Abstract
Purpose
The purpose of this study was to investigate the relationship of porcine somatic cell nuclear transfer (SCNT) embryo developmental competence with embryonic cell apoptosis and DNA methylation.
Methods
The apoptotic incidence was examined via comet assay, and the mRNA expression of genes implicated in apoptosis (Bcl-2) and DNA methylation (Dnmt1, Dnmt3a) was determined using real-time RT-PCR.
Results
Comet assay showed that the SCNT embryos exhibited significantly higher apoptotic rate at 2-cell stage (8.3% versus 2.1%, P < 0.05), 16-cell stage (27.3% versus 19.2%, P < 0.05) and morula (37.5% versus 26.9, P < 0.05) compared with IVF embryos. Compared with IVF embryos, a higher Bcl-2 mRNA expression pattern was observed in SCNT embryos before the 8-cell stage and differed significantly at 2- and 4-cell stages (P < 0.05). After the 16-stage, Bcl-2 mRNA expression pattern became significantly lower in SCNT group (P < 0.05). The relative expression level of Dnmt1 mRNA showed a higher expression level in oocytes, then sharply decreased and started to increase slightly after the 8-cell (IVF embryos) or 16-cell stage (SCNT embryos). Dnmt1 mRNA expression in IVF embryos appeared to have been lower than that of SCNT group before 16-cell stage embryos, especially at 4- and 8-cell stages (P < 0.05). Although a trend for a similar increase of Dnmt3a expression was observed in IVF and SCNT embryos after 8-cell embryos, SCNT group resulted in much higher Dnmt3a mRNA abundance compared with the IVF group, particularly after 16-cell embryos (P < 0.05).
Conclusions
The results showed that low efficiency of porcine SCNT technology may be associated with either embryonic apoptosis or incomplete reprogramming of donor nuclear caused by abnormal Dnmts mRNA expression.
Keywords: Swine, Somatic cell nuclear transfer, Apoptosis, DNA methylation
Introduction
The success of somatic cell nuclear transfer (SCNT) in swine gives promise to widespread applications such as genetically superior pig breed production, species resource preservation and xenotransplantation for humans, etc [1–3]. Since the first cloned pigs were generated using somatic cell cloning [4], substantial improvement for this technique has been realized [5, 6]. However, the efficiency of pig cloning has been lower than that of other domestic animals, with only 1%–5% of the embryos reconstructed by nuclear transfer surviving to term [7, 8]. Apoptosis, a type of programmed cell death, is a physiological process occurring spontaneously during normal preimplantation embryo development [9]. However, apoptosis also has a role in the cellular response to suboptimal developmental conditions and stress [10], and embryonic development is compromised if apoptosis surpasses a certain threshold [9]. The occurrence of apoptosis in preimplantation embryos has been considered one of the most important parameters for evaluation of embryo health [11, 12]. The process of apoptotic cell death in mammalian preimplantation embryos has been well described, and the TUNEL (terminal deoxynucleotidyl transferase-mediated dUTP nick-end labelling) assay has been widely used for detection of apoptosis. Mouse in vivo embryos usually do not exhibit any TUNEL labeling before the blastocyst stage [13], but some apoptotic morphological changes were observed already at the 8- to 16-cell-stage mouse embryos produced in vitro [9]. Apoptosis was first observed in bovine SCNT embryos at the 4-cell stage, but for in vitro fertilization (IVF) embryos at 6- to 8-cell stages using TUNEL assay [14]. In pigs, the earliest positive TUNEL labeling signals were detected in SCNT-derived blastocysts, and the percentage of cells undergoing apoptosis in the SCNT embryos was apparently higher than that of IVF-derived blastocysts [15]. DNA damage and fragmentation are commonly major characteristics of apoptosis, and the comet assay (single cell gel-electrophoresis, SCGE) appears to be a more sensitive method for assessing DNA damage to evaluate the apoptosis individual eukaryotic cell as well as in mammalian preimplantation embryos, because this method is able to distinguish DNA fragmentation present in all apoptotic stages, including early ones [16, 17]. Nevertheless, the comprehensive evaluation of DNA damage aspects of apoptosis in porcine SCNT embryos during development in vitro has not been reported to date.
