Abstract
Genome-wide association studies (GWAS) using population-based designs have identified many genetic loci associated with risk of a range of complex diseases including cancer; however, each locus exerts a very small effect and most heritability remains unexplained. Family-based pedigree studies have also suggested tentative loci linked to increased cancer risk, often characterized by pedigree-specificity. However, a comparison between the results of population-and those of family-based studies shows little concordance. Explanations for this unidentified genetic ‘dark matter’ of cancer include phenotype ascertainment issues, limited power, gene-gene and gene-environment interactions, population heterogeneity, parent-of-origin-specific effects, rare and unexplored variants. Many of these reasons converge towards the concept of genetic heterogeneity that might implicate hundreds of genetic variants in regulating cancer risk. Dissecting the dark matter is a challenging task. Further insights can be gained from both population association and pedigree studies.
Keywords: complex genetics, genetic linkage, polygenic inheritance, single nucleotide polymorphisms
Genome-wide association studies and individual risk of disease
The advent of genome-wide association studies (GWAS) has revolutionized research on genetic determinants of risk for common diseases. Hundreds of associations of common genetic variants with extremely impressive P-values have been published in past four years [1]. However, the yield of associations has varied for different diseases and phenotypes, ranging from just one association discovered for pancreatic cancer to >25 for prostate cancer. As the dust settles after the first waves of enthusiasm, it is becoming evident that for many diseases, much of the genetic risk remains unexplained, representing the so-called ‘dark matter’ of genetic risk. Cancer is a prominent example where despite many important successes in >50 GWAS published recently, this dark matter seems to remain very prominent. Even with the current exciting pace of discovery, it seems unlikely that currently available samples and genotyping platforms will be able to explain this dark matter, unless additional breakthroughs and/or amendments in the strategy of discovery are adopted. As genotyping capacity evolves towards the feasibility of full sequencing in large population samples, there is debate about how to tackle the unexplained genetic dark matter.
These new insights and evolutions prompt a re-evaluation of our concepts about the genetic architecture of cancer. Are common cancers heritable diseases? What have we learnt from GWAS studies? How extensive is the dark matter? What are possible explanations for its presence? How could the dark matter be deciphered in future research? These are important questions that we address here.
Heritability of common cancers
Inherited cancer syndromes are associated with rare and highly penetrant monogenic mutations, but genetic factors also play a role in sporadic cancer, as reported in numerous family-based studies. The contribution of inherited factors has been quantified in modeling studies among twins. Although these studies are not fully consistent regarding the heritability (see glossary) of specific common cancers, overall they suggest that at least for some cancers the heritability is considerable.
Among >90 000 Swedish, Danish and Finnish twins, statistically significant effects of heritable factors were observed for prostate, colorectal and breast cancer, with inherited genetic differences among participants (heritability) accounting for 42%, 35%, and 27% of the phenotypic variance in the respective cancers; conversely, inherited factors played no statistically significant role for other types of cancer and were not involved in cervical cancer. For thyroid cancer, no concordant monozygotic or dizygotic pairs were observed [2].
A study of 9.6 million individuals in the Swedish family-cancer database showed that the risk of thyroid cancer has the highest contribution of heritable factors (53% of the proportion of variance explained) [3]. In that study, other cancers with a high heritability included those of the endocrine system (28%), testis (25%), breast (25%) and melanoma (20%), whereas a low-to-moderate contribution of hereditary factors was estimated for cancers of colon (13%), nervous system (13%), rectum (12%), non-Hodgkin lymphoma (10%), and lung (8%); prostate cancer was excluded from the analysis [3].
In common cancers, environmental risk factors play an important role and in most, if not all, cases, supersede that of genetics [2-4]. However, the genetic component is important to decipher for estimating the individual genetic risk of cancer, improving early prevention and diagnosis, and understanding the underlying biochemical pathways as a first step to design new cancer therapies. In this regard, the advent of GWAS kindled hopes that association analyses of common variants tagging the genome with markers of high density would reveal this genetic component of cancer risk (see Box 1 for methodological aspects).
Box 1. Methodology issues in the identification of genetic factors affecting cancer risk.
Pedigree-based genetic linkage analysis
Linkage originally referred to the physical proximity of loci along the chromosome, i.e., sufficiently close (physically connected) for their alleles to co-segregate within families. Genetic linkage analysis, which is aimed at identifying the chromosomal location of loci affecting a particular phenotype, is carried out in pedigrees and is based on analysis of recombination frequency between a disease locus and marker loci. Recombination between two loci is a function of their distance. Owing to the relatively small number of generations that might be available in a given pedigree and the consequent small number of possible recombinations, related individuals tend to share large regions of the genome inherited from the same founders. Thus, linkage analysis covering the complete genome can usually be done based on genotyping <1000 informative markers. Pedigree analysis can detect rare high-risk disease alleles and can provide formal proof of the genetic modulation of a given phenotype [4].
Population-based association analysis
Association analysis is carried out at the general population level in unrelated individuals and is based on the assumption that individuals sharing a particular phenotype, e.g., cancer, also share the disease allele originating from a common ancestral founder and causing or modulating the phenotype. Mapping of loci affecting a phenotype by association is based on the existence of linkage disequilibrium (LD) between the disease allele and marker alleles. Association analysis can identify disease-risk alleles only when such alleles show strong LD with marker alleles. Because LD decays very rapidly by distance in unrelated individuals, association analysis requires a much higher marker density (i.e. >100 000) than genetic linkage analysis and it is not suited to detect the role of rare variants [4].
Discovery yield of genome-wide association studies
As of December 18, 2009, the NHGRI catalog of GWAS lists 446 GWAS for different types of diseases or common phenotypes, including 2097 GWAS-discovered associations with any P-value of 10−5 or less (http://www.genome.gov/26525384). More than 50 GWAS have evaluated cancer phenotypes. Some recurring themes are becoming clear from these studies: the few variants discovered in each GWAS, the small effect sizes of the identified variants, and the relative lack of overlap in variants discovered by different GWAS investigations on the same phenotype.
Each of the cancer GWAS has detected one or a few loci associated with the risk of the particular cancer type and most, if not all, of these putative cancer risk loci showed small effects, that is the per-allele odds ratios of most of these were ≤1.4 and usually <1% of the phenotypic variance could be attributed to a single locus.
Small genetic effects were observed not only for pathologic phenotypes with relatively low heritability, such as common cancers, but also for physiological phenotypes characterized by high heritability, such as height. As an illustrative comparison, human height, a complex trait with heritability estimated to be ~80%, has been investigated in three GWAS involving analysis of hundreds of thousands of single nucleotide polymorphisms (SNPs) in ~ 63 000 people (for a review, see Ref [5]). Although the studies identified 54 loci affecting height variation in the population, these variants combined accounted for ~5% of height heritability. Moreover, the overlap among the three GWAS was quite poor, with only four loci identified in all three studies [5]. Although the genetic effects for height can also be modulated by environmental exposures (e.g. malnutrition), for well-nourished populations, this is probably less of a concern. However, cancer risk is a much more complex phenotype than height. Beyond the possibility of incomplete penetrance of the inherited cancer susceptibility alleles, exposure to environmental risk factors could markedly modify genetic predisposition. Depending on environmental exposures, an individual at high genetic risk of developing cancer might never be affected, whereas an individual at low genetic risk for cancer might experience the disease [6].
