Abstract
Pseudomonas aeruginosa is an opportunistic pathogen that infects the lungs of patients with cystic fibrosis causing aberrant and destructive neutrophil (PMN)-dominated inflammation of airways. Interaction of P. aeruginosa with the lung epithelial barrier resulting in trans-epithelial PMN migration likely represents a key event during PMN recruitment. To investigate bacterial factors involved in interactions with lung epithelial cells, a mutant library of two-component system response regulators was evaluated to identify mutants exhibiting defects in the ability to induce PMN trans-epithelial migration. Of forty eight mutants, five reproducibly demonstrated a reduced PMN trans-epithelial migration response. All five mutants also exhibited a decreased ability to interact with lung epithelial cells. One mutant identified lacks the response regulator gene roxR, which has not previously been reported to be involved regulating factors that facilitate interactions with lung epithelial cells. This finding suggests that RoxR likely regulates genes with relevance to P. aeruginosa mediated lung disease.
Keywords: Pseudomonas aeruginosa, Neutrophils, Airway
1. Introduction
Lung disease as a consequence of bacterial infection is often associated with an over-exuberant inflammation marked by the recruitment and accumulation of polymorphonuclear leukocytes (PMNs) and their noxious products in the airway [1]. During inflammatory disease (i.e., pneumonia and cystic fibrosis) PMNs are recruited to eradicate bacterial infection, however, PMNs are less effective against certain pathogens and can be highly destructive towards host lung tissue [1, 2]. The Gram-negative bacterium Pseudomonas aeruginosa infects the lung of patients with cystic fibrosis (CF) and is capable of causing bacterial pneumonia in immune compromised patients [2, 3]. Increasing antibiotic resistance among P. aeruginosa clinical isolates has spurred significant interest in developing alternative therapeutics for treating P. aeruginosa infection, including anti-inflammatory based approaches [4]. Recruitment of PMNs from the bloodstream to the airways in response to infection is a complex multi-stage process that represents a potential anti-inflammatory target for therapeutic intervention [1]. A better understanding of the molecular mechanisms underlying P. aeruginosa interactions with lung epithelial barriers would greatly facilitate this endeavor.
P. aeruginosa utilizes an unusually large portion of its genome to encode regulatory proteins [5]. The two-component sensor/response regulator system represents a particular class of P. aeruginosa regulatory control proteins whereby a sensor histidine kinase is activated, prompting phosphorylation of a response regulator, which exerts regulatory control over target genes [5, 6]. Certain pairs of sensors and response regulators have been identified as being responsible for the orchestration of particular functional attributes of P. aeruginosa, such as motility and biofilm formation [5–10].
In this study we aim to gain further insight into molecular mechanisms underlying P. aeruginosa interactions with lung epithelial barriers, particularly as they pertain to P. aeruginosa induced PMN trans-epithelial migration [11, 12]. P. aeruginosa mutants of two-component response regulators, P. aeruginosa mutants of established virulence factors, as well as clinical isolates of P. aeruginosa, were evaluated for the ability to induce PMN migration across lung epithelial cells. Our studies underscore the importance of efficient P. aeruginosa association with the lung epithelial barrier for mediating PMN trans-epithelial migration and have revealed a novel role for the sensor response regulator pair, known as RoxS/RoxR.
2. Material and methods
2.1 Bacterial Strains
Strains listed in table 1 were grown aerobically in LB-broth overnight at 37 °C. Mutants ΔroxSR and ΔcioAB and complementation strain PAΔ4493-C were cultured with 50 μg/ml gentamicin.
Table 1.
List of Strains Used in Study
| Strain | Description | Reference |
|---|---|---|
| CF18 | P. aeruginosa clinical isolate | [19] |
| CF27 | P. aeruginosa clinical isolate | [19] |
| CF265 | P. aeruginosa clinical isolate | [19] |
| CF276 | P. aeruginosa clinical isolate | [19] |
| PAK | P. aeruginosa wild-type strain | [16, 27] |
| PAΔ1099 | P. aeruginosa (PAK) ORF mutanta (ΔfleR) (motility/attachment- regulates flagella) | [7, 9] |
| PAΔ3702 | P. aeruginosa (PAK) ORF mutant (ΔwspR) (motility/attachment/chemotaxis) | [7, 10] |
| PAΔ4547 | P. aeruginosa (PAK) ORF mutant (ΔpilR) (motility/attachment- regulates pili) | [7, 20] |
| PAΔ4493 | P. aeruginosa (PAK) ORF mutant (ΔroxR) (regulates cyanide-insensitive oxidase) | [7, 15] |
| PAΔ5261 | P. aeruginosa (PAK) ORF mutant (ΔalgR) (regulates secreted factors – toxins/alginate) | [7, 21] |
| MC1000 | E. coli K12 strain used as non-pathogenic control | [11] |
| PAK ΔfliC | P. aeruginosa mutant defective flagella | [27] |
| PAK ΔpilA | P. aeruginosa mutant defective pili | [16] |
| PAK ΔpscC | P. aeruginosa mutant defective type III secretion | [16] |
| PAΔ4493-C | P. aeruginosa complement of PA4493 (PAK Δ roxR) | This study |
| PAO1 | P. aeruginosa wild-type strain | [11] |
| PAO1 ΔroxSR | P. aeruginosa mutant: sensor/response regulator | This study |
| PAO1 ΔcioAB | P. aeruginosa mutant: cyanide insensitive oxidase | This study |
All (PAK) ORF mutants have been annotated as response regulators belonging to the two-component sensor response regulator system. PAK ORF mutants analyzed in the initial screen, but not discussed further are excluded from the body of the table, but listed here; PAΔ0034, PAΔ0173, PAΔ0179, PAΔ0463, PAΔ0601, PAΔ0928, PAΔ0929, PAΔ1179, PAΔ1243, PAΔ1437, PAΔ1456, PAΔ1637, PAΔ1799, PAΔ1978, PAΔ2523, PAΔ2424, PAΔ2686, PAΔ3192, PAΔ3204, PAΔ3346, PAΔ3462, PAΔ3604, PAΔ3714, PAΔ3879, PAΔ3947, PAΔ3948, PAΔ4101, PAΔ4112, PAΔ4196, PAΔ4381, PAΔ4396, PAΔ4726, PAΔ4776, PAΔ4843, PAΔ4856, PAΔ4885, PAΔ4938, PAΔ4982, PAΔ5166, PAΔ5360, PAΔ5364, PAΔ5483, PAΔ5511.
