configuration that optimizes interactions with internal side-chains of VEK-30 at the binding interface. Other charged residues of VEK-30, i.e., Glu19 and Arg25, which reside in the C-terminal extension of the α-helix, are oriented away from the interaction surface into the solvent. This detailed atomic-resolution structural model is supported by previous mutagenesis studies in which Lys14, Arg17, His18, and Glu20 of VEK-30 were identified as key binding determinants with respect to K2Pg. However the binding energy contributed by Asp7, which appears to engage in ionic contacts with cationic residues of the kringle binding cleft, has yet to be evaluated [14,32].
Fig. 4.
Electrostatic interactions between K2Pg and VEK-30. The K2Pg domain is shown in surface representation with color coding according to electrostatic potential (blue = positively charged, red = negatively charged). The side-chains of charged residues in VEK-30 interacting with K2Pg are represented as magenta and green sticks for positive- and negative-charged residues, respectively.
3.5. Mutational analysis of potential exosite interactions
To investigate the contribution of exosite region interactions in the stability of the VEK-30/K2Pg complex, specifically those contributed by Asp7 in tandem with Glu9 of VEK-30, we measured the binding affinities of select VEK-30 variants to K2Pg by SPR and also evaluated their α-helix contents by circular dichroism (Table 2). Replacement of Asp7 with Ala reduced the affinity of VEK-30 for K2Pg by ~7-fold relative to wild type, and decreased the α-helix content of VEK-30 in both free and kringle-bound forms. Mutation of Asp7 to Asn reduced the K2Pg binding affinity by an even greater extent (30-fold) and significantly reduced the helical propensity of the resultant peptide as evidenced by the minor increase in helicity elicited by kringle binding. Double mutations of Asp7 and Glu9 resulted in effects similar to those observed for the single mutations at Asp7. These results support an important role for Asp7 in maintaining the stability of the VEK-30/K2Pg complex, and confirm a previous mutagenesis study [14] in which it was concluded that the salt-bridge spanning Glu9 of VEK-30 and Lys39 of K2Pg does not appear to make any sizable contribution towards lowering the free energy of binding of the complex.
Table 2.
Mutational analysis of VEK-30 variants binding to K2Pg by surface plasmon resonance and circular dichroism
| Peptide | kona | koff | KD | Apo-helicityb | Maximum helicityc |
|---|---|---|---|---|---|
| M−1 s−1 | s−1 | nM | % | % | |
| VEK-30 | 4.54×105 | 2.93×10−2 | 65 | 34 | 74 |
| VEK-30[D7A] | 1.26×105 | 5.61×10−2 | 446 | 29 | 58 |
| VEK-30[D7N] | 3.62×104 | 7.04×10−2 | 1940 | 18 | 23 |
| VEK-30[D7N/E9Q] | 5.04×104 | 8.60×10−2 | 1710 | 12 | 26 |
| VEK-30[D7A/E9A] | 2.61×105 | 9.67×10−2 | 371 | 19 | 52 |
The association rate constant (kon) and dissociation rate constant (koff) were obtained by SPR and apparent equilibrium dissociation constants (KD) were calculated from the ratio of koff/kon.
% α-helix in absence of K2Pg.
% maximal attainable a-helix after titration with K2Pg.
4. Discussion
An important aspect of GAS pathogenesis is the assembly of plasmin(ogen) on the bacterial surface, in some cases mediated by Pg-binding group-A M-like protein (PAM). A region of PAM corresponding to a 30-residue internal polypeptide, VEK-30, is responsible for the interaction of PAM with Pg via the K2Pg module of Pg [33], normally a very weak lysine-binding kringle. Of great interest is the fact that VEK-30 does not contain a C-terminal Lys residue, thus suggesting that VEK-30 occupies the lysine binding site of K2Pg by adopting a pseudo-lysine conformation, i.e., a composite of peptide side-chains that is isosteric with C-terminal lysine. We have previously determined the X-ray structure of the VEK-30/K2Pg complex and found that side-chain residues, Arg17, His18, and Glu20 were positioned in the lysine binding site of K2Pg [14] in accord with the proposed pseudo-lysine topography. Beyond these interactions within the LBS, exosite contacts were also identified, namely those involving Lys14 and Glu9 of VEK-30. This crystallographic analysis, while providing a snapshot of the binding, does not account for the features that contribute to the high affinity and specificity of the binding of VEK-30 to K2Pg. Furthermore, because the asymmetric unit of the crystal is comprised of two molecules of the kringle/peptide complex, crystal packing artifacts may not be reflective of the molecularity of the solution-phase complex. Thus, we turned to modeling this binding in solution using NMR methods. Successful application of this approach allowed a comparison of the static solid state and average dynamic solution state binding interactions, and also provided a more complete description of kringle/ligand interactions.
