Abstract
Objective
Analysis of the αβ T cell receptor (TCR) repertoire in patients with myelodysplastic syndrome (MDS) using the technique of TCR β chain spectratyping has provided valuable insight into the pathophysiology of cytopenias in a subset of patients with this heterogeneous disorder. TCR β chain spectratypes are complex datasets, however, and statistical tools for their comprehensive analysis are limited. The objective of the present work was to develop a method to enable quantitative evaluation and global comparison of spectratype data from different individuals, and to study the prevalence of TCR β repertoire abnormalities in MDS patients
Patients and Methods
We developed a robust statistical method based on k-means clustering analysis, and applied this method to analysis of the αβ TCR repertoires in 50 MDS patients and 23 age-matched healthy controls.
Results
Cluster analysis identified a subset of 11 MDS patients with profoundly abnormal αβ TCR repertoires. This group of patients was characterized by advanced disease by IPSS and WHO criteria, increased expression of the WT1 oncogene, increased bone marrow myeloblast count, and older age.
Conclusions
We have developed a robust analytic algorithm that enables the comparison of αβ TCR repertoires between individuals and have shown that abnormal αβ TCR repertoire is a feature of a subset of patients with advanced MDS.
Keywords: Myelodysplastic Syndromes, TCR spectratyping
Introduction
Patients with myelodysplastic syndrome (MDS) have ineffective hematopoiesis, peripheral blood cytopenias, and are at increased risk of developing acute myeloid leukemia (AML). The pathophysiology of MDS is heterogeneous, and it is clear that MDS comprises several different conditions, rather than a single disease. A subset of MDS patients responds to immunosuppressive therapy with cyclosporine [1,2] or antithymocyte globulin (ATG) [3–10], either singly or in combination with other drugs [11], and compelling in vitro evidence suggests that autologous T lymphocytes contribute to suppression of hematopoiesis in these patients [4,12–15]. Serial analysis of the αβ TCR repertoire in a subset of patients that responded to immunessupression, using the technique of TCR β chain “spectratyping”, has identified prominent spectratype “peaks” corresponding to populations of T cells with identical complementarity-determining region 3 (CDR3) lengths and TCR β chain variable (TRBV) gene utilization, which regressed or disappeared in parallel with the clinical response to immunosuppression [4,6,15–17]. Sequencing of the CDR3 region in the cells comprising the peaks demonstrated that they were often clonal or oligoclonal [6,17], and flow cytometric analysis showed that these cells were primarily CD8+, rather than CD4+, cells, which appeared to have undergone selective proliferative expansion [15,18–21].
Although the regression of clonally expanded T cells in some MDS patients who respond to immunosuppression is consistent with a T cell-mediated autoimmune etiology, the full biological and clinical significance of such clonally expanded cells remains far from clear. A recent study observed clonally expanded T cells in 50% of MDS patients – a significantly larger fraction than would be expected to respond to immunosuppression – and did not identify any association between the presence of such cells and clinical features previously found to be predictive of a response to immunosuppression, such as IPSS score, bone marrow cellularity, and karyotype [20]. Perhaps more important, however, is the fact that clonally expanded T cells are commonly observed in healthy adults, particularly in individuals over the age of 65 years [22–25]. Thus, clonally expanded T cells per se do not identify a group of MDS patients with either a unique pathogenesis or a high likelihood of response to a specific treatment.
We hypothesized that comprehensive analysis of the αβ TCR repertoire in MDS patients might provide more useful insights into the heterogeneous pathophysiology of MDS than has to date been provided by analyses focused solely or primarily on the identification and characterization of expanded clonal populations in TCR β chain spectratypes. Comprehensive analysis of TCR diversity, however, has been limited by the complexity of spectratype data and by the lack of adequate statistical tools suitable for global comparisons between one spectratype and another. We therefore sought to develop a robust and objective statistical framework based on k-means clustering for the analysis of spectratype data that would enable both the comparison between individuals of CDR3 length distributions in specific Vβ families as well as of entire spectratypes, and applied this method to the analysis of the αβ TCR repertoires in 50 patients with MDS and 23 age-matched healthy controls.
Materials and methods
Human subjects and sample acquisition
From March 2006 to November 2007, peripheral blood and bone marrow samples from 50 patients with MDS, aged 18 to 81 years, and peripheral blood samples from 23 healthy donors, aged 50 to 81 years, were obtained with written informed consent using forms approved by the Institutional Review Board of the Fred Hutchinson Cancer Research Center (FHCRC). MDS stage was categorized according to the World Health Organization (WHO) classification [26] and prognostic stratification was assigned based on the categories of the International Prognostic Scoring System (IPSS) [27] at the time of sample acquisition. Patients with chronic myelomonocytic leukemia (CMML) were stratified according to the MD Anderson CMML scoring system into low, intermediate-1, intermediate-2, and high risk, as it has been shown to be more appropriate than the IPSS in this group of patients [28]. Early stage disease and advanced stage disease refer to WHO-defined categories with less than 5% and more than 5% bone marrow myeloblasts, respectively, and low risk disease and high risk disease refer to IPSS low / intermediate-1 and IPSS intermediate-2 / high, respectively.
Sample processing, RNA extraction, and cDNA synthesis
Peripheral blood mononuclear cells (PBMC) and bone marrow mononuclear cells (BMMC) were isolated by Ficoll-Hypaque® density gradient separation. Total RNA was extracted from 5 × 107 PBMC or BMMC using Trizol (Invitrogen), and first-strand cDNA was synthesized using oligo (dT) and SuperScript II reverse transcriptase (Invitrogen).
Analysis of WT1 expression
Quantitative WT1 RT-PCR was performed on triplicate samples using the SYBR Green/ROX PCR Master Mix (SuperArray Bioscience) and run on an ABI7900HT (Applied Biosystems) real-time PCR machine, with β-actin used as the reference gene. The primer sequences used were: WT1-F: 5’ AGCTGTCGGTGGCCAAGTTGTC 3’, and WT1-R: 5’ TGCCTGGGACACTGAACGGTC 3’, as previously described [29]. β-Actin was amplified with ACTB RT2 PCR primer set (SuperArray Bioscience). If the cycle threshold (CT) difference between the triplicates was larger than one cycle, the experiment was repeated. The calculated relative gene expression level was equal to 2−ddCT, where ddCT is the Δ Δ CT, as previously described [30]. The expression of WT1 in each sample was expressed relative to the stable level observed in the leukemic cell line HL-60, which was arbitrarily defined as 1.
T cell receptor β chain spectratyping
TCR β chain spectratyping was performed as previously described [31], with minor modifications. Complementary DNA was synthesized from RNA extracted from unfractionated PBMC and used as template for multiplex PCR amplification of the rearranged TCR β chain CDR3 region. Each multiplex reaction contained a 6-FAM-labeled antisense primer specific for the TCR β chain constant region and two to five TCR β chain variable (TRBV) gene-specific sense primers. All 23 functional Vβ families were studied. PCR reactions were carried out on a Hybaid PCR Express thermal cycler (Hybaid, Ashford, UK) under the following cycling conditions: 1 cycle at 95°C for 6 minutes, 40 cycles at 94°C for 30 seconds, 58°C for 30 seconds, and 72°C for 40 seconds, followed by 1 cycle at 72°C for 10 minutes. Each reaction contained cDNA template, 500 µM dNTPs, 2mM MgCl2 and 1 unit of AmpliTaq Gold DNA polymerase (Perkin Elmer) in AmpliTaq Gold buffer, in a final volume of 20 µl. After completion, an aliquot of the PCR product was diluted 1:50 and analyzed via capillary electrophoresis using a 3730×1 DNA Analyzer (Applied Biosystems). The output of the DNA Analyzer is a distribution of fluorescence intensity vs. time, which was converted to a distribution of fluorescence intensity vs. length by comparison with the fluorescence intensity trace of a reference sample containing known size standards.