2.2 Cell Culture
A549 and Calu-3 lung epithelial cell lines were maintained in Ham’s F-12K and MEMα medium respectively, supplemented with 10% fetal bovine serum and antibiotics. Polarized monolayers of A549 and Calu-3 were grown on the underside of 0.33 cm2 collagen coated Transwell filters to study PMN migration in the physiological basolateral to apical direction [11, 12].
2.3 PMN Isolation
PMNs were isolated from blood of healthy consenting human volunteers anti-coagulated with acid citrate/dextrose (IRB protocol #: 1999-P-007782). The buffy coat was resolved by centrifugation. Plasma and mononuclear cells were removed by aspiration, and the majority of the red blood cells (RBCs) were removed using 2% gelatin sedimentation. Residual RBCs were removed by lysis in cold NH4Cl lysis buffer [11, 12].
2.4 PMN Transmigration Assay
Transwell inserts containing A549 cell monolayers seeded on the underside was exposed for 1 hr to 25 μl of varying concentrations of bacteria as per individual experiment. After infection, PMNs (1×106) were added to the top (basolateral) chamber and incubated at 37°C for 2 hr. PMNs that fully migrated across the cell monolayer reaching the bottom (apical) chamber were quantified by the myeloperoxidase assay[11, 12]. Negative controls include uninfected A549 monolayers and A549 monolayers infected with non-pathogenic K12 E. coli (MC1000) [11, 12].
2.5 Bacterial/Epithelial Association Assay
Lung epithelial cells were grown on 24-well plates to confluence or Transwells for 9–14 days. Monolayers were infected with between 1 and 40×105 CFUs/monolayer P. aeruginosa for 3 hrs at 37°C. After infection, monolayers were washed followed by addition of 1 ml/well 1% Triton X-100. Cells were shaken at 4°C for 1 hr and plated on Pseudomonas Isolation Agar plates +/− 50 μg/ml gentamicin for colony forming unit (CFU) counting the following day.
2.6 Cell Viability/Barrier Integrity Assays
The amount of LDH released into the supernatant with and without infection of various concentrations of PAK was quantified using the LDH based In Vitro Toxicology Assay Kit (Sigma, St. Louis MO). Barrier integrity of lung epithelial monolayers grown on Transwells was assayed by the horse radish peroxidase (HRP) flux assay as previously described [11].
2.7 Swim assay
Single colonies of P. aeruginosa wild type or mutants were isolated and stabbed in the middle of a thickly poured LB-broth plate containing 0.3 % agar. Plates were incubated at 37°C overnight. The following day, the diameter of the bacterial spread on the plates was measured [13, 14].
2.8 Twitch assay
Single colonies of P. aeruginosa wild type or mutants are isolated and stabbed in the middle of a thinly poured LB-broth plate containing 1.0 % agar. Plates are incubated at 37°C for 36 to 48 hrs. Following the incubation period, the diameter of the bacterial spread under the agar surface of the plates is measured [13, 14].
2.9 Growth Assay
Optical density at 600 nm for each strain is adjusted to 0.05 in LB-broth +/− antibiotics with or without 500 μM NaN3 [15]. After shaking at 37°C for various time points, optical density at 600 nm is measured in each culture to determine the extent of growth in the presence or absence of NaN3.
2.10 Construction of ΔroxRS and ΔcioAB mutants
Internal regions of roxS and cioA were amplified by PCR from P. aeruginosa (PAO1) using 5′-GCGCGGATCCGAACAGGTCAAGCAGTGCAA-3′ and 5′-GCGCGAATTCCTTGGTGGTGAAGAACGGTTT-3′ for roxS and 5′-GCGCGGATCCGTGATCTTCAACCCGTCGTT-3′ and 5′-GCGCGAATTCACCTTGAAGCGGGTTTCCTC-3′ for cioA. Each oligonucleotide contained restriction sites for ligation of PCR product into the integration vector pEXG2 using Promega T4 DNA ligase [16].
E. coli SM10 was made competent by the T salt method and transformed by heat shock with pEXG2 containing cloned fragments [16, 17]. Transformants were plated on LB agar with 10 μg/ml gentamicin, plasmids were screened by restriction digest pattern, and identified transformants were mated with P. aeruginosa.
2.11 Construction of roxR complementation vector
The complete sequence of PA4493 (roxR) was amplified by Hi-Fi PCR using Phusion Hot Start DNA polymerase (New England Biolabs). Primer sequence specific to PA4493 was flanked by a BamH1 restriction site and a ribosomal binding site (shown in italics) on the forward primer and a HindIII site on the reverse primer; forward primer 5′-CATTCAGGATCCTGAGGAGAGATTCATGTCTGAAGAGCTGCTCTTCGAAGGGGAAGA ACA-3′ and reverse primer 5′-CATTCAAAGCTTTCAGCGCCGTACCGGCCGCTTCT-3′. The PCR product (561 bp) was ligated into expression vector pPSV35 using Promega T4 DNA ligase and used to transform Subcloning Efficiency™ DH5α™ Competent Cells (Invitrogen) via heat shock [18]. Colonies were isolated on LB agar containing 10 μg/ml gentamicin and screened by PCR. A PCR positive colony was sequenced by the MGH DNA Core Facility.
2.12 Complementation of PAΔ4493 mutant in PAK background
PAKΔ4493 was created as part of the two component response regulator mutant library as previously described [7]. Prior to movement of pPSV35-roxR into PAKΔ4493, the gentamicin resistance marker was removed by transient expression of the Flp recombinase from plasmid pFLP2 in the PAK4493 strain which was introduced by electroporation [18]. Electroporated cells were plated on Pseudomonas Isolation Agar plates containing 50 μg/ml gentamicin.
