Abstract
In several invertebrate organisms, the Sec1p/Munc18-like protein Vps45 interacts with the divalent Rab4/Rab5 effector, Rabenosyn-5 and carries out multiple functions in the endocytic/secretory pathways. In mammalian cells, Vps45 and Rabenosyn-5 also interact, but the molecular characterization of this binding, and the functional relationship between these two proteins has not been well defined. Here we identify a novel sequence within Rabenosyn-5 required for its interaction with Vps45. We demonstrate that hVps45-depletion decreases expression of Rabenosyn-5, likely resulting from Rabenosyn-5 degradation through the proteasomal pathway. Furthermore, we demonstrate that similar to Rabenosyn-5-depletion, hVps45-depletion causes impaired recycling of β1 integrins, and a subsequent delay in human fibroblast cell migration on fibronectin-coated plates. Moreover, β1 integrin recycling could be rescued by reintroduction of siRNA-resistant wild-type Rabenosyn-5, but not a mutant deficient in Vps45 binding. However, unlike Rabenosyn-5-depletion, which induces Golgi fragmentation and decreased recruitment of sorting nexin retromer subunits to the Golgi, hVps45-depletion induces Golgi condensation and accumulation of retromer subunits in the vicinity of the Golgi. In part, these phenomena could be attributed to reduced Syntaxin16 expression and altered localization of both Syntaxin16 and Syntaxin6 upon Vps45-depletion. Overall, these findings implicate hVps45 and Rabenosyn-5 in post early endosome transport, and we propose that their interaction serves as a nexus to promote bidirectional transport along the endosome-to-recycling compartment and endosome-to-Golgi axes.
Keywords: Endosome, Golgi, Integrins, Retromer
Introduction
Endocytosis is a highly conserved protein and lipid trafficking pathway in eukaryotes in which nutrients and plasma membrane proteins, including growth factor receptors and ion transporters, are internalized. Upon endocytosis, newly internalized materials are transported to early endosomes, a tubulo-vesicular sorting station usually localized to the cell periphery. From early endosomes, proteins destined for degradation are transported to late endosomes and lysosomes, whereas proteins destined for recycling are sorted to recycling endosomes, and subsequently transported back to the plasma membrane [1].
Rab proteins are involved at various steps of transport in the vesicular trafficking pathway and help determine the specificity of vesicle targeting [2]. Most of them are ubiquitously expressed in eukaryotes and function as molecular switches, cycling between GTP-bound and GDP-bound states. Rab5, a key endocytic protein, is involved in transport and tethering of vesicles to the early endosomal membrane [3]. Its function is facilitated through its binding to effector molecules, proteins that bind directly in a GTP-dependent manner and are required for various downstream functions. These effectors include Rabaptin-5, early endosomal antigen 1 (EEA1), phosphatidylinositol 3-kinase (PI3K) and the dual Rab5/Rab4 effector, Rabenosyn-5 [4].
Rabenosyn-5 is comprised of a C2H2 and a FYVE zinc finger domain both localized near its N-terminal region, a central coiled-coil region (CC), and five asparagine-proline-phenylalanine (NPF) motifs within its C-terminal region [5–7]. The function of the C2H2 zinc finger domain is still unknown, whereas the FYVE domain binds phosphatidylinositol-3-phosphate (PtdIns3P) [5, 8]. The Rab4-binding region is located within the central CC region, whereas the Rab5 binding region is at the C-terminal end of Rabenosyn-5, following the NPF motifs [6]. The first two Rabenosyn-5 NPF motifs are the most important ones for binding to the EH-domain of the C-terminal EH-domain-containing proteins (EHD proteins) [7], a family of proteins involved in various steps of intracellular protein trafficking [9].
It has been proposed that Rabenosyn-5 sequentially regulates protein recycling through its interaction with EHD1, an EHD protein that controls recycling from the endocytic recycling compartment (ERC) to the plasma membrane. Although Rabenosyn-5-depletion impairs recycling, unlike the effect of EHD1-depletion, loss of Rabenosyn-5 leads to accumulation of transferrin and major histocompatibility complex class I (MHC I) at early endosomes, causing a delay in their trafficking from early endosomes to the ERC [7]. In addition, EHD1 has been implicated in the recycling of β1 integrin, a eukaryotic receptor for adhesion to the extracellular matrix [10].
In eukaryotes, it has been demonstrated that Rabenosyn-5 (or its orthologs), interacts with Vps45 [5, 11], a member of the Sec1p/Munc18-like (SM) protein family that binds SNAREs. SM proteins are generally localized to the cytoplasm and contain ~600–700 residues which display considerable homology to one another. In a manner similar to SNAREs, SM proteins function in multiple pathways and are unlikely to be exclusively responsible for fusion specificity. SM proteins often bind directly to syntaxins, a subclass of SNAREs [12]. In yeast, Vps45p is a peripheral membrane protein whose binding to the membrane is mediated through Tlg2p, a target membrane-bound SNARE (t-SNARE) protein. A conserved hydrophobic residue within the Sec-1p like domain of Vps45p (L117), which is part of the hydrophobic binding pocket, is needed for binding to the N-terminal region of Tlg2p [13]. Mutation of Vps45p L117 to arginine leads to its inability to bind to monomeric Tlg2p, thus abolishing its membrane association [13]. Vps45p not only binds to Tlg2p in its monomeric state, but also as a part of trans- and cis-SNARE complexes through different amino acids [14].
In yeast, the interaction between Vps45p and Vps21p (the yeast ortholog of Rab5) is mediated through Vac1p, which is considered to be the yeast ortholog of Rabenosyn-5 [11]. This complex is required for carboxypeptidase Y (CPY) processing and trafficking from the Golgi to vacuoles via the pre-vacuolar compartment (PVC) [11]. In C. elegans, Vps45 and Rabenosyn-5 interact directly and Vps45 is required for receptor-mediated and fluid-phase endocytosis pathways [15]. It has also been demonstrated that both Vps45 and Rabenosyn-5 are needed for Rab5-mediated endosomal fusion [15]. Moreover, Drosophila Vps45 and Rabenosyn-5 both regulate vesicle fusion leading to early endosome formation [16].
While both the Rabenosyn-5 and Vps45 orthologs appear to coordinate endocytic regulatory functions in yeast and invertebrates, their relationship in mammalian cells is not well understood. Although these proteins have been identified as part of a complex [5], the mode of their interactions in mammalian cells and their functional inter-relationship has received considerably less attention.
In this study, we provide evidence for a direct interaction between endogenous hVps45 and Rabenosyn-5 in mammalian cells, and delineate the specific Rabenosyn-5 residues required for binding. We demonstrate that the interaction of hVps45 with Rabenosyn-5 stabilizes the latter protein, preventing its degradation that likely occurs through the proteasomal pathway. Moreover, we demonstrate that hVps45 and Rabenosyn-5 regulate recycling of β1 integrin receptors, thus controlling cell migration. Finally, we show that as observed upon Rabenosyn-5-depletion, loss of hVps45 also impairs transport from endosomes to the Golgi complex. These studies further our understanding of the complex mode by which hVps45 and Rabenosyn-5 coordinately regulate trafficking in mammalian cells.
Materials and methods
Recombinant DNA constructs
Rat Vps45 cDNA was subcloned into the mammalian pEGFP-C2 and pCMV-Myc expression vectors and the yeast two-hybrid vector pGBKT7 (BD Biosciences, Mountain View, CA). Rat Vps45 V107R L110R, siRNA-resistant rat Vps45 (Si-Vps45), HA-Rabenosyn-5 100, 101, 105-109 ala constructs and truncated Rabenosyn-5 mutant (Rabenosyn-5 50-200 95-99 ala, Rabenosyn-5 50-200 100, 100 ala, Rabenosyn-5 50-200 105-109 ala, and Rabenosyn-5 50-200 110-114 ala) constructs were generated through site-directed mutagenesis with the QuickChange kit (Stratagene, La Jolla, CA). SiRNA-resistant GFP-Rabenosyn-5 (si-GFP-R-5) clones were also generated for wild-type and mutants. Full-length HA-tagged Rabenosyn-5 and HA-Rabenosyn-5 264-784 were subcloned into the mammalian expression vector pCDNA 3.1(−) (Invitrogen, Carlsbad, CA). Truncated Rabenosyn-5 constructs (Rabenosyn-5 1-623, Rabenosyn-5 1-263, Rabenosyn-5 50-200, Rabenosyn-5 70-170, Rabenosyn-5 70-152, Rabenosyn-5 70-120, Rabenosyn-5 130-170, and Rabenosyn-5 100-170) and truncated Rabenosyn-5 mutant constructs (Rabenosyn-5 50-200 95-99 ala, Rabenosyn-5 50-200 100, 101 ala, Rabenosyn-5 50-200 102-104 ala, Rabenosyn-5 50-200 105-109 ala, and Rabenosyn-5 50-200 110-114 ala) were generated and cloned into yeast two-hybrid vector pGADT7 (BD Biosciences, Mountain View, CA). HA-Arf6-Q67L and GFP-Rab5-Q79L were kindly provided by Dr. J. Donaldson (NIH) and Dr. R. Lodge (Laval Universite), respectively.
Antibodies
Peptide-specific affinity-purified polyclonal anti-Rabenosyn-5 antibody has been previously described [16], rabbit anti-hVps45, mouse anti-Syntaxin6, and mouse anti-Syntaxin16 were purchased from Synaptic Systems (Gottingen, Germany), mouse anti-actin and goat anti-hVps35 antibodies were from Abcam (Cambridge, MA), mouse anti-Rab5 and anti-SNX1 antibodies were from Transduction Laboratories (Lexington, KY), mouse anti-human β1 integrin was from Serotec (Oxford, UK), mouse anti-γ-tubulin, mouse anti-GFP was from Roche Applied Science (Indianapolis, IN), mouse anti-HA was from Covance (Berkeley, CA), mouse anti-Myc was from Zymed Laboratories (Carlsbad, CA), and DAPI was from Molecular Probes (Carlsbad, CA). Secondary anti-mouse and anti-rabbit Alexa Fluor 488 and Alexa Fluor 568 antibodies were from Molecular Probes (Carlsbad, CA).
