Abstract
Selenoprotein H is a redox-sensing DNA binding protein that upregulates genes involved in antioxidant responses. Given the known links between oxidative stress and heavy metals, we investigated the potential for regulation of selenoprotein H by metals. In silico analysis of the selenoprotein H genes from nine species reveals multiple predicted metal response elements (MREs). To validate MRE function, we investigated the effects of zinc or cadmium addition and metal-responsive transcription factor 1 (MTF-1) knockout on selenoprotein H mRNA levels. Chromatin immunoprecipitation was used to directly assess physical binding of the transcription factor to MREs in the human and mouse selenoprotein H genes. The results reported herein show that selenoprotein H is a newly identified target for MTF-1. Further, whereas nearly all prior studies of MREs focused on those located in promoters, we demonstrate binding of MTF-1 to MREs located downstream of the transcription start sites in the human and murine selenoprotein H genes. Finally, we identified MREs in downstream sequences in 15 additional MTF-1 regulated genes lacking promoter MREs, and demonstrated MTF-1 binding in three of these genes. This regulation via sequences downstream of promoters highlights a new direction for identifying previously unrecognized target genes for MTF-1.
Keywords: selenoprotein H, MTF-1, MREs
Introduction
Human selenoprotein H ( SELH) has recently been identified as a redox-responsive member of the high mobility group DNA-binding protein family [1]. SELH contains a CxxU thioredoxin-like motif, where U indicates the amino acid, selenocysteine[ 2]. CxxU or CxxC motifs are found in enzymes with antioxidant defense or cellular redox functions. We and others have shown that SELH protects against cell death induced by exposure to UV, hydrogen peroxide or buthionine sulfoximine, an inhibitor of glutathione synthesis [1–3]. Moreover, we have also shown that SELH upregulates its own expression, and the expression of genes involved in glutathione biosynthesis and phase II detoxification in response to oxidative stress [1]. Chromatin immunoprecipitation (ChIP) revealed binding of human SELH to DNA sequences containing heat shock or stress response elements. Thus, we have identified human SELH as a redox-sensing DNA-binding protein that regulates the expression of genes involved in antioxidant protection and cellular detoxification.
Selh is moderately expressed in many human and murine tissues, with higher levels of the mRNA detected in murine brain, thymus, testes and uterus, and in early stages of embryonic development [2, 4, 5]. Analysis of human Selh expression profiles (dbEST and SAGE data sets) also suggests elevated expression in some tumors [2], possibly in response to cellular stress or damage. Currently, little is known about the mechanisms regulating Selh gene expression. Here we have investigated Selh gene regulation by performing a series of bioinformatic and experimental analyses of the Selh genes from multiple species. In silico analysis revealed numerous predicted transcription factor binding sites, including multiple metal response elements (MREs) in the Selh genes from all nine species examined. This finding is intriguing, given the redox functions of metals, the links between metals and oxidative stress, and our demonstration that SELH functions in redox sensing and protection against various stresses. We therefore focused this study on the potential regulation of Selh by metals via the putative MREs.
MREs are short cis-acting sequences that are bound by metal responsive transcription factor 1 (MTF-1). MTF-1 is a nucleocytoplasmic shuttling factor that, in the absence of metal treatment, is predominantly cytoplasmic [6-10]. Upon heavy metal exposure or cellular stress, MTF-1 translocates to the nucleus, where it regulates expression of metallothioneins and other genes [11-23]. MREs consist of a highly conserved TGCRCNC core, with variations conferring differential binding by MTF-1. MREs are categorized into 6 types, termed MREa through f. MTF-1 exhibits the strongest affinity towards MREd, decreasing in the order: d ≥ a = c > b > e > f [24]. It has been demonstrated that different Zn++ concentrations can affect MTF-1 binding affinity to different MREs [21, 25], suggesting individual MREs in a given gene may each have a specific contribution to overall MTF-1 regulatory function.
To investigate the potential regulation of Selh by metals, we tested the effects of Zn++ and Cd++ addition on endogenous Selh mRNA levels in human and murine cell lines. We further investigated regulation of Selh by individual MREs, using site-directed mutagenesis and ChIP assays. Finally, we examined the role of MTF-1 in regulation of Selh expression, using mouse embryo fibroblasts derived from wild type and MTF-1 knockout mice. Our results demonstrate that both human Selh and the murine ortholog are targets of MTF-1, and that their regulation is metal ion and cell-type specific. These findings suggest a role for SELH as a sensor of cellular stress, responding not just to oxidative damage, but to heavy metal load as well.
MATERIALS AND METHODS
Cell culture and maintenance
HEK-293 (human embryonic kidney), MSTO-211H (human mesothelioma), and WISH (human amniotic epithelium) cell lines were originally obtained from the ATCC. HEK-293 cells were used for most of our studies on human gene regulation by MTF as previous studies on MTF-1 used these cells. Mouse embryo fibroblasts, designated MEFs, were derived from wild type (w.t.) and MTF-1 knockout (MTF-1-KO) mice [37]. The MTF-1-FLAG™ cell line was created by stable transfection of MTF-1-KO cells with an expression vector for MTF-1-FLAG™[26]. The MEF, MTF-1-KO and MTF-1-FLAG™ cell lines were generous gifts from Dr. Glen Andrews (University of Kansas). Quantitation of MTF-1 expression levels in the three cell lines by western blotting showed that expression was undetectable in MTF-1-KO, and was restored in the MTF-1-FLAG cell line to ~30% of wt MEF levels. For generation and characterization of the MTF-1 levels in these cells lines please see [26, 37]. MSTO-211H cell line was maintained in RPMI-1630 media (GIBCO), supplemented with 10% FBS. HEK-293T, MEF, MTF-1-KO, and MTF-1-FLAG™ cell lines were maintained in Dulbecco’s modified Eagle’s medium (DMEM, GIBCO), supplemented with 10% FBS. WISH cells were grown in a 1:1 mixture of DMEM: F12 media (GIBCO), supplemented with 10% FBS. All cell lines were maintained at 37°C in 5% CO2. For ChIP assays, cells were grown in 100 mm dishes. For luciferase assays, cells were cultured in 24 well plates. For metal additions described below, concentrations were based on experimentally determined sensitivities of individual cell lines.