2.13 Statistics
Data displayed for each figure is presented as a representative experiment with a mean (standard deviation) of at least three independent data points/condition. Each experiment has been repeated multiple times yielding similar results. Statistical analysis was performed by Student’s t test for all comparisons.
3. Results
3.1 P. aeruginosa clinical isolates induce PMN trans-epithelial migration
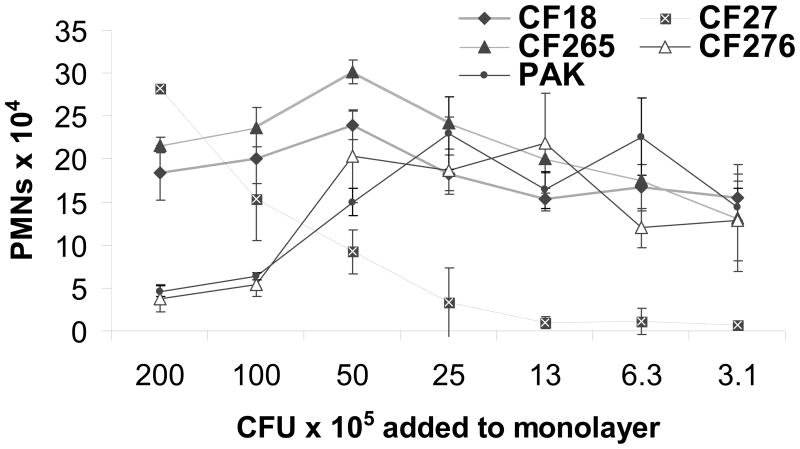
Several strains of P. aeruginosa are capable of inducing PMN migration across lung epithelial cells including, PAO1, PA14, and PA103 [11]. Four CF clinical isolates were analyzed using a range of infection concentrations (Fig. 1) [19]. All four CF isolates induced PMN trans-epithelial migration. Clinical isolate CF276 as well as pathogenic laboratory strain PAK exhibited a pattern of induced migration similar to that previously described for PAO1. A maximal numbers of PMNs migrated in response to 10 – 50 × 105 CFU/monolayer of PAK and CF276, however, reduced PMN migration occurs at higher infection concentrations (Fig. 1) [11]. A substantial PMN trans-epithelial migration response was observed at all concentrations tested for CF18 and CF265. In contrast, CF27 displayed a strong PMN transmigration response only at the highest concentrations of bacterial infection (Fig. 1). Ability to induce PMN trans-epithelial migration appears to be a common attribute amongst a diverse collection of P. aeruginosa isolates.
Figure 1.
Number of PMNs migrating across A549 monolayers grown on Transwells in response to infection with a range of concentrations (3.1 to 200 × 105 CFU/monolayers) of various P. aeruginosa clinical strains isolated from CF patients. Each data point displayed was performed in triplicate, with error bars representing standard deviation. Experiment was performed multiple times with similar results.
3.2 Mutants of two component response regulators involved in PMN trans-epithelial migration
In order to identify regulators that modulate gene expression of factor(s) involved in mediating the PMN transmigration response we employed a comprehensive mutant library encompassing targeted deletions in each of forty eight open reading frames (ORFs) representing annotated response regulators specifically of the two-component system (Table 1) [5–7]. The response regulator mutant library was developed in the background P. aeruginosa strain PAK, which displayed minimal effects on cell viability or barrier integrity at infection concentrations of 1 × 105 CFU/A549 monolayer (data not shown).
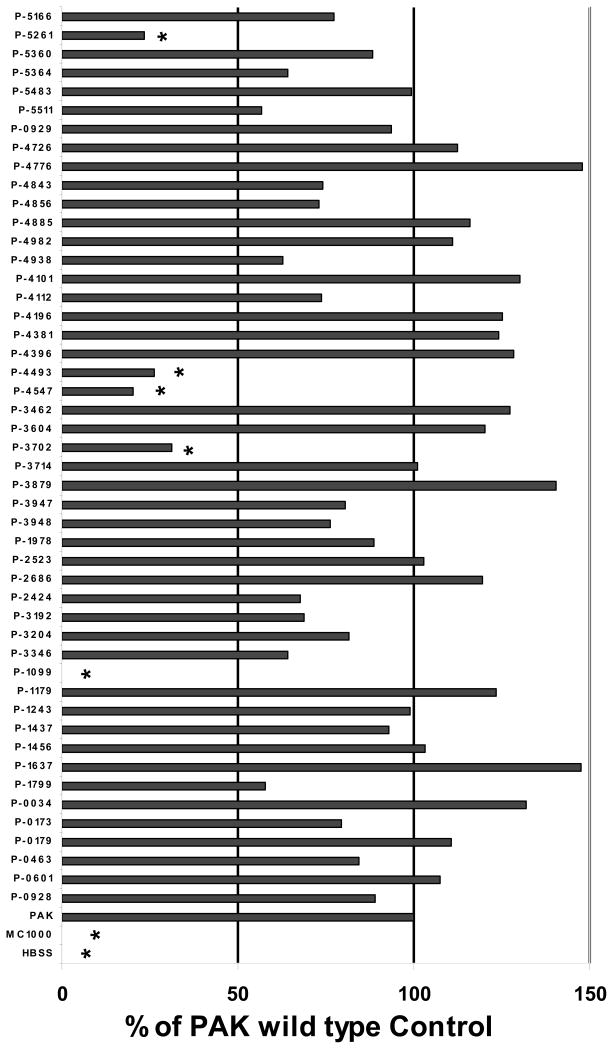
Using an infection concentration of 1 × 105 CFU/monolayer, five mutants were identified that reproducibly exhibited a greater than 50% reduction in the number of PMNs induced to migrate across the infected epithelial monolayer (PAΔ1099, PAΔ3702, PAΔ4547, PAΔ4493, and PAΔ5261 (Fig. 2). Of the five mutants, four are well-characterized as being involved in the regulation of traits important to virulence including motility and attachment to host cells (Table 1) [9, 10, 20, 21]. One of the five response regulators identified (PAΔ4493), to our knowledge, has not been shown to play a role in regulating genes involved in P. aeruginosa interaction with lung epithelial cells.