Statistical analysis and clustering of TCR β chain spectratypes
During the process of antigen-driven T cell responses, the normal Gaussian profile of the CDR3 length distribution found in antigen-naive Vβ families [32] is disrupted, as a consequence of clonal T cell expansions [33]. In order to capture the variations in the CDR3 length distribution of any given Vβ family, we chose to analyze four features that capture the most significant changes in any given distribution, as previously described [32]: (1) the number of distinct peaks, or CDR3 lengths, in the distribution, (2) the ratio of the amplitude of the highest peak in the distribution to the sum of the amplitudes of all peaks in the distribution (termed the “maximum relative height”), (3) the skewness (a measure that reflects the symmetry of a distribution above and below the mean), and (4) the kurtosis (a measure that reflects the amount of data in the tails as opposed to the central part of the distribution). Thus, the skewness captures the extent to which a CDR3 length distribution is biased toward lengths shorter or longer than the mean, whereas the kurtosis provides information about CDR3 sequences that are either very short or very long.
Feature extraction
Each TCR β chain spectratype comprises 23 individual CDR3 length distributions, one for each of the 23 Vβ families. From each digitized CDR3 length distribution all four features were extracted: A peak finding algorithm was implemented which first performed triangular smoothing of the raw data in each CDR3 length distribution, then located and measured the amplitude of the peaks in the distribution by searching for downward zero crossings in the smoothed first derivative and determining the position, height, and approximate width of each peak. Peaks whose amplitudes were less than 1.5 percent of the maximum peak amplitude were rejected as noise. In addition, if the length difference between two apparent peaks was less than three nucleotides, the two peaks were considered a single peak. Missing Vβ families were replaced by the average of the donor and patient data from the same Vβ family.
Cluster analysis
Unsupervised k-means clustering was performed on the set of four features – number of peaks, maximum relative height, skewness and kurtosis – extracted from all of the CDR3 length distributions within each Vβ family (73 distributions for each Vβ family, with one from each of the 50 MDS patients and the 23 age-matched control subjects), using the square Euclidean distance between variables as the similarity measure [34]. Optimal partitioning of the CDR3 length distributions within each Vβ family was achieved by identifying the number of clusters, k, which maximized the mean silhouette value. Partitioning the 73 CDR3 length distributions in each of the Vβ families into two variably-sized clusters maximized the silhouette value for all but three families (Vβ 1, 5, and 13), in each of which the mean silhouette values for k = 3 were marginally higher than those for k = 2. Because the difference between the mean silhouette values for k = 2 and k = 3 in these three Vβ families was small, it was elected to use k = 2 for the cluster analysis in all 23 families.
K-means clustering was also performed on the level of entire spectratypes, using composite datasets comprising the four features extracted from the CDR3 length distributions for each of the 23 Vβ families. Once again, partitioning the 73 spectratypes into two clusters maximized the mean silhouette value and therefore optimized the clustering process. To determine whether the results of the k-means cluster analysis were robust, unsupervised hierarchical clustering of the 73 spectratypes was also performed. For hierarchical clustering, the relative entropy distance Ds (f, g) between the two variables f (x) and g(x), defined as:
was used to calculate the pair-wise distance matrix D between the data. Agglomerative hierarchical clustering was applied to the spectratyping data using the pair-wise dissimilarity matrix D with the average linkage decision rule to merge two clusters.
Permutation testing
To assess the probability of all of the 23 age-matched controls being assigned to a single cluster by chance given the pair-wise distance matrixD, permutation testing was performed using the test statistic , where the indicator vector,IT = (I1,…,I73) is composed of 23 ones for the age-matched controls and 50 zeros for MDS patients. There are possible ways the 23 age-matched controls can be arranged, and each such arrangement has equal probability. In each of 107 permutations, healthy donors were randomly assigned to the indicator vector I and the test statistic s was calculated on the permuted sample. Given the observed value of statistic s̃ (all normal donors are assigned to the same group) the permutation result was considered significant if the p-value Pr(s ≤ s̃ ) calculated from random samples of a large number of permutations was less than 0.05. The simulations were done on Matlab.
Statistical analysis of clinical variables
We prospectively decided to apply statistical analysis to a limited number of clinical and laboratory features, in order to avoid multiple comparisons. Fisher’s exact test was used for the analysis of categorical variables, such as IPSS scoring, WHO classification, bone marrow cellularity, and transfusion history. The Wilcoxon rank sum test was used for the analysis of continuous variables such as age, WT1 expression, blast count, lymphocyte count, and spectratype features. A p value less than 0.05 was considered statistically significant.
Results
Cluster analysis of CDR3 length distributions in MDS patients and age-matched controls reveals two dominant clusters in each Vβ family
TCR β chain spectratyping was performed in 50 patients with MDS and 23 age-matched healthy individuals (controls). Classification of the 50 MDS patients by WHO stage and IPSS score revealed comparable numbers of patients with early stage and advanced disease (Table 1). The number of peaks, maximum relative height, skewness, and kurtosis were extracted from the 23 CDR3 length distributions comprising each TCR β chain spectratype (Figure 1). K-means clustering was applied at the Vβ family level to the extracted feature data from the CDR3 length distributions across the 73 subjects. This analysis reproducibly identified two distinct clusters within each Vβ family: a “normal” cluster characterized by a higher number of peaks, lower maximum relative height, lower skewness, and lower kurtosis, and a second “abnormal” cluster with the opposite characteristics (Figure 2A). Thus, each CDR3 length distribution was classified as normal or abnormal according to its assignment to one of these two clusters. Abnormal CDR3 length distributions were observed more commonly in the spectratypes from MDS patients than in those from the age-matched control subjects, with a mean number of abnormal CDR3 length distributions per individual of 1.6 (range, 0 to 5) for the age-matched controls and 3.7 (range, 1 to 18) (p=0.03) for the MDS patients. In both control subjects and MDS patients, the abnormal CDR3 length distributions were non-uniformly distributed across the 23 Vβ families (Figure 2B).
Table 1.