DNA methylation is a major epigenetic mark in the mammalian genome and is modulated during the reprogramming process, and the insufficient epigenetic reprogramming of the somatic donor genome might be the cause of the abnormalities and the low efficiency associated with SCNT [18, 19]. DNA methylation mechanism relies on the catalytic activity of DNA methyltransferases (Dnmts). The Dnmt1 maintains the methylation pattern during replication, whereas Dnmt3a and Dnmt3b are responsible for de novo methylation of unmethylated regions [20]. In animal cloning, the highly differentiated donor nucleus must cease its own program of gene expression and restore a particular program of embryonic expression necessary for normal development [21]. However, aberrant methylation changes of the donor genome, especially in highly repetitive sequences, were observed frequently in cloned embryos [21, 22]. In cloned bovine embryos, various genomic repeated sequences, such as satellite I, satellite II, and art-2 SINE sequences, showed aberrant methylation status in cloned blastocysts [22]. Bourc’his et al. [19] also reported that the centromeric heterochromatin in cloned bovine embryos was hypermethylated in contrast to that in IVF-derived embryos. The lack of demethylation in repetitive sequence Rsat IIE was also revealed in cloned rabbit embryos [23]. However, Kang et al. [24] using bisulfite-sequencing, demonstrated that DNA methylation changes in cloned porcine embryos appears to be normal, as both the centromeric satellite and PRE-1 sequence exhibited a typical demethylation process compared with that in control fertilized embryos. Bonk et al. [25] used a microarray-based approach but reported different results that methylation patterns of many CpG islands did differ between in vivo-generated and cloned porcine blastocysts.
These apparent conflicting results indicated different methylation status revealed by the different target sequences analyzed. Therefore, it is hard to predict the global DNA methylation patterns by evaluating individual genes and sequences. Moreover, these previous studies have focused on the DNA methylation state of repeated DNA elements, including SINE, LINE, and micro-satellite DNA [19, 24]. When studying nuclear reprogramming, however, it is essential to identify DNA methylation-related gene expression profiles; especially for the Dnmts-related genes, transcription levels changed throughout the recipient oocyte maturation process and cloned embryo development in vitro, which may provide insight into the relation between the DNA methylation state and the efficiency of procine SCNT.
The objectives of this study were to reveal the relationship of porcine SCNT embryos developmental competence with embryonic cell apoptosis and DNA methylation. The developmental competence of porcine SCNT and IVF embryos was compared, and their apoptotic incidence during development in vitro were assessed with comet assay. The expression patterns of genes implicated in apoptosis (Bcl-2) and DNA methylation (Dnmt1, Dnmt3a) were also examined using real-time RT-PCR, in order to provide insight into the causes of the low efficiency of porcine SCNT.
Materials and methods
All chemicals and reagents used in the experiments were purchased from Sigma (St. Louis, MO, USA) unless otherwise noted.
Collection and in vitro maturation of porcine oocytes
Porcine ovaries were obtained from a local abattoir and transported to the laboratory in 0.9% NaCl solution supplemented with 100 IU/ml potassium penicillin G and 100 IU/ml streptomycin sulfate (Zhong-Nuo Pharmaceutical Co., Ltd, Shijiazhuang, China) within 2 h at 37°C. Cumulus-oocyte complexes (COC) from 2–5 mm diameter follicles were aspirated to a 10-ml disposable syringe. The COC with uniform cytoplasm and at least three layers of compact cumulus cells were chosen and rinsed three times in Tyrode lactate-HEPES (TL-HEPES) containing 0.l% polyvinyl alcohol (PVA), then in vitro matured (IVM) as previously described by Wu et al.[26]. Briefly, approximately 15–20 germinal vesicle (GV) stage oocytes were placed into a 100-μl droplet of pre-equilibrated culture medium (TCM199; GIBCO BRL, Gaithersburg, MD) supplemented with 10 IU/ml PMSG (Tianjin Huafu High & New Biotech Co., Tianjin, China), 10 IU/ml hCG (Ningbo Hormonal Reagents Co., Ltd, Zhejiang, China), 69 μg/ml L-cysteine, 1 μg/ml 17β-estradiol, 10%(v/v) porcine follicular fluid (pFF, self-made), 36 mg/l sodium pyruvate, 1 ml/l sodium lactate, 10% newborn calf serum (NCS, GIBCO BRL), 70 mg/l penicillin G and 50 mg/l streptomycin, covered with mineral oil. According to the experimental design, the oocytes were cultured for various periods at 38.5°C in humidified atmosphere of 5% CO2 in air.
After maturation, cumulus cells were removed by vigorous vortexing in TL-HEPES medium supplemented with 0.1% hyaluronidase for 3 min, and the cumulus-free oocytes with uniform ooplasm, intact cytoplasmic membrane and visible first polar body (pbI) were considered to reach meiosis II (MII) stage [26].