Another generic feature of GWAS investigations is that often different GWAS discover different loci for the same phenotype. This leads to an accumulation of more discovered variants as more GWAS investigations are performed. The lack of overlap among different GWAS does not necessarily mean lack of replication. An aspect to consider is the limited power to detect small effects at the stringent required levels of genome-wide significance, even with large studies. Thus, variants that pass stringent genome-wide significance thresholds, such as P<10−8, are thought to be real [7, 8], whereas variants associated with substantially more modest P-values (e.g. 10−5−10−6) might represent false positives; not surprisingly the majority of such variants will fail to be replicated when tested in further samples. For example, in GWAS a variant with P = 10-5 is <1% likely to represent a true association [9, 10].
With these general principles in mind, we review some of the major successes of GWAS in identifying loci related to the risk of specific cancers, so as to illustrate both the extent of progress made and the remaining caveats. For illustrative purposes, we discuss two cancers where GWAS have revealed a substantial number of new loci (breast cancer and colorectal cancer) and two others where GWAS have revealed few loci, one where heritability seems to be limited (lung cancer) and another whether heritability is considered to be more prominent (thyroid cancer).
Breast cancer
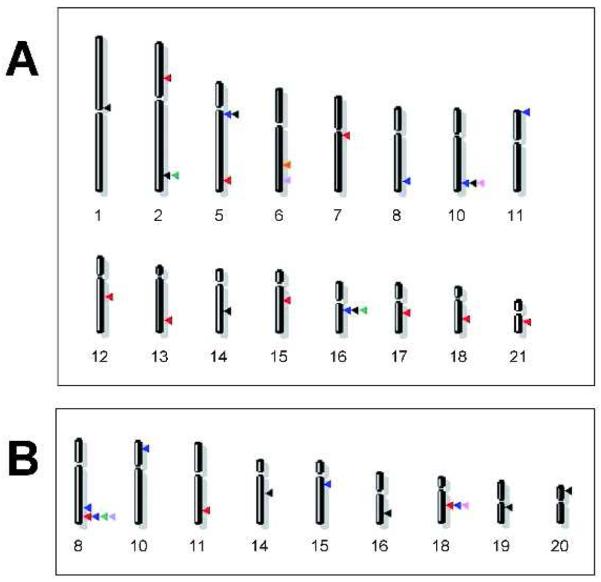
Several GWAS have identified multiple common genetic variants influencing breast cancer risk. Easton et al. [11] identified five loci mapping to 10q26, 16q12.1, 5q11.2, 8q24 and 11p15.5. Based on the NHGRI catalog, at a P<10−5 threshold, 14 additional regions were associated with breast cancer in subsequent GWAS publications [12-17]; only four of which (2q35, 5q11.2, 10q26.13 and 16q12.1) were seen in two or more GWAS [12, 14, 17] (Figure 1a). A recent study [18] found strong evidence for additional susceptibility loci on 3p and 17q by testing >800 promising associations derived from a previous GWAS [11]. Finally, combined analysis of suggestive loci using three published GWAS led to the identification of an additional locus on 5p12 [19] that seemed to be associated specifically with estrogen-receptor positive breast cancer. Claims for associations with specific subsets need even more careful replication, as subgroup differences can be spurious. For example, although the 2q35 locus was originally proposed to be associated specifically with estrogen-receptor positive breast cancer, a recent study found similar effects regardless of estrogen-receptor status [20]. All of the identified variants have small effects (per allele odds ratios <1.41, some even <1.10) and explain less of the heritability of breast cancer than the previously known breast cancer 1, early onset (BRCA1) and breast cancer 2, early onset (BRCA2) mutations can explain, perhaps with the exception of a fibroblast growth factor receptor 2 (FGFR2) common variant, where the contribution to explaining risk might be of similar magnitude as that of BRCA1/2.
Figure 1.
Chromosomal mapping of loci present in the NHGRI catalog and identified by GWAS in (a) breast or (b) colorectal cancers. Each arrow indicates the physical mapping position of a single locus. Each GWAS is associated with a different color as follows: (A) blue [11], red [13], pink [14], green [12], orange [15], violet [16], black [17]; (B) black [75], red [25], blue [26], pink [27], green [28], violet [29].
One may argue that even limited information can be informative in selected populations, where decision-making (e.g. whether mammography should be performed or not) might depend on slight modifications of risk [21, 22]. Nevertheless, currently, much of the genetic component of breast cancer risk remains uncharacterized and is thought to arise from combinations of common low-penetrance variants. These might interact with environmental exposures to cause disease risk. However, these environmental exposures remain elusive as a lot of the previously proposed environmental and lifestyle risk factors (e.g. nutrition) for breast cancer have been refuted in large studies in the last decade [23].
Colorectal cancer
Most of the heritability of colorectal cancer (CRC) cannot be explained by monogenic syndromes caused by high-risk germ-line mutations in adenomatous polyposis coli (APC) or mismatch repair genes which account for only <5% of colorectal cancer cases [24]. GWAS, carried out in large series including either sporadic colorectal cancer cases or colorectal cancer cases with a family history, have identified multiple loci at which common variants can influence the risk of developing CRC. When limited to loci included in the NHGRI catalog (threshold p<10−5), ten different susceptibility loci have been detected by GWAS [25-30] and by a meta-analysis of GWAS [31]: these include loci at 8q24.21, 18q21.1, 15q13.3, 11q23.1, 8q23.3, 10p14, 19q13.11, 20p12.3, 14q22.2 and 16q22.1 (Figure 1b). In all of these loci, the best SNP markers exhibit very modest odds ratios for colorectal cancer predisposition (range 1.10-1.26). Most likely because of the limited power to detect such modest effects, few loci (8q24 and 18q21) have been found consistently by several GWAS [27-29]. Cumulatively, these variants explain only a very small fraction of colorectal cancer risk. We are still unclear as to how these or other risk variants might interact with environmental exposures (e.g. red meat intake) to modulate colorectal cancer risk.
Lung cancer
Lung cancer is the prototype of a malignancy where environmental rather than genetic factors are apparently far more important. Three GWAS confirmed only one locus on chromosome 15q25, where nicotinic acetylcholine receptor genes map, as associated with lung cancer risk with an odds ratio of about 1.3 (for a review, see Ref [32]). However, the same locus is strongly linked to the main environmental risk factor for lung cancer (i.e. smoking), whose association with lung cancer is much greater, with odds ratios >20 for ~20 cigarettes per day [33]. It is possible that the effect of the 15q25 on lung cancer risk is mediated entirely through its influence on nicotine dependence [34]. Another two risk loci mapping to chromosomes 5p15 and 6p21 were detected by combining data of several GWAS [35-37]. The effects on cancer risk were even smaller, corresponding to odds ratios of 1.15-1.24, for the best-associated markers in these loci.