Cell culture, silencing RNA and transfection
HeLa and human foreskin fibroblast cell lines were cultured as described [19], silencing RNA (siRNA) treatment was performed for either 48 or 72 h with Oligofectamine in Optimem (Invitrogen, Carlsbad, CA) or Dharmafect1 (Dharmacon, Lafeyette, CO) in DMEM with 10% fetal calf serum using Rabenosyn-5 siRNA oligonucleotide duplexes as described [16] or On-TargetPlus SMARTpool human Vps45-siRNA oligonucleotides (Dharmacon). The addition of oligonucleotide was omitted from Mock-treated cells. FuGene HD (Roche Applied Science, Indianapolis, IN) was used for cDNA transfection of siRNA-treated cells for the final 40 h after the siRNA treatment.
Immunoprecipitations and immunoblotting
For immunoprecipitation experiments, cells were harvested and lysed for 20 min in buffer containing 50 mM Tris, pH 7.4, 150 mM NaCl, 0.5% Triton-X-100 (wt/vol), 1.8 mg/ml iodoacetamide, and Protease Inhibitor Cocktail Set I (EMD Calbiochem, Gibbstown, NJ). Insoluble matter was removed by centrifugation and the lysate supernatants were subjected to immunoprecipitations with either goat anti-HA immobilized agarose (Bethyl laboratories, Montgomery, TX) or Rabbit ExactaCruz F (immunoprecipitation matrix) (Santa Cruz Biotechnology, Santa Cruz, CA) prebound to rabbit anti-Rabenosyn-5 antibody. After 2 h incubation at 4 C, immunoprecipitates were washed three times with lysis buffer and eluted by boiling in the presence of 1% SDS. Proteins were resolved by 8% SDS-PAGE, transferred onto nitrocellulose membranes, blocked in 5% nonfat milk in phosphate-buffered saline (PBS), and immunoblotted with either rabbit anti-Rabenosyn-5, anti-Vps45 or either mouse anti-Myc or anti-HA followed by HRP-conjugated donkey anti-rabbit and goat anti-mouse (Jackson ImmunoResearch, West Grove, PA).
Membrane fractionation
Membrane fractionation assays were performed essentially as described [26]. Briefly, HeLa cells were harvested and then homogenized in ice-cold homogenization buffer (25 mM HEPES, 100 mM NaCl, 1 mM ethylenediaminetetraacetic acid, pH 7.4 and protease inhibitor cocktail at 4 °C. After removal of unbroken cells and cellular debris by centrifugation at 800 × g for 10 min, the supernatant was then ultracentrifuged at 108,000 × g for 1 h at 4 °C to yield a pellet of total cellular membranes and a supernatant containing the cytosolic fraction. The membrane pellet was dissolved in urea buffer (70 mM Tris-HCl, pH 6.8, 8 M urea, 10 mM n-ethylmaleimide, 10 mM iodoacetamide, 2.5% SDS and 0.1 M DTT). The entire membrane fraction and 24% of the cytosolic fraction were each mixed with Laemmli buffer and subjected to SDS-PAGE and immunoblot analysis.
Yeast two-hybrid analysis
Co-transformation into Saccharomyces cerevisiae strain AH109 (BD Biosciences Clontech) and colony growth assays were performed as described [27]. Briefly, co-transformation was performed by the lithium acetate procedure as described in the instructions for the MATCHMAKER two-hybrid (BD Biosciences Clontech). The co-transformants were streaked on plates lacking leucine and tryptophan and grown at 30 °C for 3 days. An average of 3–4 colonies were then chosen, suspended in water, equilibrated to the same optical density, and replated on plates lacking leucine and tryptophan (+HIS) as well as plates lacking leucine, tryptophan and histidine (−HIS).
β1 integrin recycling and migration assays
β1 integrin recyling and migration assays were done as described [19] on HeLa and human fibroblast cells, respectively. Cells were grown on coverslips, and transfected with either Rabenosyn-5- or hVps45-siRNA oligonucleotides. For migration assays, a scratch of ~900 μm was made in the confluent layer of cells on the coverslips coated with fibronectin, and migration of the cells into the wound area was monitored after 6 h by fixing the cells in 4% paraformaldehyde in PBS (v/v) and visualizing the cells by incubation with 488-Wheat Germ Agglutinin (Molecular Probes, Carlsbad, CA). Mean wound lengths were measured at several points across the scratched region using the LSM 5 Pascal marker tool.
DNA-cell cycle analysis
Mock- and hVps45-siRNA-treated cells were trypsinized, washed with PBS, fixed with 70% ethanol at 4 °C for 1h, washed with PBS, stained with Telford Reagent (0.115 mM EDTA, 13.4 mg RNAse A (93U/mg), 0.0748 mM Propidium Iodide, and 1% TX-100 in PBS) at 4 °C overnight, and subjected to flow cytometry analysis.
Immunofluorescence and quantification
HeLa cells were grown on coverslips and were either Mock-treated or treated with hVps45-siRNA or Rabenosyn-5-siRNA. In some cases, the cells were also transfected with GFP-Rab5 Q79L, HA-Arf6 Q67L, si-GFP-R-5, si-GFP-R-5 100, 101, 104-109 ala, or si-GFP-Vps45 using FuGENE-HD (Roche Diagnostics, Indianapolis, IN) for the final 48 h of siRNA treatment. After fixation with 4% paraformaldehyde in PBS (v/v), cells were incubated with primary antibodies in staining solution (0.2% saponin (w/v) and 0.5% bovine serum albumin (BSA) in PBS) for 1 h at room temperature. Then, cells were washed with PBS and incubated with the appropriate fluorochrome-conjugated secondary antibody mixture in staining solution for 30 min at room temperature. Images were acquired using Zeiss LSM 5 Pascal Confocal microscope (Carl Zeiss, Thornwood, NY), using a 63X objective with a numerical aperture of 1.4 and quantified using either LSM 5 Pascal or ImageJ software (for co-localization quantification).
Results
Interaction of endogenous Rabenosyn-5 and hVps45 in mammalian cells
GST-pulldown experiments have previously been used to demonstrate an in vitro interaction between Rabenosyn-5 and hVps45 [5]. To date, however, neither the interaction of these proteins in vivo, nor the characterization of the amino acid sequences required for such binding have been addressed. To determine if endogenous hVps45 and Rabenosyn-5 interact in cells, we performed co-immunoprecipitation experiments with HeLa cell lysates (Fig. 1A). As shown, immunoprecipitation of endogenous Rabenosyn-5 with anti-Rabenosyn-5 antibodies was highly specific as pre-immune serum did not precipitate detectable levels of Rabenosyn-5 (Fig. 1A; left panel). Moreover, this band corresponded to the ~120 kDa band observed in lysates blotted directly with the antibody (Fig. 1A; second panel). When immunopreciptation was done with Rabenosyn-5 antibodies and immunoblotting was performed with anti-Vps45 antibodies (Fig. 1A; third panel), a band corresponding to Vps45 was readily detected indicating that these two proteins interact in vivo. Control experiments showed that two other (highly expressed) endogenous proteins, Focal Adhesion Kinase (FAK) and Heat Shock 70 (Hsc70) were not immunoprecipitated with Rabenosyn-5, demonstrating the specificity of the Rabenosyn-5/Vps45 interaction (Fig. 1A; right panel).
Fig. 1.
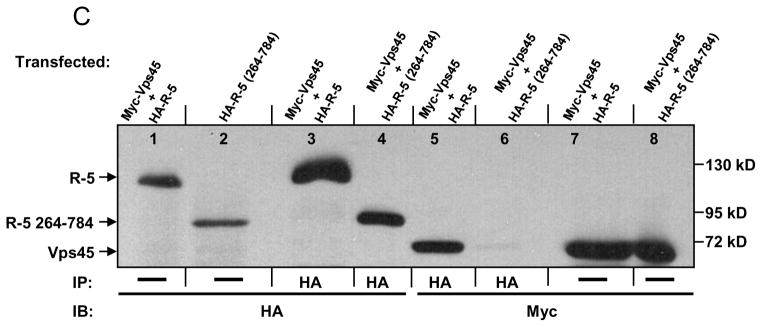
hVps45 interacts with Rabenosyn-5 in mammalian cells. (A) HeLa cells were harvested, lysed, and subjected to immunoprecipitations with either antibody against human Rabenosyn-5 (R-5) or pre-immune serum (Pre-Imm). Immunoprecipitates and lysates were resolved by 8% SDS-PAGE, transferred to nitrocellulose filters, and immunoblotted with either anti-R-5 (left panel), anti-R-5 or anti-hVps45 (center panel; nitrocellulose was cut and blotted with either antibody as indicated by the border line), and with either anti-FAK or anti-Hsc70 (far right panel; nitrocellulose was cut and blotted with either antibody as indicated by the border line) and followed by secondary anti-rabbit HRP-conjugated antibody. (B) The yeast strain AH109 was co-transformed with the following GAL4 binding domain (GAL4BD) fusion constructs: GAL4BD-Vps45 and GAL4BD-p53 (control) together with the following GAL4 transcription activation domain (GAL4AD) fusion constructs: GAL4AD-R-5 (full length), GAL4AD-R-5 1-623, GAL4AD-R-5 1-263, GAL4AD-R-5 50-200, GAL4AD-R-5 70-170, GAL4AD-R-5 70-152, GAL4AD-R-5 70-120, GAL4AD-R-5 130-170, GAL4AD-R-5 100-170, and GAL4AD-SV40 Large T-antigen (control). Co-transformants were assayed for their growth on non-selective (+HIS) and selective (−HIS) media. (C) HeLa cells were co-transfected with either Myc-Vps45 and HA-R-5 or Myc-Vps45 and HA-R-5 264-784. After 24 h, cells were harvested, lysed, and in some cases, subjected to immunoprecipitations with antibody against the HA epitope. Lysates and immunoprecipitates were resolved by 8% SDS-PAGE, transferred to nitrocellulose, and immunoblotted with either anti-HA or anti-Myc antibodies followed by anti-mouse-HRP conjugated antibody. IP=immunoprecipitation; IB=immunoblot; Ig=immunoglobulin; C2H2=zinc finger domain; FYVE=PtdIns3P-binding domain; CC=coiled-coil region; NPF=asparagine-proline-phenylalanine motif.