Isolation and in vitro culture of nuclear donor cells
Cumulus cells were used for nuclear donor cells. After maturation in vitro, COC were transferred into TL-HEPES medium supplemented with 0.1% hyaluronidase for 3 min at 38.5°C, and then equal volume of DMEM medium containing 10% NCS was added and mixed thoroughly. The cumulus cells were harvested by centrifugation at 1,500 r/min for 5 min after removal of oocytes. Cell pellets were resuspended with DMEM medium containing 10% NCS, adjusting the cell density of 5 × 105 cells/ml, and seeded into 4-well multidishes (Nunclon, Roskilde, Denmark), incubated at 38.5°C in humidified 5% CO2 in air. When reaching 80% of confluence, the cells were passed in the proportion of 1:2 for subculture. Cells at 2-5 passages were treated with 0.05% trypsin and 0.02 mM EDTA before use.
Enucleation of recipient oocytes
The enucleation of MII-stage denuded oocytes was treated in HEPES buffered TCM199 containing 7.5 μg/ml Cytochalasin B (CB). After incubation for 15 min, oocytes were enucleated blindly by aspirating pbI and 20% adjacent cytoplasm presumably containing the metaphase II plate using a 20 μm in diameter glass pipette as previously described by Boquest et al. [27]. Successful enucleation was confirmed under an inverted fluorescent microscope (TS100, Nikon, Japan) after staining with 5 μg/ml Hoechst 33342 for 10 min.
Nuclear transfer procedures
Round glossy cumulus cells were chosen as donor cells and introduced into the perivitelline space of enucleated recipient oocytes through the hole made at enucleation, and then wedged between zona pellucida and cytoplasm membrane to facilitate close membrane contact for subsequent fusion. After recovered in 0.4% BSA (bovine serum albumin) NCSU23 medium at 38.5°C for 1.5 h, the reconstructed couplets were fused and activated simultaneously with a single DC pulse of 1.5 kV/cm for 80 µs using an Electro-cell Manipulator (CRY-3, Ningbo Xinzhi Co., Ltd, Ningbo, China). The resulting reconstructed embryos were washed and cultured in 0.5 ml of overnight-equilibrated NCSU23 medium containing 4 mg/ml BSA covered with dimethylpolysiloxane at 38.5°C, 5% CO2 in air. Approximately 40–50 reconstructed embryos were cultured in each well of a 4-well multidish. Fusion results were examined under an inverted microscope at 1 h after fusion and embryos were examined daily. The percentages of cleavage and blastocyst formation were evaluated on Day 2 and Day 7 after culture, respectively.
In vitro fertilization
Boar frozen semen in 0.25 ml straw was thawed at 37°C for 30 s in a water bath, diluted in 2.25 ml ZORLESCO (ZO) solution [28] and incubated at 37°C for 5 min. After washing twice by centrifugation (1,500 r/min, 5 min) with Dulbecco’s PBS, the spermatozoa were resuspended in modified tris-buffered medium (mTBM) [29] supplemented with 200 μg/ml heparin sodium salt, adjusting the sperm concentration to 5 × 106 sperm/ml. After washing twice with mTBM, MII oocytes were transferred into 100 μl mTBM droplets and inseminated with frozen-thawed sperm for 6 h at 38.5°C. The IVF embryos were cultured in overnight-equilibrated 0.4% BSA NCSU23 medium covered with dimethylpolysiloxane at 38.5°C, 5% CO2 in air. Embryos were examined daily and evaluated on Day 2 and Day 7 for cleavage and blastocyst formation. For the comet assay and real-time RT-PCR analysis, IVF and SCNT embryos at 2-, 4-, 8-, 16-cell and morulea stages were collected at 24 h, 48 h, 72 h, 96 h and 120 h, respectively.