Patient characteristics
| Patients, N | 50 |
| Median age, y (range) | 64 (18–81) |
| Gender | 13 F / 37 M |
| WHO classification | |
| Refractory anemia | 7 |
| Refractory anemia with ringed sideroblasts | 4 |
| Refractory cytopenia with multilineage dysplasia | 6 |
| Refractory cytopenia with multilineage dysplasia and ringed sideroblasts |
2 |
| Refractory anemia with excess blasts 1 | 6 |
| Refractory anemia with excess blasts 2 | 12 |
| MDS-AML | 6 |
| Chronic myelomonocytic leukemia | 7 |
| IPSS score | |
| Low | 6 |
| Intermediate-1 | 17 |
| Intermediate-2 | 13 |
| High | 13 |
| Not available | 1 |
| Previous Treatment | |
| Growth Factor | 23 |
| Azacitidine | 10 |
| Anthi-thymocyte globulin | 3 |
| Thalidomide / lenalidomide | 8 |
| Cytotoxic Chemotherapy | 6 |
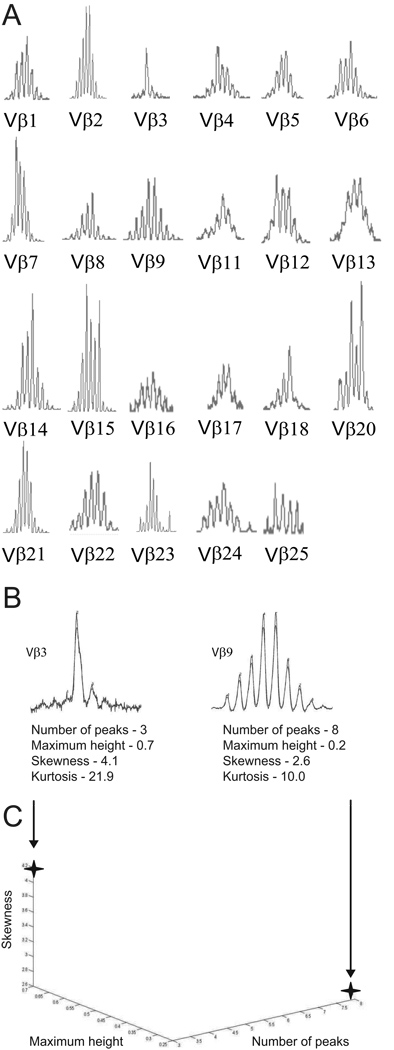
Figure 1. Extraction of features from a TCR β spectratype.
(A) The 23 constituent CDR3 length distributions comprising the TCR β spectratype of a healthy 81-year-old control subject. (B) Extraction of features from two (Vβ3 and Vβ9) of the CDR3 length distributions in (A). (C) Three-dimensional graphic representation of three of the four features – number of peaks, maximum relative height, and skewness - extracted from the two CDR3 length distributions in (B).
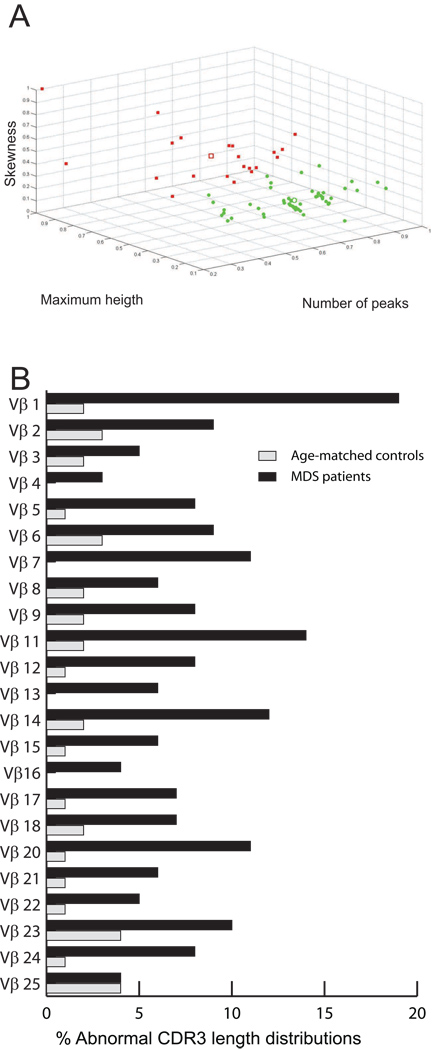
Figure 2. K-means clustering of CDR3 length distributions within Vβ families.
(A) The CDR3 length distributions for Vβ1 from 50 MDS patients and 23 age-matched controls are plotted according to their number of peaks, maximum relative height, and skewness. Two clusters – one characterized by high number of peaks, low maximum relative height, and low skewness (normal), and another with the opposite characteristics (abnormal) – are indicated by the green dots and the red squares, respectively. The open square and the open circle identify the centroids of the two clusters. (B) The percentage of CDR3 length distributions in the spectratypes of the age-matched controls and MDS patients that were assigned to the “abnormal” cluster, by Vβ family.
Cluster analysis at the spectratype level identifies a subset of MDS patients with profoundly abnormal repertoires
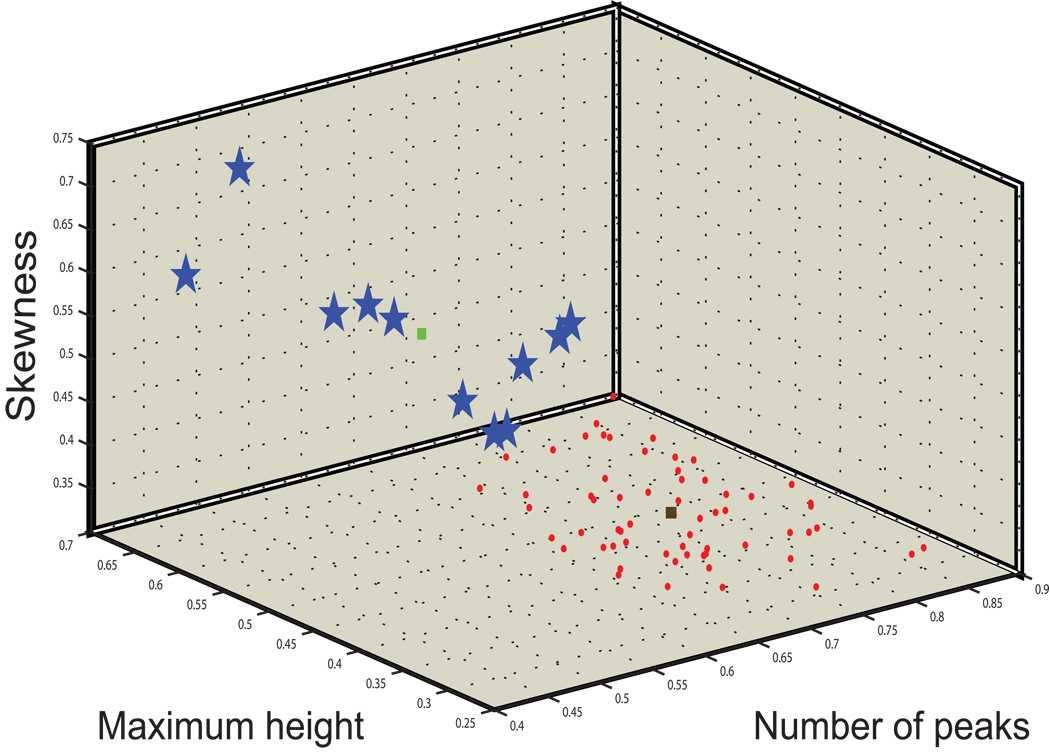
K-means clustering was also applied at the spectratype level to composite datasets consisting of the four features extracted from each of the 23 CDR3 length distributions in each individual’s spectratype. This higher-level clustering again generated 2 clusters (Figure 3). One cluster contained all of the age-matched control subjects as well as 39 MDS patients, while the other cluster contained the remaining 11 MDS patients, all of whom had profoundly abnormal TCR Vβ spectratypes (representative spectratypes from MDS patients assigned to the two clusters in Figure 4). The MDS patients assigned to the smaller, abnormal cluster were, in general, older than those assigned to the larger cluster (median age in the two clusters, 67 vs. 61 years, respectively; p=0.03).