While direct interactions between Rabenosyn-5 and Vps45 have been reported in C. elegans and mammalian cells by in vitro assays [5, 15], the nature and sequence requirements of these interactions have remained unknown to date. To map the interaction between Rabenosyn-5 and Vps45, we utilized a selective yeast two-hybrid approach. As shown in Fig. 1B, the full-length Rabenosyn-5 interacted directly with Vps45. The C2H2, FYVE, and coiled-coil (CC) domains as well as the NPF motifs of Rabenosyn-5 were all dispensable for this interaction, but a region including amino acids 70-120 was essential for the binding of Rabensoyn-5 to Vps45 (Fig. 1B).
These findings were further confirmed in vivo by co-immunoprecipitation experiments. Myc-Vps45 was co-expressed with either full-length HA-Rabenosyn-5 (HA-R-5) or HA-R-5 containing residues 264-784 in HeLa cells (Fig. 1C). Co-expression levels were validated in lanes 1, 2, 7 and 8, and successful immunoprecipitations were demonstrated (lanes 3 and 4). As shown in Fig. 1C (lane 5), the full-length HA-R-5 interacted with myc-Vps45, however, HA-R-5 264-784 was unable to do so (Fig. 1C; lane 6), indicating that the N-terminal region of Rabenosyn-5 is required for the interaction with hVps45 in vivo.
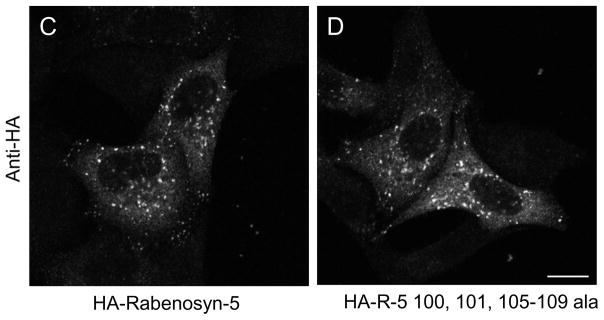
To further delineate the exact Rabenosyn-5 residues required for interacting with Vps45, we utilized alanine screening within amino acid 70-120 stretch of Rabenosyn-5. Using the ‘wild-type’ residues 50-200 as a template, we systematically mutated clusters of 2-5 residues to alanine. As demonstrated (Fig. 2A), we identified two nearby clusters of residues within the Rabenosyn-5 70-120 region (amino acids 101, 102 and 105-109) that were necessary for Rabenosyn-5 to interact with Vps45. No other sequences within the 70-120 residue stretch were required for binding to Vps45 (data not shown). To further confirm these findings in vivo, we then generated a HA-tagged Rabenosyn-5 mutant with alanine residues replacing amino acids 100, 101 and 105-109. The mutant and wild-type Rabenosyn-5 were then each co-transfected with Myc-Vps45 into HeLa cells, and immunoprecipitation experiments were performed to determine whether they interact (Fig. 2B). As demonstrated, whereas immunoprecipitation of wild-type Rabenosyn-5 co-precipitated a high level of Vps45, immunoprecipitation of the mutant Rabenosyn-5 did not co-precipitate any detectable Vps45 (Fig. 2B, right panel, last lane). Since Rabenosyn-5 is targeted to phosphatidylinositol-3-phosphate enriched early endosomes through its FYVE domain, it was not surprising that the non-interacting Rabenosyn-5 mutant displayed a normal endosomal pattern when expressed in cells (Fig. 2C and D). The data confirm the two-hybrid based alanine screening and indicate that a stretch of residues between amino acids 100-110 contained amino acids that are required for the interaction of Rabenosyn-5 with Vps45. Although Vps45 is primarily comprised of a Sec1 domain, to identify the region within Vps45 required for its interaction with Rabenosyn-5, we took advantage of studies in yeast demonstrating that L117 of Vps45p is required for its interaction with the cognate t-SNARE, Tlg2p [13]. Accordingly, we generated a double amino acid substitution within the Sec1p-like domain of mammalian Vps45 (V107R and L110R), either of which might be the ‘true’ homologous amino acid of yeast L117. Although Rabenosyn-5 does not share significant homology with Tlg2p (or its mammalian ortholog, Syntaxin 16), double Vps45 V107R L110R mutations in the mammalian Vps45 protein caused a loss of its interaction with Rabenosyn-5 (Supplemental Fig. 1).
Fig. 2.
Amino acids 100, 101, 105-109 of Rabenosyn-5 are required for its interaction with hVps45. (A) The yeast strain AH109 was co-transformed with the following GAL4BD fusion constructs: GAL4BD-Vps45 and GAL4BD-p53 (control) together with the following GAL4AD fusion constructs: GAL4AD-R-5 50-200, GAL4AD-R-5 50-200 95-99 ala, GAL4AD-R-5 50-200 100, 101 ala, GAL4AD-R-5 50-200 102-104 ala, GAL4AD-R-5 50-200 105-109 ala, GAL4AD-R-5 50-200 110-114 ala, and GAL4AD-SV40 Large T-antigen (control). Co-transformants were assayed for their growth on non-selective (+HIS) and selective (−HIS) media. (B–D), HeLa cells were either grown on 35 mm dishes (B) or coverslips (C and D) and were co-transfected with either Myc-Vps45 and HA-R-5 or Myc-Vps45 and HA-R-5 100, 101, 105-109 ala. After 24 h, cells were harvested (B), lysed and in some cases, subjected to immunoprecipitations with antibody against the HA epitope. Lysates and immunoprecipitates were resolved by 8% SDS-PAGE, transferred to nitrocellulose, and immunoblotted with either anti-HA or anti-Myc antibodies followed by anti-mouse-HRP conjugated antibody. (C and D) transfected cells were fixed and immunostained with antibody against the HA epitope followed by 568-conjugated anti-mouse antibody. Bar, 10 μm.
hVps45 stabilizes Rabenosyn-5 in vivo
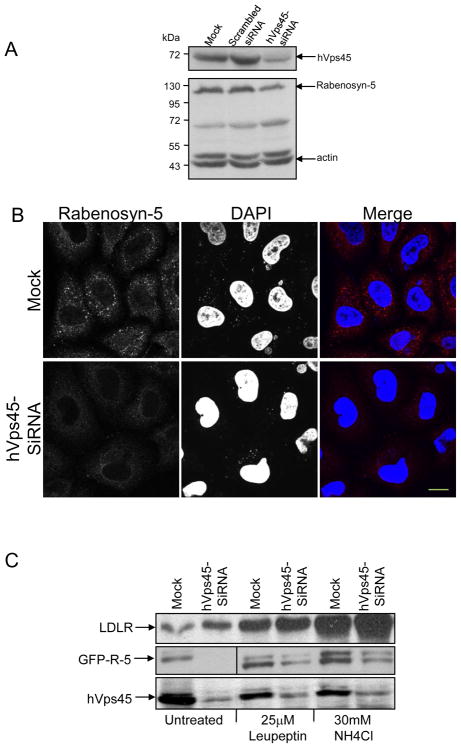
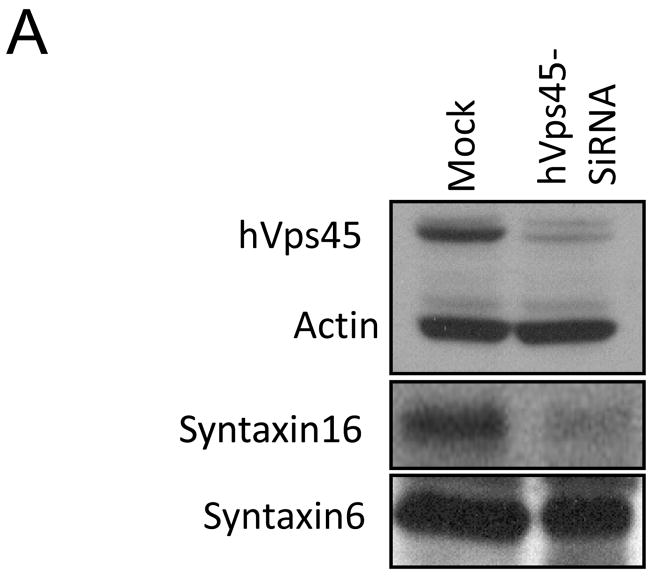
Having demonstrated binding between endogenous hVps45 and Rabenosyn-5, we next sought to determine the functional significance of this interaction. As demonstrated in Fig. 3A, oligonucleotides designed to deplete hVps45 levels (but not scrambled siRNA) were capable of reducing hVps45 levels by 80–90% in HeLa cells, with actin being used to monitor protein levels. In these partial hVps45-depleted cells, we observed that Rabenosyn-5 protein levels were typically ~50% lower than in Mock- or scrambled-siRNA-treated cells (Fig. 3A; lower panel). Indeed, following siRNA-depletion of hVps45, the level of endogenous Rabenosyn-5 observed by immunofluorescence was also considerably decreased (Fig. 3B). These results suggest that normal hVps45 expression levels are required for the stability of Rabenosyn-5.
Fig. 3.