Figure 2.
P. aeruginosa response regulator mutants induce PMN migration across A549 monolayers grown on Transwells at infection concentrations of 1 × 105 CFUs/monolayer. Each individual assay consisted of three wells per condition. Conditions for each assay include infection with twelve distinct mutants, a PAK wild type positive control, and a non-pathogenic E. coli (MC1000) infected negative control. Uninfected (HBSS) monolayers were also included in each assay serving as an additional negative control. Mutants PAΔ(ORF disrupted) are referred to as P-(ORF disrupted), for example a mutant of ORF PA1099 referred to in the text as PAΔ1099 is depicted in this figure as P-1099. All forty eight mutants were screened over the course of at least eight separate assays with each individual mutant tested on at least two separate occasions. The PMN transmigration response was internally controlled in each assay by setting the number of PMNs migrating in response to PAK to 100% and reporting each mutant as a percentage of the PAK wild type control. Data from all individual assays has been combined and presented herein. Mutants that fall below the 50% response threshold and reach statistical significance by Student’s T-test when compared with the response to PAK are reflected by the symbol (*) above the data bar (p < 0.05).
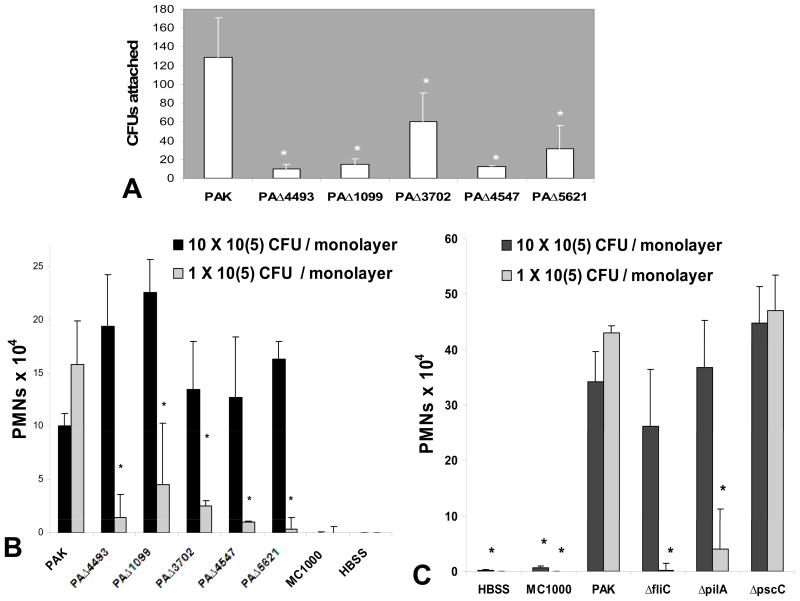
As a compromised ability to associate with lung epithelial monolayers might underlie a reduced ability to stimulate PMN trans-epithelial migration, we investigated whether any of the five mutants identified displayed a reduced ability to interact with A549 monolayers. All five mutants, including PAΔ4493, exhibit significantly reduced ability to interact with A549 monolayers when compared with the wild type PAK strain (Fig. 3A). We next tested whether infecting with 10 fold more of each of the mutant PAK strains could reconstitute the PMN trans-epithelial migration response, and found that all five mutants were capable of inducing PMN trans-epithelial migration that was quantitatively equivalent to PAK infection when 10 fold more mutant bacteria was used (Fig. 3B). Thus, a deficiency in the ability of P. aeruginosa to interact with lung epithelial cells likely exerts an effect on the ability to trigger PMN trans-epithelial migration. This can likely be overcome if a sufficient threshold of bacteria is present to engage the epithelial cell to produce key PMN chemo-attractants [11].
Figure 3.
(A) Number of colony forming units recovered from A549 monolayers grown on Transwells after an initial infection with an equivalent concentration of PAK or selected mutant strains. The symbol (*) represent a statistically significant decrease compared to PAK using the Student’s T-test (p < 0.05). (B) PMNs migrate across A549 monolayers grown on Transwells in response to infection with a concentration of either 1 (gray bars) or 10 (black bars) × 105 CFU/monolayer of P. aeruginosa strain PAK or isogenic mutants; PAΔ4493 (ΔroxR). PAΔ1099 (ΔfleR), PAΔ3702 (ΔwspR), PAΔ4547 (ΔpilR), and PAΔ5621 (ΔalgR). Uninfected A549 monolayers (HBSS) and monolayers infected with non-pathogenic E. coli MC1000 serve as negative controls for the PMN transmigration response. (C) PMNs migrate across A549 monolayers grown on Transwells in response to infection with a concentration of either 1 (gray bars) or 10 (black bars) × 105 CFU/monolayer of P. aeruginosa strain PAK or isogenic mutants; ΔfliC, ΔpilA, or ΔpscC. Uninfected A549 monolayers (HBSS) and monolayers infected with non-pathogenic E. coli MC1000 serve as negative controls for the PMN transmigration response. The symbol (*) represents a statistically significant decrease by Student’s T-test in the number of PMNs migrating compared to induction by 1 × 105 CFUs/monolayer PAK infection (p < 0.05). Data displayed is a representative experiment with each data point performed in triplicate. Experiment was performed on at least three separate occasions yielding similar results.