4.1. Solution-phase molecularity and general structural features of the VEK-30/K2Pg complex
The crystal structure of the VEK-30/K2Pg complex reveals that the dimer interface occurs between the outer face of the VEK-30 helices, consistent with the tendency of M and M-like proteins to dimerize as coiled-coils on the bacterial surface [34]. However, the Mw,app obtained for VEK-30 reflects the monomeric form of the peptide, and can be reconciled with evidence suggesting that the hypervariable, N-terminal segments of M and M-like proteins (wherein the VEK-30 sequence resides) are less prone to dimerization [35]. With respect to the VEK-30/K2Pg complex, the NMR-derived global correlation time and Mw,app value are also consistent with a monomeric species. We conclude that the functional unit of the complex is defined by one molecule of VEK-30 bound to one molecule of K2Pg.
The NMR-derived solution structure of the VEK-30/K2Pg complex confirms the most salient aspect of the crystallographic model, i.e., that the side-chains of Arg17, His18, and Glu20 are involved in numerous hydrogen bonding and electrostatic interactions within the LBS of K2Pg. In previous mutagenesis studies, when the centrally-positioned Lys14 in VEK-30 was replaced by Ala, the binding affinity of the resultant VEK-30 variant for K2Pg was reduced by over 20-fold, demonstrating that interactions of Lys14 with K2Pg are important to the macroscopic binding event [14]. In our solution structure, Lys14 is in close contact with the anionic center of K2Pg, and also forms hydrogen bonds with the carbonyl groups of Asp54 and Arg55, respectively, of K2Pg. In this study, as well as in the analysis of the crystal structure of VEK-30/K2Pg, the proximity of the carboxylate of Glu9 of VEK-30 to the ε-amino group of Lys39 of K2Pg mitigates for the existence of a salt-bridge between these two moieties. However, substitution of Glu9, either individually[14], or in tandem with substitutions at Asp7 (this study), is without significant effect on the binding constant of the peptide for K2Pg. In the absence of any structural information related to the complex formed between a Glu9-substituted version of VEK-30 and K2Pg, it is difficult to speculate on the molecular basis for the “non-effect” of the Glu9 replacement. Because neither an Ala nor a Gln replacement modifies the binding affinity, it is unlikely that a compensatory interaction occurs between the residue at position 9 and the kringle, given the size and polarity differences between Ala and Gln. It is doubtful that two disparate residues could offset the elimination of the Glu9-Lys39 salt-bridge through another type of contact. It is more likely that a compensating interaction takes place distal to sequence position 9 of VEK-30 in the variant peptides. Alternatively, the unfavorable enthalpy that most likely results from the elimination of the salt-bridge may be counterbalanced by favorable entropic effects that prevail in the complexation of the variant peptides with K2Pg.
The effects of replacing Asp7 of VEK-30 can be interpreted in a more straightforward manner and point to the importance of the ionic contacts between this residue and Arg55 and Lys43 of K2Pg in augmenting the stability of the peptide/kringle complex. Upon analysis of the solution structure details of VEK-30/K2Pg, it is seen that Asp7 is positioned at the N-terminus of the binding interface and is flanked by cationic residues, Arg55 and Lys43 of K2Pg, forming a sandwiched electrostatic field interaction (Fig. 4). This arrangement results in two favorable factors for complex binding. Firstly, it enables the aliphatic atoms of Lys43 to make numerous hydrophobic contacts with the side-chain of Leu10 of VEK-30, thus stabilizing the N-terminus of the binding interface and facilitating α-helix formation. Secondly, the electrostatic interaction between Asp7 and Arg55 abrogates the salt-bridge between Arg55 and Asp54 of K2Pg by attracting the side-chain of Arg55 to be in closer proximity to VEK-30. This allows Asp54 to flip into the LBS, a feature consistent with that observed for the VEK-30/angiostatin complex [15]. The proposed stabilizing interactions attributed to Asp7 are born out by the SPR-monitored binding data of Asp7 variants of VEK-30. Substitution of Asp7 with Ala raises the free energy of binding of the complex by 1.1 kcal/mol while the Asn replacement is disfavorable by 2.0 kcal/mol. This latter substitution effects an approximately 20% diminution in the total free energy of binding compared to that of the parent complex. However, it is unclear if the reduced binding affinity resulting from the Asn replacement is due to steric considerations or because the helical content of this VEK-30 variant, compared to the VEK-30 parent, is significantly compromised in both apo and kringle-bound forms. In short, upon binding to K2Pg, VEK-30 mimics free lysine ligands such that the carboxylate and protonated amino groups of lysine are supplanted by the side-chains of Lys14, Arg17, His18, and Glu20. This intricate network of intermolecular hydrophobic and electrostatic interactions, with support from contacts made by Asp7, accounts for the high binding affinity of VEK-30 to K2Pg (Kd ~ 40 nM) and, in tandem with mutagenesis studies, demonstrate that these residues are crucial for the interaction of VEK-30 with K2Pg in solution.