Thyroid cancer
Although thyroid cancer displays the highest heritability among solid tumors (up to 53% in [3]), little is known about genetic variants affecting the risk of this cancer. A GWAS for thyroid cancer detected only two loci (9q22.33 and 14q13.3) where SNPs reached genome-wide significance [38]. The per allele odds ratios were greater than those seen for most other common variants in association with cancer risk (1.75 and 1.37, respectively), but with only two variants available, the proportion of the variance explained is still limited. The GWAS used only 192 cases in the discovery stage, and thus the power is extremely limited despite the availability of over 30 000 controls; one might speculate that several more variants would be discovered, if the sample size could be enlarged.
What is missing in GWAS?
Although we know that genetics-related determinants of risk exist, we cannot explain the majority of this risk through specific identified genetic variants. Numerous hypothetical arguments have been proposed to explain that dark matter, as summarized in Table 1 [1, 1, 39, 40].
Table 1.
What accounts for the genetic ‘dark matter’ in cancer studies?a
| Reason | Ability of the different study designs to address these reasons |
||
|---|---|---|---|
| Population-based case-control studies |
Family-based association studies |
Genetic linkage studies in pedigrees |
|
| Biased phenotype definition and ascertainment |
Modest and/or Poor b |
Good and/or Modest b |
Good and/or Modest b |
| Insufficient sample size resulting in low power |
Good | Modest | Poor |
| Epistatic (gene–gene) interactions |
Modest c | Modest c | Good and/or Modest c |
| Gene–environment interactions |
Poor | Modest c | Good and/or Modest c |
| Differential effects in different populations |
Poor | Poor | Good |
| Incomplete genome coverage for common variants |
Good | Good | Good and/or Modest d |
| Effects of rare alleles | Poor | Poor | Good |
| Parent-of-origin specific effects |
Poor | Good and/or Poor | Good |
Potential reasons explaining the genetic dark matter and the ability of different study designs to address them.
Dependent on the type of cancer and on clinical/pathological ascertainments that are carried out; some types of cancers, e.g., liver cancer, are particularly prone to misclassification if histology of the tumor samples are not available, since the liver is a common site of cancer.
Dependent on the strength of the interaction, here for relatively strong interactions.
In pedigree studies, the reduced numbers of genetic recombinations determine a high number of redundant co-segregating genetic markers and, thus, high-density coverage by SNP-arrays does not allow fine mapping.
The ability of GWAS to detect associations with common SNPs can be reduced if the phenotypes are poorly or inconsistently defined and ascertained, and/or if controls are also suboptimally screened for exclusion of disease. Even with large sample sizes of several thousands cases and controls, there is usually limited power to detect alleles of modest effect sizes (odds ratios of 1.20), and minimal power to detect risk allele odds ratios of <1.10 even for very common variants. Power is also limited to detect epistatic interactions of multiple modest effect genes. The detection of gene-environment interactions is hampered not only by limited power, but also by the lack of concurrent availability of both genetic and high-quality, standardized, and consistently collected environmental exposure data [6]. Residual population stratification or genotyping error can also lead to the attrition of some associations, although current GWAS investigations have dramatically improved study performance on these fronts. The genetic architecture can differ substantially across different populations, and most GWAS to-date have targeted European-descent populations [41], while there is some evidence that different loci can emerge and the implicated haplotype blocks and strength of association can vary when populations of other ancestry are examined [42]. Moreover, despite generally good coverage of the whole genome in currently used genotyping platforms, some areas of the genome are still imperfectly covered and thus variants lurking in these areas would remain undiscovered. This is a more of a concern for African-descent populations than those of European descent. Finally, parent-of-origin-specific genetic effects would have been largely missed with the current mode of association analysis in most GWAS investigations. For example, the association of rs157935[T] at 7q32 with the risk of cutaneous basal cell carcinoma seems to be dependent to the parent-of-origin of the risk allele, estimated in silico [43].
In particular, the hypothesis of insufficient sample size, a potential limiting factor in the statistical power to detect weak genetic loci, has gathered considerable supporting evidence based on GWAS conducted to-date and deserves some more elaboration. The general rule has been that the larger the sample size, the greater the yield of new discovered loci. The pattern of discoveries to-date in terms of the minor allele frequency and odds ratio of the risk alleles that emerge could be largely explained based on power considerations alone. This is leading GWAS research groups worldwide to assemble huge numbers of patients and controls to conduct GWAS on increasingly large series of cases and controls. As the technology is becoming cheaper, instead of multi-stage designs, performance of GWAS using the whole genotyping platform on very large samples (e.g. 100 000 cases and as many controls) might become feasible and cost-efficient [5]. However, the practical issues in accumulating such a huge number of cases and controls are not easy to solve. For the most common cancers such as breast cancer and colorectal cancer, consortia with sample sizes in the range of up to 30 000 cases and as many controls are already in place and further enrollment of additional teams will be able to increase this further. For less common cancers, it will be a challenge to obtain such numbers, even if new large population cohorts are established [44]. Moreover, it is possible that even with 100 000 cases and 100 000 controls, the proportion of variance explained by the discovered variants, cumulatively, might still not exceed 20%. However, the use ever larger cohorts providing the main solution to the identification of the elusive dark matter is a topic of debate in the community. One view is that many additional common variants are unlikely to be discovered or are not worth discovering, and that most genetic control is caused by variants that are not represented at all in the current studies [45]. This is an interesting speculation, but there are still no data to support it or refute it.
Rare variants are an obvious contender for the source of the missing heritability. By default, usually variants with frequency <5-10% are excluded from current GWAS analyses. Many rare variants (especially those with minor allele frequencies of 0.5-5%) would be possible to capture using full-sequencing, if a sufficient number of individuals is genotyped. Indeed, the 1000 Genomes Project (http://www.1000genomes.org/page.php) is aimed to the identification of the rare variants and thus facilitating association studies. However, unless the effects that they convey are large, the power to detect them would be practically zero. Various analytical approaches have been proposed to improve power, typically generating composite scores by merging many rare variants together that might share common function [46]. However, such merging usually provides speculative results and no clear functional evidence exists to perform the grouping of variants with certainty.
Structural variants also need to be considered as an underlying cause of cancer. Several investigators have already performed association studies that capture copy number variants (CNVs), which can correspond to either common or rare alleles. So far, strong associations with common CNV are limited, although common CNV can be in strong LD with common SNPs (frequencies of the minor allele >0.1) [47], suggesting that the detection power for common CNVs is probably adequate. Conversely, a considerable number of rare CNV have been proposed to be associated with various neuropsychiatric traits, including schizophrenia, mental retardation, autism, and epilepsy [48]. In all of these cases, the CNV have been seen with a frequency of 0.2-1%, although they are exceedingly rare in the general population (generally ≤0.03%). No CNVs have yet been associated robustly with cancer phenotypes, but no large studies have yet been published in cancer patients to pursue this avenue.
Power considerations are important also for the detection of CNV associations in association studies, and those that can currently be detected for rare CNV are those with very large effect sizes.
Pedigree studies in familial cancer
Genetic linkage analysis in pedigrees containing multiple affected members can complement association analyses. Such studies have been traditionally hampered by their relatively small size and much lower markers density as compared with the recent GWAS. Despite the relatively low power, results of pedigree analyses can provide strong and convincing indications of genetic effects, since they are based on genetic transmission of disease alleles within a family and thus do not have to make the population assumptions of association analyses. How do results for family-linkage studies compare with those of GWAS?