Figure 3. K-means clustering of TCR β spectratypes.
The spectratypes from 50 MDS patients and 23 age-matched controls were clustered on the basis of their number of peaks, maximum relative height, skewness, and kurtosis. Two clusters are evident – one (red squares) characterized by high number of peaks, low maximum relative height, low skewness, and low kurtosis (kurtosis is not plotted in this three-dimensional representation), and another cluster with the opposite characteristics (blue stars). The brown and green squares identify the centroids of the normal and abnormal clusters, respectively. The “normal” cluster contains the spectratypes of all the age-matched controls and 39 MDS patients, while the abnormal cluster contains the spectratypes of 11 MDS patients.
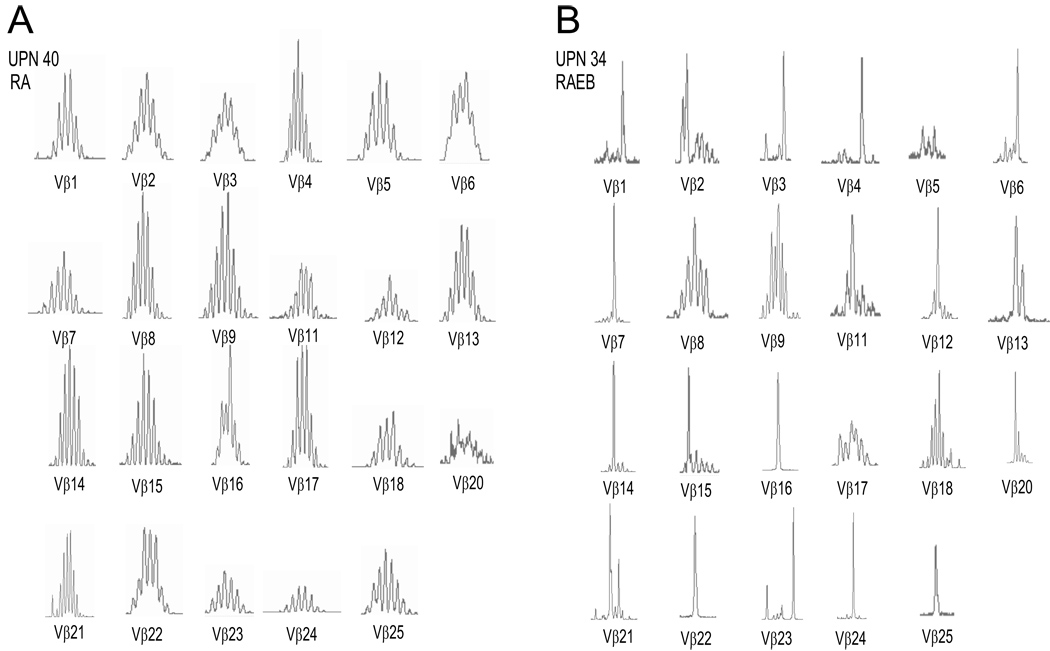
Figure 4. Representative TCR β spectratypes from the normal and abnormal clusters.
TCR β spectratypes from two representative MDS patients, one belonging to the normal cluster (A) and another belonging to the abnormal cluster (B).
To assess the power of cluster analysis at the spectratype level to identify two groups with significantly different αβ TCR repertoires, we compared the distributions in the two groups of the four features on which the clustering was based (number of peaks, maximum relative height, skewness, and kurtosis) as well as the average number of abnormal CDR3 length distributions (Table 2). All four of the features extracted from each CDR3 length distribution were significantly differently distributed between the two groups. Moreover, the average number of abnormal CDR3 length distributions was 2.1 for MDS patients who were assigned to the larger cluster containing all of the control subjects, and 9.4 for the 11 MDS patients assigned to the smaller cluster, confirming that the k-means clustering technique is an effective tool for the identification of patients with an abnormal αβ TCR repertoire.
Table 2.
Comparison of spectratype features between normal and abnormal clusters
| Normal cluster |
Abnormal cluster |
p value |
|
|---|---|---|---|
| Number of peaks | 6.81 | 5.61 | 1×10−4 |
| Maximum height | 0.33 | 0.47 | 8×10−7 |
| Skewness | 2.39 | 3.41 | 7×10−7 |
| Kurtosis | 9.55 | 18.42 | 9×10−7 |
| Median number of abnormal Vβ families | 2 | 9 | 0.02 |
Abnormal TCR repertoire is not attributable to lymphopenia, red cell transfusion, or infections
The two groups of MDS patients defined by cluster assignment were evaluated for potential differences in three variables that could potentially confound the analysis of TCR β chain spectratyping: peripheral blood lymphopenia [35], active infection, and a history of transfusion (Table 3). The median peripheral blood lymphocyte count (1310 / µl versus 1410 / µl, p=0.37), the incidence of MDS-related infection, as defined by a viral, fungal or bacterial infection identified after the diagnosis of MDS but before sample acquisition (27% versus 21%, p=0.69), and a history of transfusion (70% versus 70%; Table 3), did not differ significantly between the MDS patients in the abnormal and normal clusters, respectively.
Table 3.
Comparison of clinical and laboratorial data between the normal and abnormal clusters
| Normal cluster |
Abnormal cluster |
p value |
|
|---|---|---|---|
| Number of patients | 39 | 11 | |
| IPSS score | |||
| Low / Intermediate-1 | 21 (55%) | 2 (18%) | |
| Intermediate-2 / High / MDS-AML | 17 (45%) | 9 (82%) | 0.031 |
| NA | 1 | ||
| WHO classification | |||
| <5% bone marrow blasts | 18 (46%) | 1 (9%) | |
| >5% bone marrow blasts | 16 (41%) | 8 (73%) | 0.027 |
| Chronic myelomonocytic leukemia | 5 (13%) | 2 (18%) | |
| Infection | 8 (21%) | 3 (27%) | 0.69 |
| History of transfusion | |||
| Yes | 24 (70%) | 7 (70%) | 0.67 |
| No | 10 (30%) | 3 (30%) | |
| NA | 5 | 1 | |
| Peripheral blood lymphocyte count (median) | 1410 | 1310 | 0.37 |
| WT1 expression (median) | 0.0062 | 0.034 | 0.047 |
| Bone marrow blasts at sample (median) | 2 | 10 | 0.056 |
| Peripheral blood lymphocyte count (median) | 1410 | 1310 | 0.37 |
| Bone marrow cellularity | |||
| Hypocellular | 6 (15%) | 3 (27%) | 0.4 |
| Normocellular / Hypercellular | 28 (71%) | 7 (63%) | |
| NA | 5 | 1 | |
| Previous Treatment | |||
| Growth Factor | 20 | 3 | 0.189 |
| 5’ Azacytidine | 4 | 6 | 0.004 |
| Anthi-thymocyte globulin | 3 | 0 | 0.4 |
| Thalidomide / lenalidomide | 7 | 1 | 0.66 |
| Cytotoxic Chemotherapy | 4 | 2 | 0.6 |
Abnormal TCR αβ repertoire correlates with features of advanced MDS
The MDS patients assigned to the two clusters were compared to determine if an abnormal αβ TCR repertoire correlated with disease characteristics or stage (Table 3). By IPSS criteria, 82% of the patients in the abnormal cluster had high-risk disease (IPSS intermediate-2 and high), compared to only 45% of the MDS patients in the other cluster (p=0.03), and 73% of patients in the abnormal cluster had advanced disease according to WHO criteria (>5% marrow blasts) as opposed to 41% of those in the other cluster (p=0.027). The 11 MDS patients in the abnormal cluster also had a higher median expression level of the Wilms’ tumor-1 (WT1) gene, as determined by quantitative RT-PCR in peripheral blood (0.034 versus 0.0062, p=0.047), and a higher median bone marrow myeloblast count (10% versus 2%, p=0.056) (Table 3). There were no differences in bone marrow cellularity between the groups. Multivariate analysis was not performed because IPSS score, WHO disease stage, and bone marrow myeloblast count are not, by definition, independent variables. Although patients in the abnormal cluster were characterized by high risk disease, the prevalence of patients with MDS-AML was not different between the 2 clusters (27% in the abnormal and 8.3% in the normal cluster, p = 0.11). The distribution of CMML patients was also similar in both clusters (13% and 18%, respectively).