4.2. Comparison of the solution and crystal structures of the VEK-30/K2Pg complex
The previously determined crystal structure of VEK-30 bound to K2Pg revealed an asymmetric unit containing two molecules of the VEK-30/K2Pg complex [14]. Superimposition of the lowest energy NMR structure and the two molecules obtained from the crystallographic data (PDB entry 1I5K) is presented in Fig. 5. The structural alignment of the backbone shows that all three complexes exhibit similar overall folding (Fig. 5A), with minor differences at the N-termini of both VEK-30 and the kringle. Comparisons of the conformations of side-chain residues in the interacting interface of the crystal structure with those of the solution structure show that most superimpose well, with some differences in the charged residues, Lys39, Lys43, Arg55 and Arg69 of K2Pg, and Asp7, Glu9, Lys14, Glu16, His18 and Glu20 of VEK-30 (Fig. 5B). In particular, the sandwiched ion pairs, Lys43 and Arg55 of K2Pg, are hydrogen-bonded to the carboxylate of Asp7 of VEK-30 in 11 of 20 and 9 of 20 lowest energy structures, respectively, indicating that these ion pairs make closer contacts in the solution structure than in the crystal structure. This is likely due to a dynamic conformational equilibrium that exists among these ion pairs. This is reflected by the line-broadening of Asp7 of VEK-30 that occurs upon complexation with K2Pg (Fig. 2B). There are also some differences in the orientation of the carboxyl group of Asp7 of VEK-30, and especially that of the side-chain of Arg55 of K2Pg, that occur between the two molecules of the asymmetric unit, indicating flexibility of these side-chains even in the more static crystalline state. Minor differences also exist in the orientation of side-chains of the ion pair, Lys39 of K2Pg and Glu9 of VEK-30, as well as the side-chains Lys14 of the peptide, suggesting that some mobility of these residues persists in the complexed state. A more notable difference among these three complexes involves Arg69 of K2Pg and Glu20 of VEK-30. The two structures in the asymmetric unit of the crystal manifest different orientations of the guanidinium group of Arg69, resulting in different binding modes. Specifically, molecule 1 of the crystal structure appears to have a lone hydrogen bond with little or no salt bridge component, while in molecule 2 of the crystal structure, the ion pair interaction is stronger because the carboxylate of Glu20 of VEK-30 is somewhat closer to Arg69 of K2Pg. This slightly different orientation additionally leads to hydrogen bonds with Arg69-NH1. Also, the lowest energy NMR-derived structures show that the guanidinium group of Arg69 of K2Pg is located in close proximity to the carboxylate group of Glu16 of VEK-30, indicating that a doubly H-bonded salt bridge exists between Arg69 of K2Pg and Glu16/Glu20 of VEK-30. Additionally, in the C-terminus of the binding interface, the ion pair interaction between Lys68 of K2Pg and Glu24 of VEK-30 differs slightly among the three complex molecules. In molecule 1 of the crystal structure, only a long-range ionic interaction exists between this ion pair. In molecule 2, this electrostatic interaction is assisted further by an H-bond. Of the 20 refined solution structures, 4 exhibited this H-bond. The imidazole ring of His18 of VEK-30 has a somewhat different orientation in three of the complexes.
Fig. 5.
