Abstract
Aromatase is a unique cytochrome P450 that catalyzes the removal of the 19-methyl group and aromatization of the A-ring of androgens for the synthesis of estrogens. All human estrogens are synthesized via this enzymatic aromatization pathway. Aromatase inhibitors thus constitute a frontline therapy for estrogen-dependent breast cancer. Despite decades of intense investigation, this enzyme of the endoplasmic reticulum membrane has eluded all structure determination efforts. We have determined the crystal structure of the highly active aromatase purified from human placenta, in complex with its natural substrate androstenedione. The structure shows the binding mode of androstenedione in the catalytically active oxidized high-spin ferric state of the enzyme. Hydrogen bond forming interactions and tight packing hydrophobic side chains that complement the puckering of the steroid backbone provide the molecular basis for the exclusive androgenic specificity of aromatase. Locations of catalytic residues and water molecules shed new light on the mechanism of the aromatization step. The structure also suggests a membrane integration model indicative of the passage of steroids through the lipid bilayer.
1. Introduction
Cytochrome P450 aromatase (CYP19A1) uses with high specificity androstenedione, testosterone, and 16α-hydroxytestosterone (all with the same androgen backbone) as substrates converting them to estrone, 17β-estradiol, and 17β,16α-estriol (all with the same estrogen backbone), respectively [1–3]. The reaction is a three-step process, each requiring one mol of O2, one mol of NADPH, and coupling with its redox partner cytochrome P450 reductase for the transfer of all important electrons. Aromatase is the only known enzyme in vertebrates capable of catalyzing the aromatization of a six-membered ring. The functional human enzyme is monomeric, comprised of a heme group and a single polypeptide chain of 503 amino-acid residues. It is an integral membrane protein of the endoplasmic reticulum, anchored to the membrane by an amino terminal transmembrane domain [4,5], in addition to other membrane-associating regions. Many soluble bacterial cytochrome P450s including p450cam [6], P450BM-3 [7], p450terp [8], and p450eryf [9], have been crystallized and structures determined by X-ray crystallography. More recently, crystal structures of several recombinant, microsomal human cytochrome P450s (PDB ID codes: 1A2, 2A6, 2A13, 2C8, 2C9, 2D6, 2R1 and 3A4) have been determined [10–14 and references therein]. Many of these microsomal P450s catalyze metabolism of a wide variety of endogenous and xenobiotic compounds and drugs with low substrate specificities.
Aromatase has been the subject of intense biochemical and biophysical investigations for the past 35 years [1–5,15–34]. Nevertheless, many aspects of the aromatase-catalyzed reaction, especially the third aromatization step, remain poorly understood. Lack of a crystal structure of aromatase prompted a number of homology models based on other experimental P450 structures and site-directed mutagenesis data, and generated proposals for androgen-binding scenarios as well as models for the enzyme’s mechanism of action [2,5,15,20–26,28–33]. However, validation of all these results necessitated an experimental three-dimensional model of the enzyme showing the binding mode of the steroidal substrate and its interactions with active side amino acids. Additionally, because inhibition of estrogen biosynthesis by aromatase inhibitors (AIs) constitutes one of the modern therapies for postmenopausal estrogen-dependent breast cancer [35,36], details of the substrates and inhibitor binding interactions at the active site have become increasingly critical information for the development of next generation AIs.
The major impediments to aromatase crystallization have been its strong hydrophobic character, and susceptibility to rapid denaturation in the absence of the protective lipid bilayer. A number of laboratories have reported purification of aromatase from human placenta [16–19] and recombinant expression systems [27,33]. Nevertheless, attempts to crystallize either the placental or a recombinant and modified aromatase have been unsuccessful. Using term human placenta as a rich source of aromatase and a purification technique that employs a highly specific monoclonal antibody-based affinity chromatography [34], we have been able to purify large quantities of the enzyme in the active form under suitable detergent conditions that has permitted the growth of diffraction-quality single crystals and elucidation of the crystal structure [37].
2. Materials and Methods
2.1. Purification of aromatase from human placenta
Aromatase was purified from microsomal fraction of human term placenta as previously described [34,37]. Briefly, an immunoaffinity chromatography column made with the highly specific monoclonal antibody mAb 3-2C2 was used for the purification of aromatase. Emulgen913 was used as the detergent during the purification process. All buffer solutions contained 0.5µM androstenedione. Hydroxyapetite (HA) chromatography was employed to exchange emulgen with β-D-dodecyl maltopyranoside (BDM), a small-molecule detergent suitable for crystallization as well as for the retention of aromatase activity for a prolonged period at 4 °C.
2.2. Crystallization, data collection and structure solution
Details have already been published [37]. Freshly purified aromatase in 100mm phosphate buffer, ph 7.4, containing 20% glycerol, 0.1mm androstenedione, 20mm dithiothreitol and 1mm BDM was mixed with the reservoir cocktails of 24 to 30% polyethylene glycol 4000 and vapor diffused in sealed 24-well sitting drop plates against corresponding reservoir solution. The purification and crystallization experiments were all conducted at 4 °C.