Comet assay
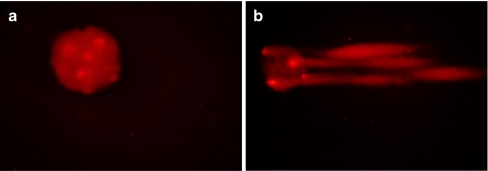
DNA damage of the embryos was assessed with the comet assay as previously described by Fabian et al. [16] with minor modifications. Prior to comet assay, the zona pellucida was digested by 0.2% protease and polar body was removed by microsurgery to ensure that only embryonic cell DNA was evaluated [30]. Briefly, 10–15 embryos were randomly selected from each group, put into 50 μl 1% low-melting agarose, and then transferred to a glass slide pre-coated with 100 μl 1% normal-melting agarose. The slide was sealed with a cover slip to solidify the agarose at 4°C for 10 min. After gently removing the cover slip, the third layer of 75 μl 1% normal-melting agarose was spread and sealed again with a cover slip to solidify at 4°C for another 10 min. After carefully removing the coverslip, slide with adhered embryo-embedded agarose was immersed in lysis solution (10 mM Tris, 100 mM EDTA, 1% SDS, 2.5 mM NaCl and 1% Triton X-100) at 4°C for 1 h to dissolve the cells and permit DNA unfolding and placed on a horizontal gel electrophoresis unit and equilibrated for 15 min in Tris-botate-EDTA buffer (TBE). Electrophoresis was conducted at 50 V, 300 mA for 15 min. Following electrophoresis, DNA of embryos was stained with 250 μg/ml propidium iodide (PI, Boster) by 20 min incubation. Stained embryos were examined under a fluorescence microscope. Fluorescence-stained images were analyzed according to the method used by Fabian et al. [16], the embryos with a full head but no tail or a very short tail no longer than diameter of head were assigned to undamaged embryos (Fig. 1a); contrarily, those that showed a half-empty head (due to emigration of high levels of fragmented DNA) with a long and intensive tail were classified into apoptotic embryos (Fig. 1b).
Fig. 1.
Apoptosis of embryos evaluated by comet assay (200×). a Undamaged embryo without tail. b Apoptotic embryo with a half-empty head together with long and intensive tails
Real-time RT-PCR
To compare the relative abundance of mRNA transcripts of Bcl-2, Dnmt1 and Dnmt3a genes in the oocytes or the embryos at various developmental stages from each group, real-time RT-PCR was performed. Total RNA was extracted from 30 oocytes or embryos from each group using TRIzol reagent (Invitrogen), according to the manufacturer’s instruction. A total of 0.5 μg RNA from each sample was converted to cDNA by using ReverTra Ace®qPCR RT Kit (TOYOBO, Co., Japan) in a 10 μl reaction mixture containing 2 μl of 5 × RT Buffer, 0.5 μl of RT Enzyme Mix, 0.5 μl of Primer Mix, 0.5 μg of RNA, and nuclease-free water added to 10 μl. Briefly, the RNA solution was incubated at 65°C for 5 min, then immediately placed on ice, after which the other components were added, incubated at 37°C for 15 min and then at 98°C for 5 min. Finally, the reacted solution was stored at −20°C. As negative controls, tubes were prepared in which RNA or reverse transcriptase was omitted during the reverse-transcription reaction.
Real-time PCR was performed on a LightCycler PCR (MJ ResearchTM, Mont., USA) by using SYBR® Green Real-time PCR Master Mix (TOYOBO), according to the manufacturer’s protocol. Primer sequences, the size of amplified products, and the GenBank accession numbers are shown in Table 1. The total volume of 20 μl real time RT-PCR reaction mixture contained 10 μl of SYBR® Green Real-time PCR Master Mix, 0.4 μM each of forward and reverse primers, 2 μl of cDNA and 6.4 μl of nuclease-free water. The program used for all genes consisted of a denaturing cycle of 30 s at 95°C, 45 cycles of PCR (95°C for 5 s, 57°C for 10 s, and 72°C for 15 s), a melting cycle consisting of 95°C for 0 s, 72°C for 5 min, and a step cycle starting at 65°C with a 0.2°C/s transition rate to 95°C. The specificity of the real-time RT-PCR product was confirmed by melting curve analysis. The PCR product sizes were confirmed by submarine agarose gel electrophoresis and staining with ethidium bromide. Three replications were performed, and the mRNA level of each sample was normalized to that of β-Actin mRNA level. The calibrator was the RNA from GV stage oocytes in each real-time amplification. Results of real-time PCR were analyzed using 2−ΔΔ CT method [31] to compare the relative transcriptional levels of the target genes in each sample.
Table 1.