DNA sequence analysis
Putative transcription factor binding sites were identified using Genomatix (www.genomatix.de), TFSEARCH (http://transfac.gbf.de) and MOTIF (http://www.motif.genome.ad.jp) software. Genomatix Software was also used to extract promoters from database sequences and to assign the positions of extracted fragments for cloning and analysis. Zebrafish and mosquito Selh gene sequences were provided by Drs. Alexsei Lobanov and Vadim Gladyshev (University of Nebraska). The accession numbers (GenBank™) for the selenoprotein genes sequences analyzed are as follows: Selh in different species H. sapiens (3516960), P. troglodytes (114642405), M. mulata (109106230), M. musculus (82546880), R. norvegicus (109470184), B. taurus (119907899), D. rerio (31342376), D. melanogaster (14718671), A. gambiae (27645579); mouse Txnrd2 (NM_013711; GI:145309318); drosophila SelK1 (CG1844). The accession numbers for the non selenoprotein metal responsive genes are as follows: sulfotransferase family 5A, member 1(Sult5a1, NM_020564.2, GI:142377411); Ubiquitin specific protease 24b (Usp24b, XM_001481281.1, GI:149252835), granzyme B (Gzmb, NM_013542.2, GI:133892516), pyruvate kinase, muscle (Pkm2, NM_011099.2, GI:31981561), armadillo repeat containing, X-linked 2 (Armcx2, NM_026139.3, GI:142372774), Fc receptor, IgG, high affinity, (Fcgr1, NM_010186.3, GI:142356991), coactosin-like 1 (Cotl1, NM_028071.2, GI:142383258), interleukin 7 (IL7, NM_008371.3, GI:142372734), retinoid X receptor gamma (Rxrg, NM_009107.2, GI:42476335), Rab7, member RAS oncogene family-like 1 (Rab7l,1 NM_144875.1, GI:21450114), glutathione S-transferase omega 1 (Gsto,1 NM_010362.2, GI:133892764), Membrane progestin receptor alpha (Mrpa, NM_027995.2, GI:142388621), Ubc (XM_001471699.1, GI:149254583), Lipoprotein lipase (Lpl, NM_008509.2, GI:126723005). For MRE identification, we used a modified Transfac matrix. The original matrix is derived from MREs in vertebrate metallothionein genes, and was modified based on information provided in published studies on additional MREs [25, 27–30], which resulted in a more sensitive but still highly specific matrix. Our matrix for MRE search did not include the “G” at 8th position following the MRE core sequence. While we expect 0.30 matches per 1000 nucleotides with the original pattern, our core matrix results in about 0.73 chance matches per 1000 nucleotides.
Human Selh gene promoter constructs
The fragment -641/+4 relative to the human Selh translation start site was amplified by PCR with DeepVent exo minus DNA polymerase (NE Biolabs) and cloned into the pGL3-basic plasmid (Promega) upstream of the firefly luciferase reporter gene to generate the pGL-645 construct (wt MRE1). The mutant, mut MRE1, was generated by introducing three point mutations into the MRE1 core sequence (TGCGCTC → ATCGCTA) of the pGL-645 construct. Constructs were verified by sequencing. The primer sequences are given in Supplemental Table 1.
Luciferase assays
Luciferase assays were performed using Dual luciferase assay kits from Promega and a Turner Designs Luminometer Model TD-20/20. For transient transfections, we used the TransIT-LT1 Reagent (Mirus-Bio Company) following the manufacturer’s instructions. Cells were cultured in complete media until 90% confluent, and co-transfected with 300ng of human wt MRE1 or mut MRE1 reporter plasmid and 30ng of pRL-SV40 expression vector per well. To evaluate the short-term effects of different metals on human Selh promoter activity, 4 h post transfection cells were supplemented with serum-free media containing ZnCl2 or CdCl2, at concentrations indicated in Table 1. After 24 h of incubation, metal treatment was stopped by washing the cells with 1× PBS. Cells were then harvested and assayed for luciferase activity. At least three independent transfections were carried out, with triplicate samples in each.
Table 1.
Treatment conditions in RT-PCR, ChIP and luciferase assays.
| Cell type | Supplementation or treatment | Concentrations used | Time of treatment | Experiment or assay |
|---|---|---|---|---|
| HEK-293T | CdCl2 ZnCl2 |
10; 20μM 50; 100μM |
20 h 20 h |
Luciferase |
| HEK 293T MSTO-211H |
CdCl2 ZnCl2 |
20μM 100μM |
1h* | ChIP, mRNA |
| WISH | CdCl2 ZnCl2 |
20μM 100μM |
1h* | mRNA |
| MTF-1 FLAG MTF-1-KO |
CdCl2 ZnCl2 |
10μM 50μM |
30 min* | mRNA |
After the indicated time of treatment media was exchanged to media without metals and incubated for an additional 2 hours prior to RNA isolation. For ChIP assays this step was omitted.
Chromatin Immunoprecipitation
Cells were cultured in complete media until 90% confluent. Media was then exchanged for serum-free media and cells were incubated for 24h. Cells were stimulated with metals at concentrations and durations indicated in Table 1. DNA-protein complexes were cross-linked for 10 minutes at room temperature using 1% formaldehyde, quenched with 0.125 M glycine and washed with ice cold 1x PBS buffer. Chromatin was sheared into 100 to 700 bp fragments. ChIP assays were performed according to standard protocols [31] using 2 μg anti-MTF polyclonal antibodies (Santa Cruz Biotechnology). For negative controls, an aliquot of cross-linked chromatin was immunoprecipitated with normal goat serum instead of anti-MTF antibody (referred to as “no Ab” fraction). Immunoprecipitated DNAs were treated with RNAse A and crosslinks were reversed by incubation at 65oC for 5 hours, followed by proteinase K treatment. DNA was phenol/chloroform purified, ethanol precipitated and specific enrichment was measured by quantitative PCR analysis. Quantification of co-immunoprecipitated DNA fragments was based on Chakrabarti et al. [32]. Samples from three independent immunoprecipitations were quantified by real time PCR in triplicate. A melting curve analysis was performed for each sample after PCR amplification to ensure that a single product was obtained. If primer dimers were observed for particular primer sets, those samples were analyzed by 4% TBE-PAGE gel electrophoresis and the corresponding band of specific amplification product quantified using Adobe Photoshop software.
Quantitative PCR (qPCR)
For evaluation of mRNA levels, cells were incubated with metals at concentrations and durations indicated in Table 1. Cells were cultured in complete media until 90% confluent. Media was then exchanged with serum free media and cells were further incubated for 24h. Cells were stimulated with metals at concentrations and durations indicated in Table 1. After the indicated time of treatment cells were washed with 1x PBS and media was exchanged to media without metals and incubated for an additional 2 hours prior to RNA isolation. Total RNA was isolated from cells using the Qiagen RNeasy purification kit and treated with DNase I. First strand cDNA was synthesized using a Superscript III reverse transcriptase kit primed with oligo-dT (Invitrogen). PCR reactions were carried out using Power SYBR green mix (Applied Biosystems) in an Applied Biosystems 7900HT Real-Time PCR System. Intron-spanning primers specific to the human Selh and mouse Selh genes, or specific to the MREs in the 5′ flanking or coding regions of the human Selh gene, were used for amplification (Supplemental Table 1). Each reaction was performed in triplicate with default conditions: 50°C for 2 min, 95°C for 10 min, and 40 cycles of 95°C for 15 s, and 55–60°C for 1 min followed by a dissociation cycle.
Statistical Analysis of Gene Expression
Statistical differences were evaluated by Paired Student’s t-test. P values below the nominal 0.05 were deemed significant.