Initially, diffraction data sets to about 3.3Å resolution were collected at the A-1 station of the Cornell High Energy Synchrotron Source (CHESS). Additionally, two data sets, each to about 4Å resolution, were measured at the CHESS F-2 station by tuning the beam to the peak and the inflexion point of the iron absorption edge. Finally, a 2.9Å diffraction dataset used for solution and refinement of the structure was gathered at the beamline SBC 19-ID-D of the Advanced Photon Source, Argonne National Laboratory, Argonne, IL. The data was processed with HKL3000 software package [38]. The space group is P3221 and the unit cell parameters are a=b=140.208Å, c=119.266Å, α=β=90°, γ=120°. There is one aromatase molecule in the asymmetric unit, with a solvent content of about 79%. A total of 184,295 diffraction intensities were measured yielding 30,371 unique reflections. The diffraction data was 99.4% complete to 2.90Å resolution with an overall Intensity to sigma ratio of 31.1 and Rmerge of 0.067. The intensity to sigma ratio was 2.8 in the highest resolution shell.
The structure was solved by molecular replacement method using CCP4 package of software [39], coupled with Bijvoet difference Fourier synthesis utilizing the Fe-absorption edge datasets. Model building and refinement were performed with Coot [40] and Refmac5 [41] routines, respectively. The final model contained 452 amino acid residues, a heme group, one androstenedione molecule, 35 solvent waters, and 2 phosphate ions (3792 total atoms). Models for 44 amino- and 7 carboxyl-terminal residues could not be built because of weakness of the their electron densities. Apart from these residues, the electron density for the rest of the molecule was mostly well defined, except for two short loop/turn regions. In the space of missing amino-terminal residues, an isolated patch of weak electron density was identifiable and could be fitted to a four-turn helix. However, owing to weakness of the density, this region could not be unambiguously modeled. Additionally, electron densities appropriate for 2 detergent molecules were located near the transmembrane region of the enzyme, but were not included in the final refinement owing to the possibility of multiple orientations of the sugar moiety or the alkyl chain. A small piece of residual electron density adjacent to the protein chain near the heme proximal site was also not modeled. The final R factor for all reflections between 37.77Å and 2.90Å resolutions was 0.214 and the R-free value was 0.244. The root-mean squared deviations of bond-lengths and angles from ideal values were 0.009Å and 1.32°, respectively. The average isotropic thermal factor (B) for all atoms was 77.3Å2, whereas the Wilson plot B-value was 94.5Å2. There were only 2 violations in the backbone torsion angle Ramachandran plot, both in weaker loop regions. Chimera [42] was used for visualization and analysis of the structural results and preparation of illustrations (Figure 2 and Figure 3).
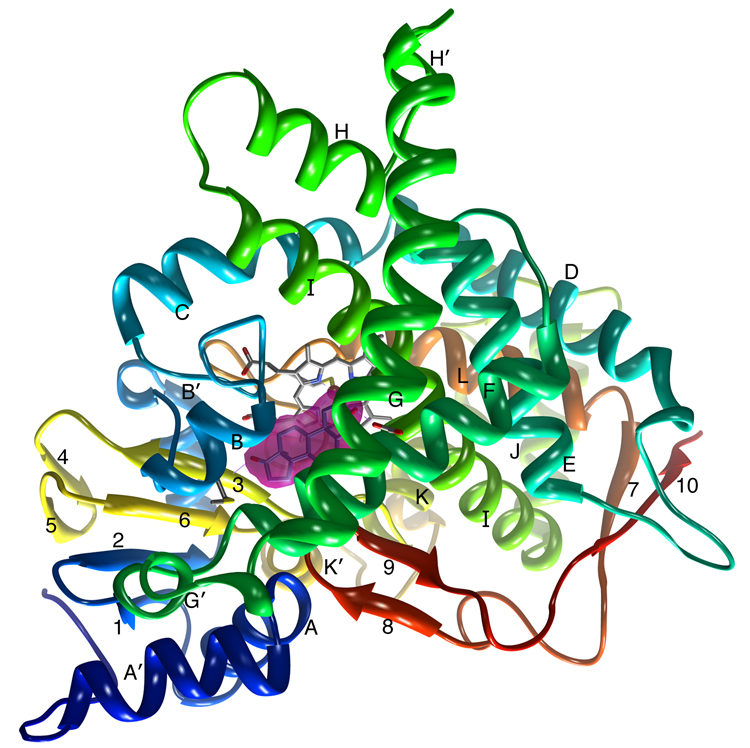
Figure 2.
A ribbon diagram showing the overall structure of human placental aromatase. The amino terminus, starting at residue 45, is colored dark blue and the carboxyl terminus ending at residue 496 is colored red. The α-helices are labeled from A to L and β-strands are numbered from 1 to 10. The heme group and the bound androstenedione molecule at the active site are shown. The bound androstenedione molecule is shown within its unbiased electron density surface contoured in magenta at ~4.5 times the standard deviation. The hydrogen bond-forming interactions at the 3-keto and 17-keto end with Asp309 side chain and Met374 backbone amide, respectively, are also indicated. All 3D illustrations are prepared with Chimera [35].