Primer sequences used for real-time RT-PCR
| Gene | Accession number | Primer sequence 5′- 3′ | Product size(bp) |
|---|---|---|---|
| β-Actin | SSU07786 | F-CTCGATCATGAAGTGCGACGT | 114 |
| R-GTGATCTCCTTCTGCATCCTGTC | |||
| Bcl-2 | NM_214285 | F-GAAACCCCTAGTGCCATCAA | 196 |
| R-GGGACGTCAGGTCACTGAAT | |||
| Dnmt1 | NM_001032355 | F-TCGAACCAAAACGGCAGTAGT | 215 |
| R-CGGTCAGTTTGTGTTGGAGAAG | |||
| Dnmt3a | NM_001097437 | F-CTGAGAAGCCCAAGGTCAAG | 238 |
| R-CAGCAGATGGTGCAGTAGGA |
Experimental design
Developmental competence of porcine SCNT and IVF embryos
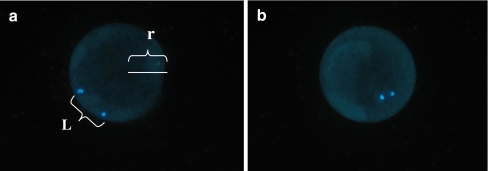
This experiment mainly focused on the optimization of production of porcine SCNT and IVF embryos. Optimal procedures reported previously were adopted directly in the present study [26, 32]. Porcine MII stage oocytes were randomly allocated to two groups: IVF and SCNT. Cleavage and blastocyst formation in culture were evaluated on Day 2 and Day 7 after reconstruction or IVF. Optimization of enucleation of recipient oocytes was emphasized here in the paper. The relative location between the metaphase plate and pbI of a cumulus-free oocyte was occasionally found to be associated with maturation culture time of COC (culture for 42 h, 44 h and 46 h, respectively). After blind enucleation, successful enucleation of the oocytes was determined by staining with 5 μg/ml Hoechst 33342 for 10 min under an inverted fluorescent microscope. According to the relative distance between metaphase plate and pbI (Fig. 2), oocytes were divided into two groups: (1) Adjacent. The length of the distance between metaphase plate and pbI was less than 1/3 radius of the oocyte (L<1/3r). (2) Displaced. The distance between metaphase plate and pbI was more than 1/3 radius of the oocyte (L>1/3r). The dynamic alterations of pbI extrusion rate, enucleation rate and relative location between pbI and nucleolus of an oocyte were evaluated, respectively, at different maturation culture time (42 h, 44 h and 46 h).
Comparison of apoptotic incidence of porcine IVF and SCNT embryos during development in vitro
Fig. 2.
The relative location between pbI and nucleus of procine ooctyes (400×). L. The length of the distance between the metaphase plate and the pbI, r. The radius of the porcine oocytes. a Displacement: the length of the distance between the metaphase plate and the pbI was more than 1/3 radius of the oocytes(L > 1/3 r). b Adjacent: the length of the distance between the metaphase plate and the pbI was less than 1/3 radius of the oocytes (L < 1/3 r)
For evaluation of apoptotic incidence, porcine IVF and SCNT embryos were randomly divided into five groups: 2-, 4-, 8-, 16-cell stage embryos and morula derived from IVF and SCNT, respectively. Embryos from each group were used for comet assay separately to assess apoptotic incidence of each group.
Relative expression level of Bcl-2 in porcine embryos produced in vitro
To compare relative expression level of anti-apoptotic gene (Bcl-2), embryos derived from IVF and SCNT were randomly divided into five groups: 2-, 4-, 8-, 16-cell stage embryos and morula. Thirty embryos were pooled from each group and directly conserved in liquid nitrogen, preparing for real-time RT- PCR analysis. All embryos samples were analyzed in triplicate for every gene.
Relative expression levels of Dnmt1 and Dnmt3a mRNA in porcine oocytes and embryos
To compare relative expression levels of DNA methyltransferases genes (Dnmt1, Dnmt3a), oocytes or embryos were randomly divided into seven groups: GV and MII oocytes, 2-, 4-, 8-, 16-cell stage embryos and morula. Thirty oocytes or embryos were pooled from each group and directly conserved in liquid nitrogen, preparing for real-time RT- PCR analysis. All oocytes and embryos samples were analyzed in triplicate for every gene.
Statistical analysis
Experiments were replicated at least three times, the percentages were subjected to an arc-sine transformation, and the transformed values were analyzed by ANOVA. Differences at P < 0.05 were considered significant.
Results
Developmental competence of porcine SCNT and IVF embryos
Table 2 summarizes the developmental competence of porcine IVF and SCNT embryos using the optimal protocols. There was no significant difference for cleavage rate between groups (P > 0.05), but the developmental percentage to blastocyst stage of SCNT group was significantly lower than that of IVF group (20.4% versus 29.8%, P < 0.05; Fig. 3).
Table 2.