Superimposition of X-ray crystallographic and NMR solution structures of the VEK-30/K2Pg. (A). The backbone trace of two molecules (molecule-1 and molecule-2) in the unit cell of the crystal structure (PDB entry 115K), and the lowest energy NMR structure of VEK-30/K2Pg are overlaid and represented as blue, green and red lines, respectively. (B). Overlay of side-chain residues involved in the binding interactions. The intermolecular side-chain interactions of K2Pg and VEK-30 of molecule-1 (blue) and molecule-2 (green) of the X-ray crystal structure and the lowest energy NMR structure (red) are compared with side-chain orientations (represented as sticks). The main-chain of VEK-30 in the three structures is depicted as a ribbon diagram.
4.3. Comparison of K2Pg in complex with different ligands
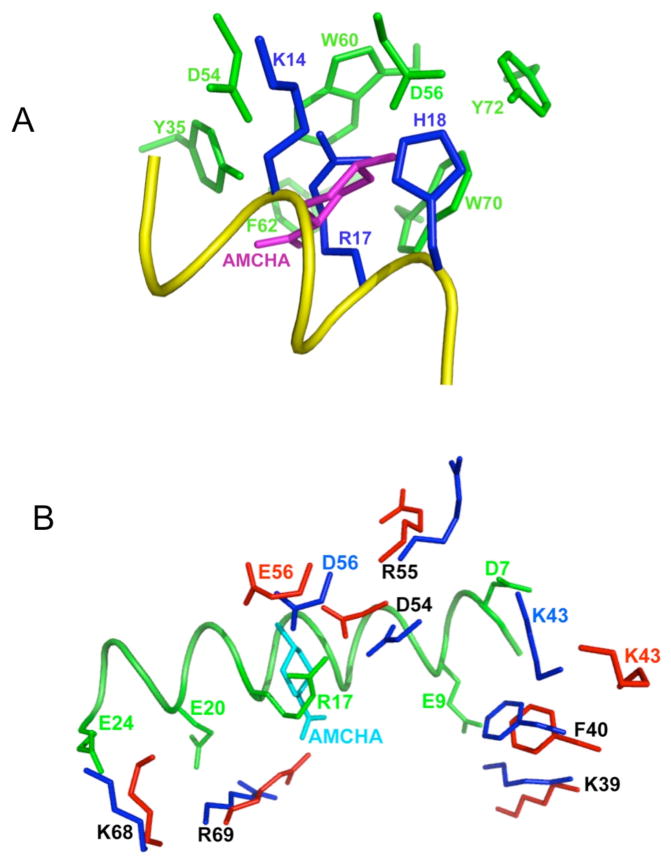
Our solution structure of the K2Pg domain in the VEK-30/K2Pg complex is similar to of the solution structure of K2Pg bound to the lysine analog, t-aminomethyl-cyclohexane-1-carboxylic acid (AMCHA) [36]. When the structures of these two complexes are overlaid and all K2Pg atoms eliminated from the depiction, it is clear that AMCHA resides in the VEK-30/K2Pg binding site with its amino group ideally positioned to interact with Asp54 and Asp56 (Fig. 6A). The hydrophobic sandwich of Trp60 and Trp70 is in close proximity to the cyclohexane ring of AMCHA, providing additional binding energy for AMCHA to the kringle. Comparison of the K2Pg moieties of the AMCHA/K2Pg and VEK-30/K2Pg solution structures shows clear differences in the side-chain geometries of several LBS residues (Fig. 6B). Firstly, in the N-terminal region of the kringle binding domains, two cationic residues, Lys39 and Lys43, exhibit distinctly different side-chain orientations in the two complexes. Both side-chains of these residues in AMCHA/K2Pg are far removed from the ligand, in contrast to Lys39 and Lys43 in VEK-30/K2Pg, which make ionic contacts with Asp7 and Glu9 of VEK-30. Secondly, it is noteworthy that some modulation of ligand binding can occur because of the presence of the guanidinium group of Arg55, which is located between the two essential anionic groups of Asp54 and Asp56. In the solution structure of AMCHA/K2Pg, the guanidinium group of Arg55 of K2Pg does not appear to directly interfere with the binding of the ligand [36]. However, in the VEK-30/K2Pg complex, this side-chain interacts electrostatically with Asp7 of VEK-30. Analysis of the ensemble of lowest-energy conformers shows that 12 out of 20 have an H-bonded electrostatic interaction involving Asp7 of VEK-30 and Arg55 of K2Pg. Finally, there are some differences in the orientations of side-chains of Lys68 and Arg69, which are located in the C-terminal region of the K2Pg binding domain. In the complex of AMCHA and wt-K2Pg, the side-chain of Arg69 is located closer to the ligand-binding center, in proximity to the carboxylate group of AMCHA. Also, the side-chain of Lys68 in VEK-30/K2Pg adopts a more open conformation relative to its orientation in AMCHA/K2Pg, facilitating its interaction with Glu24 of VEK-30.