Four of the five regulatory mutants identified modulate genes known to directly interact with lung epithelial cells (Table 1). For example, structural components of flagella and pili are involved in bacterial motility and have been shown to facilitate interactions with the lung epithelial cell, including stimulating the release of chemokines [13, 22–25]. PilR (encoded by ORF PA4547) and FleR (PA1099) serve as regulators controlling expression of pili and flagella respectively. To evaluate whether mutation of the structural components of flagella and pili are compromised in ability to induce PMN trans-epithelial migration, we infected lung epithelial monolayers with wild type pathogenic P. aeruginosa strain PAK and isogenic mutants; ΔfliC (lacking flagella) and ΔpilA (lacking pili). Both ΔfliC and ΔpilA were substantially less able to induce PMN transmigration at an infection concentration of 1 × 105 CFU/monolayer (Fig. 3C, gray bars). When A549 monolayers were treated with a 10-fold higher infection concentration of either ΔfliC or ΔpilA, no significant differences in the induction of PMN trans-epithelial migration was observed between wild type strain PAK and either of these two mutants, suggesting that although flagella or pili facilitate interactions with lung epithelial cells that promote PMN trans-epithelial migration, neither structure is required for P. aeruginosa to induce PMN trans-epithelial migration (Fig. 3C, black bars). P. aeruginosa possesses a type three secretion system (TTSS) that allows injection of bacterial toxins into lung cells, which can directly modulate host cell signaling [18, 26]. PscC is an outer membrane transport protein that serves as a crucial component of the TTSS and a pscC mutant lacks a functional TTSS [16, 27]. No significant differences are observed when the cells were infected with PAK ΔpscC at either concentration, suggesting that the TTSS does not play a role in the process by which P. aeruginosa triggers PMN trans-epithelial migration (Fig. 3C).
3.3 Characterization of ΔroxR interaction with lung epithelial cells
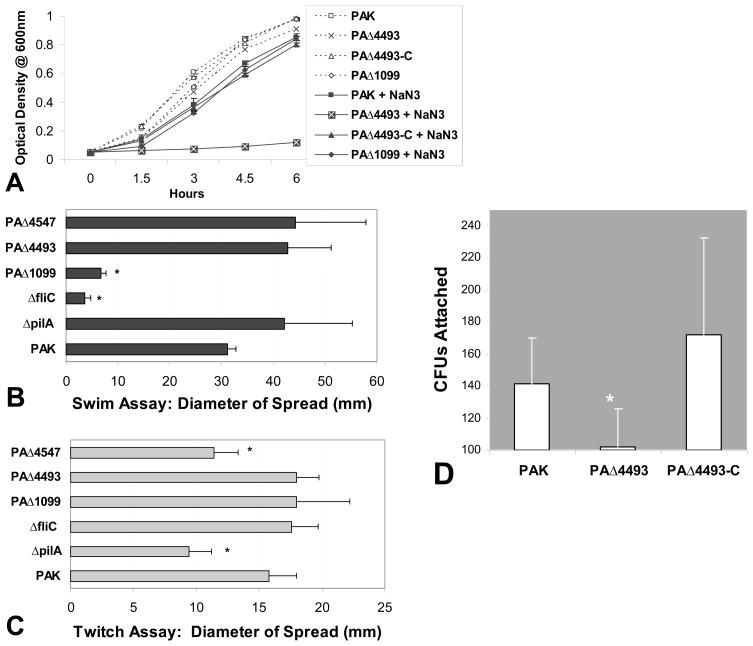
The ORF PA4493 encodes the response regulator RoxR, located downstream of and co-transcribed with PA4494 (roxS) a gene encoding its cognate sensor histidine kinase [15]. RoxS/RoxR induce expression of cyanide insensitive oxidase (cioAB) [15]. CioAB exhibits enzymatic activity that is insensitive to oxidase inhibitors such as cyanide and sodium azide (NaN3) [15]. As shown in Fig. 4A, wild type PAK experienced only a slight decrease in culture density in the presence of NaN3 over a six hour time course. Growth of PAΔ4493 (ΔroxR) in LB was indistinguishable from the wild type, but in the presence of NaN3, this mutant failed to grow and the cell density after 6 hrs remained on slightly above that of the inoculum. PAΔ1099 (ΔfleR), a response regulatory mutant identified as defective in our PMN transmigration assay, but not thought to be involved in regulating cioAB expression, grew comparable to wild type in the presence of NaN3 (Fig. 4A).
Figure 4.
(A) P. aeruginosa culture densities; PAK, PAΔ4493 (roxR mutant), PAΔ4493-C (complemented roxR mutant), and PAΔ1099 (fleR mutant) after incubation for up to 6 hrs in the absence (dotted lines, empty symbols) or presence (solid lines, filled symbols) of 500 μM NaN3. (B) Flagella based motility, Swim Assay. Representative of the degree of spread from an initial inoculation of PAK or mutant strains [ΔpilA, ΔfliC, PAΔ1099(ΔfleR-regulates flagella assembly), PAΔ4547(ΔpilR)-regulates pili formation, and PAΔ4493(ΔroxR-regulates CioAB)] in the center of a low concentration thickly poured LB agar plate. (C) Pili based motility, Twitch Assay. Representative of the degree of spread from an initial inoculation of PAK or mutant strains [ΔpilA, ΔfliC, PAΔ1099(ΔfleR-regulates flagella assembly), PAΔ4547(ΔpilR)-regulates pili formation, and PAΔ4493(ΔroxR-regulates CioAB)] in the center of a high concentration thinly poured LB agar plate. (D) Colony counts recovered from Calu-3 monolayers grown on Transwells after an initial infection with an equally concentrated inoculum of PAK, PAΔ4493(roxR mutant), and PAΔ4493-C (complemented roxR mutant). The symbol (*) represent a statistically significant decrease compared to PAK using the Student’s T-test (p < 0.05). Each above experiment was performed on at least three separate occasions yielding similar results.
Little is known regarding regulatory targets of RoxR that might explain the reduced ability of PAΔ4493 (ΔroxR) to associate with lung epithelial cellls. To determine if RoxR contributes to expression of genes required for flagella or pili function, we subjected PAΔ4493 (ΔroxR) to motility assays. The swim assay was utilized to evaluate flagella based motility [13, 14]. PAΔ4493 (ΔroxR) displayed a spreading ability similar to wild type PAK indicating the presence of a fully functional flagella (Fig. 4B). The twitch motility assay was next employed to assess whether PAΔ4493 (ΔroxR) possessed functional pili [13, 14]. PAΔ4493 (ΔroxR) displayed a normal pili phenotype, as no reduction in spread compared to PAK wild type was observed (Fig. 4C).