RESULTS
In silico analysis identifies multiple MREs in Selh genes
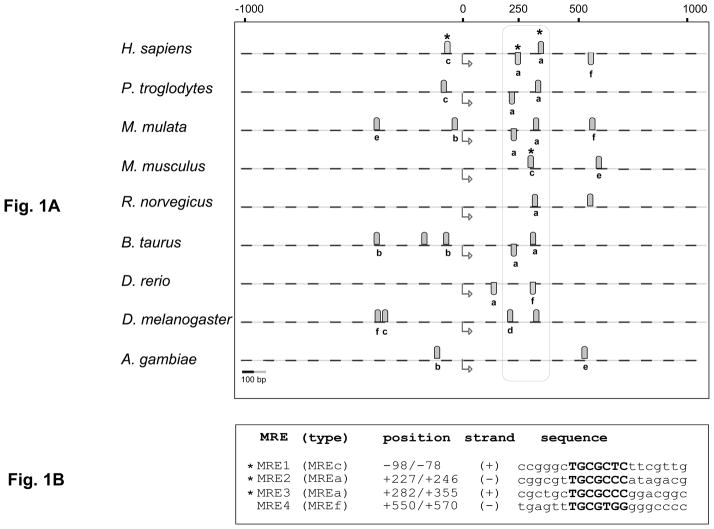
Selh orthologs are found in species ranging from insects to humans. To investigate potential metal responsiveness of Selh gene expression, we carried out in silico analysis of the Selh genes in nine species, spanning the range of organisms in which the gene is present. Previous studies of MREs have focused on those found in promoters, and we found candidate MREs in the promoters of 4 of the 9 Selh genes (Fig. 1A). Intriguingly, computational analysis predicts putative MREs located 200–350 bp downstream of the transcription start site (TrSS) in the Selh genes in all nine species examined (Fig. 1A, encircled by dotted line and Supplementary material Table 2). Nine of the 13 MREs in this region share an identical core sequence with the high affinity MREa element located in the mouse metallothionein promoter. Thus, the MREs in this region have sequence contexts characteristic of high affinity binding to MTF-1. In contrast, MREs located in the 5′UTR (MREb, e and f) and in the region ~550bp downstream of the TrSS (MREe and f) have sequence contexts characteristic of the lower binding affinity to MTF-1 (Fig. 1A).
Fig. 1. Metal response elements in Selh genes from nine species.
A. Position of MREs predicted in the Selh genes from nine species. Genomic DNA from the indicated species (dashed lines) was analyzed over the region spanning 1000bp upstream and downstream from the translational start (TrSS) sites, indicated by bent arrows below the dashed lines. Gray ovals above and below the dashed lines represent predicted MREs on the plus and minus strand, respectively. Experimentally verified MTF-1 binding sites are marked by asterisks. MRE type a through f indicates the mouse metallothionein MRE with which the given MRE shows identity in the core sequence. MREx indicates that those sequences do not have a corresponding counterpart among the mouse metallothionein MREs. The conserved MREs located 200–350bp downstream of the TSS are encircled in the vertical box. Scale is shown at the lower left. B. Predicted MRE sequences in the human Selh gene. MRE1 through 4 predicted in the human Selh gene share identical core sequences (in bold) with MREs from the mouse metallothionein gene. MRE types are given in parentheses.
The conserved core sequence in MREs is often followed by a semi-conserved GC-rich region [28–30]. We noted that in the Selh genes the GC-rich region was in some cases located 5′ to the core MREs sequences, rather than 3′. For example, MRE1 and MRE2 in the human Selh gene contain the GC-rich sequence 5′ to the MRE core sequence (Fig. 1B), whereas MRE3 and MRE4 have the GC-rich region at the 3′ end.
Metal regulation of human Selh mRNA
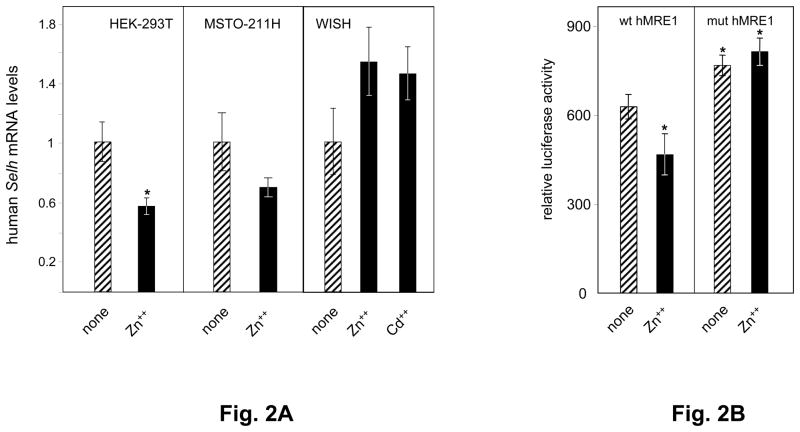
Expression of endogenous human Selh mRNA was evaluated in three human cell lines incubated in the absence or presence of 100 μM Zn++ or 20 μM Cd++. In HEK-293T cells, Zn++ treatment resulted in a significant 43% decrease in human Selh mRNA level, while in MSTO-211H a non-significant 32% decrease was seen (Fig. 2A). Cd++ treatment exerted minimal effects on human Selh mRNA levels in HEK-293 and MSTO-2H11 cells (data not shown). In contrast, in WISH cells, Zn++ or Cd++ treatment increased human Selh mRNA levels by 54 and 45%, respectively, but these changes were also not statistically significant. These results demonstrate that regulation of human Selh mRNA levels is both heavy metal and cell type dependent.
Fig. 2. Human Selh expression is regulated by metals.
A. Human Selh mRNA expression was assessed in HEK-293T, MSTO-211H, and WISH cell lines in the absence (striped bars) or presence (black bars) of heavy metal treatment, as described in the text and in Table 1. mRNA levels relative to Hprt were analyzed by real time PCR (n=3) and are plotted as mean ± SD from 3 experiments. Student’s paired t test was used for statistical evaluation; (*) indicates P<0.05. B. Wt and mut human MRE1 luciferase expression vectors introduced into HEK-293T cells were used to evaluate Selh promoter activity in the absence (striped bars) or presence (black bars) of Zn++. Relative luciferase units are represented as ratio of firefly luciferase activity driven by Selh promoter fragment to Renilla luciferase activity from co-transfected control plasmid (pRLSV40). Each value represents the mean ± standard deviation of four independent transfection experiments, each performed in triplicate. Asterisks indicate values below the nominal P<0.05 compared to untreated wt MRE1 construct.
Mutagenesis of human MRE1 confirms its function
The MRE identified in the 5′UTR of the human Selh gene is defined herein as human MRE1. To evaluate its contribution to Selh regulation, we performed site-directed mutagenesis, replacing three nucleotides in the core MRE1 sequence (TGCGCTC → ATCGCTA). The effect of human MRE1 mutations was tested by comparing wild type (wt human MRE1) and mutant (mut human MRE1) luciferase expression constructs in HEK-293T cells, and the effects of Zn++ addition, as indicated in Table 1. In the absence of added Zn++, the expression from the wt human MRE1 construct was 79% of that from the mut human MRE1 construct (Fig. 2B, compare striped bars in both panels). Treatment with 100 μM Zn++ decreased luciferase expression from the wt human MRE1 construct by a further 21% (Fig. 2B, left panel). The decrease caused by Zn++ treatment was abolished with the mut human MRE1 luciferase expression construct (Fig. 2B, right panel), demonstrating the importance of human MRE1 in human Selh regulation. This result demonstrates that MRE-1 is involved in repression of Selh expression by Zn++.