Fig. 6.
Close-up views of the intermolecular interactions comparing VEK-30/K2Pg and K2Pg/AMCHA interactions as derived from their solution structures. (A). Side-chain atoms in the hydrophobic groove (green-formed by Y35, F40, W60, W70 and Y72) and anionic center (formed by D54 and D56) of K2pg that interact with side-chain atoms from three critical amino acids of VEK-30 (blue-K14, R17, and H18) are shown. A portion of the VEK-30 helix is shown in yellow. An overlay of the K2Pg/AMCHA structure on the K2Pg/VEK-30 structure was performed and all atoms undisplayed, except for AMCHA (magenta), thus providing the relationships between the pseudo-lysine of VEK-30 and AMCHA in the K2Pg lysine binding site. (B). The binding of K2Pg to VEK-30 and K2Pg to AMCHA are overlaid for comparison and selected residues are displayed. The side-chains of K2Pg residues in the VEK-30/K2Pg complex and in the AMCHA/K2Pg complex are shown as dark blue and red sticks, respectively. The ligand, AMCHA, is colored cyan and the ribbon of VEK-30 and side-chains in VEK-30 are both colored green.
4.4. Ligand specificity of K2Pg
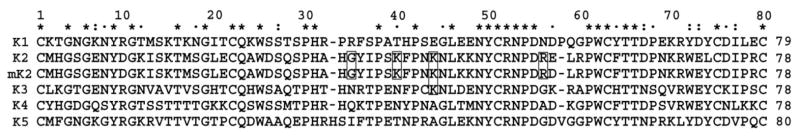
The binding modes between the five individual plasminogen kringle domains and lysine-like ligands have been the subject of numerous structural and mutagenesis-based studies. From these studies it has been concluded that the architecture of the canonical kringle LBS involves a bipolar cleft that harbors cationic and anionic loci that stabilize the carboxyl and amino groups, respectively, of bound C-terminal lysine and its analogs [2–5,14,37–39]. Despite the common structural features shared among the lysine-binding kringles of Pg, VEK-30 specifically binds K2Pg, exhibiting no measurable affinity for any of the other isolated Pg kringle domains [5,33]. It is apparent from the sequence alignment of K2Pg and the five human Pg kringles (Fig. 7) that high homology, including many strictly conserved residues, exists among these domains. This suggests that the specificity of VEK-30 for K2Pg arises from the interactions of VEK-30 with non-conserved kringle residues. Both the present study and a previous crystallographic analysis [14] have confirmed that the VEK-30 helix forms extensive hydrophobic and ionic interactions when complexed with K2Pg. In K2Pg, the conserved hydrophobic core (formed by aromatic residues Tyr35, Phe40, Trp60, Phe62, Trp70, and Y72), anionic (Asp54 and Asp56) and cationic (Arg69) centers interact with VEK-30 in a similar fashion as lysine analogs, resulting in a basic level of binding affinity. However, while EACA and AMCHA bind K2Pg with Kd values of 60 μM and 7 μM, respectively [5], the approximately 65 nM Kd value that describes the VEK-30/K2Pg complex mitigates for intermolecular contacts beyond those that involve the pseudo-lysine arrangement of VEK-30 side-chains in the LBS pocket, and suggests that exosites in the interface function in binding mode differently of kringles bound to simple lysine analogs. Hence, the additional binding energy for tight complex formation must derive from exosite interactions in the kringle/peptide complex. These include contacts between the positively charged residues Lys39, Lys43, and Arg55 of K2Pg, which are not conserved in kringle modules and are unique to K2Pg, and the corresponding negatively charged residues (Asp7, Glu9) in the N-terminus of the VEK-30 helix. These complementary electrostatic interactions likely play important roles in the docking of VEK-30 to K2Pg. Another non-conserved K2Pg residue, Gly34, is positioned at the kringle/peptide interface and, based on NOE data, also participates in intermolecular interactions. Analysis of the VEK-30/K2Pg structure revealed that a small side-chain is required at this position because of steric constraints imposed by the proximity of Leu13 and Glu9 of VEK-30. The sequence alignments show that bulky side-chain residues occupy this position in all other kringle module sequences, suggesting a basis for their null interactions with VEK-30. Thus, although the various kringle modules exhibit similar global folds, variations in their primary structure and in the nature of binding residues are likely to contribute to their ligand-binding specificities and distinct biochemical roles. These different intermolecular contacts provide a structural basis for specificity, since these interactions assist in the recognition of target peptide by placing and orienting VEK-30 in the lysine binding site of K2Pg and/or by inducing the formation of the helix, which is necessary in providing the steric arrangement of VEK-30 side-chains needed for interactions with the lysine binding site of K2Pg. These specific differences in primary sequence may explain why only the K2Pg domain binds to VEK-30. Thus, although the various kringle modules exhibit similar global folds, variations in their primary structure and in the nature of binding residues are likely to contribute to their ligand-binding specificities and distinct biochemical roles.