Decreased expression of Rabenosyn-5 upon hVps45-depletion. (A) HeLa cells were either Mock-treated, treated with scrambled-siRNA, or treated with hVps45-siRNA for 72 h. Cells were harvested and lysates were resolved by 8% SDS-PAGE, transferred to nitrocellulose, and immunoblotted with anti-hVps45, anti-Rabenosyn-5, and anti-actin antibodies followed by anti-mouse-HRP conjugated and anti-rabbit-HRP conjugated antibodies. (B) Cells were grown on coverslips and were either Mock-treated (upper panels) or treated with hVps45-siRNA (lower panels) for 72 h. Cells were then fixed and immunostained with anti-Rabenosyn-5 antibody followed by 568 conjugated anti-mouse antibody and DAPI. (C) Cells were either Mock-treated or treated with hVps45-siRNA. After 48 h, cells were left untreated or subjected to the lysosomal inhibitors NH4Cl or leupeptin for 24 h. Cells were harvested, lysed, resolved by 8% SDS-PAGE, transferred to nitrocellulose, and immunoblotted with anti-Rabenosyn-5, anti-LDL receptor (LDL-R, control), anti-hVps45, and anti-actin (control) antibodies followed by anti-mouse-HRP and anti-rabbit-HRP conjugated antibodies. Bar, 10 μm.
Based on the interaction between hVps45 and Rabenosyn-5, we hypothesized that in the absence of sufficient levels of hVps45, Rabenosyn-5 is likely destabilized and undergoes degradation. To test whether depletion of hVps45 leads to degradation through the lysosomal pathway, HeLa cells were treated with hVps45-siRNA for 3 days. In the last 24 h, the lysosomal inhibitors leupeptin or ammonium chloride (NH4Cl) were added to the media. Control cells were left without inhibitors (Fig. 3C, untreated). When the lysosomal inhibitors were added to either Mock- or hVps45-siRNA-treated cells, we did not observe significant accumulation of Rabenosyn-5 compared to the untreated cells (Fig. 3C, middle panel). On the other hand, as a positive control, we did observe enhanced levels of LDL receptor (LDL-R) upon treatment with lysosomal inhibitors Fig. 3C, upper panel), suggesting that Rabenosyn-5 is not degraded through the lysosomal pathway upon reduction of hVps45 levels.
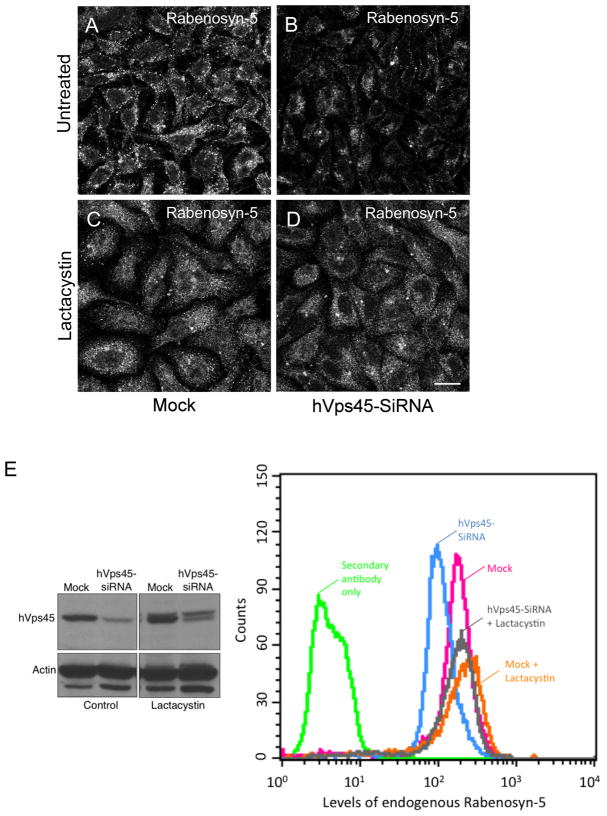
We next attempted to determine whether proteasomal degradation is responsible for the loss of Rabenosyn-5 upon depletion of hVps45, by using the proteasomal inhibitor lactacystin. As depicted in the micrographs in Fig. 4A–D, reduction of Rabenosyn-5 levels was evident in cells partially depleted of hVps45 (compare Fig. 4B and 4A). Upon treatment with the proteasomal inhibitor lactacystin, the level of Rabenosyn-5 expression was similar for either Mock- or hVps45-siRNA-treated cells (Fig. 4, compare D and C). However, immunoblotting analysis showed that whereas depletion of hVps45 was significant in untreated cells, the hVps45-siRNA depletion was considerably less marked in the lactacystin-treated cells (Fig. 4E, upper right panel). Indeed, quantitative analysis by flow cytometry indicated that total Rabenosyn-5 protein levels were returned to near normal levels upon lactacystin treatment (Fig. 4E, compare gray to blue peaks in graph). Accordingly, hVps45 itself appears to be targeted to the proteasomal degradation pathway, and its degradation is delayed under these conditions. Taken together, the stabilization of Rabenosyn-5 upon proteasomal inhibition likely resulted from: (i) the propensity of Rabenosyn-5 to undergo degradation primarily via the proteasome, and (ii) the rescue of hVps45 levels upon inhibition of its own degradation
Fig. 4.
Inhibition of proteasomal degradation stabilizes Rabenosyn-5 and hVps45 expression. HeLa cells were either grown on coverslips (A–D) or 35 mm dishes (E) and either Mock-treated (A, C, and E) or treated with hVps45-siRNA (B, D, and E). After 36 h, cells were either left untreated (control in A, B, and E) or subjected to treatment with 5 μM of the proteasomal inhibitor lactacystin (C, D, and E) for an additional 36 h. (A–D) Cells were fixed and immunostained with anti-Rabenosyn-5 antibody followed by 568-conjugated anti-rabbit antibody. (E) Cells were harvested, lysed, resolved by 8% SDS-PAGE, transferred to nitrocellulose, and immunoblotted with anti-hVps45 and anti-actin antibodies followed by anti-mouse HRP and anti-rabbit HRP conjugated antibodies (left panels), or trypsinized, fixed with 4% paraformaldyhide, immunostained with anti-Rabenosyn-5 antibody followed by 488-conjugated anti-rabbit antibody, and subjected to flow cytometry analysis (right panel). Bar, 10 μm.
Rabenosyn-5 and hVps45 regulate β1 integrin recycling and cell migration
In both C. elegans and D. melanogaster, it has been demonstrated that Vps45 plays multiple and complex regulatory roles, controlling the internalization of plasma membrane proteins and early endosome formation, respectively [15, 16]. In addition, Vps45 controls Rab5 activation in C. elegans [15]. Initially, we proposed to test the requirement of hVps45 and its interaction partner, Rabenosyn-5, for Rab5-induced endosome fusion (Supplemental Fig. 2). As depicted, although depletion of hVps45 and Rabenosyn-5 were ~80% and ~90% effective respectively, (Supplemental Figure 2D), the loss of either protein had no bearing on the ability of the ‘GTP-locked’ active Rab5 mutant (Q79L) to induce hyperfused early endosomes (Supplemental Figure 2; compare B and C with A). Indeed, membrane fractionation experiments showed that the ratio of active, membrane-bound Rab5 to inactive, cytosolic Rab5 remained similar upon hVps45-depletion. (Supplemental Figure 2E). Moreover, no change in the distribution of the early endosomal protein EEA1 was elicited upon hVps45-depletion (Supplemental Figure 2F and G). This implies that in mammalian cells, the primary roles of hVps45 may lie downstream of Rab5 function at the early endosome.
It has been previously demonstrated that Arf6 regulates early endocytic internalization events including macropinocytosis [19] as well as recycling of receptors internalized through clathrin-independent mechanisms [19, 20]. To determine whether early fusion events of newly incoming vesicles supported by Arf6 are affected by depletion of hVps45, we transfected the HA-Arf6 Q67L (GTP-hydrolysis resistant mutant) to Mock-treated and hVps45-siRNA-treated cells. This constitutively active mutant of Arf6 elicits rigorous fusion of newly internalized vesicles, forming large clusters of fused membranes [20]. As shown, vacuoles were formed that contained HA-Arf6 Q67L in both Mock-treated and hVps45-siRNA-treated cells (Supplemental Fig. 2H and I, respectively), suggesting that hVps45 is not involved in the formation of Arf6-containing membrane structures in mammalian cells.
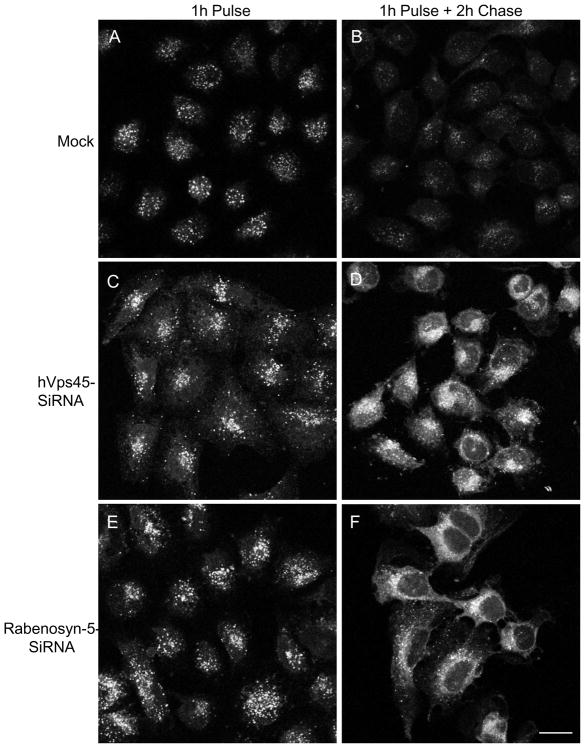
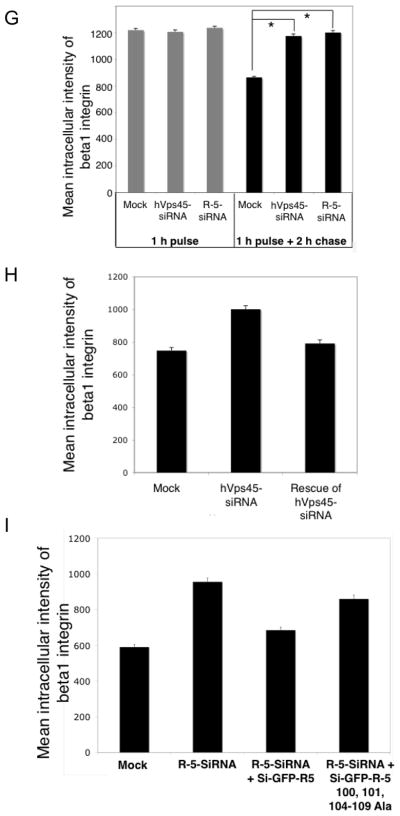
To assess the role of hVps45 in post-early endosomal trafficking pathways, we followed the itinerary of β1 integrins (Fig. 5), a protein that we have previously shown is regulated by the Rabenosyn-5 interaction partner, EHD1 [7, 10]. To this aim, we first pulsed Mock-treated HeLa cells (Fig. 5A), hVps45-siRNA-treated cells (Fig. 5C), and Rabenosyn-5-siRNA-treated cells (Fig. 5E) with a β1 integrin antibody for 1 hour at 37°C (followed by stripping of non-internalized antibody) and monitored the level of internalized β1 integrins by confocal microscopy. During the course of internalization, no significant differences in uptake were observed (Fig. 5A, C, E, and quantified in G), further decreasing the likelihood of hVps45 involvement in the process of internalization. To assess the recycling of β1 integrins, following the internalization period, the cells were ‘chased’ in complete media for 2 h to allow recycling of internalized integrins (Fig. 5B, D, and F). Following the chase period, anti-β1 integrin antibodies were stripped from integrins that returned to the cell surface by a brief acid strip rendering the cells exclusively with an intracellular pool of β1 integrin-antibody complexes. In Mock-treated cells, the β1 integrin signal remaining within the cells was significantly reduced compared to the pulse alone, indicating that it recycled back to the plasma membrane (Fig. 5B). However, upon depletion of hVps45 (Fig. 5D) or Rabenosyn-5 (Fig. 5F), we observed accumulation of internalized β1 integrins within endocytic compartments (quantified in Fig. 5G).