Comparison between k-means clustering and hierarchical clustering
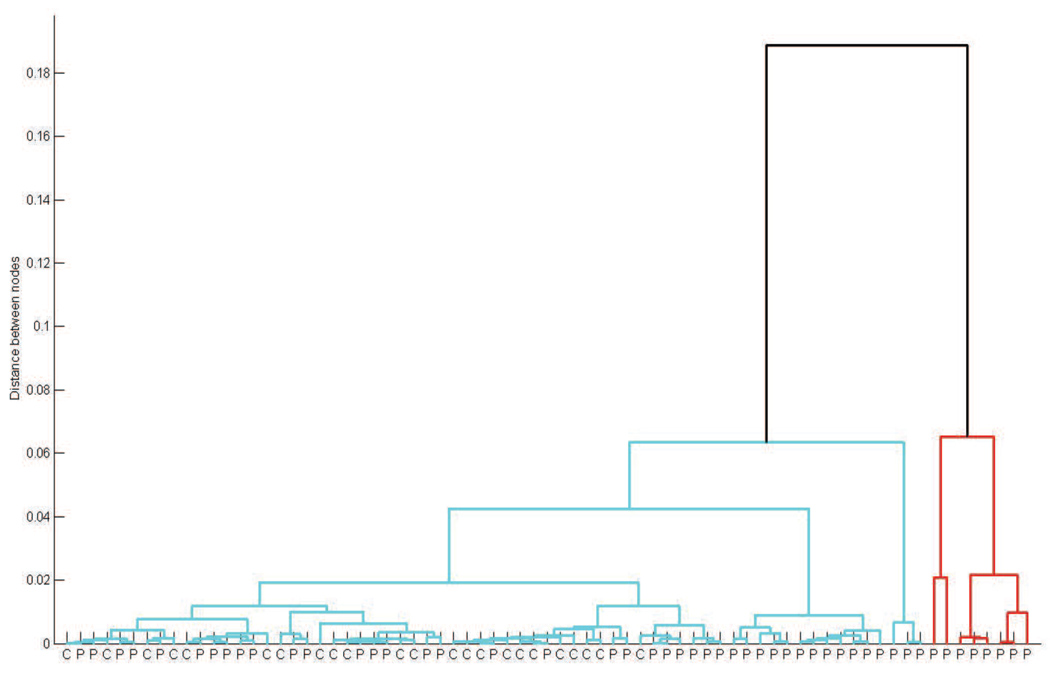
To assess whether the partitioning of the TCR β chain spectratypes from the 50 MDS patients and 23 age-matched controls by k-means clustering was robust, the 73 spectratypes were clustered using a different algorithm and a different distance measure. Unsupervised hierarchical clustering using relative entropy as the measure of dissimilarity reproduced all of the essential features of k-means clustering (Figure 5). The 23 age-matched controls clustered closely with 27 of the MDS patients, all of whom had been assigned to the normal cluster defined by k-means analysis. Permutation testing demonstrated that the assignment of all 23 age-matched controls to the same cluster was highly significant (p<1×10−6). Three additional clusters consisting solely of 12, 3, and 8 MDS patients, respectively, were also identified, which included all of the 11 MDS patients assigned to the abnormal cluster defined by k-means analysis (Figure 5). In particular, 8 of these patients comprised the most distantly related cluster (red branch in dendrogram of Figure 5).
Figure 5. Hierarchical clustering of TCR β spectratypes.
Dendrogram depicting the results of hierarchical clustering of TCR β spectratypes from the 50 MDS patients and 23 age-matched controls. Each branch in the lowest level of the dendrogram represents the spectratype of a single patient (P) or control (C).
Stability of TCR β spectratype over time and during therapy
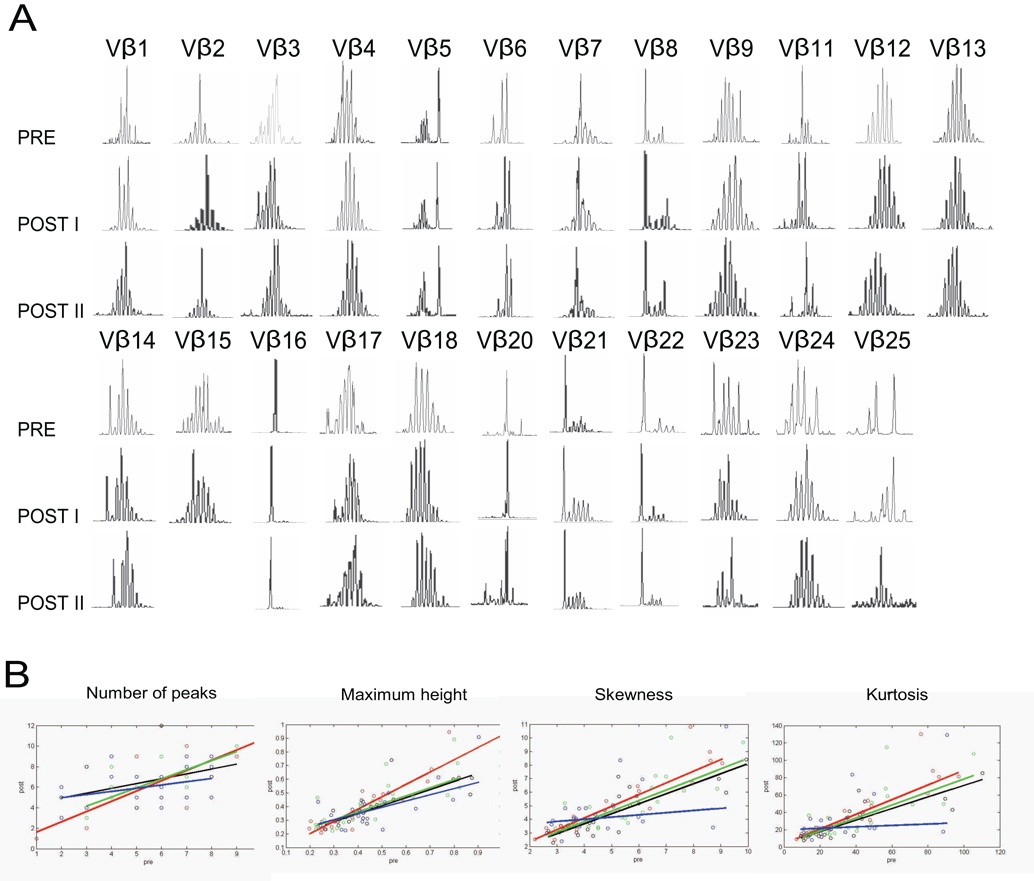
Comparison of the treatment history of the subsets of MDS patients with normal and abnormal spectratyping revealed that prior treatment with azacitidine was significantly more prevalent in the group with abnormal spectratyping (Table 3). To address a possible causal relationship between treatment with azacitidine and abnormalities in the TCRβ spectratype, we analyzed serial bone marrow mononuclear cells samples (BMMC), before and after treatment with azacitidine and etanercept, in 4 MDS patients, as it has been shown that in MDS patients, the analysis of BMMC is more sensitive for detection of abnormal Vβ families than PBMC [20]. All four patients had abnormal TCRβ spectratypes before treatment. In all cases, the spectratypes remained stably abnormal over months of observation, during which time two patients achieved partial and two patients achieved complete remission of their disease (Figure 6, and data not shown).