Familial breast cancer
Inherited mutations in the two major susceptibility genes for breast cancer, namely, BRCA1 and BRCA2, lead to a high risk of breast cancer, but account for only about 20% of familial breast cancer [49]. Other genes that have fairly robust evidence for conferring susceptibility to familial breast cancer through inherited mutations include ataxia telangiectasia mutated (ATM) and tumor protein p53 (TP53); moreover, a 1100delC variant in CHK2 checkpoint homolog (Schizosaccharomyces pombe) (CHEK2) with minor allele frequency close to 1% has strong evidence for increasing breast cancer risk, and even more so familial cancer risk [50, 51]. All these known mutations still explain the minority of familial breast cancer and probably <5% of all breast cancer risk.
Several linkage studies have reported candidate regions containing breast cancer susceptibility genes. However, the logarithm of the odds ratio (LOD) score values obtained for these regions were not significant or of borderline statistical significance, and the percentage of families putatively linked to each region was low. In a recent linkage study in Spanish breast cancer families, three regions of interest, located on 3q25, 6q24, and 21q22, were observed [52]. Overall, 20 distinct putative breast cancer susceptibility loci have been proposed, but these do not overlap among studies and independent loci cluster in each family [52] (Table 2).
Table 2.
Genetic heterogeneity in breast or colorectal cancer families a
| Disease | Chromosomal regions linked to cancer |
LOD score b |
Family ID | Refs |
|---|---|---|---|---|
| Breast cancer |
2p21 | 1.67 | 699003 | [76] |
| 4p14-12; 20q13 | 1.84; 1.80 |
2191 c | ||
| 3p14; 11p13 | 1.52; 1.59 |
MAYO 151 | ||
| 11q14 | 1.67 | 153 d | ||
| 15q14 | 1.5 | EUR 60a | ||
| 4q13.1-13.2; 22q13.2 | 1.91; 2.62 |
EUR 60b | ||
| 2q32 | 3.2 | 178, 261, 277, 381, 437, 477, 574, 3004, 3008, 3009, 3011 |
[77] | |
| 10q23; 19q13; 17p13 | 1.66; 1.52; 1.51 |
14 | [78] | |
| 16p13.2-12.1; 19p13.3-12 |
1.91; 2.10 |
1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13 |
||
| 2p22.3 | 1.92 | 3395 | [79] | |
| 4p14-12 | 1.8 | 2191 c | ||
| 11q13.5-14.3; 14q21.1-21.3 |
2.2; 2.2 | 153 d | ||
| 3q25 | 3.01 | 3, 5, 10, 21, 24, 27 | [52] | |
| 6q24 | 2.26 | 2, 6, 31, 33, 35 | ||
| 21q22 | 3.55 | 8, 9, 18, 24, 41 | ||
| Colorectal cancer |
3q13.31-27.1 | 4.1 | 70, 119, 125, 197, 242, 256, 309, 409 |
[58] |
| 6q22.31 | 2.3 | 1 to 30 | ||
| 9q22.32–31.1 | 2.4 | 24 | [59] e | |
| 9q22.2-31.2 | 3.3 | 1 to 53 | [57] | |
| 11q13.2–13.4; 11q22.1–23.1 |
1.57; 1.55 |
70, 100, 161, 197, 201, 244 |
[80] | |
| 14q23.1–24.1 | 1.4 | 53, 70, 101, 161, 201, 202 | ||
| 9q22.33 | 1.44 | 1 to 57 | [81] e | |
| 3q29; 4q31.3; 7q31.31 |
2.61; 2.13; 3.08 |
1 to 83 | [82] | |
| 3q21–24 | 3.4 | 1 to 38 | [83] |
Based on the results of genetic linkage studies in pedigrees of breast or colorectal cancer families; genetic heterogeneity evidenced by mapping of multiple study-specific and pedigree-specific loci.
The highest LOD score values reported in the paper, independent of type of analysis (parametric or non-parametric).
Same family.
Same family.
Replication study with a candidate locus design.
One potential explanation for these results is genetic heterogeneity, with several putative breast cancer susceptibility genes playing an important role in the genetic risk of breast cancer but relevant only in a small number of families. The effects can be large in the specific families, but they would be completely lost once diluted in a large association population sample.
Familial colorectal cancer
Hereditary non-polyposis colorectal cancer (HNPCC), or Lynch syndrome, is a hereditary condition that predisposes to colorectal cancer. Inherited mutations causing defects in the DNA mismatch repair machinery and occurring at different genes, such as mutL homolog 1, colon cancer, nonpolyposis type 2 (E. coli) (MLH1), mutS homolog 2, colon cancer, nonpolyposis type 1 (E. coli) (MSH2), mutS homolog 6 (E. coli) (MSH6), PMS2 postmeiotic segregation increased 2 (S. cerevisiae) (PMS2) and possibly mutL homolog 3 (E. coli) (MLH3) and PMS1 postmeiotic segregation increased 1 (S. cerevisiae) (PMS1) [53], are the main cause of HNPCC. However, microsatellite instability or alterations in expression of DNA mismatch repair proteins (i.e., markers of inactivating mutations at these genes) are not identified in ~ 60% of HNPCC families fulfilling the Amsterdam criteria of HNPCC. This suggests the existence of genetic variations at as yet unidentified genes, leading to a possible family-specific autosomal dominant trait of HNPCC risk [54]. In addition, many families with multiple CRC cases but not completely fulfilling the Amsterdam criteria have been described [55, 56], suggesting a wider spectrum of familial disease.
Linkage studies in colorectal cancer families with no evidence of deficiency in DNA mismatch repair genes have mapped putative loci on several chromosomes (Table 2). Among these loci, most have very modest LOD scores and might well represent false-positives, but those on chromosome 3q21.1-q26.2 and 9q22.32-31.1 have shown the most consistent findings [57-59] (Table 2). Moreover, specific and different loci have been detected not only in different studies but also in single pedigrees within the same study (Table 2), providing another example of genetic heterogeneity.
Lung cancer
Lung cancer occurs mostly in a sporadic form, although several familial clusters have been reported. Genetic linkage analysis of families with multiple cases of lung cancer detected a locus influencing lung cancer risk on chromosome 6q23-25 [60]. Statistically significant linkage (LOD = 4.26) was observed in a subset of 23 families with five or more affected individuals in two or more generations, out of a total of 52 families. Linkage heterogeneity was detected, as in 14 families with only three affected relatives no linkage was found. No other loci were detected, suggesting that a single, relatively weak locus might affect familial lung cancer [60].
Thyroid cancer
Estimates indicate that ~ 5% of non-medullary thyroid carcinoma (NMTC) is familial. Familial NMTC is often characterized by an earlier age of onset, higher aggressiveness, and more frequent multifocal disease and recurrence as compared to sporadic NMTC. Linkage studies in pedigrees of different geographical origin identified loci on chromosomes 1q21, 2q21, 8p23.1-p22, and 19p13.2 [61-64]. Typically, a single locus was detected in each pedigree, and LOD scores were quite low, ranging from 3.01 to 4.41, pointing to weak effects and to the possibility of genetic heterogeneity.