Fig. 5.
hVps45 and Rabenosyn-5 regulate β1 integrin recycling. HeLa cells were grown on coverslips and either Mock-treated (A and B), treated with hVps45-siRNA (C and D), or treated with Rabenosyn-5-siRNA (E and F) for 48 h. Cells were incubated in starvation media (DMEM + 0.5% BSA) for 1 h, “pulsed” with anti-β1 integrin antibody in starvation media for 1 h at 37°C (A, C, and E), and “chased” in complete media (DMEM + 10% fetal calf serum) for 2 h, and then acid-stripped to remove β1 integrin that had returned to the plasma membrane (B, D, and F). After fixation and permeabilization, the cells were immunostained with 568-conjugated anti-mouse antibody. (G) Quantification of the experiments performed in A–F. Images of 150 cells from each treatment were taken using a Zeiss LSM5 PASCAL confocal microscope, and the mean intracellular signal intensity of non-recycled β1 integrin was analyzed by using LSM5 PASCAL software. (H) Cells were grown on coverslips and either Mock-treated or treated with hVps45-siRNA. After 6 h, the cells were transfected with a GFP-Vps45 construct resistant to hVps45-siRNA oligonucleotides. 66 h later, cells were incubated in starvation media for 1 h, “pulsed” with β1 integrin antibody in starvation media for 1 h at 37°C, “chased” in complete media for 3 h, stripped, fixed, and immunostained with 568 conjugated anti-mouse antibody. Images of cells from each treatment (180 cells in H) were analyzed. (I) Cells were grown on coverslips and either Mock-treated or treated with Rabenosyn-5-siRNA. After 24 h, cells were transfected with either siRNA-resistant GFP-Rabenosyn-5 (si-GFP-R-5) or the siRNA-resistant Rabenosyn-5 mutant that is incapable of Vps45-binding (si-GFP-R-5 100, 101, 104-109 ala), for an additional 48 h. Then, cells were subjected to β1 integrin recycling assay as described in H and images of cells from each treatment (150 cells) were analyzed. Error bars display standard error and asterisks denote p-values less than 0.01 utilizing the t-Test for independent samples (in G). Results were derived from 3 independent experiments. Bar, 10 μm.
To highlight the specificity of hVps45-siRNA on β1 integrin recycling, we designed an siRNA-resistant wild-type GFP-Vps45 construct. Utilizing the same β1 integrin recycling assay, we again demonstrated that loss of hVps45 caused an accumulation of β1 integrins in the cells (Fig. 5H). However, upon transfection of the hVps45-siRNA-treated cells with the siRNA-resistant wild-type GFP-Vps45 construct, recycling was rescued and we observed a marked (and statistically significant) decrease in the level of internalized β1 integrins that accumulated within the cell, further illustrating that hVps45 regulates β1 integrin recycling.
To analyze the significance of Rabenosyn-5/Vps45 interactions for β1 integrin recycling, we re-introduced either wild-type Rabenosyn-5 or the Vps45-binding deficient rabenosyn-5 mutant back into cells depleted of endogenous Rabenosyn-5 and then analyzed β1 integrin recycling (Fig. 5I). Whereas the wild-type Rabenosyn-5 significantly expedited recycling to near normal levels, the mutant Rabenosyn-5 displayed significantly less ability to ‘rescue’ the recycling defect. These data suggest that the interaction between Rabenosyn-5 and Vps45 are necessary for optimal β1 integrin transport to the endocytic recycling compartment and return to the plasma membrane.
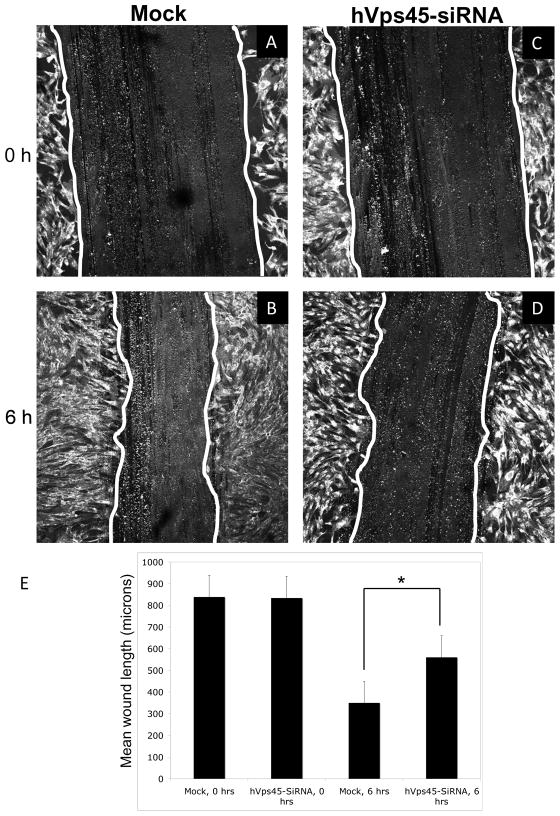
We have previously demonstrated that delayed recycling of β1 integrin results in impaired cell motility upon depletion of the endocytic regulatory protein, EHD1 [10]. We therefore hypothesized that hVps45 might also indirectly regulate cell motility, since its depletion leads to delayed recycling of β1 integrin. To this aim, human foreskin fibroblast cells grown on a confluent monolayer on fibronectin-coated cover-slides were treated with Mock- or hVps45-siRNA oligonucleotides for 48 h, and a ~850 μm wound (scratch) was applied to the monolayer (Fig. 6A and C). After 6 h, the Mock-treated cells migrated significantly more towards the wounded area compared to the hVps45-siRNA-treated cells (Fig. 6; compare B to D; quantified in E). To validate these results and ensure that siRNA-treated cells do not display altered levels of quiescent cells that might affect motility/migration, we performed cell cycle analysis for Mock- and SiRNA-treated cells (Supplemental Fig. 3A–C). As shown, the percentages of cells in G1, G2 and S phase for each treatment were virtually identical. Collectively, these results indicate that hVps45-depletion leads to impaired integrin recycling, and as a consequence, decreased cell migration.
Fig. 6.
hVps45 regulates cell migration. (A–E) Human foreskin fibroblast cells were grown on coverslips coated with fibronectin. Cells were either Mock-treated or treated with hVps45-siRNA for 48h. Scratches (wounds) approximately 850 μm wide were introduced onto the coverslips using a pipet tip. Fields of cells were fixed immediately after introducing the scratch (A, C), whereas the cells shown in B and D were fixed 6 h later. Cells were immunostained with 488-conjugated anti-wheat germ agglutinin antibody, which marked the outlines of the cells. (E) Quantification of three independent experiments as performed in A–D. Error bars display standard error and asterisk denotes p-values less than 0.05.