Figure 6. Serial TCR β spectratype analysis of bone marrow mononuclear cells from MDS patients undergoing treatment with azacitidine and etanercept.
(A) CDR3 length distributions from all 23 Vβ families from one patient achieving complete remission after treatment with azacitidine and etanercept, measured before (PRE), three months (POST I), and six months (POST II) after treatment. (B) Correlation plots comparing each feature (number of peaks, maximum relative height, skewness and kurtosis) by Vβ family before and after (POST II) treatment with azacitidine and etanercept in four MDS patients, two of whom achieved complete remission and two, partial remission. Pre-treatment data are plotted on the x-axis and post-treatment data on the y-axis. Different colors are used to distinguish the data from the four patients.
Discussion
The technique of TCR β spectratyping has been used by numerous groups to study the αβ TCR repertoire in patients with various bone marrow failure syndromes, including MDS [4–6,15,16,19,36]. The interpretation of spectratyping studies in MDS patients has been complicated by the significant heterogeneity of pathogenesis, prognosis, and treatment response in this disease, as well as by the heterogeneity in the statistical methods used for the analysis of spectratype data in different studies. Spectratype data are complex, and the development of comprehensive analytic methods for global comparison of spectratypes from MDS patients and healthy subjects has proved very challenging [37]. In the present study we developed a robust statistical approach to the analysis and comparison of spectratype data from multiple individuals that is based on the extraction of four quantitative features from each of the 23 CDR3 length distributions that comprise each spectratype, followed by k-means clustering of the extracted feature data. Application of this method to the analysis of spectratypes from 50 MDS patients and 23 age-matched controls assigned all age-matched controls, along with a large majority of the MDS patients to a single cluster. However, the cluster analysis also identified a subset of MDS patients with advanced disease and highly abnormal αβ TCR repertoires. Identification of this distinct subset of patients by cluster analysis was robust, as it was reproduced using an entirely different clustering algorithm and similarity metric.
The results of the current study show that the spectrum of abnormalities in the αβ TCR repertoire in patients with MDS has likely been underestimated. Several previous studies have focused on the identification of expanded populations of clonal or oligoclonal T cells [4,6,15,16,19,20,36]. Similar expanded T cell populations have also been commonly observed in healthy adults, particularly those in their seventh decade of life and beyond [22–25], in whom the incidence of MDS is the highest [38,39]. Our results confirm that abnormal CDR3 length distributions characterized by a small number of dominant peaks are observed in the spectratypes of most healthy elderly adults and MDS patients. The most abnormal αβ TCR repertoires, however, are observed in a minority of patients identified by cluster analysis, most of whom had advanced disease. The spectratypes of these patients were characterized by a large number of abnormal CDR3 length distributions with a small number of peaks and high maximum relative height, skewness, and kurtosis, which collectively indicate that these repertoires have severely decreased αβ TCR diversity when compared with healthy elderly adults and the majority of MDS patients.
Further studies will be required to determine the etiology and define the clinical significance of such profoundly abnormal αβ TCR repertoires. The 11 MDS patients with the most abnormal αβ TCR repertoires were slightly older, as a group, than the rest of the cohort. Diversity of the αβ TCR repertoire declines precipitously during the seventh and eighth decades of life [24], and it is thus conceivable that age may, at least in part, account for the more pronounced abnormalities observed in the smaller cluster. Although previous studies have demonstrated that the neoplastic clone does not contribute significantly to the T cell compartment in MDS patients [40], factors produced by the neoplastic cells could potentially disrupt normal lymphoid homeostasis and thereby impair maintenance of a diverse αβ T cell repertoire. The neoplastic clone could also shape the T cell repertoire through expression of antigens that trigger autologous T cell responses. For example, the MDS patients in the abnormal cluster expressed significantly higher levels of WT1, the Wilms’ tumor oncogene, which has been shown to be correlated with disease progression in MDS [41,42]. The protein product of WT1 is a frequent auto-antigen in MDS patients that elicits both autologous humoral [43–45] and cellular [46] immune responses. Although WT1-specific immune responses were not examined in our cohort, our data suggest that future studies should address whether such responses are found more frequently in the subset of patients with the most abnormal TCR repertoires.
Serial TCR β spectratyping of bone marrow mononuclear cells from four MDS patients treated with azacitidine and etanercept showed that the spectratypes remained stably abnormal over intervals ranging from 6 to 18 months, during which time two of the patients achieved complete and two achieved partial remissions of their disease. The cellular and molecular mechanisms underlying responses to azacitidine and etanercept are unknown [47]. Limited data from patients treated with decitabine suggest that the efficacy of demethylating agents in MDS and CMML may in part be attributable to immunologic mechanisms [48,49]. If so, our data suggest that these immunologic mechanisms are not reflected by significant changes in the bone marrow TCR β spectratype.
Our studies demonstrate the value of TCR β spectratyping as a relatively simple tool for analysis of αβ TCR repertoire in diseases such as MDS, but also highlight some of its limitations. Although spectratyping provides useful information on the magnitude of TCR diversity in populations of αβ T cells, this information is more qualitative than quantitative. Thus, spectratyping cannot provide precise estimates of the relative frequency of each unique T cell receptor β chain in a population of T cells. The advent of high throughput, massively parallel, single molecule DNA sequencing [50] now offers the prospect of determining all of the uniquely rearranged TCR β chain CDR3 sequences, and the relative frequency of each CDR3 sequence, in any defined sample of αβ T lymphocytes. The application of these new sequencing technologies to comprehensive definition of the αβ TCR repertoire is the focus of current research in our laboratory.
Acknowledgements
The authors wish to thank Brent L. Wood and Denise A. Wells for their assistance with flow cytometric analysis, Nobuharu Fujii for assistance with RT-PCR, and Colette Chaney for assistance with sample acquisition.
Supported by NIH grants R21 CA119599 (to HJD) and R01 CA106512 (to EHW).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest disclosure
No financial interest/relationships with financial interest relating to the topic of this article have been declared.