Suitability and feasibility of different types of studies
Although larger population-based GWAS studies will continue to be an important avenue to pursue in identifying more risk variants, large family-based case-control studies might represent an alternative design that incorporates the advantages of studying sporadic cancer, of avoiding problems with population structure, and of analyzing hundred thousands of genetic markers as in the case of population-based GWAS. However, collecting family-based samples might be more difficult than collecting a series of unrelated cases and controls and the matching of transmitted with nontransmitted alleles from the same family might reduce statistical power (Table 1). However, if properly collected, this information would be valuable, especially if effect sizes are much stronger within pedigrees. This could cut sample size requirements considerably.
Pedigree and family-based studies might be best suited to detect gene-gene or gene-environment interactions, because the ability of such detection depends not only on the size of the study population and the number of examined polymorphisms but even more so on the accuracy of the phenotype and of the environmental exposure measurements. Evaluation of environmental exposures might be more uniform and standardized in the setting of pedigrees where subjects have been born and have lived in the same location (Table 1).
Parent-of-origin-specific effects can suitably be dissected by pedigree studies, because no parental genotypes or family structure need be taken into account in population-based studies. In family-based association studies, parent-of-origin effects can also be detected when the design of the study includes genetic information from the parents (Table 1).
If rare variants are a hallmark of cancer risk, pedigrees but not family-based association studies would be expected to be particularly enriched in such rare variants. Also, pedigree studies do not suffer of problems related to different populations or ethnicity (Table 1).
Pedigree-based and population-based GWAS studies can also be combined (e.g. by weighing loci in GWAS differentially based on pedigree-derived signals). This would reduce the stringency of the required P-values. Such approaches would benefit from verification of their efficiency and discovery yield with appropriate replication studies.
An emerging model of genetic heterogeneity
Overall, comparison of the results from GWAS and pedigree studies shows hardly any overlap for breast, colon, lung and thyroid cancer (Figure 1 and Table 2). The relative success of discovery of GWAS versus loci discovered with familial approaches varies across cancers. Going beyond the examples of the four cancers that we discussed in detail above, the cancer for which we currently have the largest number of discovered loci through GWAS is prostate cancer, with a large number of GWAS identifying up to more than 25 independent loci [65-67], albeit all of them with very small effects. Conversely, for prostate cancer there have been no genes with strong evidence identified through familial approaches and our knowledge of environmental and lifestyle determinants also remains very rudimentary. At the other end of the spectrum, for testicular cancer, GWAS have identified fewer genes, but several of them have relatively strong effects [68, 69], although family studies demonstrated no statistically significant regions of linkage [70].
To some extent, the lack of overlap might simply mean that associations emerging from GWAS with modest P-values and modest LOD scores emerging from family-based linkage studies could well represent false-positives. However, false-positives are unlikely to represent the whole story. Numerous GWAS signals are definitely genuine, and several of the linkage signals have considerable credibility. One possibility is that many of the loci identified by pedigree analysis would not be detectable in population-based studies, if they are specific for each pedigree. Thus genetic heterogeneity might play an important role in genetic predisposition to different cancer types.
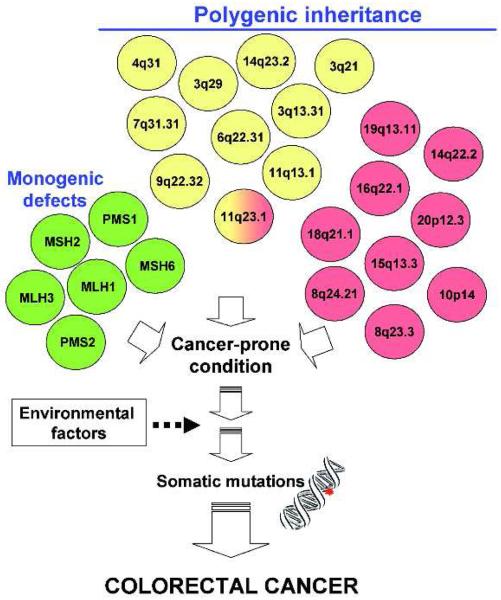
Although the idea that genetic heterogeneity can hamper the detection of loci relevant for complex disease is not new, insights from recent studies allow us to have a better grasp of its possible manifestations and implications for cancer heritability and to speculate on the extent of its complexity. The accumulating data suggest that we can exclude with confidence the scenario where one or a few loci alone can explain the majority of the genetic risk for any common cancer. Regardless of frequency (i.e. common, rare, very rare) and type (i.e. SNP or CNV), it is likely that hundreds or even thousands of genetic variants are implicated in cancer risk. One could envision a model, based on the example of colorectal cancer, in which genetic heterogeneity plays a major role in both familial cancer due to monogenic mutations and in polygenic inheritance of sporadic cancer, modulated by a complex architecture of a multiplicity of genetic loci (Figure 2). The relative contribution of low-penetrance low-risk common variants and high-penetrance, high-risk uncommon/rare variants is unknown and it might vary from one type of cancer to another and between sporadic and familial disease (acknowledging that in some cases, the exact boundaries of the definition of familial disease might still be unclear).
Figure 2.
Proposed role of genetic heterogeneity in individual predisposition to cancer. Colorectal cancer is given as an example. Several causative mutations at single genes (monogenic syndromes, in green), as well as variations at several loci (polygenic control) detected by either pedigree analysis (in yellow) or GWAS (in red), increase the risk of cancer. Only the 11q23.1 locus was common to pedigree and GWA studies. Either the single genetic defects or the polygenic conditions produce a cancer-prone condition in the normal tissue; individual risk of cancer might be further modulated by environmental factors, leading to somatic mutations and, ultimately, to cancer.
Moreover, it is possible that in some situations low-penetrance, low-risk common variants identified through GWAS could simply be markers of high-penetrance, high-risk uncommon/rare variants to which they are in linkage disequilibrium. These linked variants might be in the same genetic locus, but linkage sometimes can extent to very distant positions [40]. The coexistence of mutations and common markers of risk in the same gene has been extensively documented in GWAS studies for some metabolic traits such as lipid levels [71]. The extent of this coexistence in cancer is less well documented, given the relatively smaller number of bona-fide known mutated genes with familial risk that have been conclusively identified to-date, but some evidence already exists (e.g. for pigmentation-related genes and melanoma) [72].
Concluding remarks and future perspectives
Overall, the recognition of genetic heterogeneity should be seen as an opportunity rather than a problem in genetic epidemiology. Indeed, the analysis of candidate genes identified in population-based and in pedigree studies could allow tracing possible biochemical pathways affecting a specific type of cancer. This could reduce the extreme multi-dimensionality of the genetic architecture to fewer pathways. For example, although several independent loci cause the Lynch syndrome, the known germ-line mutations are observed in genes all involved in the control of DNA repair. Moreover, given the complexity of regulation of gene expression, variants of many different independent loci can bind to the same regulatory elements and affect expression of the same target gene.
The identification of common pathways underlying genetic heterogeneity might result in the design of new tests for early diagnosis that are based on biochemical targets of the causal loci modulating the disease. In addition, identification of such pathways could enable the design of new therapeutic strategies to control cancer outcome.