hVps45 regulates trafficking between the Golgi and early endosomes
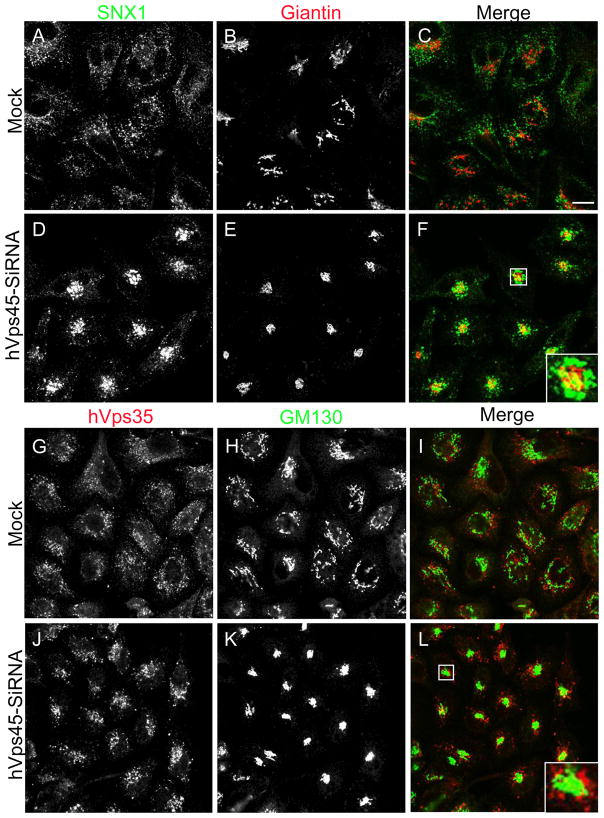
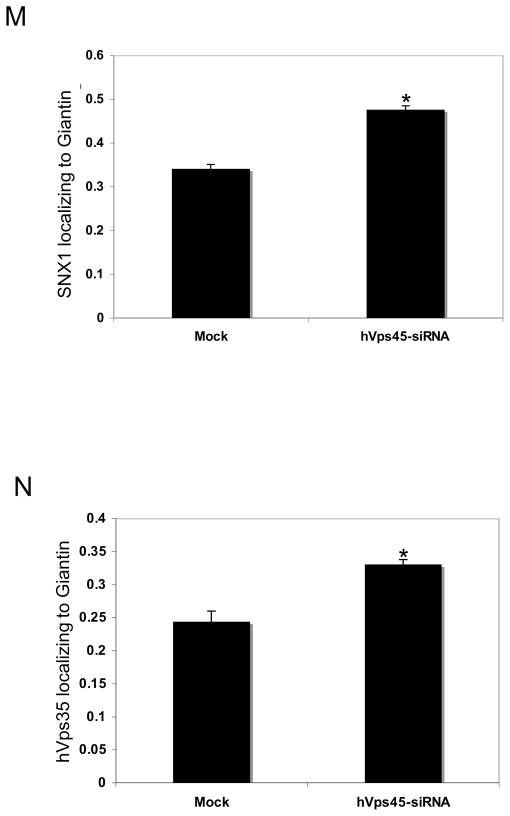
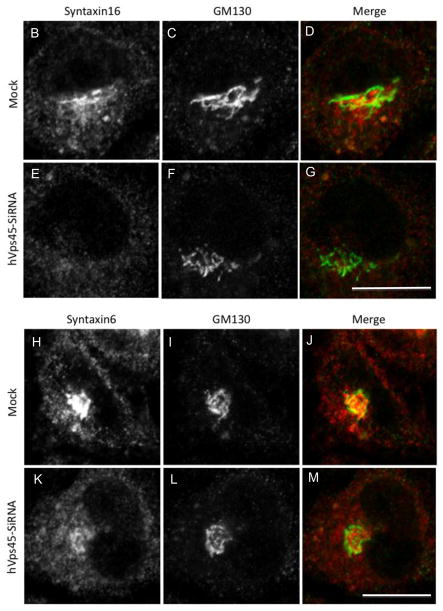
In yeast, another role of Vps45p is regulating the transport of carboxypeptidase Y (CPY) from the Golgi to the pre-vacuolar compartment [21]. We thus aimed to determine whether this function is conserved in mammalian cells. Accordingly, we chose to monitor the subcellular localization of retromer subunit proteins SNX1 and hVps35, which normally cycle between endosomes and the Golgi [22]. We observed that depletion of hVps45 induced the accumulation of vesicles containing both SNX1 (Fig. 7A–F, see inset; quantified in M) and hVps35 (Fig. 7G–L, see inset; quantified in N) adjacent to the Golgi region (stained with Giantin or GM130), but not coinciding with the Golgi apparatus. Moreover, the absence of hVps45 had a dramatic effect on Golgi morphology, causing it to become condensed into a tightly packed structure (Fig. 7; compare E to B and K to H). These affects might in part be due to the reduction in expression levels of Syntaxin16 (Fig. 8A) and the more diffused subcellular localization of both Syntaxin16 (Fig. 8B–G) and Syntaxin6 (Figure 8H–M), both of which interact with Vps45 [23–26]. These results provide an additional role for hVps45 in regulating vesicular trafficking between the Golgi and early endosomes, both dependently and independently of Rabenosyn5, thereby affecting Golgi morphology.
Fig. 7.
Depletion of hVps45 affects retromer localization. HeLa cells were grown on coverslips and were Mock-treated or treated with hVps45-siRNA for 72 hours prior to fixation. Cells were then co-immunostained with anti-SNX1 and anti-Giantin (A–F and M), and with anti-hVps35 and anti-GM130 (G–L and N) followed by 568 conjugated anti-mouse and 488 conjugated anti-rabbit or 488 conjugated anti-goat antibodies. (M and N) Co-localization between SNX1 and Giantin (M), and between hVps35 and GM130 (N), respectively, were quantified using ImageJ software. Error bars display standard error and asterisks denote p-values less than 0.01 utilizing the t-Test for independent samples. Results were derived from 3 independent experiments by counting 100 cells from each experiment. Insets (F and L) depict the co-staining in a single cell. Bar, 10 μm.
Fig. 8.
hVps45-depletion affects the subcellular distribution of both Syntaxin6 and Syntaxin16 and reduces Syntaxin16 protein levels. (A) Cells were either Mock-treated or treated with hVps45-siRNA for 72 h, harvested, lysed, resolved by 8% SDS-PAGE, immunoblotted with either anti-hVps45, anti-actin, anti-Syntaxin6 or anti-Syntaxin16 antibodies followed by appropriate HRP-conjugated antibodies. (B–M) Cells were grown on coverslips and either Mock-treated (B–D and H–J) or treated with hVps45-siRNA (E–G and K–M) for 72 h. Cells were then immunostained with either Syntaxin16 and GM130 antibodies (B–G) or Syntaxin6 and GM130 antibodies (H–M) followed by 568-conjugated goat anti-mouse and 488-conjugated goat anti-rabbit antibodies.
Discussion
Vesicle docking and fusion events involve several families of conserved proteins, such as Rab proteins, their molecular effectors, SM-family and SNARE proteins. It has been demonstrated in yeast that one member of the SM protein family, Vps45p, is involved in the transport of CPY hydrolase from the Golgi to the pre-vacuolar compartment or early endosomes [11, 27]. This occurs via the Vps45p interaction with Vac1p [11] and also with its cognate SNAREs, Tlg2p and Pep12p [13, 14, 23, 28–30]. Loss of Vps45p function leads to accumulation of vesicles adjacent to the vacuole [31]. This indicates that Vps45p functions at a vesicle docking and/or fusion event. This is consistent with recent reports indicating that the Arabidopsis thaliana ortholog, AtVps45, functions in transport to the vacuole [32]. In apparent contrast, in C. elegans, Vps45 is essential for receptor-mediated and fluid-phase endocytosis pathways and its interaction with Rabenosyn-5 is required for Rab5-mediated endosomal fusion [15]. D. melanogaster Vps45, Rabenosyn-5, Rab5, and Avl (an ortholog of the SNARE Syntaxin7) are needed for endosome formation as well [16], indicating that Vps45 plays multiple roles in regulating endocytic and secretory pathways.
Although in HeLa cells hVps45 cycles on and off membranes during vesicle transport [30], overexpression of Vps45 in Chinese hamster ovary (CHO) cells displays a Golgi-like pattern [33]. It binds specifically to its cognate t-SNAREs, Syntaxin6 and Syntaxin16 [23, 33], the mammalian orthologs of Pep12p and Tlg2p, respectively. In addition, Nielsen and coworkers demonstrated that hVps45 is also capable of interacting directly with the early endosomal Rab4/5 effector, Rabenosyn-5, suggesting the evolution of multiple and complex functions for this SM protein [5]. However, the nature and functional roles of the Vps45-Rabenosyn-5 interaction in mammalian cells remains largely unexplored to date.
In this study, we have: (1) identified a unique region of Rabenosyn-5 that specifically interacts with Vps45 in mammalian cells, (2) demonstrated that Vps45 depletion leads to proteasome-mediated Rabenosyn-5 degradation, reduced expression of Syntaxin16, and altered subcellular localization of Syntaxin6 and Syntaxin16, (3) shown that both Rabenosyn-5 and Vps45 are required for recycling of β1 integrins to the plasma membrane, and that the Vps45-Rabenosyn-5 interaction is necessary for optimal recycling to occur. In addition to its role in recycling, Vps45 is also involved in the endosome-to-Golgi retromer-based trafficking pathway.
In the course of mapping the Rabenosyn-5 region required for interaction with hVps45, we identified an important stretch of ~50 residues (70–120) required for binding. By a systematic mutational approach, we identified a series of residues between amino acids 100-110 that are necessary for binding to Vps45. This region was highly conserved in other species, displaying 82%, 64% and 54% homology in chicken, zebrafish and yeast, respectively. Indeed, despite showing 54% homology to a sequence in the yeast protein, the overall level of homology of yeast Vac1 (the Rabenosyn-5 homolog) to human Rabenosyn-5 was only ~20%, indicating that this binding region is selectively conserved throughout evolution. Although this region did not yield close homology to the sequence of any known human protein, it did show a loose homology with a region of Vps52, a member of the mammalian Golgi-associated retrograde protein (GARP) complex. However, attempts to immunoprecipitate Vps52 with Vps45 were unsuccessful (data not shown).
Vps45, on the other hand, is comprised largely of a Sec1 domain. Substituting amino acids V107 and L110 of Vps45 to arginine residues induced a loss of interaction with Rabenosyn-5. Alignment of hVps45 with yeast Vps45p showed that these two hydrophobic amino acids are each potentially homologous to amino acid L117 of yeast Vps45p, which is positioned within a hydrophobic pocket of the Sec1 domain. Indeed, mutating amino acid L117 of the yeast Vps45p to arginine abolished its interaction with monomeric Tlg2p [13]. While the mode of interaction between Vps45 and Tlg2p may be similar to that of Vps45 and Rabenosyn-5, we were unable to identify a conserved motif between Rabenosyn-5 and Tlg2p (or its mammalian ortholog, Syntaxin16). Since the same region of Vps45 is capable of binding both Rabenosyn-5 and Syntaxin16, we speculate that Vps45 binds each protein sequentially, although we cannot rule out the formation of oligomeric complexes that enable Vps45 to interact with both proteins simultaneously.