References
- 1.Jonasova A, Neuwirtova R, Cermak J, et al. Cyclosporin A therapy in hypoplastic MDS patients and certain refractory anaemias without hypoplastic bone marrow. Br J Haematol. 1998;100:304–309. doi: 10.1046/j.1365-2141.1998.00551.x. [DOI] [PubMed] [Google Scholar]
- 2.Shimamoto T, Tohyama K, Okamoto T, et al. Cyclosporin A therapy for patients with myelodysplastic syndrome: multicenter pilot studies in Japan. Leuk Res. 2003;27:783–788. doi: 10.1016/s0145-2126(03)00008-0. [DOI] [PubMed] [Google Scholar]
- 3.Molldrem JJ, Caples M, Mavroudis D, Plante M, Young NS, Barrett AJ. Antithymocyte globulin for patients with myelodysplastic syndrome. Br J Haematol. 1997;99:699–705. doi: 10.1046/j.1365-2141.1997.4423249.x. [DOI] [PubMed] [Google Scholar]
- 4.Molldrem JJ, Jiang YZ, Stetler-Stevenson M, Mavroudis D, Hensel N, Barrett AJ. Haematological response of patients with myelodysplastic syndrome to antithymocyte globulin is associated with a loss of lymphocyte-mediated inhibition of CFU-GM and alterations in T-cell receptor Vbeta profiles. Br J Haematol. 1998;102:1314–1322. doi: 10.1046/j.1365-2141.1998.00920.x. [DOI] [PubMed] [Google Scholar]
- 5.Saunthararajah Y, Nakamura R, Nam JM, et al. HLA-DR15 (DR2) is overrepresented in myelodysplastic syndrome and aplastic anemia and predicts a response to immunosuppression in myelodysplastic syndrome. Blood. 2002;100:1570–1574. [PubMed] [Google Scholar]
- 6.Kochenderfer JN, Kobayashi S, Wieder ED, Su C, Molldrem JJ. Loss of T-lymphocyte clonal dominance in patients with myelodysplastic syndrome responsive to immunosuppression. Blood. 2002;100:3639–3645. doi: 10.1182/blood-2002-01-0155. [DOI] [PubMed] [Google Scholar]
- 7.Killick SB, Mufti G, Cavenagh JD, et al. A pilot study of antithymocyte globulin (ATG) in the treatment of patients with 'low-risk' myelodysplasia. Br J Haematol. 2003;120:679–684. doi: 10.1046/j.1365-2141.2003.04136.x. [DOI] [PubMed] [Google Scholar]
- 8.Yazji S, Giles FJ, Tsimberidou AM, et al. Antithymocyte globulin (ATG)-based therapy in patients with myelodysplastic syndromes. Leukemia. 2003;17:2101–2106. doi: 10.1038/sj.leu.2403124. [DOI] [PubMed] [Google Scholar]
- 9.Stadler M, Germing U, Kliche KO, et al. A prospective, randomised, phase II study of horse antithymocyte globulin vs rabbit antithymocyte globulin as immune-modulating therapy in patients with low-risk myelodysplastic syndromes. Leukemia. 2004;18:460–465. doi: 10.1038/sj.leu.2403239. [DOI] [PubMed] [Google Scholar]
- 10.Broliden PA, Dahl IM, Hast R, et al. Antithymocyte globulin and cyclosporine A as combination therapy for low-risk non-sideroblastic myelodysplastic syndromes. Haematologica. 2006;91:667–670. [PubMed] [Google Scholar]
- 11.Deeg HJ, Jiang PY, Holmberg LA, Scott B, Petersdorf EW, Appelbaum FR. Hematologic responses of patients with MDS to antithymocyte globulin plus etanercept correlate with improved flow scores of marrow cells. Leuk Res. 2004;28:1177–1180. doi: 10.1016/j.leukres.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 12.Smith MA, Smith JG. The occurrence subtype and significance of haemopoietic inhibitory T cells (HIT cells) in myelodysplasia: an in vitro study. Leuk Res. 1991;15:597–601. doi: 10.1016/0145-2126(91)90028-r. [DOI] [PubMed] [Google Scholar]
- 13.Sugawara T, Endo K, Shishido T, et al. T cell-mediated inhibition of erythropoiesis in myelodysplastic syndromes. Am J Hematol. 1992;41:304–305. doi: 10.1002/ajh.2830410425. [DOI] [PubMed] [Google Scholar]
- 14.Baumann I, Scheid C, Koref MS, Swindell R, Stern P, Testa NG. Autologous lymphocytes inhibit hemopoiesis in long-term culture in patients with myelodysplastic syndrome. Exp Hematol. 2002;30:1405–1411. doi: 10.1016/s0301-472x(02)00968-2. [DOI] [PubMed] [Google Scholar]
- 15.Sloand EM, Mainwaring L, Fuhrer M, et al. Preferential suppression of trisomy 8 compared with normal hematopoietic cell growth by autologous lymphocytes in patients with trisomy 8 myelodysplastic syndrome. Blood. 2005;106:841–851. doi: 10.1182/blood-2004-05-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Epperson DE, Nakamura R, Saunthararajah Y, Melenhorst J, Barrett AJ. Oligoclonal T cell expansion in myelodysplastic syndrome: evidence for an autoimmune process. Leuk Res. 2001;25:1075–1083. doi: 10.1016/s0145-2126(01)00083-2. [DOI] [PubMed] [Google Scholar]
- 17.Wlodarski MW, Gondek LP, Nearman ZP, et al. Molecular strategies for detection and quantitation of clonal cytotoxic T-cell responses in aplastic anemia and myelodysplastic syndrome. Blood. 2006;108:2632–2641. doi: 10.1182/blood-2005-09-3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kook H, Zeng W, Guibin C, Kirby M, Young NS, Maciejewski JP. Increased cytotoxic T cells with effector phenotype in aplastic anemia and myelodysplasia. Exp Hematol. 2001;29:1270–1277. doi: 10.1016/s0301-472x(01)00736-6. [DOI] [PubMed] [Google Scholar]
- 19.Melenhorst JJ, Eniafe R, Follmann D, Nakamura R, Kirby M, Barrett AJ. Molecular and flow cytometric characterization of the CD4 and CD8 T-cell repertoire in patients with myelodysplastic syndrome. Br J Haematol. 2002;119:97–105. doi: 10.1046/j.1365-2141.2002.03802.x. [DOI] [PubMed] [Google Scholar]
- 20.Epling-Burnette PK, Painter JS, Rollison DE, et al. Prevalence and clinical association of clonal T-cell expansions in Myelodysplastic Syndrome. Leukemia. 2007;21:659–667. doi: 10.1038/sj.leu.2404590. [DOI] [PubMed] [Google Scholar]
- 21.Fozza C, Contini S, Galleu A, et al. Patients with myelodysplastic syndromes display several T-cell expansions, which are mostly polyclonal in the CD4(+) subset and oligoclonal in the CD8(+) subset. Exp Hematol. 2009;37:947–955. doi: 10.1016/j.exphem.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 22.Posnett DN, Sinha R, Kabak S, Russo C. Clonal populations of T cells in normal elderly humans: the T cell equivalent to "benign monoclonal gammapathy". J Exp Med. 1994;179:609–618. doi: 10.1084/jem.179.2.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schwab R, Szabo P, Manavalan JS, et al. Expanded CD4+ and CD8+ T cell clones in elderly humans. J Immunol. 1997;158:4493–4499. [PubMed] [Google Scholar]