Improvements on the sequencing front might also change our understanding of the genetic architecture. Family trees for example can be successfully analyzed with exome sequencing, such as the approach used recently for understanding the genetics of Miller syndrome, a rare mendelian disorder [73]. It is interesting to see to what extent, if any, various cancers might be a conglomerate of a large number of rare mendelian syndromes that can be dissected with a similar sequencing approach.
However, while such advances are desirable, the difficulties of achieving them should not be understated. If genetic heterogeneity is extreme, then cancer will become an example of ‘private epidemiology’ [74], where what happens to one individual with cancer is not applicable to others. As we add more, even if incremental, discoveries, we should be able to gain increasing insights about the complex puzzle of cancer genetic architecture.
Acknowledgments
This work was funded in part by grants from Associazione and Fondazione Italiana Ricerca Cancro (AIRC and FIRC), and Fondo Investimenti Ricerca di Base (FIRB), Italy, to TAD. Scientific support for this project to JPAI was provided through the Tufts Clinical and Translational Science Institute (Tufts CTSI) under funding from the National Institute of Health/National Center for Research Resources (UL1 RR025752). Points of view or opinions in this paper are those of the authors and do not necessarily represent the official position or policies of the Tufts CTSI.
Glossary
- Genetic heterogeneity
is a common phenomenon that occurs when a disease might be caused by any of multiple and unlinked susceptibility loci, each of which by itself or in subgroups affecting individual disease risk [84, 85]. The term was introduced by Harris in 1953 and Fraser in 1956, although it has been somehow recognized by Waardenburg in 1935 [86]. As natural consequence of genetic heterogeneity, pedigree-specific linkages to different chromosomal regions can be detected, due to the causation of the same disease by different loci in different families. Taking the polycystic kidney disease as an example of monogenic disease with genetic heterogeneity, the same disease is caused in certain individuals by mutations in the PKD1 gene (chromosome 16p13.3) whereas in other individuals by mutations in the PKD2 gene (4q21) [87].
- Heritability
is the proportion of the phenotypic variation that is attributable to genetic inheritance, in a given population and for a specific phenotype.
- Linkage disequilibrium
occurs when random formation of haplotypes from alleles based on their frequencies is not respected, thus resulting in a statistically significant combination of alleles at two or more loci. Linkage disequilirium at the same pair of loci can vary in different populations and tends to disappear over time at a rate depending on the mendelian recombination frequency, in the absence of interfering evolutionary forces.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Manolio TA, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lichtenstein P, et al. Environmental and heritable factors in the causation of cancer--analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med. 2000;343:78–85. doi: 10.1056/NEJM200007133430201. [DOI] [PubMed] [Google Scholar]
- 3.Czene K, et al. Environmental and heritable causes of cancer among 9.6 million individuals in the Swedish Family-Cancer Database. Int J Cancer. 2002;99:260–266. doi: 10.1002/ijc.10332. [DOI] [PubMed] [Google Scholar]
- 4.Borecki IB, Province MA. Linkage and association: basic concepts. Adv Genet. 2008;60:51–74. doi: 10.1016/S0065-2660(07)00403-8. [DOI] [PubMed] [Google Scholar]
- 5.Weedon MN, Frayling TM. Reaching new heights: insights into the genetics of human stature. Trends Genet. 2008;24:595–603. doi: 10.1016/j.tig.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 6.Ioannidis JPA, et al. Researching Genetic Versus Nongenetic Determinants of Disease: A Comparison and Proposed Unification. Science Translational Medicine. 2009;1:7ps8–7ps8. doi: 10.1126/scitranslmed.3000247. [DOI] [PubMed] [Google Scholar]
- 7.Hoggart CJ, et al. Genome-wide significance for dense SNP and resequencing data. Genet Epidemiol. 2008;32:179–185. doi: 10.1002/gepi.20292. [DOI] [PubMed] [Google Scholar]
- 8.Ioannidis JP, et al. Replication validity of genetic association studies. Nat Genet. 2001;29:306–309. doi: 10.1038/ng749. [DOI] [PubMed] [Google Scholar]
- 9.Khoury MJ, et al. Genome-wide association studies, field synopses, and the development of the knowledge base on genetic variation and human diseases. Am J Epidemiol. 2009;170:269–279. doi: 10.1093/aje/kwp119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ioannidis JP. Calibration of credibility of agnostic genome-wide associations. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:964–972. doi: 10.1002/ajmg.b.30721. [DOI] [PubMed] [Google Scholar]
- 11.Easton DF, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–1093. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stacey SN, et al. Common variants on chromosomes 2q35 and 16q12 confer susceptibility to estrogen receptor-positive breast cancer. Nat Genet. 2007;39:865–869. doi: 10.1038/ng2064. [DOI] [PubMed] [Google Scholar]
- 13.Murabito JM, et al. A genome-wide association study of breast and prostate cancer in the NHLBI's Framingham Heart Study. BMC Med Genet. 2007;8(Suppl 1):S6. doi: 10.1186/1471-2350-8-S1-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hunter DJ, et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet. 2007;39:870–874. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gold B, et al. Genome-wide association study provides evidence for a breast cancer risk locus at 6q22.33. Proc Natl Acad Sci U S A. 2008;105:4340–4345. doi: 10.1073/pnas.0800441105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng W, et al. Genome-wide association study identifies a new breast cancer susceptibility locus at 6q25.1. Nat Genet. 2009;41:324–328. doi: 10.1038/ng.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thomas G, et al. A multistage genome-wide association study in breast cancer identifies two new risk alleles at 1p11.2 and 14q24.1 (RAD51L1) Nat Genet. 2009;41:579–584. doi: 10.1038/ng.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ahmed S, et al. Newly discovered breast cancer susceptibility loci on 3p24 and 17q23.2. Nat Genet. 2009;41:585–590. doi: 10.1038/ng.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stacey SN, et al. Common variants on chromosome 5p12 confer susceptibility to estrogen receptor-positive breast cancer. Nat Genet. 2008;40:703–706. doi: 10.1038/ng.131. [DOI] [PubMed] [Google Scholar]
- 20.Milne RL, et al. Risk of estrogen receptor-positive and -negative breast cancer and single-nucleotide polymorphism 2q35-rs13387042. J Natl Cancer Inst. 2009;101:1012–1018. doi: 10.1093/jnci/djp167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pharoah PD, et al. Polygenes, risk prediction, and targeted prevention of breast cancer. N Engl J Med. 2008;358:2796–2803. doi: 10.1056/NEJMsa0708739. [DOI] [PubMed] [Google Scholar]