Chromatin immunoprecipitation confirms physical binding of MTF1 to human Selh MRE1
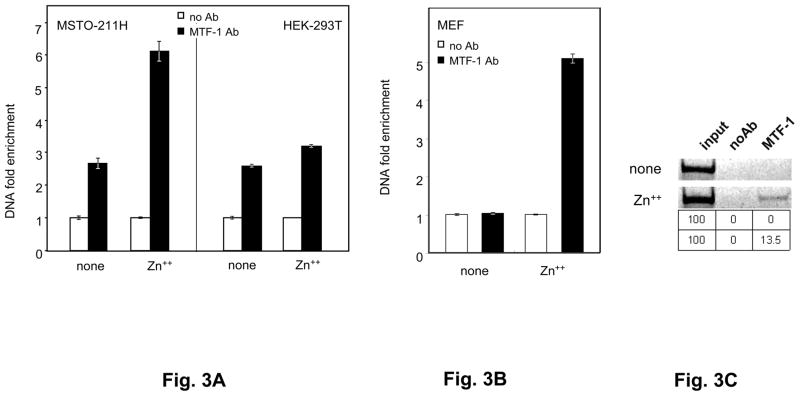
To directly assess MTF-1 binding to hMRE1 in the human Selh gene in the presence or absence of metals, we performed ChIP assays coupled to RT-PCR quantitation of binding. Real-time PCR quantitation of human MRE1 bound by MTF-1 in the absence of added metals showed a ~2.8 fold increase over background (no Ab) binding in both MSTO-211H and HEK-293T (Fig. 3A). This presumably reflects MTF-1 activated by endogenous metals, consistent with the results in Fig. 2B. Zn++ addition resulted in a 3-fold increase in binding over the background level in MSTO-211H cells, but no increase in HEK-293T cells. In contrast, after Cd++ addition, we did not observe MTF-1 binding (data not shown), These results further support a role for MTF-1 in human Selh gene regulation and responsiveness to zinc, through physical binding of MTF-1 to human MRE1.
Fig. 3. Mouse and human Selh genes are new targets of MTF-1.
A. MTF-1 binding to the MRE-1 located in the 5′ UTR of the human Selh gene. ChIP assays were performed on MSTO-211H and HEK-293T cell extracts in the absence or presence of heavy metal treatment. Black bars represent immunoprecipitation with MTF-1 specific antibody (MTF-1 Ab) and white bars represent negative control (normal goat serum), indicated as no Ab. DNA fold enrichment units are normalized against input of total DNA used for immunoprecipitation and no Ab control. Three independent immunoprecipitations were carried out, immunoprecipitated MRE-containing DNA was quantified by real time PCR (n=3) and data are plotted as mean ± SD. B. MTF-1 binding to MREs of mouse Selh coding region was assessed in MEF cells, as described in the legend for Fig 3A. Quantitation of MTF-1 binding for mouse MRE1 was evaluated by real time PCR (n=3) and data are plotted as mean ± SD. C. Gel electrophoresis analysis (4% polyacrylamide -TBE) of the amplification products from HEK-293T cell ChIP assays in the absence or presence Zn++ addition. Band intensities from the gel were quantified using Adobe Photoshop software. Percent MTF-1 binding (signal minus background (no Ab) relative to input) is shown in the table below the gel pictures. Three independent immunoprecipitations were carried out and a representative of the output data is shown in this figure.
Effects of MTF-1 and metals on murine Selh mRNA levels in mouse cell lines
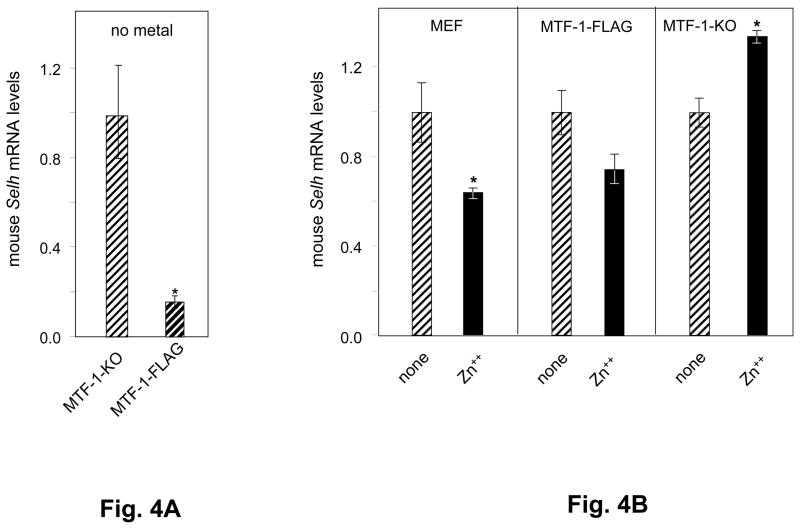
In two species, mouse and rat, putative MREs were identified in the coding regions only, providing a model to investigate whether MTF-1 regulatory functions extend to MREs located downstream of the translation start site (TrSS). To investigate the role of MTF-1 in mouse Selh transcriptional regulation, we analyzed mouse Selh mRNA expression levels in mouse embryo fibroblast cell lines derived from MTF-1-knockout (MTF-1-KO) mice, and from a cell line created by transfection of MTF-1-KO cells with an expression vector for MTF-1-FLAG™ (MTF-1-FLAG), in an attempt to restore MTF-1 expression [26]. Western blotting analysis showed that the MTF-1-FLAG cell line expressed MTF-1 at ~30% of w.t. levels. Quantitation of mouse Selh mRNA levels in the absence of added metals revealed ~73% lower levels in MTF-1-FLAG than in MTF-1-KO cells (Fig. 4A), indicating that MTF-1 expression significantly decreased mouse Selh mRNA levels. This suggests the existence of an activated nuclear pool of MTF-1 in the MTF-1-FLAG cells that is capable of repressing mouse Selh mRNA expression in the absence of added metals.
Fig. 4. Murine Selh mRNA expression is regulated by metals via MTF-1.
A. Mouse Selh mRNA expression was analyzed in MTF-1-KO and MTF-1-FLAG cells in the absence of added metal. mRNA levels in MTF-1-KO assigned a level of 1.0 and the levels in MTF-1-FLAG cells were calculated accordingly. B. Mouse Selh expression was assessed in wild type (MEF) and MTF-1-KO cells in the absence (striped bars) or presence (black bars) of heavy metal treatment, as described in the text and in Table 1. mRNA levels in untreated cells were assigned as 1.0 and the levels in Zn++ treatments were calculated accordingly. Three independent experiments were carried out. Real time PCR data (n=3) are plotted as mean ± SD. Means of two groups were compared using a student. s t-test and asterisks indicate values below the nominal P<0.05 compared to no metal addition.
Mouse Selh mRNA levels were next quantitated following addition of 50 μM Zn++ or 10 μM Cd++, as described in Table 1 and the corresponding legend. In wild type MEF cells, Zn++ addition decreased mouse Selh mRNA levels by ~35% (Fig. 4B, left panel). In MTF-1-FLAG cells, Zn++ addition resulted in a non-statistically significant 25% decrease (Fig. 4B, center panel). In contrast, in the MTF-1-KO cell line, Zn++ addition increased mouse Selh mRNA levels by 35% (Fig. 4B, right panel). These results demonstrate that Zn++ represses mouse Selh mRNA levels in an MTF-1-dependent manner. The absence of MTF-1 in the MTF-1KO cells indicates Zn++ activation via an MTF-1-independent pathway, possibly through other Zn++ dependent transcription factors. The MTF-1-dependent Zn++ effect is further indicated by the difference between the expression levels in MTF-1-KO with Zn++ addition (35% increase) and MEF with Zn++ addition (35% decrease), an ~70% change. Finally, addition of 10 μM Cd++ exerted minimal effects on mouse Selh transcript levels (data not shown), consistent with the pattern for human Selh in HEK-293T and MSTO-211H cells.