Developmental competence of porcine IVF and NT embryos in vitro
| Group | No. embryos cultured | Cleavage rate (%) | Blastocyst rate(%) |
|---|---|---|---|
| IVF | 131 | 79.4 (104/131)a | 29.8 (31/104)a |
| SCNT | 127 | 77.2 (98/127)a | 20.4 (20/98)b |
a,bValues with different letters within columns are significantly different, P < 0.05
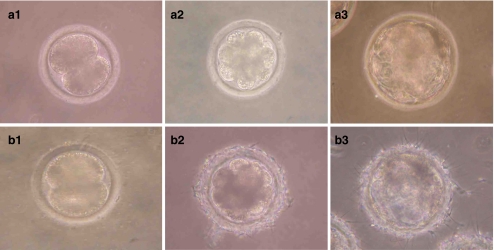
Fig. 3.
Development in vitro of porcine SCNT (a) and IVF (b) embryo (200×). a1 and b1. 2-cell stage embryo. a2 and b2. 8-cell stage. a3 and b3. blastocyst stage
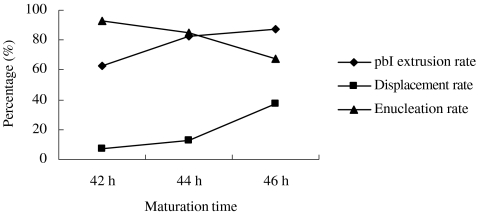
Interestingly, during blind enucleation of recipient oocytes in the present experiment, it was found that pbI extrusion rate appeared to rise with the maturation culture period (Fig. 4), whereas enucleation rate of recipient oocytes decreased gradually. The relative location between pbI and nucleus dramatically varied with the culture period, and the displacement percentage of pbI increased significantly as the oocytes aged during continuous culture. Under the present experimental condition, maturation culture for 44 h in vitro was the optimal time-point for enucleation of porcine oocytes with relatively higher pbI extrusion rate (82.5%, 33/40) as well as acceptable enucleation rate (85.0%, 34/40).
Fig. 4.
The dynamic alterations of pbI extrusion rate, enucleation rate and the relative location between pbI and nucleus of porcine oocytes during culture in vitro. The pbI extrusion rate is the percentage of the number of oocytes with visible pbI. Displacement rate means the percentage of the number of those oocytes that length of distance between metaphase plate and pbI was more than 1/3 radius of the oocyte (L>1/3 r). Enucleation rate is the percentage of the number of oocytes with successful enucleation
Comparison of apoptotic incidence and relative abundance of Bcl-2 mRNA transcript in porcine IVF and SCNT embryos during development in vitro
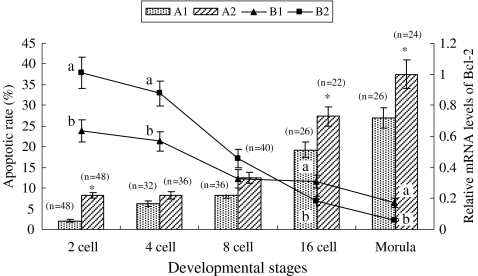
Comet assay showed that the apoptotic rate in both IVF and SCNT embryos tended to increase similarly with embryonic developmental stage (Fig. 5). However, SCNT embryos exhibited significantly higher apoptotic rate at the 2-cell stage (8.3% versus 2.1%, P < 0.05), 16-cell stage (27.3% versus 19.2%, P < 0.05) and morula (37.5% versus 26.9%, P < 0.05) compared with IVF embryos.
Fig. 5.
The apoptosis and relative expression level of Bcl-2 mRNA in porcine IVF and SCNT embryos during development in vitro. A1 and A2 shown the apoptotic rate of IVF and SCNT embryos, respectively; B1 and B2 shown the relative Bcl-2 mRNA levels of IVF and SCNT embryos, respectively. The experiment was replicated three times. dates are presented as mean ± SEM. a,b Values with different letter superscripts are significant difference(P < 0.05) in the relative Bcl-2 transcript abundance between in IVF and SCNT embryos at given stage of development; Bars with an asterisk(*) are significantly difference(P < 0.05) in apoptotic rate between in IVF and SCNT embryos at given stage of development; ‘n’ is the number of embryos tested by comet assay
On the other hand, as shown in Fig. 5, the relative level of Bcl-2 mRNA in both IVF and SCNT embryos decreased with embryonic development. Compared with IVF embryos, a higher Bcl-2 mRNA expression pattern was observed in SCNT embryos before the 8-cell stage and significantly differed at 2- and 4-cell stages (P < 0.05). After the 16-cell stage, Bcl-2 mRNA expression pattern became significantly lower in the SCNT group (P < 0.05).