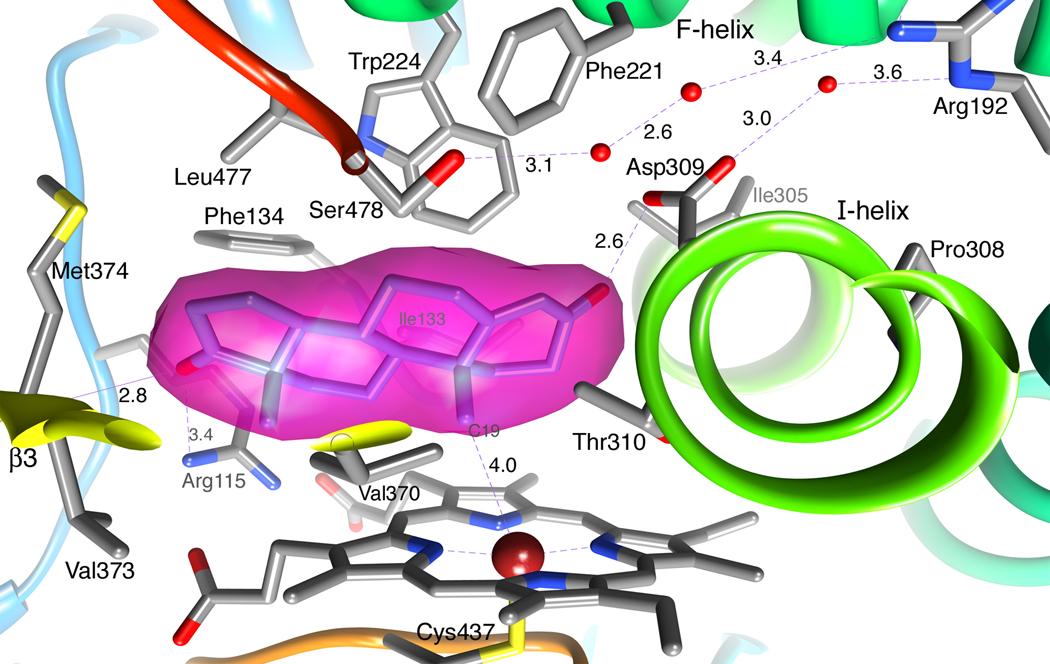
Figure 3.
A close up view of the human placental aromatase active site showing the bound androstenedione molecule within its unbiased electron density surface. Important side chains, heme and water molecules are depicted in the following element colors: gray (carbon), blue (nitrogen), red (oxygen), yellow (sulfur) and firebrick (Fe). The carbon atoms of androstenedione are colored in cornflower blue. The color code is maintained for all figures throughout the manuscript. The distances are in Å.
3. Results and Discussion
3.1. Purification and crystallization
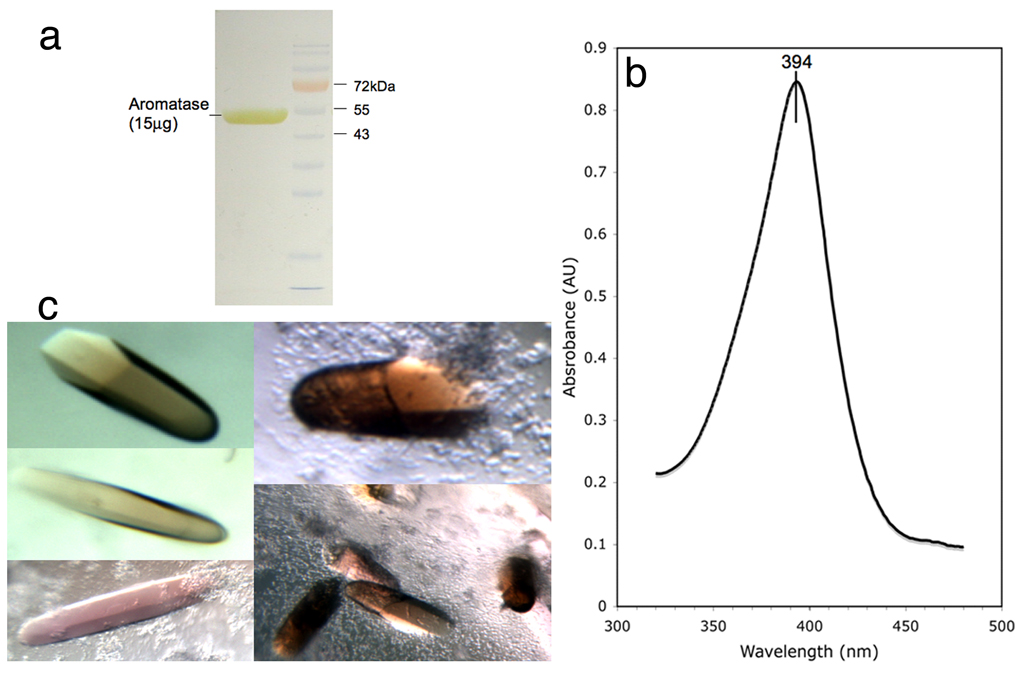
One placenta typically yielded 5 to 10mg of aromatase purified essentially to homogeneity (Figure 1a). The purified enzyme was highly active – the specific activity with androstenedione as the substrate ranged between ~10 and ~100nmol/min/mg over a large number of purification experiments. The absorption spectrum of the androstenedione complex exhibited a Soret band at 394nm (Figure 1b), characteristic of oxidized high-spin ferric (Fe3+) state of the heme iron, suggestive of the formation of the androstenedione-complex. Reddish-brown color hexagonal rod-shaped crystals appeared in 7–10 days and continued to grow up to 14–16 days. Typically, the crystals are about 0.05mm to 0.50mm in lengths, and have a hexagonal cross-section of about 0.01mm to 0.12mm (Figure 1c).
Figure 1.
(a) A sodium dodecyl sulfate polyacrylamide gel electrophoretic analysis of purified and concentrated human placental aromatase. (b) Optical spectrum of purified aromatase (1.6mg/ml) showing the Soret peak at 394 nm, characteristic of oxidized high spin ferric (Fe3+)-androstenedione complex. (c) Single crystals of aromatase-androstenedione complex from various crystallization experiments. Typically, the crystals are 0.1–0.5mm long.