Fig. 7.
Sequence alignment of the 5 human Pg kringles. Strictly conserved residues are designated by *. Several important non-conserved residues presented in K2Pg and are boxed in the sequences. In K2Pg, C4, E56, and L72 were mutated to C, D, and Y (mK2Pg), respectively to bring the canonical LBS of K2Pg into conformity with other lysine binding kringles.
5. Conclusions
In conclusion, two notable conundrums had existed in the binding of VEK-30 to K2Pg, i.e., the high specificity of VEK-30 for K2Pg and the much higher affinity of VEK-30 for K2Pg, as compared with small molecule lysine analogs. Neither of these is adequately addressed by invoking the formation of a pseudo-lysine residue via optimized side-chains of VEK-30. The intermolecular interactions between the pseudo-lysine-motif, composed of Lys14, Arg17, His18, and Glu20 and the LBS of K2Pg are indispensable, but also insufficient, for the tight complex formation observed between VEK-30 and K2Pg. The electrostatic exosite interactions, in concert with numerous hydrophobic associations, augment the affinity and facilitate the docking of the bacterial peptide to the K2Pg domain by modulating ligand recognition and binding specificity.
Acknowledgments
This work was supported by grant HL013423 from the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sottrup-Jensen L, Claeys H, Zajdel M, Petersen TE, Magnusson S. The primary structure of human plasminogen: isolation of two lysine-binding fragments and one “mini” plasminogen (MW, 38000) by elastase-catalyzed-specific limited proteolysis. Prog Chem Fibrinolysis Thrombolysis. 1978;3:191–209. [Google Scholar]
- 2.Menhart N, Sehl LC, Kelley RF, Castellino FJ. Construction, expression and purification of recombinant kringle 1 of human plasminogen and analysis of its interaction with ω-amino acids. Biochemistry. 1991;30:1948–1957. doi: 10.1021/bi00221a031. [DOI] [PubMed] [Google Scholar]
- 3.Hoover GJ, Menhart N, Martin A, Warder S, Castellino FJ. Amino acids of the recombinant kringle 1 domain of human plasminogen that stabilize its interaction with ω-amino acids. Biochemistry. 1993;32:10936–10943. doi: 10.1021/bi00092a002. [DOI] [PubMed] [Google Scholar]
- 4.McCance SG, Menhart N, Castellino FJ. Amino acid residues of the kringle-4 and kringle-5 domains of human plasminogen that stabilize their interactions with omega-amino acid ligands. J Biol Chem. 1994;269:32405–32410. [PubMed] [Google Scholar]
- 5.Nilsen SL, Prorok M, Castellino FJ. Enhancement through mutagenesis of the binding of the isolated kringle 2 domain of human plasminogen to omega-amino acid ligands and to an internal sequence of a Streptococcal surface protein. J Biol Chem. 1999;274:22380–22386. doi: 10.1074/jbc.274.32.22380. [DOI] [PubMed] [Google Scholar]
- 6.Fleury V, Gurewich V, Angles-Cano E. A study of the activation of fibrin-bound plasminogen by tissue-type plasminogen activator, single chain urokinase and sequential combinations of activators. Fibrinolysis. 1993;7:87–96. [Google Scholar]
- 7.Fleury V, Loyau S, Lijnen HR, Nieuwenhuizen W, Angles-Cano E. Molecular assembly of plasminogen and tissue-type plasminogen activator on an evolving fibrin surface. Eur J Biochem. 1993;216:549–556. doi: 10.1111/j.1432-1033.1993.tb18173.x. [DOI] [PubMed] [Google Scholar]
- 8.Miles LA, Dahlberg CM, Plow EF. The cell binding domains of plasminogen and their function in plasma. J Biol Chem. 1988;263:11928–11934. [PubMed] [Google Scholar]