ChIP assay confirms physical binding of MTF-1 to MREs in Selh coding region
Putative MREs were identified in the promoter and coding regions of the Selh gene in multiple species (Fig. 1A). In the murine and rat genes, MREs were identified only in the coding region, which allows investigating if those MREs are involved in Selh gene regulation by MTF1.
To assess MTF-1 binding to mouse MRE1, located in exon 2 of the mouse Selh coding region, we performed ChIP assays in the presence or absence of Zn++. In the absence of metal addition, MTF-1 binding was not detected over background levels, whereas addition of Zn++ increased binding ~5-fold above background (Fig. 3B). This result confirms physical binding of MTF-1 to an MRE located in the coding region of mouse Selh, and stimulation of this binding by Zn++.
Human MRE2 and MRE3 are located in the second exon of human Selh, in close proximity to each other (Fig. 1A). MTF-1 binding to these sites in the absence of added Zn++ was not detected over background levels in HEK-293T cells (Fig. 3C). Addition of Zn++ resulted in significant MTF-1 binding.
Txnrd2 gene is a new target of MTF-1
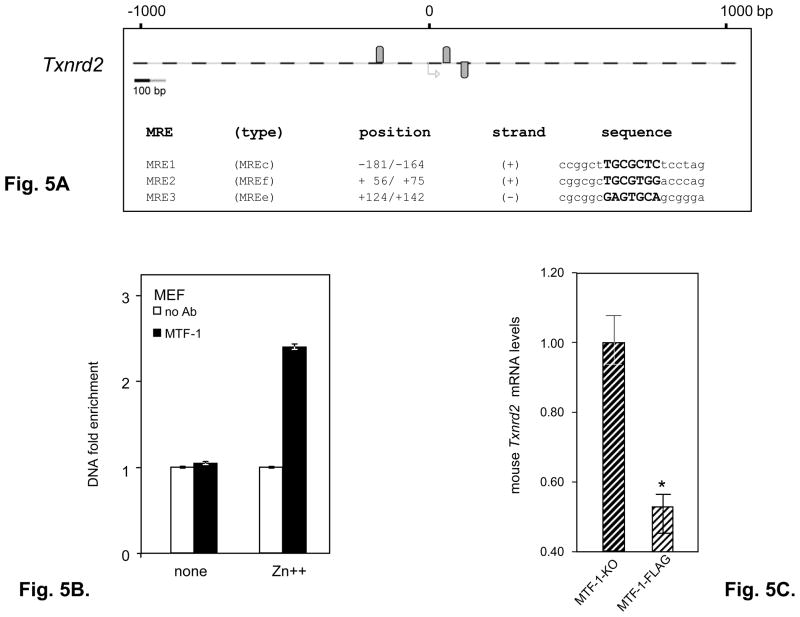
Our in-silico analysis of the human and murine selenoprotein gene promoters identified putative MREs in the coding regions and introns of other selenoprotein genes as well ([33] and our unpublished data). We identified 3 putative MREs for the mouse Txnrd2 gene (Fig. 5A). To investigate if mouse Txnrd2 is also a target of the MTF-1 transcription factor we examined mRNA levels of the mouse Txnrd2 in MTF-FLAG and MTF-KO cell lines and performed ChIP assay in MEF cells with primers specific for the MRE2 and 3 located in the coding region. In the absence of metal addition, MTF-1 binding was not detected over background levels, whereas addition of Zn++ increased binding to 2.4 fold above background (Fig. 5B), consistent with the data observed for mouse and human Selh MREs located in the coding region.
Fig. 5. Mouse Txnrd2 gene is a new target of MTF-1.
A. In-silico analysis of the mouse selenoprotein gene Txnrd2 identified 3 putative MRE sequences. We analyzed the region spanning 1000bp upstream and downstream from the translational start site. Gray ovals above and below the dashed lines represent predicted MREs on the plus and minus strand, respectively. MRE1 through 3 share identical core sequences (in bold) with MREs from the mouse metallothionein gene. The MRE types are given in parentheses. B. MTF-1 binding was assessed in MEF cells using primers specific for MRE2+3 in the absence or presence of heavy metal treatment as described in the text and in the legend for Fig. 3A. Three independent immunoprecipitations were carried. Immunoprecipitated MRE-containing DNA was quantified by real time PCR (n=3) and data are plotted as mean ± SD. C. Mouse Txnrd2 mRNA expression was analyzed in MTF-1-KO and MTF-1FLAG cells in the absence of added metal. Three independent experiments were carried out. mRNAs levels relative to Hprt were analyzed by real time PCR (n = 3) and are plotted as mean ± SD. Student’s t-test was used to evaluate statistical significance (indicated by asterisk) between the two groups.
Thus MRE sequences of mouse Selh, human Selh and mouse Txnrd2 located in the coding region may play differential roles in MTF-1 regulation compared to human MRE1 located in the 5′ UTR, which is constitutively bound by MTF-1 and this binding is further stabilized by Zn++ addition.
Identification of MREs in the coding regions or introns of other MTF-1 responsive genes
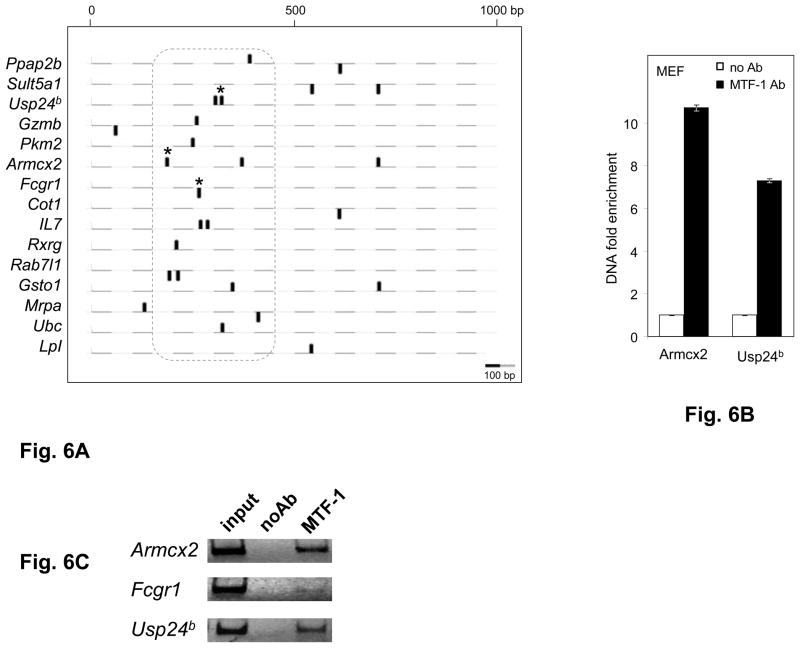
Our finding of functional MREs in the coding region of the mouse Selh, human Selh and mouse Txnrd2 genes suggests that MTF-1 regulatory function may also occur through MREs located in the downstream regions of other genes. A recent study examined the effects of liver-specific MTF-1 knockout on mRNA expression and response to Cd++ exposure [34]. The study identified 55 genes that were significantly up or down-regulated, and of these, 20 did not contain MREs in the promoter regions. We examined the sequences of these 20 genes, and found putative MREs in sequences downstream of the TSS (Fig. 6A) in 15 of the 20, with multiple MREs in 7 genes. These include 5 in 5′UTRs, 17 in introns, and 3 in coding regions (Table 2, and supporting material Table 3). Most of the in silico identified MREs are located in the region +200 /+400bp relative to the TSS encompassed by the dotted line (Fig. 6A).