We next sought to determine whether decreased cell association is observed when interacting with other lung epithelial cell lines and whether we could complement this defect by transforming PAΔ4493 with a plasmid expressing roxR. As shown in Fig. 4A, PAΔ4493 transformed with pPSV35-roxR regains the ability to grow in the presence of NaN3, indicating RoxR is expressed and functional in PAΔ4493-C. Furthermore, PAΔ4493 is defective at interacting with not only A549 lung epithelial cells (Fig. 3A), but also Calu-3 bronchial epithelial cells (Fig. 4D). The total number of PAΔ4493 CFUs recovered from Calu-3 monolayers was significantly less than the number of CFUs recovered upon infection with wild type PAK (Fig. 4D). When infecting Calu-3 cells with PAΔ4493-C, the number of CFUs recovered is similar to that of wild type PAK (Fig. 4D).
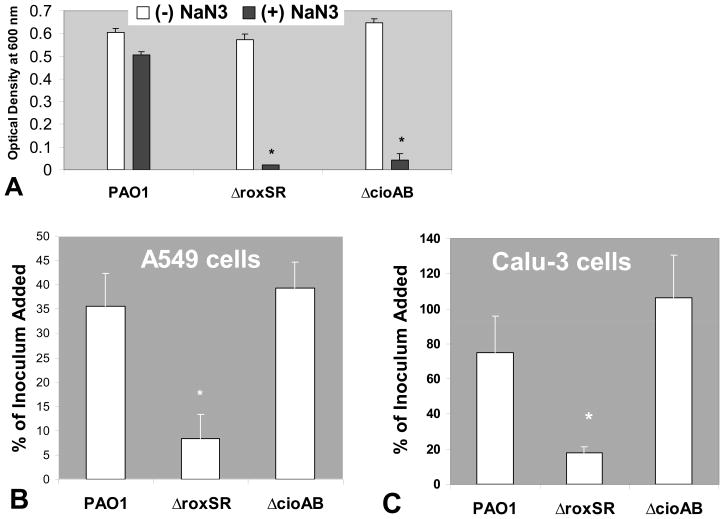
To our knowledge, the only reported gene modulated by RoxS/RoxR in Pseudomonas aeruginosa is cioAB [15]. A roxSR and cioAB mutant in the PAO1 background strain was constructed to address the question of whether P. aeruginosa lacking cioAB expression are also defective at associating with lung epithelial cells [16]. Both mutants grew normally in LB, but neither was capable of growing in the presence of 500 μM NaN3 unlike wild type PAO1 (Fig. 5A). Having confirmed the previously described phenotype of both mutants, we next sought to determine if either ΔroxSR or ΔcioAB were defective at associating with lung epithelial cells. The number of CFUs recovered from lung epithelial cells as a percentage of the initial infection concentration of ΔroxSR was significantly decreased compared to percent recovery of an identical inoculum of wild type PAO1 (Fig. 5B and 5C). In contrast, the recovery of the ΔcioAB was similar to that observed for wild type PAO1 from both A549 and Calu-3 lung epithelial cells (Fig. 5B and 5C).
Figure 5.
(A) P. aeruginosa culture densities (PAO1 wild type and mutants) after incubation for 5 hr in the absence (white bars) or presence (black bars) of 500 μM NaN3. (B) Representative of the percentage of P. aeruginosa CFUs recovered from confluent A549 cells grown on 24-well plates after an initial infection with an equivalent concentration of PAO1, ΔroxSR, or ΔcioAB. (C) Representative of the percentage of P. aeruginosa CFUs recovered from confluent Calu-3 cells grown on 24-well plates after an initial infection with an equivalent concentration of PAO1, ΔroxSR, or ΔcioAB. Percent of the inoculum added refers to the number of CFUs recovered divided by the number of bacteria initially added to the lung epithelial cells. This value is then multiplied by 100 to give percentage. The symbol (*) represent a statistically significant decrease compared to PAO1 by Student’s T-test (p < 0.05). Each above experiment was performed on at least three separate occasions yielding similar results.
4. Discussion
Infection with P. aeruginosa is extremely detrimental for immuno-compromised and cystic fibrosis patients [2]. P. aeruginosa is particularly suited for occupying the lung of these patients where infection instigates a destructive inflammatory response culminating in large numbers of PMNs emigrating from the bloodstream to the airway lumen [2, 3]. Recent evidence has revealed that P. aeruginosa stimulates lung epithelial cell release of hepoxilin A3, which serves as a PMN chemo-attractant directing trans-epithelial migration [11, 12]. Little is known regarding how P. aeruginosa triggers this inflammatory pathway.
P. aeruginosa exhibits tremendous adaptability, as nearly ten percent of the coding portion of its genome is allocated to genes encoding regulatory proteins [5]. One class of regulatory proteins, known as the two-component system, consists of paired sensors and response regulators [5, 6]. Numerous activities of P. aeruginosa are controlled by this mechanism, including swimming and twitching motiliy, metabolic processes, and biofilm formation [10, 15, 16, 27]. We evaluated a mutant library consisting of a P. aeruginosa mutant for each of forty eight annotated response regulators of the two-component system and identified five mutants that reproducibly displayed a defect in the ability to induce PMN transmigration. Four of the five have been well characterized and previously shown to modulate genes important to pathogenesis, including ΔfleR, which regulates flagella assembly, ΔwspR, which regulates biofilm formation, ΔpilR which regulates pili production, and ΔalgR which regulates exopolysaccharides [9, 10, 20, 21, 23, 27, 28]. Each of these mutants also displayed significantly less ability to associate with lung epithelial monolayers in the absence of PMNs, consistent with previous observations [10, 23, 24, 28, 29].