- 22.Gulcher J, Stefansson K. Genetic risk information for common diseases may indeed be already useful for prevention and early detection. Eur J Clin Invest. 2010;40:56–63. doi: 10.1111/j.1365-2362.2009.02233.x. [DOI] [PubMed] [Google Scholar]
- 23.Boyle P, et al. Diet, nutrition and cancer: public, media and scientific confusion. Ann Oncol. 2008;19:1665–1667. doi: 10.1093/annonc/mdn561. [DOI] [PubMed] [Google Scholar]
- 24.Green RC, et al. Prediction of Lynch syndrome in consecutive patients with colorectal cancer. J Natl Cancer Inst. 2009;101:331–340. doi: 10.1093/jnci/djn499. [DOI] [PubMed] [Google Scholar]
- 25.Tenesa A, et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on 11q23 and replicates risk loci at 8q24 and 18q21. Nat Genet. 2008;40:631–637. doi: 10.1038/ng.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tomlinson IP, et al. A genome-wide association study identifies colorectal cancer susceptibility loci on chromosomes 10p14 and 8q23.3. Nat Genet. 2008;40:623–630. doi: 10.1038/ng.111. [DOI] [PubMed] [Google Scholar]
- 27.Broderick P, et al. A genome-wide association study shows that common alleles of SMAD7 influence colorectal cancer risk. Nat Genet. 2007;39:1315–1317. doi: 10.1038/ng.2007.18. [DOI] [PubMed] [Google Scholar]
- 28.Tomlinson I, et al. A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24.21. Nat Genet. 2007;39:984–988. doi: 10.1038/ng2085. [DOI] [PubMed] [Google Scholar]
- 29.Zanke BW, et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on chromosome 8q24. Nat Genet. 2007;39:989–994. doi: 10.1038/ng2089. [DOI] [PubMed] [Google Scholar]
- 30.Pittman AM, et al. Refinement of the basis and impact of common 11q23.1 variation to the risk of developing colorectal cancer. Hum Mol Genet. 2008;17:3720–3727. doi: 10.1093/hmg/ddn267. [DOI] [PubMed] [Google Scholar]
- 31.Houlston RS, et al. Meta-analysis of genome-wide association data identifies four new susceptibility loci for colorectal cancer. Nat Genet. 2008;40:1426–1435. doi: 10.1038/ng.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chanock SJ, Hunter DJ. Genomics: when the smoke clears. Nature. 2008;452:537–538. doi: 10.1038/452537a. [DOI] [PubMed] [Google Scholar]
- 33.Freedman ND, et al. Cigarette smoking and subsequent risk of lung cancer in men and women: analysis of a prospective cohort study. Lancet Oncol. 2008;9:649–656. doi: 10.1016/S1470-2045(08)70154-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Galvan A, Dragani TA. Nicotine dependence may link the 15q25 locus to lung cancer risk. Carcinogenesis. 2009 doi: 10.1093/carcin/bgp282. [DOI] [PubMed] [Google Scholar]
- 35.Wang Y, et al. Common 5p15.33 and 6p21.33 variants influence lung cancer risk. Nat Genet. 2008;40:1407–1409. doi: 10.1038/ng.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Broderick P, et al. Deciphering the impact of common genetic variation on lung cancer risk: a genome-wide association study. Cancer Res. 2009;69:6633–6641. doi: 10.1158/0008-5472.CAN-09-0680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Landi MT, et al. A genome-wide association study of lung cancer identifies a region of chromosome 5p15 associated with risk for adenocarcinoma. Am J Hum Genet. 2009;85:679–691. doi: 10.1016/j.ajhg.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gudmundsson J, et al. Common variants on 9q22.33 and 14q13.3 predispose to thyroid cancer in European populations. Nat Genet. 2009;41:460–464. doi: 10.1038/ng.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu YJ, et al. Is replication the gold standard for validating genome-wide association findings? PLoS ONE. 2008;3:e4037. doi: 10.1371/journal.pone.0004037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ioannidis JP, et al. Validating, augmenting and refining genome-wide association signals. Nat Rev Genet. 2009;10:318–329. doi: 10.1038/nrg2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Need AC, Goldstein DB. Next generation disparities in human genomics: concerns and remedies. Trends Genet. 2009;25:489–494. doi: 10.1016/j.tig.2009.09.012. [DOI] [PubMed] [Google Scholar]
- 42.Ioannidis JP. Population-wide generalizability of genome-wide discovered associations. J Natl Cancer Inst. 2009;101:1297–1299. doi: 10.1093/jnci/djp298. [DOI] [PubMed] [Google Scholar]
- 43.Stacey SN, et al. New common variants affecting susceptibility to basal cell carcinoma. Nat Genet. 2009;41:909–914. doi: 10.1038/ng.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Collins FS, Manolio TA. Merging and emerging cohorts: necessary but not sufficient. Nature. 2007;445:259. doi: 10.1038/445259a. [DOI] [PubMed] [Google Scholar]
- 45.Goldstein DB. Common genetic variation and human traits. N Engl J Med. 2009;360:1696–1698. doi: 10.1056/NEJMp0806284. [DOI] [PubMed] [Google Scholar]
- 46.Morris AP, Zeggini E. An evaluation of statistical approaches to rare variant analysis in genetic association studies. Genet Epidemiol. 2009 doi: 10.1002/gepi.20450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McCarroll SA, et al. Integrated detection and population-genetic analysis of SNPs and copy number variation. Nat Genet. 2008;40:1166–1174. doi: 10.1038/ng.238. [DOI] [PubMed] [Google Scholar]
- 48.Williams HJ, et al. Schizophrenia genetics: new insights from new approaches. Br Med Bull. 2009;91:61–74. doi: 10.1093/bmb/ldp017. [DOI] [PubMed] [Google Scholar]
- 49.Stratton MR, Rahman N. The emerging landscape of breast cancer susceptibility. Nat Genet. 2008;40:17–22. doi: 10.1038/ng.2007.53. [DOI] [PubMed] [Google Scholar]
- 50.CHEK2 Breast Cancer Case-Control Consortium CHEK2*1100delC and susceptibility to breast cancer: a collaborative analysis involving 10,860 breast cancer cases and 9,065 controls from 10 studies. Am J Hum Genet. 2004;74:1175–1182. doi: 10.1086/421251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Meijers-Heijboer H, et al. Low-penetrance susceptibility to breast cancer due to CHEK2(*)1100delC in noncarriers of BRCA1 or BRCA2 mutations. Nat Genet. 2002;31:55–59. doi: 10.1038/ng879. [DOI] [PubMed] [Google Scholar]
- 52.Rosa-Rosa JM, et al. Genome-wide linkage scan reveals three putative breast-cancer-susceptibility loci. Am J Hum Genet. 2009;84:115–122. doi: 10.1016/j.ajhg.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Peltomaki P. Lynch syndrome genes. Fam Cancer. 2005;4:227–232. doi: 10.1007/s10689-004-7993-0. [DOI] [PubMed] [Google Scholar]
- 54.Llor X, et al. Differential features of colorectal cancers fulfilling Amsterdam criteria without involvement of the mutator pathway. Clin Cancer Res. 2005;11:7304–7310. doi: 10.1158/1078-0432.CCR-05-0965. [DOI] [PubMed] [Google Scholar]
- 55.Kievit W, et al. Current clinical selection strategies for identification of hereditary non-polyposis colorectal cancer families are inadequate: a meta-analysis. Clin Genet. 2004;65:308–316. doi: 10.1111/j.1399-0004.2004.00220.x. [DOI] [PubMed] [Google Scholar]