Fig. 6. Downstream MREs are functional in other genes.
A. Positions of MREs predicted in 15 non-selenoprotein genes known as targets of MTF-1. Genomic DNA was analyzed over the region spanning 1000bp downstream from the transcriptional start sites (TSS). Black ovals above and below the dashed lines represent predicted MREs on the plus and minus strand, respectively. Experimentally verified MTF-1 binding sites are marked by asterisks. The dotted line encompasses the MREs located 150–450bp downstream of the TSS. Scale is shown at the lower left. B. MTF-1 binding to predicted MREs in the murine Armcx2, Fcgr1 and Usp24b genes was assessed in MEF cells in the absence of heavy metal treatment. Black bars represent immunoprecipitation with MTF-1 specific antibody (MTF-1 Ab) and white bars represent negative control (normal goat serum), indicated as no Ab. DNA fold enrichment units for immunoprecipitation and no Ab control are normalized against input of total DNA used. Three independent immunoprecipitations were carried out, immunoprecipitated MRE-containing DNA was quantified by real time PCR (n=3) and data are plotted as mean ± SD. C. Gel electrophoresis analysis (4% polyacrylamide -TBE) of the amplification products of a representative PCR reaction.
Table 2.
Novel MREs identified in genes shown to be regulated by MTF-1.
| Gene | number of MREs | Location |
|---|---|---|
| Ppap2b | 2 | 5′UTR, 1st intron, 5′UTR+CR |
| Sult5a1 | 2 | 1stintron, CR |
| Usp24b | 2 | CR |
| Gzmb | 2 | 5′UTR, 1st intron |
| Pkm2 | 1 | 1st intron |
| Armcx2 | 2 | 5′UTR, 1st intron |
| Fcgr1 | 1 | 1st intron |
| Cotl1 | 1 | 2nd intron |
| IL7 | 2 | 1st intron |
| Rxrg | 1 | 5′UTR |
| Rab7l1 | 2 | 1st intron |
| Gsto1 | 1 | 2nd intron |
| Mrpa | 1 | 1st intron |
| Ubc | 1 | 1st intron |
| Lpl | 1 | 1st intron |
MRE sequences and additional information are given in Supporting Material Table 3.
To investigate whether MTF-1 interacts with these putative downstream MREs, we carried out ChIP analysis of three of the genes, Armcx2, Fcgr1 and Usp24b. Strong MTF-1 binding was seen for the MRE in Armcx2, a high affinity MRE of the MREd type, and for the region in Usp24b gene containing two adjacent MREs, of the MREc and MREn40 types, while weak but specific detectable binding was observed with the Fcgr1 MRE, of the MREn40 type. Melting curve analysis following the real time PCR amplification of Fcgr1 indicated formation of primer dimers, therefore PCR reactions were analyzed by polyacryalamide gel elecrtrophoresis (Fig. 6B and 6C). Images of the gel, indicating strong binding to Armcx2 and Usp24b and faint but specific product for the Fcgr1 PCR amplification, are shown in the middle panel.
Sequence analysis of the MREs identified in silico in the 15 MTF-1 responsive genes (Supporting material, Table 3) reveals that 10 of the genes had core sequences identical to MREn40, suggesting its role in MTF-1 regulation. This sequence was previously reported in a set of sequences that does not require addition of Zn++ for MTF-1 binding [25].
Thus, regulation of expression by MREs downstream of TSSs appears to be a potentially widespread mechanism of MTF-1 regulation.
DISCUSSION
In the present study, we demonstrate that human Selh, mouse Selh and mouse Txnrd2 are new targets of MTF-1. Our expression studies show that human Selh mRNA levels were reduced in Zn++ treated HEK-293T and MSTO-211H cells. Interestingly, expression increased in Zn++ treated WISH cells, indicating cell-type specific regulation of human Selh by Zn++. Mutational analysis of hMRE1 supported our hypothesis that MTF-1 regulates human Selh gene expression. We show that mouse Selh mRNA levels decrease dramatically following introduction of MTF-1 into MTF-1-KO cells. Moreover, following Zn++ treatment, mouse Selh mRNA levels were reduced in MEF and increased in MTF-1-KO knockout cells, demonstrating that MTF-1 is involved in regulation of mouse Selh gene expression. Finally, ChIP assays demonstrate that MTF-1 physically binds to human Selh, mouse Selh and Txnrd2 MREs.
MREs have previously been identified in the promoter regions of glutathione peroxidase 3 and selenoprotein W, suggesting a role for MTF-1 in regulating expression of these two selenoprotein genes [22, 34, 35]. Selenoprotein W expression is upregulated by metals and downregulated in MTF-1-KO mice, and MTF-1 has been shown to bind the selenoprotein W promoter MRE [34]. In this study, we found that MTF-1 represses Selh mRNA levels in response to metal in several cell lines. Thus MTF-1 could serve as either an activator or repressor of selenoprotein gene expression.
Previous investigations of MTF-1 target genes focused on validating MREs located predominantly in promoter regions [18, 36, 37]. Exceptions to this are the reports of MTF-1 repressing the mouse Slc39a10 and its fly homolog Zip10 genes via MREs located downstream of the TSS [34, 38]. Herein, we report that multiple MREs are located up to 650bp downstream from the TrSS of the Selh genes in nine species and in the TXNRD2 gene. Further, our demonstration of functional MREs in the coding regions of the hSelh, mSelh and Txnrd2 genes, and application of this concept to the other MTF-1 regulated genes, revealed MTF-1 binding to MREs in the coding regions or introns in Armcx2, Fcgr1 and Usp24b.
Given the role of SELH in sensing and responding to cellular stress, we might have predicted that its expression would be regulated in response to metals. The repression of Selh mRNA levels by Zn++ in MEF, HEK-293 and MSTO-211H cells, the upregulation of Selh mRNA levels by both Zn++ and Cd++ in WISH cells, and the disparate effects of Zn++ and Cd++ in the ChIP assays suggests that metal regulation of Selh is complex. It is likely that this regulation is dependent on the cellular redox status or oxidant/antioxidant balance of the cell line. In addition, nucleocytoplasmic distribution of MTF-1 in a given cell line under basal conditions will certainly influence metal regulation. Clearly, further investigation will be required to elucidate the mechanistic details of Selh regulation in response to metals and the contributions of the individual MREs to Txnrd2 gene regulation by MTF-1.
Rate limiting steps for MTF-1 regulation of target genes include metal-induced nuclear translocation, activation of DNA binding, and interaction with MREs to form stable MTF-1-chromatin complexes [26]. In contrast, regulation of gene expression by the RNA Pol II stalling mechanism provides rapid transcriptional reactivation in response to stimuli. Heat shock and developmental control genes are well known to be regulated by this mechanism [39-42]. Like heat shock genes, selenoproteins are also involved in multiple cell defense pathways, and regulation by Pol II pausing would be beneficial for rapid responses. Given that we showed MTF-1 regulation via MREs located downstream of the TSS, one can speculate that this regulation might be linked to the RNA Pol II stalling mechanism where MTF-1 binding would possibly contribute to the Pol II stalling.