One mutant identified, PA4493 (ΔroxR), presented a less obvious explanation for inducing a defective PMN transmigration response. RoxR is a homologue of the response regulator PrrA/RegA of the Rhodobacter species, which is responsible for regulating enzymes involved in energy generating and energy utilizing systems [15]. RoxR in P. aeruginosa has been shown control expression of cyanide insensitive oxidase (CioAB), an energy generating enzyme that allow P. aeruginosa to withstand the presence of oxidase inhibitors, cyanide or azide [15]. We confirm these results herein by demonstrating that neither PAK (ΔroxR mutant) nor PAO1 (ΔroxSR mutant) are capable of growing in the presence of NaN3. We also present a novel role for the response regulator RoxR as our screen revealed that ΔroxR was defective at inducing PMN trans-epithelial migration. Reduced ability of ΔroxR to induce PMN transmigration is likely a consequence of ineffective interaction with the lung epithelial monolayer, however, an explanation for why P. aeruginosa strains carrying a mutant RoxR are unable to associate properly with lung epithelial cells remains elusive. The association defect of the RoxR mutant does not appear to involve its regulatory target CioAB, as PAO1 (ΔcioAB) displays no interaction defect. Furthermore, ΔroxR does not exhibit reduced survival in HBSS in the absence of lung epithelial cells nor does ΔroxR exhibit a reduced growth rate in LB grown under static or oxygen limiting conditions (data not shown). Results presented herein suggest that RoxR regulates genes other than cioAB that play an important role in P. aeruginosa interaction with lung epithelial cells. Since RoxR mutants behaved normally in multiple motility assays, genes involved in flagella or pili function are likely not regulatory targets of RoxR.
A recent study has identified the RoxS/RoxR homologues in the common soil bacterium Pseudomonas putida, which regulates genes not only involved in oxidase activity, but also density dependent genes, genes involved in redox signaling, and genes involved in sugar and amino acid metabolism [30]. Interestingly, the RoxR mutant of Pseudomonas putida exhibited a significantly reduced ability to adhere to corn seeds [30]. It remains unclear why RoxR mutants are less able to associate with host surfaces and whether defective interaction with lung epithelial cells for P. aeruginosa RoxR mutants and defective adhesion to corn seeds for P. putida RoxR mutants share any mechanistic similarities.
In summary, P. aeruginosa clinical isolates from CF patients are capable of stimulating PMN migration across lung epithelial cell monolayers, suggesting that the ability to induce PMN trans-epithelial migration is a widely shared attribute amongst a diverse collection of P. aeruginosa strains [11]. Components of flagella, pili, or TTSS that have previously been shown to induce inflammatory responses in epithelial cells such as the secretion of CXCL8 [22–26], are not directly involved in the ability to induce PMN transmigration, although flagella and pili exert and indirect effect by virtue of their ability to facilitate association with lung epithelial cells. It remains unclear which P. aeruginosa factor(s) are necessary and sufficient for stimulating PMN trans-epithelial migration, but P. aeruginosa lung epithelial cell association is clearly important to facilitate this process. To this end, we have identified a previously unrecognized role for the two component system response regulator RoxR. RoxR regulates genes important for proper association of P. aeruginosa with lung epithelial cells. Identification of such genes may reveal novel insight into the pathogenic process of P. aeruginosa infection of the human lung and proteins encoded by such genes may serve as useful targets to exploit for therapeutic intervention or vaccine design.
Acknowledgments
This work is supported by the Cystic Fibrosis Foundation and through a Research Scholarship Development Award (K22_AI065425-01A2) from the National Institute of Health/National Institute of Allergy and Infectious Disease. Dr. Beth A. McCormick is supported by NIH/NIDDK (R01_DK56754). Work in Stephen Lory’s laboratory was supported by the grant AI21451 from the NIH.
We thank Dr. W. Allan Walker MD, Chief of the Mucosal Immunology Laboratory at Massachusetts General Hospital for continued support. We also thank Waheed Pirzai, Jeffrey D. Bien, and Michael A. Pazos for technical assistance.
Abbreviations
- PMNs
Neutrophils
- CF
Cystic Fibrosis
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Burns AR, Smith CW, Walker DC. Unique structural features that influence neutrophil emigration into the lung. Physiol Rev. 2003;83:309–336. doi: 10.1152/physrev.00023.2002. [DOI] [PubMed] [Google Scholar]
- 2.Lyczak JB, Cannon CL, Pier GB. Lung infections associated with cystic fibrosis. Clin Microbiol Rev. 2002;15:194–222. doi: 10.1128/CMR.15.2.194-222.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McIntosh K. Community-acquired pneumonia in children. N Engl J Med. 2002;346:429–437. doi: 10.1056/NEJMra011994. [DOI] [PubMed] [Google Scholar]
- 4.Prescott WA, Jr, Johnson CE. Antiinflammatory therapies for cystic fibrosis: past, present, and future. Pharmacotherapy. 2005;25:555–573. doi: 10.1592/phco.25.4.555.61025. [DOI] [PubMed] [Google Scholar]
- 5.Goodman AL, Lory S. Analysis of regulatory networks in Pseudomonas aeruginosa by genomewide transcriptional profiling. Curr Opin Microbiol. 2004;7:39–44. doi: 10.1016/j.mib.2003.12.009. [DOI] [PubMed] [Google Scholar]
- 6.Rodrigue A, Quentin Y, Lazdunski A, Mejean V, Foglino M. Two-component systems in Pseudomonas aeruginosa: why so many? Trends Microbiol. 2000;8:498–504. doi: 10.1016/s0966-842x(00)01833-3. [DOI] [PubMed] [Google Scholar]