One intriguing finding is that hVps45 regulates the protein level of Rabenosyn-5, as depletion of hVps45 induces reduced the level of Rabenosyn-5 as observed by immunoblot and immunofluorescence analysis. It has previously been demonstrated that loss of functional Vps45p leads to rapid degradation of Tlg2p through the proteasomal pathway in yeast [30], and loss of the Arabidopsis thaliana AtVps45 protein similarly destabilizes a complex containing Syp41, a plant ortholog of Tlg2p [32]. Therefore we hypothesized that depletion of hVps45 might similarly destabilize Rabenosyn-5 at a post-translational level. Indeed, when we applied lysosomal inhibitors to cells depleted for hVps45, we did not observe an accumulation of Rabenosyn-5. On the other hand, treatment of cells with a proteasomal inhibitor, lactacystin, led to increased protein levels of both Rabenosyn-5 and hVps45. As expression of both interaction partners is enhanced upon proteasomal inhibition, we cannot discern whether the lactacystin directly prevents Rabenosyn-5 degradation, or does so by preventing degradation of hVps45. However, the presence of a ubiquitin interacting motif in Rabenosyn-5 could signal its degradation via the proteasome.
Another intriguing finding from this study is that unlike its C. elegans and D. melanogaster homologs, hVps45 does not appear to be involved in internalization and early endosome formation. As shown in Supplemental Figure 2, both hVps45 and Rabenosyn-5 do not affect Rab5 function in endosomal fusion and depletion of hVps45 does not significantly alter the membrane localization of Rab5, suggesting that hVps45 may function in a pathway(s) downstream of the formation of early endosomes. To further investigate this possibility, we examined whether hVps45 and its interacting partner, Rabenosyn-5, are involved in the regulation of receptor recycling back to the plasma membrane. We observed that β1 integrin receptors accumulate in early endosomes and in the vicinity of the recycling compartment upon depletion of Rabenosyn-5 and hVps45, respectively. The difference in the subcellular point of accumulation of β1 integrin receptors between cells lacking Rabenosyn-5 and hVps45 likely reflects subtle differences in the function of these two endocytic regulatory proteins. Rabenosyn-5, as a Rab5 effector, mediates specificity of vesicle targeting, while hVps45, after recruitment by its cognate t-SNARE to the target membrane, functions in vesicle-docking events. Accordingly, the interaction between Rabenosyn-5 and hVps45 is required for vesicle docking and subsequently, vesicle fusion. Indeed, upon Rabenosyn-5-depletion, rescue expriments with wild-type Rabensyn-5 restore normal β1 integrin recycling. However, re-introduction of a mutant Rabenopsyn-5 that is incapable of interacting with Vps45 does not similarly restore recycling capacity, highlighting the significance of the interaction between the two proteins.
Although Vps45-depletion causes a loss of 50% of the Rabenosyn-5 expression, it is unlikely that this relatively modest reduction significantly impacts β1 integrin receptor recycling. Indeed, cells treated with hVps45-siRNA did not display the pattern of β1 integrin receptor accumulation typically observed in peripheral endosomes upon direct and efficient Rabenosyn-5 depletion. However, the delay in β1 integrin receptor recycling observed upon hVps45-depletion led to impaired cell motility, consistent with observations for depletion of EHD1, another regulator of β1 integrin recycling [10].
Our observations also describe a function for hVps45 in vesicle docking at the Golgi. hVps45 depletion causes accumulation of vesicles containing the retromer complex in the vicinity of the Golgi area [32]. Consistent with these findings, the plant AtVps45 also affects retromer localization and pre-vacuolar compartment-to-Golgi transport [32]. In contrast, depletion of Rabenosyn-5 leads to accumulation of the retromer complex in or at early endosomes [34]. This likely reflects differences in the primary sites of function of these two proteins. Rabenosyn-5 mediates delivery of vesicles containing the retromer complex to the Golgi from early endosomes, while hVps45, recruited to the Golgi membrane by its cognate t-SNARE (possibly Syntaxin6 or Syntaxin16), interacts with Rabenosyn-5 during the docking event. Differences in the functions of Rabenosyn-5 and hVps45 may also indirectly influence the morphology of the Golgi. In cells treated with Rabenosyn-5-siRNA, the Golgi became more fragmented and dispersed compared to the Mock-treated cells [34]. However, in cells depleted for hVps45, the Golgi appeared more compact rather than dispersed. While endosome-to-Golgi and retromer transport of Golgi constituents is clearly implicated in these phenomena, they remain poorly understood.
Overall, we demonstrate that the interaction between hVps45 and Rabenosyn-5 plays an important role in multiple trafficking pathways. Both proteins are required for β1 integrin receptor recycling as well as trafficking of vesicles between the Golgi and early endosomes. Although the effects of depleting either hVps45 or Rabenosyn-5 are somewhat different due to their distinctive but overlapping functions, their interaction is required for the same process, which is vesicle docking and subsequently, vesicle fusion.
Supplementary Material
SUPPLEMENTAL FIG. 1- Amino acids 107 and 110 of Vps45 are required for its interaction with Rabenosyn-5. The yeast strain AH109 was co-transformed with the following GAL4 binding domain (GAL4BD) fusion constructs: GAL4BD-Vps45, GAL4BD-Vps45 V107R L110R (val 107 to arg and leu 110 to arg), and GAL4BD-p53 (control) together with the following GAL4 transcription activation domain (GAL4AD) fusion constructs: GAL4AD-R-5 and GAL4AD-SV40 Large T-antigen (control). Co-transformants were assayed for their growth on non-selective (+HIS) and selective (−HIS) media.
SUPPLEMENTAL FIG. 2- hVps45 acts downstream of Rab5. HeLa cells were either grown on coverslips (A–C, F–I) or on 35 mm dishes (D and E). They were Mock-treated (A, D, E, F and H), treated with hVps45-siRNA (B, D, E, G and I), or Rabenosyn-5-siRNA (C and D). After 24 h, cells were transfected with either GFP-Rab5 Q79L (A–C), or HA-Arf6 Q67L (H and I) for 48 h prior to fixation (A–C, F–I) or harvesting (D and E). D, cells were lysed, resolved by 8% SDS-PAGE, and immunoblotted with anti-Rabenosyn-5, anti-hVps45, and anti-actin antibodies followed by appropriate HRP-conjugated antibodies. (E) Cells were homogenized in detergent-free homogenization buffer and subjected to ultracentrifugation to separate the membrane pellets and the cytosolic fractions. Both fractions were then resolved by 10% SDS-PAGE, and immunoblotted with anti-Rab5, anti-transferrin receptor, and anti-γ-tubulin antibodies followed by anti-mouse HRP conjugated antibodies. (A–C, F–I) Cells were fixed and immunostained with anti-EEA1 antibody (F and G), or antibody against the HA-epitope (H and I) followed by 568-conjugated anti-mouse antibody (F–I). Bars, 10 μm.
SUPPLEMENTAL FIG. 3- Depletion of hVps45 does not alter cell cycle. Human foreskin fibroblasts were either Mock-treated (A) or treated with hVps45-siRNA (B) for 72 h, fixed with 70% ethanol, stained with Telford reagent, and subjected to flow cytometry assay. The percentage of cells in diploid G1 (Dip G1), diploid G2 (Dip G2), and diploid S (Dip S) are shown in C.
Acknowledgments
This work was supported by National Institutes of Health Grants GM074876 (SC) and P20 RR16469 from the National Center for Research Resources (NN). We are grateful to Dr. P. Mehta for his discussions. We also thank Dr. J. Donaldson and Dr. R. Lodge for kindly providing constructs.