Recently published data on a Drosophila genome-wide screening for genes regulated by the Pol II stalling mechanism reveal that two of the three Drosophila selenoproteins, SELK and SPS2, showed Pol II promoter-proximal enrichment, suggesting that those genes may be regulated by Pol II stalling [43]. In silico analysis of the Drosophila Selk gene reveals two putative MREs, one in the intron and one in the coding region, suggesting that Drosophila Selk is a potential target of MTF-1 regulation as well. Mouse SelK mRNA levels are reduced by 50% in MTF-KO cells (our unpublished data) supporting involvement of MTF-1 in regulation of mouse Selk gene expression. In this respect, MTF-1 regulation via the MREs located downstream of the TSS may contribute to Pol II pausing and serve as a mechanism to reactivate transcription of Selh and Selk.
Finally, the findings reported herein highlight a new avenue for genome-wide searches to identify previously unrecognized target genes for MTF-1.
Supplementary Material
Acknowledgments
FUNDING. This work was supported by the National Institutes of Health [DK47320, DK52963, NS40302 to MJB] and the Hawaii Community Foundation [No. 20071388 to ZS].
We thank Dr. Glen Andrews (University of Kansas) for providing us with MEF, MTF-1-KO and MTF-1-FLAG cell lines, and for helpful discussions and advice. We thank Dr. Claire Wright (University of Hawaii) for providing us with the WISH cell line, and for helpful discussions. We are grateful to Drs. Vadim Gladyshev, Alexei Lobanov and Perry Ridge (University of Nebraska) for assembling and generously providing us with the Selh gene sequences of Danio rerio and Anopheles gambiae. We also thank Drs. Matthias Hentze and Ralph Shohet for critical reading of the manuscript and helpful suggestions.
ABBREVIATIONS
- MRE
metal responsive element
- MTF-1
metal element binding transcription factor 1
- TrSS
translation start site
- TSS
transcription start site
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Panee J, Stoytcheva ZR, Liu W, Berry MJ. Selenoprotein H is a redox-sensing high mobility group family DNA-binding protein that up-regulates genes involved in glutathione synthesis and phase II detoxification. J Biol Chem. 2007;282:23759–23765. doi: 10.1074/jbc.M702267200. [DOI] [PubMed] [Google Scholar]
- 2.Novoselov SV, Kryukov GV, Xu XM, Carlson BA, Hatfield DL, Gladyshev VN. Selenoprotein H is a nucleolar thioredoxin-like protein with a unique expression pattern. J Biol Chem. 2007;282:11960–11968. doi: 10.1074/jbc.M701605200. [DOI] [PubMed] [Google Scholar]
- 3.Ben Jilani KE, Panee J, He Q, Berry MJ, Li PA. Overexpression of selenoprotein H reduces Ht22 neuronal cell death after UVB irradiation by preventing superoxide formation. Int J Biol Sci. 2007;3:198–204. doi: 10.7150/ijbs.3.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thisse C, Degrave A, Kryukov GV, Gladyshev VN, Obrecht-Pflumio S, Krol A, Thisse B, Lescure A. Spatial and temporal expression patterns of selenoprotein genes during embryogenesis in zebrafish. Gene Expr Patterns. 2003;3:525–532. doi: 10.1016/s1567-133x(03)00054-1. [DOI] [PubMed] [Google Scholar]
- 5.Amsterdam A, Nissen RM, Sun Z, Swindell EC, Farrington S, Hopkins N. Identification of 315 genes essential for early zebrafish development. Proc Natl Acad Sci U S A. 2004;101:12792–12797. doi: 10.1073/pnas.0403929101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Radtke F, Georgiev O, Muller HP, Brugnera E, Schaffner W. Functional domains of the heavy metal-responsive transcription regulator MTF-1. Nucleic Acids Res. 1995;23:2277–2286. doi: 10.1093/nar/23.12.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smirnova IV, Bittel DC, Ravindra R, Jiang H, Andrews GK. Zinc and cadmium can promote rapid nuclear translocation of metal response element-binding transcription factor-1. J Biol Chem. 2000;275:9377–9384. doi: 10.1074/jbc.275.13.9377. [DOI] [PubMed] [Google Scholar]
- 8.Andrews GK. Cellular zinc sensors: MTF-1 regulation of gene expression. Biometals. 2001;14:223–237. doi: 10.1023/a:1012932712483. [DOI] [PubMed] [Google Scholar]
- 9.Saydam N, Georgiev O, Nakano MY, Greber UF, Schaffner W. Nucleo-cytoplasmic trafficking of metal-regulatory transcription factor 1 is regulated by diverse stress signals. J Biol Chem. 2001;276:25487–25495. doi: 10.1074/jbc.M009154200. [DOI] [PubMed] [Google Scholar]
- 10.Laity JH, Andrews GK. Understanding the mechanisms of zinc-sensing by metal-response element binding transcription factor-1 (MTF-1) Arch Biochem Biophys. 2007;463:201–210. doi: 10.1016/j.abb.2007.03.019. [DOI] [PubMed] [Google Scholar]
- 11.Zhang B, Georgiev O, Hagmann M, Gunes C, Cramer M, Faller P, Vasak M, Schaffner W. Activity of metal-responsive transcription factor 1 by toxic heavy metals and H2O2 in vitro is modulated by metallothionein. Mol Cell Biol. 2003;23:8471–8485. doi: 10.1128/MCB.23.23.8471-8485.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Selvaraj A, Balamurugan K, Yepiskoposyan H, Zhou H, Egli D, Georgiev O, Thiele DJ, Schaffner W. Metal-responsive transcription factor (MTF-1) handles both extremes, copper load and copper starvation, by activating different genes. Genes Dev. 2005;19:891–896. doi: 10.1101/gad.1301805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lichtlen P, Schaffner W. The “metal transcription factor” MTF-1: biological facts and medical implications. Swiss Med Wkly. 2001;131:647–652. doi: 10.4414/smw.2001.09672. [DOI] [PubMed] [Google Scholar]
- 14.Tamura Y, Maruyama M, Mishima Y, Fujisawa H, Obata M, Kodama Y, Yoshikai Y, Aoyagi Y, Niwa O, Schaffner W, Kominami R. Predisposition to mouse thymic lymphomas in response to ionizing radiation depends on variant alleles encoding metal-responsive transcription factor-1 (Mtf-1) Oncogene. 2005;24:399–406. doi: 10.1038/sj.onc.1208197. [DOI] [PubMed] [Google Scholar]
- 15.Ran R, Xu H, Lu A, Bernaudin M, Sharp FR. Hypoxia preconditioning in the brain. Dev Neurosci. 2005;27:87–92. doi: 10.1159/000085979. [DOI] [PubMed] [Google Scholar]
- 16.Murphy BJ, Sato BG, Dalton TP, Laderoute KR. The metal-responsive transcription factor-1 contributes to HIF-1 activation during hypoxic stress. Biochem Biophys Res Commun. 2005;337:860–867. doi: 10.1016/j.bbrc.2005.09.124. [DOI] [PubMed] [Google Scholar]