3.2. The overall structure of aromatase
A ribbon diagram of the overall crystal structure of human placental aromatase is shown in Figure 2. The tertiary structure consists of 12 major α-helices (labeled A through L) and 10 β-strands (numbered 1 through 10) distributed into 1 major and 3 minor sheets, and follows the characteristic cytochrome P450 fold. The bound androstenedione molecule at the heme distal site, the active site of the enzyme, and shown within its unbiased electron density, makes two hydrogen bond-forming contacts - the 3-keto and 17-keto oxygens with Asp309 side chain and Met374 backbone amide, respectively (Figure 2). The major β-sheet is a mixed 4-stranded sheet that begins near the amino terminus (first two strands are β1:83–88 and β2:93–97) but ends in two strands from the carboxyl terminal half of the polypeptide chain (β3:373–376 and β6:393–396). A feature somewhat special to the aromatase structure is that the amino-terminal residues 47–50, which makes one backbone hydrogen bond with β1, adds an extra β-strand-like element to this sheet. Each of the three minor sheets consists of two anti-parallel strands scattered over the polypeptide chain (sheet2: β4:381–383 and β5:386–388; sheet3: β8:473–475 and β9:479–481; sheet4: β7:458–461 and β10:491–494). Of the 12 major helices, the lengths, locations and orientations of helices I (293–324), F (210–227), G (242–267), H (278–287), C (138–152), D (155–174), E (187–205), J (326–341), K (354–366) and L (440–455) are similar to those found in most of the cytochrome p450s. Other helices, namely A’ (57–68), A (69–80), B (100–109), B’ (119–126), G’ (232–236), H’ (271–274), J’ (346–349), K’ (398–404), and K” (414–418) are 1 to 4 turns long and have more variability among P450s in terms of their locations, lengths and orientations. For instance, when compared with two human P450s 3A4 and 2D6 that aromatase has the closest resemblance to (~14–18% sequence identity), the helix A’ in aromatase is longer than a similar one in 3A4 and is not seen in 2D6. The other notable difference in the secondary structures between aromatase and 3A4 is that the helix F in 3A4 is separated into two shorter helices by a stretch of polypeptide. As discussed below, this region of the structure contributes significantly to the constitution of the active site. Another difference is that the G-helix in aromatase is at least one turn longer than those in 3A4 and 2D6. The F-helix-loop-G-helix region, in general, appears to be different in different p450s. With the helix G’ in the middle, the loop is tighter in aromatase than in either 3A4 or 2D6, both of which have longer intervening loops.
A striking feature of the tertiary aromatase structure is that long loops interconnect well-defined secondary structure elements, in general agreement with other P450 structures. One example is the polypeptide between the 2-turn helix K’ and helix L. This stretch of 35 residue polypeptide (405–439), devoid of much secondary structure except the 1-turn helix K”, contributes the all important cysteine ligand (Cys437) to the heme iron. Other examples of long loops are between helices B’ and C, β7 and β8, and β9 and β10, all of which either contribute active site residues or have roles in the scaffolding of functionally important elements. Although these loops have little intra-loop interactions through hydrogen bonding, they stabilize by interacting with other structural elements and are, thus, well defined in the electron density map. Another feature common to all cytochrome P450s is the ligation of the heme group via its propionate moieties by arginine and tryptophan side chains through ionic and hydrogen bonding interactions. These side residues in aromatase are Arg115, Trp141, Arg145, Arg375, and Arg435, homologous to those in 3A4, 2D6 and others.
3.3. Androstenedione snugly fits in the active site cavity
The active site of aromatase is the distal cavity of the heme-binding pocket and the heme iron is the reaction center of the enzyme. The active site, thus, is buried deep within the roughly spherical molecule near its geometrical center. Androstenedione binds with its β-face oriented towards the heme group and C19 of the methyl group positioned at a distance 4.0Å from the Fe-atom (Figure 2 and Figure 3). The refined Fe-position is displaced roughly 0.2Å away from the heme plane towards the ligand Cys437. The 17-keto oxygen of the substrate is ideally situated at a distance 2.8Å from the backbone amide nitrogen of Met374 to accept a proton and make a hydrogen bond. The 3-keto oxygen O1 at the other end is at 2.6Å from the carboxylate Oδ2 of the Asp309 side chain, suggesting protonation of the carboxylate moiety and the formation of a hydrogen bond. The geometries of these two hydrogen bonds are such that 3-keto O1 and the water oxygen atom lie roughly in the carboxylate plane. Furthermore, this water molecule is situated at 3.6Å from the guanidinium group of the Arg192 side chain, which is salt-bridged to Glu483 (not sown). Two water molecules were found hydrogen-bonded to each other and also to the Ser478 side chain OH, which in turn donates its proton to His480 Nδ1 (Ser478OH---Nδ1His480: 2.9Å; not shown), further away from the active site. Moreover, the Ser478 side chain is linked via these two water molecules to Arg192 by a weak hydrogen bond of length 3.4Å (Figure 3).