- 24.Naylor K, Li G, Vallejo AN, et al. The influence of age on T cell generation and TCR diversity. J Immunol. 2005;174:7446–7452. doi: 10.4049/jimmunol.174.11.7446. [DOI] [PubMed] [Google Scholar]
- 25.Khan N, Shariff N, Cobbold M, et al. Cytomegalovirus seropositivity drives the CD8 T cell repertoire toward greater clonality in healthy elderly individuals. J Immunol. 2002;169:1984–1992. doi: 10.4049/jimmunol.169.4.1984. [DOI] [PubMed] [Google Scholar]
- 26.Vardiman JW, Harris NL, Brunning RD. The World Health Organization (WHO) classification of the myeloid neoplasms. Blood. 2002;100:2292–2302. doi: 10.1182/blood-2002-04-1199. [DOI] [PubMed] [Google Scholar]
- 27.Greenberg P, Cox C, LeBeau MM, et al. International scoring system for evaluating prognosis in myelodysplastic syndromes. Blood. 1997;89:2079–2088. [PubMed] [Google Scholar]
- 28.Onida F, Kantarjian HM, Smith TL, et al. Prognostic factors and scoring systems in chronic myelomonocytic leukemia: a retrospective analysis of 213 patients. Blood. 2002;99:840–849. doi: 10.1182/blood.v99.3.840. [DOI] [PubMed] [Google Scholar]
- 29.Barragan E, Cervera J, Bolufer P, et al. Prognostic implications of Wilms' tumor gene (WT1) expression in patients with de novo acute myeloid leukemia. Haematologica. 2004;89:926–933. [PubMed] [Google Scholar]
- 30.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 31.Akatsuka Y, Martin EG, Madonik A, Barsoukov AA, Hansen JA. Rapid screening of T-cell receptor (TCR) variable gene usage by multiplex PCR: application for assessment of clonal composition. Tissue Antigens. 1999;53:122–134. doi: 10.1034/j.1399-0039.1999.530202.x. [DOI] [PubMed] [Google Scholar]
- 32.Peggs KS, Verfuerth S, D'Sa S, Yong K, Mackinnon S. Assessing diversity: immune reconstitution and T-cell receptor BV spectratype analysis following stem cell transplantation. Br J Haematol. 2003;120:154–165. doi: 10.1046/j.1365-2141.2003.04036.x. [DOI] [PubMed] [Google Scholar]
- 33.McHeyzer-Williams MG, Davis MM. Antigen-specific development of primary and memory T cells in vivo. Science. 1995;268:106–111. doi: 10.1126/science.7535476. [DOI] [PubMed] [Google Scholar]
- 34.Xu R, Wunsch D., 2nd Survey of clustering algorithms. IEEE Trans Neural Netw. 2005;16:645–678. doi: 10.1109/TNN.2005.845141. [DOI] [PubMed] [Google Scholar]
- 35.Kook H, Risitano AM, Zeng W, et al. Changes in T-cell receptor VB repertoire in aplastic anemia: effects of different immunosuppressive regimens. Blood. 2002;99:3668–3675. doi: 10.1182/blood.v99.10.3668. [DOI] [PubMed] [Google Scholar]
- 36.Sloand EM, Kim S, Fuhrer M, et al. Fas-mediated apoptosis is important in regulating cell replication and death in trisomy 8 hematopoietic cells but not in cells with other cytogenetic abnormalities. Blood. 2002;100:4427–4432. doi: 10.1182/blood-2002-01-0096. [DOI] [PubMed] [Google Scholar]
- 37.Miqueu P, Guillet M, Degauque N, Dore JC, Soulillou JP, Brouard S. Statistical analysis of CDR3 length distributions for the assessment of T and B cell repertoire biases. Mol Immunol. 2007;44:1057–1064. doi: 10.1016/j.molimm.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 38.Ma X, Does M, Raza A, Mayne ST. Myelodysplastic syndromes: incidence and survival in the United States. Cancer. 2007;109:1536–1542. doi: 10.1002/cncr.22570. [DOI] [PubMed] [Google Scholar]
- 39.Rollison DE, Howlader N, Smith MT, et al. Epidemiology of myelodysplastic syndromes and chronic myeloproliferative disorders in the United States, 2001–2004, using data from the NAACCR and SEER programs. Blood. 2008;112:45–52. doi: 10.1182/blood-2008-01-134858. [DOI] [PubMed] [Google Scholar]
- 40.Bernell P, Jacobsson B, Nordgren A, Hast R. Clonal cell lineage involvement in myelodysplastic syndromes studied by fluorescence in situ hybridization and morphology. Leukemia. 1996;10:662–668. [PubMed] [Google Scholar]
- 41.Tamaki H, Ogawa H, Ohyashiki K, et al. The Wilms' tumor gene WT1 is a good marker for diagnosis of disease progression of myelodysplastic syndromes. Leukemia. 1999;13:393–399. doi: 10.1038/sj.leu.2401341. [DOI] [PubMed] [Google Scholar]
- 42.Cilloni D, Gottardi E, Messa F, et al. Significant correlation between the degree of WT1 expression and the International Prognostic Scoring System Score in patients with myelodysplastic syndromes. J Clin Oncol. 2003;21:1988–1995. doi: 10.1200/JCO.2003.10.503. [DOI] [PubMed] [Google Scholar]
- 43.Elisseeva OA, Oka Y, Tsuboi A, et al. Humoral immune responses against Wilms tumor gene WT1 product in patients with hematopoietic malignancies. Blood. 2002;99:3272–3279. doi: 10.1182/blood.v99.9.3272. [DOI] [PubMed] [Google Scholar]
- 44.Gaiger A, Reese V, Disis ML, Cheever MA. Immunity to WT1 in the animal model and in patients with acute myeloid leukemia. Blood. 2000;96:1480–1489. [PubMed] [Google Scholar]
- 45.Wu F, Oka Y, Tsuboi A, et al. Th1-biased humoral immune responses against Wilms tumor gene WT1 product in the patients with hematopoietic malignancies. Leukemia. 2005;19:268–274. doi: 10.1038/sj.leu.2403539. [DOI] [PubMed] [Google Scholar]
- 46.Rezvani K, Yong AS, Mielke S, et al. Leukemia-associated antigen-specific T-cell responses following combined PR1 and WT1 peptide vaccination in patients with myeloid malignancies. Blood. 2008;111:236–242. doi: 10.1182/blood-2007-08-108241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Holsinger AL, Ramakrishnan A, Storer B, et al. Therapy of Myelodysplastic Syndrome (MDS) with Azacitidine Given in Combination with Etanercept: A Phase II Study. Blood. 2007;110 (ASH Annual Meeting Abstracts) Abstract 1452. [Google Scholar]
- 48.Sigalotti L, Altomonte M, Colizzi F, et al. 5-Aza-2'-deoxycytidine (decitabine) treatment of hematopoietic malignancies: a multimechanism therapeutic approach? Blood. 2003;101:4644–4646. doi: 10.1182/blood-2002-11-3458. [DOI] [PubMed] [Google Scholar]
- 49.Oki Y, Jelinek J, Shen L, Kantarjian HM, Issa JP. Induction of hypomethylation and molecular response after decitabine therapy in patients with chronic myelomonocytic leukemia. Blood. 2008;111:2382–2384. doi: 10.1182/blood-2007-07-103960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Holt RA, Jones SJ. The new paradigm of flow cell sequencing. Genome Res. 2008;18:839–846. doi: 10.1101/gr.073262.107. [DOI] [PubMed] [Google Scholar]