- 9.DeVries TJ, Vanmuijen GNP, Ruiter DJ. The plasminogen activation system in tumour invasion and metastasis. Pathol Res Pract. 1996;192:718–733. doi: 10.1016/S0344-0338(96)80094-X. [DOI] [PubMed] [Google Scholar]
- 10.Lottenberg R, Broder CC, Boyle MD. Identification of a specific receptor for plasmin on a group A streptococcus. Infect Immun. 1987;55:1914–1918. doi: 10.1128/iai.55.8.1914-1918.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carapetis JR, Steer AC, Mulholland EK, Weber M. The global burden of group A streptococcal diseases. Lancet Infect Dis. 2005;5:685–694. doi: 10.1016/S1473-3099(05)70267-X. [DOI] [PubMed] [Google Scholar]
- 12.Sodeinde OA, Subrahmanyam YV, Stark K, Quan T, Bao Y, Goguen JD. A surface protease and the invasive character of plague. Science. 1992:1004–1007. doi: 10.1126/science.1439793. [DOI] [PubMed] [Google Scholar]
- 13.Wistedt AC, Ringdahl U, Mulleresterl W, Sjobring U. Identification of a plasminogen-binding motif in PAM, a bacterial surface protein. Mol Microbiol. 1995;18:569–578. doi: 10.1111/j.1365-2958.1995.mmi_18030569.x. [DOI] [PubMed] [Google Scholar]
- 14.Rios-Steiner JL, Schenone M, Mochalkin I, Tulinsky A, Castellino FJ. Structure and binding determinants of the recombinant kringle-2 domain of human plasminogen to an internal peptide from a group A Streptococcal surface protein. J Mol Biol. 2001;308:705–719. doi: 10.1006/jmbi.2001.4646. [DOI] [PubMed] [Google Scholar]
- 15.Cnudde SE, Prorok M, Castellino FJ, Geiger JH. X-ray crystallographic structure of the angiogenesis inhibitor, angiostatin, bound to a peptide from the group A streptococcal surface protein PAM. Biochemistry. 2006;45:11052–11060. doi: 10.1021/bi060914j. [DOI] [PubMed] [Google Scholar]
- 16.Cheng Y, Patel DJ. An efficient system for small protein expression and refolding. Biochem Biophys Res Commun. 2004;317:401–405. doi: 10.1016/j.bbrc.2004.03.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen YH, Yang JT, Martiner HM. Determination of secondary structure of proteins by circular dichroism and optical rotary dispersion. Biochemistry. 1972;11:4120–4131. doi: 10.1021/bi00772a015. [DOI] [PubMed] [Google Scholar]
- 18.Markley JL, Bax A, Arata Y, Hilbers CW, Kaptein R, Sykes BD, Wright PE, Wüthrich K. Recommendations for the presentation of NMR structures of proteins and nucleic acids. IUPAC-IUBMB-IUPAB Inter-Union Task Group on the Standardization of Data Bases of Protein and Nucleic Acid Structures Determined by NMR Spectroscopy. J Biomol NMR. 1998;12:1–23. doi: 10.1023/a:1008290618449. [DOI] [PubMed] [Google Scholar]
- 19.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;63277–293:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 20.Johnson BA. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol Biol. 2004;278:313–352. doi: 10.1385/1-59259-809-9:313. [DOI] [PubMed] [Google Scholar]
- 21.Sattler M, Schleucher J, Griesinger C. Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Prog NMR Spectrosc. 1999;34:93–158. [Google Scholar]
- 22.Zwahlen C, Legault P, Vincent SJF, Greenblatt J, Konrat R, Kay LE. Methods for measurement of intermolecular NOEs by multinuclear NMR spectroscopy: Application to a bacteriophage N-peptide/boxB RNA complex. J Am Chem Soc. 1997;119:6711–6721. [Google Scholar]
- 23.Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 24.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;154:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 25.Linge JP, Williams MA, Spronk CA, Bonvin AM, Nilges M. Refinement of protein structures in explicit solvent. Proteins. 2003;15:496–506. doi: 10.1002/prot.10299. [DOI] [PubMed] [Google Scholar]