Abbreviations
- SNARE
soluble N-ethylmaleimide-sensitive factor attachment protein receptors
- Vps
vacuole protein sorting
- SiRNA
silencing RNA
- PtdIns
phosphatidylinositol
- PtdIns3P
phosphatidylinositol 3-phosphate
- GST
glutathione S-transferase
- HRP
horseradish peroxidase
- GTP
guanine triphosphate
- BSA
bovine serum albumin
- Arf
ADP ribosylation factor
- SNX
sorting nexin
- MHC I
major histocompatibility complex class I
- PtdIns
phosphatidylinositol
- SM
Sec1p/Munc18-like
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Maxfield FR, McGraw TE. Endocytic recycling. Nat Rev Mol Cell Biol. 2004;5:121–132. doi: 10.1038/nrm1315. [DOI] [PubMed] [Google Scholar]
- 2.Grosshans BL, Ortiz D, Novick P. Rabs and their effectors: achieving specificity in membrane traffic. Proc Natl Acad Sci U S A. 2006;103:11821–11827. doi: 10.1073/pnas.0601617103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McBride HM, Rybin V, Murphy C, Giner A, Teasdale R, Zerial M. Oligomeric complexes link Rab5 effectors with NSF and drive membrane fusion via interactions between EEA1 and syntaxin 13. Cell. 1999;98:377–386. doi: 10.1016/s0092-8674(00)81966-2. [DOI] [PubMed] [Google Scholar]
- 4.Deneka M, van der Sluijs P. ‘Rab’ing up endosomal membrane transport. Nat Cell Biol. 2002;4:E33–35. doi: 10.1038/ncb0202-e33. [DOI] [PubMed] [Google Scholar]
- 5.Nielsen E, Christoforidis S, Uttenweiler-Joseph S, Miaczynska M, Dewitte F, Wilm M, Hoflack B, Zerial M. Rabenosyn-5, a novel Rab5 effector, is complexed with hVPS45 and recruited to endosomes through a FYVE finger domain. J Cell Biol. 2000;151:601–612. doi: 10.1083/jcb.151.3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eathiraj S, Pan X, Ritacco C, Lambright DG. Structural basis of family-wide Rab GTPase recognition by rabenosyn-5. Nature. 2005;436:415–419. doi: 10.1038/nature03798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Naslavsky N, Boehm M, Backlund PS, Jr, Caplan S. Rabenosyn-5 and EHD1 Interact and Sequentially Regulate Protein Recycling to the Plasma Membrane. Mol Biol Cell. 2004;15:2410–2422. doi: 10.1091/mbc.E03-10-0733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burd CG, Emr SD. Phosphatidylinositol(3)-phosphate signaling mediated by specific binding to RING FYVE domains. Mol Cell. 1998;2:157–162. doi: 10.1016/s1097-2765(00)80125-2. [DOI] [PubMed] [Google Scholar]
- 9.Grant BD, Caplan S. Mechanisms of EHD/RME-1 Protein Function in Endocytic Transport. Traffic. 2008 doi: 10.1111/j.1600-0854.2008.00834.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jovic M, Naslavsky N, Rapaport D, Horowitz M, Caplan S. EHD1 regulates beta1 integrin endosomal transport: effects on focal adhesions, cell spreading and migration. J Cell Sci. 2007;120:802–814. doi: 10.1242/jcs.03383. [DOI] [PubMed] [Google Scholar]
- 11.Tall GG, Hama H, DeWald DB, Horazdovsky BF. The phosphatidylinositol 3-phosphate binding protein Vac1p interacts with a Rab GTPase and a Sec1p homologue to facilitate vesicle-mediated vacuolar protein sorting. Mol Biol Cell. 1999;10:1873–1889. doi: 10.1091/mbc.10.6.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Toonen RF, Verhage M. Vesicle trafficking: pleasure and pain from SM genes. Trends Cell Biol. 2003;13:177–186. doi: 10.1016/s0962-8924(03)00031-x. [DOI] [PubMed] [Google Scholar]
- 13.Carpp LN, Ciufo LF, Shanks SG, Boyd A, Bryant NJ. The Sec1p/Munc18 protein Vps45p binds its cognate SNARE proteins via two distinct modes. J Cell Biol. 2006;173:927–936. doi: 10.1083/jcb.200512024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bryant NJ, James DE. The Sec1p/Munc18 (SM) protein, Vps45p, cycles on and off membranes during vesicle transport. J Cell Biol. 2003;161:691–696. doi: 10.1083/jcb.200212078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gengyo-Ando K, Kuroyanagi H, Kobayashi T, Murate M, Fujimoto K, Okabe S, Mitani S. The SM protein VPS-45 is required for RAB-5-dependent endocytic transport in Caenorhabditis elegans. EMBO Rep. 2007;8:152–157. doi: 10.1038/sj.embor.7400882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morrison HA, Dionne H, Rusten TE, Brech A, Fisher WW, Pfeiffer BD, Celniker SE, Stenmark H, Bilder D. Regulation of early endosomal entry by the Drosophila tumor suppressors Rabenosyn and Vps45. Mol Biol Cell. 2008;19:4167–4176. doi: 10.1091/mbc.E08-07-0716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sharma M, Naslavsky N, Caplan S. A role for EHD4 in the regulation of early endosomal transport. Traffic. 2008;9:995–1018. doi: 10.1111/j.1600-0854.2008.00732.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Naslavsky N, Rahajeng J, Sharma M, Jovic M, Caplan S. Interactions between EHD Proteins and Rab11-FIP2: A Role for EHD3 in Early Endosomal Transport. Mol Biol Cell. 2006;17:163–177. doi: 10.1091/mbc.E05-05-0466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Radhakrishna H, Donaldson JG. ADP-ribosylation factor 6 regulates a novel plasma membrane recycling pathway. J Cell Biol. 1997;139:49–61. doi: 10.1083/jcb.139.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brown FD, Rozelle AL, Yin HL, Balla T, Donaldson JG. Phosphatidylinositol 4,5-bisphosphate and Arf6-regulated membrane traffic. J Cell Biol. 2001;154:1007–1017. doi: 10.1083/jcb.200103107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abeliovich H, Darsow T, Emr SD. Cytoplasm to vacuole trafficking of aminopeptidase I requires a t-SNARE-Sec1p complex composed of Tlg2p and Vps45p. EMBO J. 1999;18:6005–6016. doi: 10.1093/emboj/18.21.6005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seaman MN, McCaffery JM, Emr SD. A membrane coat complex essential for endosome-to-Golgi retrograde transport in yeast. J Cell Biol. 1998;142:665–681. doi: 10.1083/jcb.142.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dulubova I, Yamaguchi T, Gao Y, Min SW, Huryeva I, Sudhof TC, Rizo J. How Tlg2p/syntaxin 16 ‘snares’ Vps45. EMBO J. 2002;21:3620–3631. doi: 10.1093/emboj/cdf381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simonsen A, Bremnes B, Ronning E, Aasland R, Stenmark H. Syntaxin-16, a putative Golgi t-SNARE. Eur J Cell Biol. 1998;75:223–231. doi: 10.1016/S0171-9335(98)80116-7. [DOI] [PubMed] [Google Scholar]
- 25.Xu H, Boulianne GL, Trimble WS. Drosophila syntaxin 16 is a Q-SNARE implicated in Golgi dynamics. J Cell Sci. 2002;115:4447–4455. doi: 10.1242/jcs.00139. [DOI] [PubMed] [Google Scholar]
- 26.Bock JB, Klumperman J, Davanger S, Scheller RH. Syntaxin 6 functions in trans-Golgi network vesicle trafficking. Mol Biol Cell. 1997;8:1261–1271. doi: 10.1091/mbc.8.7.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bryant NJ, Piper RC, Gerrard SR, Stevens TH. Traffic into the prevacuolar/endosomal compartment of Saccharomyces cerevisiae: a VPS45-dependent intracellular route and a VPS45-independent, endocytic route. Eur J Cell Biol. 1998;76:43–52. doi: 10.1016/S0171-9335(98)80016-2. [DOI] [PubMed] [Google Scholar]
- 28.Nichols BJ, Holthuis JC, Pelham HR. The Sec1p homologue Vps45p binds to the syntaxin Tlg2p. Eur J Cell Biol. 1998;77:263–268. doi: 10.1016/s0171-9335(98)80084-8. [DOI] [PubMed] [Google Scholar]
- 29.Gerrard SR, Levi BP, Stevens TH. Pep12p is a multifunctional yeast syntaxin that controls entry of biosynthetic, endocytic and retrograde traffic into the prevacuolar compartment. Traffic. 2000;1:259–269. doi: 10.1034/j.1600-0854.2000.010308.x. [DOI] [PubMed] [Google Scholar]
- 30.Bryant NJ, James DE. Vps45p stabilizes the syntaxin homologue Tlg2p and positively regulates SNARE complex formation. Embo J. 2001;20:3380–3388. doi: 10.1093/emboj/20.13.3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cowles CR, Emr SD, Horazdovsky BF. Mutations in the VPS45 gene, a SEC1 homologue, result in vacuolar protein sorting defects and accumulation of membrane vesicles. J Cell Sci. 1994;107(Pt 12):3449–3459. doi: 10.1242/jcs.107.12.3449. [DOI] [PubMed] [Google Scholar]
- 32.Zouhar J, Rojo E, Bassham DC. AtVPS45 Is a Positive Regulator of the SYP41/SYP61/VTI12 SNARE Complex Involved in Trafficking of Vacuolar Cargo. Plant Physiol. 2009;149:1668–1678. doi: 10.1104/pp.108.134361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tellam JT, James DE, Stevens TH, Piper RC. Identification of a mammalian Golgi Sec1p-like protein, mVps45. J Biol Chem. 1997;272:6187–6193. doi: 10.1074/jbc.272.10.6187. [DOI] [PubMed] [Google Scholar]
- 34.Naslavsky N, McKenzie J, Altan-Bonnet N, Sheff D, Caplan S. EHD3 regulates early-endosome-to-Golgi transport and preserves Golgi morphology. J Cell Sci. 2009;122:389–400. doi: 10.1242/jcs.037051. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SUPPLEMENTAL FIG. 1- Amino acids 107 and 110 of Vps45 are required for its interaction with Rabenosyn-5. The yeast strain AH109 was co-transformed with the following GAL4 binding domain (GAL4BD) fusion constructs: GAL4BD-Vps45, GAL4BD-Vps45 V107R L110R (val 107 to arg and leu 110 to arg), and GAL4BD-p53 (control) together with the following GAL4 transcription activation domain (GAL4AD) fusion constructs: GAL4AD-R-5 and GAL4AD-SV40 Large T-antigen (control). Co-transformants were assayed for their growth on non-selective (+HIS) and selective (−HIS) media.
SUPPLEMENTAL FIG. 2- hVps45 acts downstream of Rab5. HeLa cells were either grown on coverslips (A–C, F–I) or on 35 mm dishes (D and E). They were Mock-treated (A, D, E, F and H), treated with hVps45-siRNA (B, D, E, G and I), or Rabenosyn-5-siRNA (C and D). After 24 h, cells were transfected with either GFP-Rab5 Q79L (A–C), or HA-Arf6 Q67L (H and I) for 48 h prior to fixation (A–C, F–I) or harvesting (D and E). D, cells were lysed, resolved by 8% SDS-PAGE, and immunoblotted with anti-Rabenosyn-5, anti-hVps45, and anti-actin antibodies followed by appropriate HRP-conjugated antibodies. (E) Cells were homogenized in detergent-free homogenization buffer and subjected to ultracentrifugation to separate the membrane pellets and the cytosolic fractions. Both fractions were then resolved by 10% SDS-PAGE, and immunoblotted with anti-Rab5, anti-transferrin receptor, and anti-γ-tubulin antibodies followed by anti-mouse HRP conjugated antibodies. (A–C, F–I) Cells were fixed and immunostained with anti-EEA1 antibody (F and G), or antibody against the HA-epitope (H and I) followed by 568-conjugated anti-mouse antibody (F–I). Bars, 10 μm.
SUPPLEMENTAL FIG. 3- Depletion of hVps45 does not alter cell cycle. Human foreskin fibroblasts were either Mock-treated (A) or treated with hVps45-siRNA (B) for 72 h, fixed with 70% ethanol, stained with Telford reagent, and subjected to flow cytometry assay. The percentage of cells in diploid G1 (Dip G1), diploid G2 (Dip G2), and diploid S (Dip S) are shown in C.