The steroid-binding pocket that snuggly fits an androstenedione molecule is delineated by a confluence of tight packing hydrophobic residues from various parts of the polypeptide chain. The residues contributing to the catalytic cleft are Ile305, Ala306, Asp309 and Thr310 from I-helix, Phe221 and Trp224 from F-helix, Ile133 and Phe134 from the B’-C loop, Val370, Leu372 and Val373 from the K-β3 loop, Met374 from β3, and Leu477 and Ser478 from the β8–β9 loop. Additionally, 4 arginine and 1 tryptophan side chains are involved in binding polar interactions with the heme propionate moieties, as described earlier. Of these, the NH1 atom of Arg115 approaches O2 (of 17-keto) of androsenedione from a distance of 3.4Å (Figure 3). The hydrophobic residues and porphyrin rings of heme surround and pack against the steroid backbone from all sides at van der waals distances. Side chains of residues Arg115, Ile133, Phe134, Phe221, Trp224, Ala306, Thr310, Val370, Val373, Met374 and Leu477 have direct van der waals contacts with the bound androstenedione molecule. Among these, Ile133, Phe134, Phe221, Trp224 and Leu477 approach the substrate from the α-face of androstenedione and follow the contour and puckering of the steroid backbone, while Arg115, Ala306, and Met374 make contacts at its edge and Thr310, Val370, and Val373 on its β-face. The combined surface creates a pocket just right in volume and shape to tightly enclose androstenedione [37]. This is particularly true for the α-face of the ligand where the van der waals surfaces of the large hydrophobics Phe134, Phe221, and Trp224, dictate the shape. The volume of the binding pocket is estimated to be no more than 400Å3, considerably smaller than those in 3A4 and 2D6 [11,12].
The only significant opening to the pocket is at the bottom right corner where 3 water molecules, having hydrogen bonds to Asp309 and Ser478, are located (Figure 3). This leads to a channel that exits to the exterior of the protein surface. The salt bridging Arg192-Glu483 side chain pair as well as those of Asp309 and Ser478 line the channel, right underneath the surface [37]. It is possible that this channel, although too narrow at points to let steroids through, permits the passage of the substrate and the product, owing to the flexibility of the surrounding tertiary structure.
Pro308 plays a key role in the architecture of the active site. The first 5 amino-terminal turns (Arg293 to Ala306) of I-helix in aromatase have a slightly different orientation – in a direction away from the active site. Pro308 causes a bend in the backbone and a shift in the helix axis, bringing it back more towards the active site to follow its normal course. As a result, some extra space, where the 5th turn of the helix (Met303 to Ala307) otherwise would have been, is created accommodating the 3-keto end of the A-ring. In a normal course of I-helix, the 5th turn would have a steric clash with this end of the substrate [37]. Furthermore, as previously discussed, in addition to the bulky hydrophobic residues and differences in the F-helix that greatly restrict the available space, the catalytic cleft in aromatase is further constricted by the altered location and length of the β8–β9 loop that contributes important residues Ser478 and Leu477 to the active site (Figure 3).
3.4. Structural perspective on the mechanism of aromatization
In a three-step catalytic process that requires 3 mol of O2, 3 mol of NADPH, and the flavoprotein CPR for transferring electrons from NADPH, aromatase converts androgens to estrogens. There is a general agreement among previously proposed models that the first two steps are typical P450 hydroxylation reactions. The mechanism of the third step, the aromatization step unique to aromatase, is not yet clear. Some of the residues derived by homology modeling and implicated in these proposed mechanisms are, in fact, present in the active site. For instance, several studies proposed Asp309 to be a catalytically important residue [2,23,28–30,33]. However, unlike the proposed models, Asp309 appears to be involved not only in catalysis but also in substrate binding. On the other hand, Thr310 is highly conserved in the P450 superfamily and thus was postulated to have the same generic role in hydroxylation as in other P450s. Novelties of the active site also include the strong substrate anchoring interaction at the 17-keto end as well as the specificity-defining hydrophobic interactions that drive the critically important orientation and positioning of the 19-methyl group with respect to the heme Fe.
A plausible scenario for H2β abstraction 2,3-enolization for the aromatization step based on the structural data has already been described [37]. Briefly, H2β of the A-ring of the bound androstenedione that gets abstracted in the aromatization step is close to the Ala306CO---HOγThr310 pair (C=O-----H2β-C2: 3.7Å and C2-H2β----OγH: 3.8Å). In the dioxygen complex of P450cam, the residue pair Thr252-Gly248 carbonyl, and two catalytic water molecules are involved in the activation of ferrous-dioxygen to the hydroxylating Fe(IV)=O species by providing two protons [43]. We propose that in aromatase a similar hydroxylation mechanism involving the corresponding Thr310-Ala306 carbonyl pair, and catalytic water molecules (the binding of which could be promoted by dioxygen binding as in P450cam) are responsible not only for the first two steps, but also for the H2β abstraction of the 2,3-enolization processes of the aromatization reaction. The H2β abstraction and 2,3-enolization by tautomerization could be achieved by a nucleophilic attack on H2β-C by the Ala306CO----HOγThr310 moiety (perhaps along with a water) and a concerted electrophilic attack on the C3-keto oxygen by a protonated Asp309 side chain [37]. Asp309 thus appears to have a direct participation in enolization, unlike the indirect roles of Asp251 and Glu244 in hydroxylation by P450cam and P450eryF, respectively [43,44]. The 1β hydrogen is probably removed following the Fe-peroxy nucleophilic attack on 19-aldehyde as previously proposed [15,21,37].