- 26.Nederveen AJ, Doreleijers JF, Vranken W, Miller Z, Spronk CA, Nabuurs SB, Güntert P, Livny M, Markley JL, Nilges M, Ulrich EL, Kaptein R, Bonvin AM. RECOORD: a recalculated coordinate database of 500+ proteins from the PDB using restraints from the BioMagResBank. Proteins. 2005;59:662–672. doi: 10.1002/prot.20408. [DOI] [PubMed] [Google Scholar]
- 27.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHEK: a program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;26:283–291. [Google Scholar]
- 28.Vriend G. WHAT IF: a molecular modeling and drug design program. J Mol Graph. 1990;8:52–56. doi: 10.1016/0263-7855(90)80070-v. [DOI] [PubMed] [Google Scholar]
- 29.Koradi R, Billeter M, Wüthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J Mol Graph. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 30.Byeon IJ, Llinas M. Solution structure of the tissue-type plasminogen activator kringle 2 domain complexed to 6-aminohexanoic acid an antifibrinolytic drug. J Mol Biol. 1991;222:1035–1051. doi: 10.1016/0022-2836(91)90592-t. [DOI] [PubMed] [Google Scholar]
- 31.Abad MC, Arni RK, Grella DK, Castellino FJ, Tulinsky A, Geiger JH. The X-ray crystallographic structure of the angiogenesis inhibitor angiostatin. J Mol Biol. 2002;318:1009–1017. doi: 10.1016/S0022-2836(02)00211-5. [DOI] [PubMed] [Google Scholar]
- 32.Schenone MM, Warder SE, Martin JA, Prorok M, Castellino FJ. An internal histidine residue from the bacterial surface protein, PAM, mediates its binding to the kringle-2 domain of human plasminogen. J Pept Res. 2000;56:438–445. doi: 10.1034/j.1399-3011.2000.00810.x. [DOI] [PubMed] [Google Scholar]
- 33.Wistedt AC, Kotarsky H, Marti D, Ringdahl U, Castellino FJ, Schaller J, Sjobring U. Kringle 2 mediates high affinity binding of plasminogen to an internal sequence in streptococcal surface protein PAM. J Biol Chem. 1998;273:24420–24424. doi: 10.1074/jbc.273.38.24420. [DOI] [PubMed] [Google Scholar]
- 34.Phillips GN, Flicker PF, Cohen C, Manjula BN, Fischetti VA. Streptococcal M protein: alpha-helical coiled-coil structure and arrangement on the cell surface. Proc Natl Acad Sci USA. 1981;78:4689–4993. doi: 10.1073/pnas.78.8.4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.André I, Persson J, Blom AM, Nilsson H, Drakenberg T, Lindahl G, Linse S. Streptococcal M protein: structural studies of the hypervariable region, free and bound to human C4BP. Biochemistry. 2006;45:4559–4568. doi: 10.1021/bi052455c. [DOI] [PubMed] [Google Scholar]
- 36.Marti DN, Schaller J, Llinas M. Solution structure and dynamics of the plasminogen kringle 2-AMCHA complex: 3(1)-helix in homologous domains. Biochemistry. 1999;38:15741–15755. doi: 10.1021/bi9917378. [DOI] [PubMed] [Google Scholar]
- 37.Wu TP, Padmanabhan K, Tulinsky A, Mulichak AM. The refined structure of the ε-aminocaproic acid complex of human plasminogen kringle 4. Biochemistry. 1991;30:10589–10594. doi: 10.1021/bi00107a030. [DOI] [PubMed] [Google Scholar]
- 38.Mathews II, Vanderhoff-Hanaver P, Castellino FJ, Tulinsky A. Crystal structures of the recombinant kringle 1 domain of human plasminogen in complexes with the ligands ε-aminocaproic acid and trans-4-(aminomethyl)cyclohexane-1-carboxylic acid. Biochemistry. 1996;35:2567–2576. doi: 10.1021/bi9521351. [DOI] [PubMed] [Google Scholar]
- 39.Chang Y, Mochalkin I, McCance SG, Cheng BS, Tulinsky A, Castellino FJ. Structure and ligand binding determinants of the recombinant kringle 5 domain of human plasminogen. Biochemistry. 1998;37:3258–3271. doi: 10.1021/bi972284e. [DOI] [PubMed] [Google Scholar]