Relative abundance of Dnmt1 and Dnmt3a mRNA transcripts in porcine oocytes and embryos
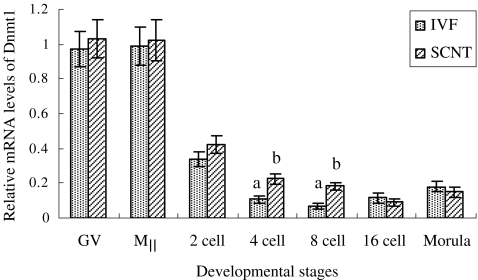
The relative expression level of Dnmt1 mRNA seemed to be similar in either GV or MII stage oocytes but sharply decreased before the 8-cell stage in embryos produced in vitro (Fig. 6), behaving similar Dnmt1 mRNA expression trend in two groups throughout development. The Dnmt1 mRNA showed a higher expression level in oocytes, then sharply decreased and started to increase slightly after 8-cell (IVF embryos) or 16-cell stage (SCNT embryos). Dnmt1 mRNA expression in IVF embryos appeared to have been lower than that of SCNT group before 16-cell stage embryos, especially at 4- and 8-cell stage (P < 0.05).
Fig. 6.
The relative expression level of Dnmt1 mRNA in porcine oocytes and embryos. The reaction was conducted in triplicate for each sample. Date are presented as mean ± SEM(bars). a,b Values with different letter superscripts on bars are significant difference( P < 0.05) in the relative Dnmt1 transcript abundance between in IVF and SCNT embryos at given stage of development
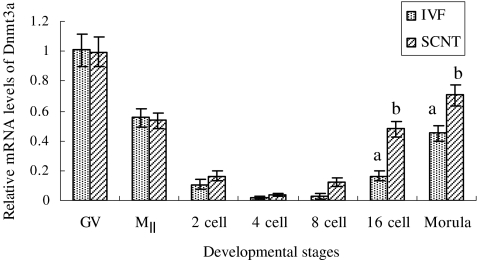
The relative expression level of Dnmt3a mRNA quickly decreased from GV-stage oocytes to 4-cell embryos produced in vitro (Fig. 7) and displayed a very low level from 2- to 8-cell stage in both groups. Although similar increase pattern of Dnmt3a expression was observed in IVF and SCNT embryos after 8-cell embryos, the SCNT group resulted in much higher Dnmt3a mRNA abundance compared with the IVF group, particularly after 16-cell embryos (P < 0.05).
Fig. 7.
The relative expression level of Dnmt3a mRNA in porcine oocytes and embryos. The reaction was conducted in triplicate for each sample. Date are presented as mean ± SEM(bars). a,b Values with different letter superscripts on bars are significant difference( P < 0.05) in the relative Dnmt3a transcript abundance between in IVF and SCNT embryos at given stage of development
Discussion
Effects of relative location between metaphase plate and pbI on enucleation efficiency of recipient oocytes
For preparation of recipient oocytes, enucleation is necessary to remove genetic material from the recipient cytoplasm. Efficient enucleation is a crucial step to avoid such problems as aneuploidy abnormalities and possibly parthenogenetic activation [33]. However, using a blind enucleation method could affect enucleation rate, which is influenced by a number of factors. The pbI extrusion rate appeared to show a rising trend, whereas enucleation rate of the recipient oocytes decreased gradually as in vitro maturation period increased. Therefore, it is important to identify a cross-point for maturation culture period and enucleation rate. At this point, both pbI extrusion and enucleation rate maximized. Both the nucleus and pbI of oocytes move during the maturation process, resulting in the distance between them changing and severely impacting the enucleation rate. In the present experiment, maturation culture for 44 h of porcine oocytes appeared to be the optimal time-point for enucleation.
In vitro development and apoptosis of porcine SCNT embryos
DNA damage and fragmentation are the major hallmarks of apoptosis, and accurate assessment is crucial when the biological significance of apoptosis is investigated. Although the TUNEL assay is frequently used as a method enabling in situ detection of apoptotic cells by labeling specific DNA degradation, its specificity is relatively low since the nuclei of cells undergoing necrosis are also labeled [34, 35]. The comet assay has been widely accepted as a simple, sensitive, and rapid tool for assessing DNA damage in individual eukaryotic as well as in preimplantation embryos, and can be used for the detailed analysis of DNA breaks during apoptosis [16]. Hao et al. [15] used TUNEL assay on porcine SCNT embryos and found that the earliest positive TUNEL signals happened in blastocysts. In this study, comet assay was used to analyze apoptosis in porcine cloned embryos, showing that the damaged embryos containing apoptotic cells could be detected in all developmental stages before blastocyst. The reason for the difference of these results may be attributed to the more sensitivity of comet assay than TUNEL assay for the determination of early cell apoptosis. Additionally, the rate of blastocyst formation by porcine SCNT embryos in the present experiment was significantly lower than that of IVF embryos (20.4% versus 29.8%, P < 0.05), whereas the apoptotic incidence of porcine SCNT embryos was higher than that of IVF embryos at all early developmental stages (Fig. 5). Hao et al. [15] reported a higher apoptotic incidence by TUNEL assay of porcine SCNT blastocysts compared with IVF ones. Gjørret et al. [36] also found that the incidence of apoptosis is higher in bovine blastocysts produced by SCNT than that of blastocysts derived in vivo. In general, all these results indicate that the high level of apoptotic incidence during development in vitro may be associated with the low efficiency of porcine SCNT.