3.5. Membrane integration and its implication
A hydrophobicity plot of aromatase suggests lipid bilayer integration for residues 21–42 and 49–71, with the possibility of partial lipid association for the regions 43–48, 72–80, and 450–470. The model proposed for membrane integration of aromatase shows that these amino terminal segments penetrate the membrane and the loop 450–470 near the carboxyl terminus touches the interior of the bilayer [37]. The model is in agreement with previous proposals [4,5] that the first 20 residues at the amino terminus of aromatase, having a glycosylation site at Asn12, reside in the luminal space. This scenario also supports the notion that the residues 21–42 are truly trans membrane, since the bulk of aromatase, with 7 free sulfahydryl groups, 1 cysteine ligand to the heme iron, and no disulfide, must rest in the reducing environment of the cytoplasm. Furthermore, since a helix has to be at least 7–8 turns and 28–30 residues long to span the entire lipid bilayer, it is likely the trans-membrane residues 21–42 traverses the lipid bilayer as an extended polypeptide devoid of secondary or tertiary structures, and hence are conformationally flexible once removed from the lipid bilayer. This scenario is consistent with our finding that electron density of the polypeptide chain is weak beyond residue 45 towards the amino terminus. Interestingly, endoplasmic reticulum membrane-spanning 43 residues at the carboxyl terminus of human placental and recombinant estrogenic 17β-hydroxysteroid dehydrogenase were similarly found to be disordered in the crystal structure and could not be traced [45].
Based on the structural evidence, we propose that the hydrophobic helix A’ (residues 57–68) and at least partially helix A (residues 69–80), are embedded into the membrane, thereby positioning several arginine (Arg64, Arg79 and Arg86) and tryptophan (Trp67 and Trp88, as well as Trp239 from the F-G loop) residues at the lipid-protein interface, a telltale sign of lipid integration of proteins [46 and references therein]. Furthermore, electron densities for at least 2 detergent molecules were identified in the proximity of helix A’, near the Trp67 side chain. Thus, the lipid integration of aromatase possibly begins with these helices, as the amino terminus traverses further into the bilayer towards the lumen side. Such a membrane integration model positions the entrance to the active site access channel on the membrane surface. This arrangement allows steroids to enter and/or exit the aromatase active site directly from/to the lipid bilayer, traveling between and across the F-G loop and the β8–β9 loop [37], as well as a P450 reductase molecule to couple at the heme proximal site [47] for the transfer of electrons. The structure of aromatase thus provides a rationale for its membrane integration.
Acknowledgement
The authors wish to thank Dr. Yoshio Osawa, who pioneered aromatase research and its purification from human placenta at the institute, for many helpful discussions and encouragement. The authors also thank Women’s and Children’s Hospital of Buffalo for providing the placenta used in this work. The research is supported in part by grants GM062794 and GM086893 (to DG) from the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference
- 1.Thompson EA, Siiteri PK. Utilization of oxygen and reduced nicotinamide adenine dinucleotide phosphate by human placental microsomes during aromatization of androstenedione. J. Biol. Chem. 1974;249:5364–5372. [PubMed] [Google Scholar]
- 2.Simpson ER, Mahendroo MS, Means GD, Kilgore MW, Hinshelwood MM, Graham-Lorence S, Amarneh B, Ito Y, Fisher CR, Michael MD, Meldenson CR, Bulun SE. Aromatase cytochrome P450, the enzyme responsible for estrogen biosynthesis. Endocrine Reviews. 1994;15:342–355. doi: 10.1210/edrv-15-3-342. [DOI] [PubMed] [Google Scholar]
- 3.O’Neal Johnston J. Aromatase inhibitors. Critical Rev. Biochem. Mol. Biol. 1998;33:375–405. [PubMed] [Google Scholar]
- 4.Shimozawa O, Sakaguchi M, Ogawa H, Harada N, Mihara K, Omura T. Core glycosylation of cytochrome P-450(arom). Evidence for localization of N terminus of microsomal cytochrome P-450 in the lumen. J Biol Chem. 1993;268:21399–21402. [PubMed] [Google Scholar]
- 5.Amarneh B, Corbin CJ, Peterson JA, Simpson ER, Graham-Lorence S. Functional domains of human aromatase cytochrome P450 characterized by linear alignment and site-directed mutagenesis. Mol Endocrinol. 1993;7:1617–1624. doi: 10.1210/mend.7.12.8145767. [DOI] [PubMed] [Google Scholar]
- 6.Poulos TL, Finzel BC, Howard AJ. High-resolution Crystal Structure of Cytochrome p450cam. J. Mol. Biol. 1987;195:697–700. doi: 10.1016/0022-2836(87)90190-2. [DOI] [PubMed] [Google Scholar]