- 56.Pinol V, et al. Frequency of hereditary non-polyposis colorectal cancer and other colorectal cancer familial forms in Spain: a multicentre, prospective, nationwide study. Eur J Gastroenterol Hepatol. 2004;16:39–45. doi: 10.1097/00042737-200401000-00007. [DOI] [PubMed] [Google Scholar]
- 57.Wiesner GL, et al. A subset of familial colorectal neoplasia kindreds linked to chromosome 9q22.2-31.2. Proc Natl Acad Sci U S A. 2003;100:12961–12965. doi: 10.1073/pnas.2132286100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Picelli S, et al. Genome-wide linkage scan for colorectal cancer susceptibility genes supports linkage to chromosome 3q. BMC Cancer. 2008;8:87. doi: 10.1186/1471-2407-8-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Skoglund J, et al. Linkage analysis in a large Swedish family supports the presence of a susceptibility locus for adenoma and colorectal cancer on chromosome 9q22.32-31.1. J Med Genet. 2006;43:e7. doi: 10.1136/jmg.2005.033928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bailey-Wilson JE, et al. A major lung cancer susceptibility locus maps to chromosome 6q23-25. Am J Hum Genet. 2004;75:460–474. doi: 10.1086/423857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Canzian F, et al. A gene predisposing to familial thyroid tumors with cell oxyphilia maps to chromosome 19p13.2. Am J Hum Genet. 1998;63:1743–1748. doi: 10.1086/302164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cavaco BM, et al. Mapping a new familial thyroid epithelial neoplasia susceptibility locus to chromosome 8p23.1-p22 by high-density single-nucleotide polymorphism genome-wide linkage analysis. J Clin Endocrinol Metab. 2008;93:4426–4430. doi: 10.1210/jc.2008-0449. [DOI] [PubMed] [Google Scholar]
- 63.Malchoff CD, et al. Papillary thyroid carcinoma associated with papillary renal neoplasia: genetic linkage analysis of a distinct heritable tumor syndrome. J Clin Endocrinol Metab. 2000;85:1758–1764. doi: 10.1210/jcem.85.5.6557. [DOI] [PubMed] [Google Scholar]
- 64.McKay JD, et al. Localization of a susceptibility gene for familial nonmedullary thyroid carcinoma to chromosome 2q21. Am J Hum Genet. 2001;69:440–446. doi: 10.1086/321979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Eeles RA, et al. Multiple newly identified loci associated with prostate cancer susceptibility. Nat Genet. 2008;40:316–321. doi: 10.1038/ng.90. [DOI] [PubMed] [Google Scholar]
- 66.Thomas G, et al. Multiple loci identified in a genome-wide association study of prostate cancer. Nat Genet. 2008;40:310–315. doi: 10.1038/ng.91. [DOI] [PubMed] [Google Scholar]
- 67.Gudmundsson J, et al. Genome-wide association and replication studies identify four variants associated with prostate cancer susceptibility. Nat Genet. 2009;41:1122–1126. doi: 10.1038/ng.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kanetsky PA, et al. Common variation in KITLG and at 5q31.3 predisposes to testicular germ cell cancer. Nat Genet. 2009;41:811–815. doi: 10.1038/ng.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rapley EA, et al. A genome-wide association study of testicular germ cell tumor. Nat Genet. 2009;41:807–810. doi: 10.1038/ng.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Crockford GP, et al. Genome-wide linkage screen for testicular germ cell tumour susceptibility loci. Hum Mol Genet. 2006;15:443–451. doi: 10.1093/hmg/ddi459. [DOI] [PubMed] [Google Scholar]
- 71.Wang J, et al. Polygenic determinants of severe hypertriglyceridemia. Hum Mol Genet. 2008;17:2894–2899. doi: 10.1093/hmg/ddn188. [DOI] [PubMed] [Google Scholar]
- 72.Bishop DT, et al. Genome-wide association study identifies three loci associated with melanoma risk. Nat Genet. 2009;41:920–925. doi: 10.1038/ng.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ng SB, et al. Exome sequencing identifies the cause of a mendelian disorder. Nat Genet. 2009 doi: 10.1038/ng.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ioannidis JP. Commentary: grading the credibility of molecular evidence for complex diseases. Int J Epidemiol. 2006;35:572–577. doi: 10.1093/ije/dyl003. [DOI] [PubMed] [Google Scholar]
- 75.Houlston RS, et al. Meta-analysis of genome-wide association data identifies four new susceptibility loci for colorectal cancer. Nat Genet. 2008;40:1426–1435. doi: 10.1038/ng.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Smith P, et al. A genome wide linkage search for breast cancer susceptibility genes. Genes Chromosomes Cancer. 2006;45:646–655. doi: 10.1002/gcc.20330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Huusko P, et al. Genome-wide scanning for linkage in Finnish breast cancer families. Eur J Hum Genet. 2004;12:98–104. doi: 10.1038/sj.ejhg.5201091. [DOI] [PubMed] [Google Scholar]
- 78.Bergman A, et al. Genome-wide linkage scan for breast cancer susceptibility loci in Swedish hereditary non-BRCA1/2 families: suggestive linkage to 10q23.32-q25.3. Genes Chromosomes Cancer. 2007;46:302–309. doi: 10.1002/gcc.20405. [DOI] [PubMed] [Google Scholar]
- 79.Gonzalez-Neira A, et al. Genomewide high-density SNP linkage analysis of non-BRCA1/2 breast cancer families identifies various candidate regions and has greater power than microsatellite studies. BMC Genomics. 2007;8:299. doi: 10.1186/1471-2164-8-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Djureinovic T, et al. A genome wide linkage analysis in Swedish families with hereditary non-familial adenomatous polyposis/non-hereditary non-polyposis colorectal cancer. Gut. 2006;55:362–366. doi: 10.1136/gut.2005.075333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kemp ZE, et al. Evidence of linkage to chromosome 9q22.33 in colorectal cancer kindreds from the United Kingdom. Cancer Res. 2006;66:5003–5006. doi: 10.1158/0008-5472.CAN-05-4074. [DOI] [PubMed] [Google Scholar]
- 82.Neklason DW, et al. Common familial colorectal cancer linked to chromosome 7q31: a genome-wide analysis. Cancer Res. 2008;68:8993–8997. doi: 10.1158/0008-5472.CAN-08-1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kemp Z, et al. Evidence for a colorectal cancer susceptibility locus on chromosome 3q21-q24 from a high-density SNP genome-wide linkage scan. Hum Mol Genet. 2006;15:2903–2910. doi: 10.1093/hmg/ddl231. [DOI] [PubMed] [Google Scholar]
- 84.Ahmed ZM, et al. The molecular genetics of Usher syndrome. Clin Genet. 2003;63:431–444. doi: 10.1034/j.1399-0004.2003.00109.x. [DOI] [PubMed] [Google Scholar]
- 85.Zhang XJ, et al. The genetic concept of vitiligo. J Dermatol Sci. 2005;39:137–146. doi: 10.1016/j.jdermsci.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 86.Passarge E. Letter: Genetic heterogeneity recognized by Waardenburgh in 1935. Am J Hum Genet. 1976;28:300–301. [PMC free article] [PubMed] [Google Scholar]
- 87.Torres VE, Harris PC. Autosomal dominant polycystic kidney disease: the last 3 years. Kidney Int. 2009;76:149–168. doi: 10.1038/ki.2009.128. [DOI] [PMC free article] [PubMed] [Google Scholar]