- 7.Goodman AL, Kulasekara B, Rietsch A, Boyd D, Smith RS, Lory S. A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in Pseudomonas aeruginosa. Dev Cell. 2004;7:745–754. doi: 10.1016/j.devcel.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 8.Kulasekara HD, Ventre I, Kulasekara BR, Lazdunski A, Filloux A, Lory S. A novel two-component system controls the expression of Pseudomonas aeruginosa fimbrial cup genes. Mol Microbiol. 2005;55:368–380. doi: 10.1111/j.1365-2958.2004.04402.x. [DOI] [PubMed] [Google Scholar]
- 9.Ritchings BW, Almira EC, Lory S, Ramphal R. Cloning and phenotypic characterization of fleS and fleR, new response regulators of Pseudomonas aeruginosa which regulate motility and adhesion to mucin. Infect Immun. 1995;63:4868–4876. doi: 10.1128/iai.63.12.4868-4876.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hickman JW, Tifrea DF, Harwood CS. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc Natl Acad Sci U S A. 2005;102:14422–14427. doi: 10.1073/pnas.0507170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hurley BP, Siccardi D, Mrsny RJ, McCormick BA. Polymorphonuclear cell transmigration induced by Pseudomonas aeruginosa requires the eicosanoid hepoxilin A3. J Immunol. 2004;173:5712–5720. doi: 10.4049/jimmunol.173.9.5712. [DOI] [PubMed] [Google Scholar]
- 12.Hurley BP, Sin A, McCormick BA. Adhesion molecules involved in hepoxilin A3-mediated neutrophil transepithelial migration. Clin Exp Immunol. 2008;151:297–305. doi: 10.1111/j.1365-2249.2007.03551.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harshey RM. Bacterial motility on a surface: many ways to a common goal. Annu Rev Microbiol. 2003;57:249–273. doi: 10.1146/annurev.micro.57.030502.091014. [DOI] [PubMed] [Google Scholar]
- 14.Rashid MH, Rao NN, Kornberg A. Inorganic polyphosphate is required for motility of bacterial pathogens. J Bacteriol. 2000;182:225–227. doi: 10.1128/jb.182.1.225-227.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Comolli JC, Donohue TJ. Pseudomonas aeruginosa RoxR, a response regulator related to Rhodobacter sphaeroides PrrA, activates expression of the cyanide-insensitive terminal oxidase. Mol Microbiol. 2002;45:755–768. doi: 10.1046/j.1365-2958.2002.03046.x. [DOI] [PubMed] [Google Scholar]
- 16.Wolfgang MC, Lee VT, Gilmore ME, Lory S. Coordinate regulation of bacterial virulence genes by a novel adenylate cyclase-dependent signaling pathway. Dev Cell. 2003;4:253–263. doi: 10.1016/s1534-5807(03)00019-4. [DOI] [PubMed] [Google Scholar]
- 17.Cohen SN, Chang AC, Hsu L. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc Natl Acad Sci U S A. 1972;69:2110–2114. doi: 10.1073/pnas.69.8.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vance RE, Rietsch A, Mekalanos JJ. Role of the type III secreted exoenzymes S, T, and Y in systemic spread of Pseudomonas aeruginosa PAO1 in vivo. Infect Immun. 2005;73:1706–1713. doi: 10.1128/IAI.73.3.1706-1713.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wolfgang MC, Kulasekara BR, Liang X, Boyd D, Wu K, Yang Q, Miyada CG, Lory S. Conservation of genome content and virulence determinants among clinical and environmental isolates of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 2003;100:8484–8489. doi: 10.1073/pnas.0832438100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ishimoto KS, Lory S. Identification of pilR, which encodes a transcriptional activator of the Pseudomonas aeruginosa pilin gene. J Bacteriol. 1992;174:3514–3521. doi: 10.1128/jb.174.11.3514-3521.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Whitchurch CB, Alm RA, Mattick JS. The alginate regulator AlgR and an associated sensor FimS are required for twitching motility in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 1996;93:9839–9843. doi: 10.1073/pnas.93.18.9839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.DiMango E, Ratner AJ, Bryan R, Tabibi S, Prince A. Activation of NF-kappaB by adherent Pseudomonas aeruginosa in normal and cystic fibrosis respiratory epithelial cells. J Clin Invest. 1998;101:2598–2605. doi: 10.1172/JCI2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hahn HP. The type-4 pilus is the major virulence-associated adhesin of Pseudomonas aeruginosa--a review. Gene. 1997;192:99–108. doi: 10.1016/s0378-1119(97)00116-9. [DOI] [PubMed] [Google Scholar]
- 24.Prince A. Flagellar activation of epithelial signaling. Am J Respir Cell Mol Biol. 2006;34:548–551. doi: 10.1165/rcmb.2006-0022SF. [DOI] [PubMed] [Google Scholar]
- 25.Verma A, Arora SK, Kuravi SK, Ramphal R. Roles of specific amino acids in the N terminus of Pseudomonas aeruginosa flagellin and of flagellin glycosylation in the innate immune response. Infect Immun. 2005;73:8237–8246. doi: 10.1128/IAI.73.12.8237-8246.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cuzick A, Stirling FR, Lindsay SL, Evans TJ. The type III pseudomonal exotoxin U activates the c-Jun NH2-terminal kinase pathway and increases human epithelial interleukin-8 production. Infect Immun. 2006;74:4104–4113. doi: 10.1128/IAI.02045-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dasgupta N, Wolfgang MC, Goodman AL, Arora SK, Jyot J, Lory S, Ramphal R. A four-tiered transcriptional regulatory circuit controls flagellar biogenesis in Pseudomonas aeruginosa. Mol Microbiol. 2003;50:809–824. doi: 10.1046/j.1365-2958.2003.03740.x. [DOI] [PubMed] [Google Scholar]
- 28.Lizewski SE, Lundberg DS, Schurr MJ. The transcriptional regulator AlgR is essential for Pseudomonas aeruginosa pathogenesis. Infect Immun. 2002;70:6083–6093. doi: 10.1128/IAI.70.11.6083-6093.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feldman M, Bryan R, Rajan S, Scheffler L, Brunnert S, Tang H, Prince A. Role of flagella in pathogenesis of Pseudomonas aeruginosa pulmonary infection. Infect Immun. 1998;66:43–51. doi: 10.1128/iai.66.1.43-51.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fernandez-Pinar R, Ramos JL, Rodriguez-Herva JJ, Espinosa-Urgel M. A two-component regulatory system integrates redox state and population density sensing in Pseudomonas putida. J Bacteriol. 2008;190:7666–7674. doi: 10.1128/JB.00868-08. [DOI] [PMC free article] [PubMed] [Google Scholar]