- 7.Ravichandran KG, Boddupalli SS, Haserman CA, Peterson JA, Deisenhofer J. Crystal Structure of Hemoprotein Domain of P450BM-3, a Prototype for Microsomal p450s. Science. 1993;261:731–736. doi: 10.1126/science.8342039. [DOI] [PubMed] [Google Scholar]
- 8.Hasemann CA, Ravichandran KG, Peterson JA, Deisenhofer J. Crystal Structure and Refinement of p450terp at 2.3Å Resolution. J. Mol. Biol. 1994;236:1169–1185. doi: 10.1016/0022-2836(94)90019-1. [DOI] [PubMed] [Google Scholar]
- 9.Cupp-Vickery J, Poulos TL. Structure of Cytochrome p450eryf: an Enzyme Involved in Erythromycin Biosynthesis. Nat. Struct. Biol. 1995;2:144–153. doi: 10.1038/nsb0295-144. [DOI] [PubMed] [Google Scholar]
- 10.Williams PA, Cosme J, Ward A, Angove HC, Vinković DM, Jhoti H. Crystal structure of human cytochrome P450 2C9 with bound warfarin. Nature. 2003;424:464–468. doi: 10.1038/nature01862. [DOI] [PubMed] [Google Scholar]
- 11.Williams PA, Cosme J, Vinkovic DM, Ward A, Angove HC, Day PJ, Vonrhein C, Tickle IJ, Jhoti H. Crystal structures of human cytochrome P450 3A4 bound to metyrapone and progesterone. Science. 2004;305:683–686. doi: 10.1126/science.1099736. [DOI] [PubMed] [Google Scholar]
- 12.Rowland P, Blaney FE, Smyth MG, Jones JJ, Leydon VR, Oxbrow AK, Lewis CJ, Tennant MG, Modi S, Eggleston DS, Chenery RJ, Bridges AM. Crystal structure of human cytochrome P450 2D6. J. Biol. Chem. 2006;281:7614–7622. doi: 10.1074/jbc.M511232200. [DOI] [PubMed] [Google Scholar]
- 13.Sansen S, Yano JK, Reynald RL, Schoch GA, Griffin KJ, Stout CD, Johnson EF. Adaptations for the Oxidation of Polycyclic Aromatic Hydrocarbons Exhibited by the Structure of Human P450 1A2. J Biol Chem. 2007;282:14348–14355. doi: 10.1074/jbc.M611692200. [DOI] [PubMed] [Google Scholar]
- 14.Smith BD, Sanders JL, Porubsky PR, Lushington GH, Stout CD, Scott EE. Structure of the human lung cytochrome P450 2A13. J Biol Chem. 2007;282:17306–17313. doi: 10.1074/jbc.M702361200. [DOI] [PubMed] [Google Scholar]
- 15.Akhtar M, Calder DL, Corina DL, Wright JN. Mechanistic studies on C19-demethylation in oestrogen biosynthesis. Biochem. J. 1982;201:569–580. doi: 10.1042/bj2010569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakajin S, Shinoda M, Hall PF. Purification to homogeneity of aromatase from human placenta. Biochem Biophys Res Commun. 1986;134:704–710. doi: 10.1016/s0006-291x(86)80477-6. [DOI] [PubMed] [Google Scholar]
- 17.Kellis JT, Vickery LE. Purification and characterization of human placental aromatase cytochrome P-450. J. Biol. Chem. 1987;262:4413–4420. [PubMed] [Google Scholar]
- 18.Osawa Y, Higashiyama T, Fronckowiak M, Yoshida N, Yarborough C. Aromatase. J Steroid Biochem. 1987;27:781–789. doi: 10.1016/0022-4731(87)90150-6. [DOI] [PubMed] [Google Scholar]
- 19.Yoshida N, Osawa Y. Purification of Human Placental aromatase Cytochrome P-450 with Monoclonal Antibody and Its Characterization. Biochemistry. 1991;30:3003–3010. doi: 10.1021/bi00226a004. [DOI] [PubMed] [Google Scholar]
- 20.Zhou DJ, Pompon D, Chen S. Structure-function studies of human aromatase by site-directed mutagenesis: kinetic properties of mutants Pro-308----Phe, Tyr-361----Phe, Tyr-361----Leu, and Phe-406----Arg. Proc. Natl. Acad. Sci. U. S. A. 1991;88:410–414. doi: 10.1073/pnas.88.2.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Akhtar M, Njar VC, Wright JN. Mechanistic studies on aromatase and related C-C bond cleaving P-450 enzymes. J. Steroid Biochem. Mol. Biol. 1993;44:375–387. doi: 10.1016/0960-0760(93)90241-n. [DOI] [PubMed] [Google Scholar]
- 22.Laughton CA, Zvelebil MJ, Neidle S. A detailed molecular model for human aromatase. J. Steroid Biochem. Mol. Biol. 1993;44:399–407. doi: 10.1016/0960-0760(93)90243-p. [DOI] [PubMed] [Google Scholar]
- 23.Chen S, Zhou D, Swiderek KM, Kadohama N, Osawa Y, Hall PF. Structure-function studies of human aromatase. J Steroid Biochem. Mol. Biol. 1993;44:347–356. doi: 10.1016/0960-0760(93)90238-r. [DOI] [PubMed] [Google Scholar]
- 24.Oh SS, Robinson CH. Mechanism of human placental aromatase: a new active site model. J. Steroid Biochem. Molec. Biol. 1993;44:389–397. doi: 10.1016/0960-0760(93)90242-o. [DOI] [PubMed] [Google Scholar]
- 25.Kadohama N, Zhou D, Chen S, Osawa Y. Catalytic efficiency of expressed aromatase following site-directed mutagenesis. Biochim. Biophys. Acta. 1993;1163:195–200. doi: 10.1016/0167-4838(93)90181-p. [DOI] [PubMed] [Google Scholar]
- 26.Zhou D, Cam LL, Laughton CA, Korzekwa KR, Chen S. Mutagenesis study at a postulated hydrophobic region near the active site of aromatase cytochrome P450. J Biol. Chem. 1994;269:19501–19508. [PubMed] [Google Scholar]
- 27.Amarneh B, Simpson ER. Expression of a recombinant derivative of human aromatase P450 in insect cells utilizing the baculovirus vector system. Mol. Cell. Endocrinol. 1995;109:R1–R5. doi: 10.1016/0303-7207(95)03524-b. [DOI] [PubMed] [Google Scholar]