- 17.Heuchel R, Radtke F, Georgiev O, Stark G, Aguet M, Schaffner W. The transcription factor MTF-1 is essential for basal and heavy metal-induced metallothionein gene expression. Embo J. 1994;13:2870–2875. doi: 10.1002/j.1460-2075.1994.tb06581.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lichtlen P, Wang Y, Belser T, Georgiev O, Certa U, Sack R, Schaffner W. Target gene search for the metal-responsive transcription factor MTF-1. Nucleic Acids Res. 2001;29:1514–1523. doi: 10.1093/nar/29.7.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Westin G, Schaffner W. A zinc-responsive factor interacts with a metal-regulated enhancer element (MRE) of the mouse metallothionein-I gene. Embo J. 1988;7:3763–3770. doi: 10.1002/j.1460-2075.1988.tb03260.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Radtke F, Heuchel R, Georgiev O, Hergersberg M, Gariglio M, Dembic Z, Schaffner W. Cloned transcription factor MTF-1 activates the mouse metallothionein I promoter. Embo J. 1993;12:1355–1362. doi: 10.1002/j.1460-2075.1993.tb05780.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stuart GW, Searle PF, Chen HY, Brinster RL, Palmiter RD. A 12-base-pair DNA motif that is repeated several times in metallothionein gene promoters confers metal regulation to a heterologous gene. Proc Natl Acad Sci U S A. 1984;81:7318–7322. doi: 10.1073/pnas.81.23.7318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Amantana A, Vorachek WR, Butler JA, Costa ND, Whanger PD. Effect of copper, zinc and cadmium on the promoter of selenoprotein W in glial and myoblast cells. J Inorg Biochem. 2002;91:356–362. doi: 10.1016/s0162-0134(02)00453-1. [DOI] [PubMed] [Google Scholar]
- 23.Li Y, Kimura T, Huyck RW, Laity JH, Andrews GK. Zinc-induced formation of a coactivator complex containing the zinc-sensing transcription factor MTF-1, p300/CBP, and Sp1. Mol Cell Biol. 2008;28:4275–4284. doi: 10.1128/MCB.00369-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Labbe S, Prevost J, Remondelli P, Leone A, Seguin C. A nuclear factor binds to the metal regulatory elements of the mouse gene encoding metallothionein-I. Nucleic Acids Res. 1991;19:4225–4231. doi: 10.1093/nar/19.15.4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y, Lorenzi I, Georgiev O, Schaffner W. Metal-responsive transcription factor-1 (MTF-1) selects different types of metal response elements at low vs. high zinc concentration. Biol Chem. 2004;385:623–632. doi: 10.1515/BC.2004.077. [DOI] [PubMed] [Google Scholar]
- 26.Jiang H, Daniels PJ, Andrews GK. Putative zinc-sensing zinc fingers of metal-response element-binding transcription factor-1 stabilize a metal-dependent chromatin complex on the endogenous metallothionein-I promoter. J Biol Chem. 2003;278:30394–30402. doi: 10.1074/jbc.M303598200. [DOI] [PubMed] [Google Scholar]
- 27.Kille P, Kay J, Sweeney GE. Molecular biology and environmental induction of piscine metallothionein. Marine Environmental Research. 1995;39:107–110. [Google Scholar]
- 28.Culotta VC, Hamer DH. Fine mapping of a mouse metallothionein gene metal response element. Mol Cell Biol. 1989;9:1376–1380. doi: 10.1128/mcb.9.3.1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stuart GW, Searle PF, Palmiter RD. Identification of multiple metal regulatory elements in mouse metallothionein-I promoter by assaying synthetic sequences. Nature. 1985;317:828–831. doi: 10.1038/317828a0. [DOI] [PubMed] [Google Scholar]
- 30.Koizumi S, Suzuki K, Ogra Y, Yamada H, Otsuka F. Transcriptional activity and regulatory protein binding of metal-responsive elements of the human metallothionein-IIA gene. Eur J Biochem. 1999;259:635–642. doi: 10.1046/j.1432-1327.1999.00069.x. [DOI] [PubMed] [Google Scholar]
- 31.Kuo MH, Allis CD. In vivo cross-linking and immunoprecipitation for studying dynamic Protein:DNA associations in a chromatin environment. Methods. 1999;19:425–433. doi: 10.1006/meth.1999.0879. [DOI] [PubMed] [Google Scholar]
- 32.Chakrabarti SK, James JC, Mirmira RG. Quantitative assessment of gene targeting in vitro and in vivo by the pancreatic transcription factor, Pdx1. Importance of chromatin structure in directing promoter binding. J Biol Chem. 2002;277:13286–13293. doi: 10.1074/jbc.M111857200. [DOI] [PubMed] [Google Scholar]
- 33.Stoytcheva ZR, Berry MJ. Transcriptional regulation of mammalian selenoprotein expression. Biochim Biophys Acta. 2009;1790:1429–1440. doi: 10.1016/j.bbagen.2009.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wimmer U, Wang Y, Georgiev O, Schaffner W. Two major branches of anti-cadmium defense in the mouse: MTF-1/metallothioneins and glutathione. Nucleic Acids Res. 2005;33:5715–5727. doi: 10.1093/nar/gki881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bierl C, Voetsch B, Jin RC, Handy DE, Loscalzo J. Determinants of human plasma glutathione peroxidase (GPx-3) expression. J Biol Chem. 2004;279:26839–26845. doi: 10.1074/jbc.M401907200. [DOI] [PubMed] [Google Scholar]
- 36.Green CJ, Lichtlen P, Huynh NT, Yanovsky M, Laderoute KR, Schaffner W, Murphy BJ. Placenta growth factor gene expression is induced by hypoxia in fibroblasts: a central role for metal transcription factor-1. Cancer Res. 2001;61:2696–2703. [PubMed] [Google Scholar]
- 37.Gunes C, Heuchel R, Georgiev O, Muller KH, Lichtlen P, Bluthmann H, Marino S, Aguzzi A, Schaffner W. Embryonic lethality and liver degeneration in mice lacking the metal-responsive transcriptional activator MTF-1. Embo J. 1998;17:2846–2854. doi: 10.1093/emboj/17.10.2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zheng D, Feeney GP, Kille P, Hogstrand C. Regulation of ZIP and ZnT zinc transporters in zebrafish gill: zinc repression of ZIP10 transcription by an intronic MRE cluster. Physiol Genomics. 2008;34:205–214. doi: 10.1152/physiolgenomics.90206.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zeitlinger J, Stark A, Kellis M, Hong JW, Nechaev S, Adelman K, Levine M, Young RA. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat Genet. 2007;39:1512–1516. doi: 10.1038/ng.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lis J. Promoter-associated pausing in promoter architecture and postinitiation transcriptional regulation. Cold Spring Harb Symp Quant Biol. 1998;63:347–356. doi: 10.1101/sqb.1998.63.347. [DOI] [PubMed] [Google Scholar]
- 41.Xiao H, Lis JT. Heat shock and developmental regulation of the Drosophila melanogaster hsp83 gene. Mol Cell Biol. 1989;9:1746–1753. doi: 10.1128/mcb.9.4.1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Core LJ, Lis JT. Transcription regulation through promoter-proximal pausing of RNA polymerase II. Science. 2008;319:1791–1792. doi: 10.1126/science.1150843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, Zeitlinger J, Adelman K. RNA polymerase is poised for activation across the genome. Nat Genet. 2007;39:1507–1511. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.