- 28.Graham-Lorence S, Amarneh B, White RE, Peterson JA, Simpson ER. A three-dimensional model of aromatase cytochrome P450. Protein Sci. 1995;4:1065–1080. doi: 10.1002/pro.5560040605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kao YC, Korzekwa KR, Laughton CA, Chen S. Evaluation of the mechanism of aromatase cytochrome P450. A site-directed mutagenesis study. Eur. J. Biochem. 2001;268:243–251. doi: 10.1046/j.1432-1033.2001.01886.x. [DOI] [PubMed] [Google Scholar]
- 30.Chen S, Zhang F, Sherman MA, Kijima I, Cho M, Yuan YC, Toma Y, Osawa Y, Zhou D, Eng ET. Structure-function studies of aromatase and its inhibitors: a progress report. J. Steroid Biochem. Mol. Biol. 2003;86:231–237. doi: 10.1016/s0960-0760(03)00361-3. [DOI] [PubMed] [Google Scholar]
- 31.Hong Y, Yu B, Sherman M, Yuan YC, Zhou D, Chen S. Molecular basis for the aromatization reaction and exemestane-mediated irreversible inhibition of human aromatase. Mol Endocrinol. 2007;21:401–414. doi: 10.1210/me.2006-0281. [DOI] [PubMed] [Google Scholar]
- 32.Karkola S, Höltje H-D, Wähälä K. A three-dimensional model of CYP19 aromatase for structure-based drug design. J. Steroid Biochem. Mol. Biol. 2007;105:63–70. doi: 10.1016/j.jsbmb.2006.11.023. [DOI] [PubMed] [Google Scholar]
- 33.Hong Y, Cho M, Yuan YC, Chen S. Molecular basis for the interaction of four different classes of substrates and inhibitors with human aromatase. Biochem Pharmacol. 2008;75:1161–1169. doi: 10.1016/j.bcp.2007.11.010. [DOI] [PubMed] [Google Scholar]
- 34.Lala P, Higashiyama T, Erman M, Griswold J, Wagner T, Osawa Y, Ghosh D. Suppression of human cytochrome P450 aromatase activity by monoclonal and recombinant antibody fragments and identification of their stable antigenic complex. J. Steroid Biochem. Mol. Biol. 2004;88:235–245. doi: 10.1016/j.jsbmb.2003.12.010. [DOI] [PubMed] [Google Scholar]
- 35.Brueggemeier RW. Update on the use of aromatase inhibitors in breast cancer. Expert Opin Pharmacother. 2006;7:1919–1930. doi: 10.1517/14656566.7.14.1919. [DOI] [PubMed] [Google Scholar]
- 36.Eisen A, Trudeau M, Shelley W, Messersmith H, Pritchard KI. Aromatase inhibitors in adjuvant therapy for hormone receptor positive breast cancer: A systematic review. Cancer Treat Rev. 2008;34:157–174. doi: 10.1016/j.ctrv.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 37.Ghosh D, Griswold J, Erman M, Pangborn W. Structural basis for androgen specificity and oestrogen biosynthesis in human aromatase. Nature. 2009;457:219–223. doi: 10.1038/nature07614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Otninowski Z, Minor W. HKL Manual. New Haven, CT: Yale University; 1995. The HKL Program Suit. [Google Scholar]
- 39.Collaborative Computational Project. Number 4, The CCP4 Suite: Programs for Protein Crystallography. Acta Crystallogr. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 40.Emsley P, Cowtan K. Coot: model building tools for molecular graphics. Acta Crystallogr. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 41.Murshudov GN, Vagin AA, Dodson EJ. Refinement of Macromolecular Structures by the Maximum-Likelihood Method. Acta Crystallogr. 1997;D53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 42.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera - A Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 43.Nagano S, Poulos TL. Crystallographic study on the dioxygen complex of wild-type and mutant cytochrome P450cam. Implications for the dioxygen activation mechanism. J. Biol. Chem. 2005;280:31659–31663. doi: 10.1074/jbc.M505261200. [DOI] [PubMed] [Google Scholar]
- 44.Nagano S, Cupp-Vickery JR, Poulos TL. Crystal structures of the ferrous dioxygen complex of wild-type cytochrome P450eryF and its mutants, A245S and A245T: investigation of the proton transfer system in P450eryF. J. Biol. Chem. 2005;280:22102–22107. doi: 10.1074/jbc.M501732200. [DOI] [PubMed] [Google Scholar]
- 45.Ghosh D, Pletnev VZ, Zhu DW, Wawrzak Z, Duax WL, Pangborn W, Labrie F, Lin SX. Structure of human estrogenic 17 beta-hydroxysteroid dehydrogenase at 2.20 A resolution. Structure. 1995;3:503–513. doi: 10.1016/s0969-2126(01)00183-6. [DOI] [PubMed] [Google Scholar]
- 46.Hernandez-Guzman FG, Higashiyama T, Pangborn W, Osawa Y, Ghosh D. Structure of human estrone sulfatase suggests functional roles of membrane association. J. Biol. Chem. 2003;278:22989–22997. doi: 10.1074/jbc.M211497200. [DOI] [PubMed] [Google Scholar]
- 47.Sevrioukova IF, Li H, Zhang H, Peterson JA, Poulos TL. Structure of a cytochrome P450-redox partner electron-transfer complex. Proc. Natl. Acad. Sci. U. S. A. 1999;96:1863–1868. doi: 10.1073/pnas.96.5.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]