Comet assay is an appropriate method for studying apoptosis in preimplantation embryos, however, it is essential to note that the degenerating polar bodies are also present in early stage embryos, such as in two to four cell stage embryos. The mechanism of polar body disintegration is very close to apoptosis and it is accompanied by the DNA fragmentation which also results in the formation of the comet tail in the comet assay [16]. In the present study, the zona pellucida was digested by 0.2% protease prior to comet assay, and the polar bodies were removed by microsurgery, which ensure that the only embryonic cells DNA were evaluated.
Another interesting finding from this experiment was that both apoptotic incidence by comet assay and expression level of Bcl-2 mRNA of SCNT embryos were significantly higher than those of IVF embryos at 2-cell stage. Bcl-2 is considered to be an anti-apoptotic gene, and its expression level should be low when apoptotic incidence is high. The higher apoptotic incidence of porcine 2-cell stage SCNT embryos than IVF group may be attributed to the in vitro micromanipulation[6]. The in vitro micromanipulation could cause cellular mechanical damage to some extent, resulting in higher apoptotic rate of porcine SCNT reconstructed embryos compared with IVF embryos. This could be another demonstration that in vitro manipulation may reduce embryo development, and increase the incidence of apoptosis [6]. On the other hand, the higher bcl-2 mRNA expression of SCNT embryos at 2-and 4-cell stages may be the active response of SCNT embryos to deal with the manipulation stress in vitro, suggesting that the embryos have a higher capability of response to the mechanical stimulin of in vitro manipulation at early developmental stages. However, the capability of response gradually decreased along with the embryonic development. The Bcl-2 gene family includes pro-apoptotic (Bax, Bad, Bak, Bokl, Bcl-xs, Bik, and Bid) and anti-apoptotic (Bcl-2, Bcl-xl, Mcl-1, and Bcl-w) subgroups and plays an important role in regulation of cell apoptosis during embryo development [37]. In our study, IVF embryos yielded significantly higher expression of Bcl-2 mRNA compared to SCNT embryos after 16-cell stage. Correspondingly, the apoptotic incidence of IVF embryos after 16-cell stage is significantly lower than that of SCNT embryos.
Relative abundance of Dnmt1 and Dnmt3a mRNA transcripts in porcine oocytes and embryos
During normal embryonic development, reprogramming of genomic DNA modifications, such as DNA methylation, is observed shortly before and after the formation of the zygote. Shortly after fertilization, the global pattern of genomic DNA methylation is high in male and female gametes. In most species, such as mouse, rat, cattle, human and pig, a similar remodeling process has been observed that the paternal DNA is actively and rapidly demethylated immediately within 4 h after fertilization, whilst the maternal DNA becomes passively demethylated during subsequent cleavages [22, 38–41]. The genome-wide demethylation process is a general phenomenon in mammals and may play an important role for normal development of early embryos [21]. Remethylation or remodeling of the genome appears to occur around the time of implantation and is maintained in somatic tissues.
Successful cloning requires epigenetic reprogramming of donor nuclei [21]. DNA methylation, a major mechanism of this process, involves addition of a methyl group to the cytosine residues within the CpG dinucleotide, and generally associated with gene silencing. The correct pattern of cytosine methylation in CpG dinucleotides is required for normal mammalian development [42]. Nevertheless, cloned embryos have been found frequently to possess aberrant DNA methylation changes [21, 22, 25]. Genome-wide aberrant demethylation of the donor genome has been found in cloned bovine embryos during preimplantation development [43]. The lack of demethylation in repetitive sequence Rsat IIE was also observed in cloned rabbit embryos [23]. To date, several studies have also focused on the methylation changes during development of cloned pig embryos. It is of interest that a gradual demethylation pattern, which is similar to the endogenous demethylation process in fertilized embryos, was observed in cloned porcine embryos by bisulfite-sequencing technology when using PRE-1 and centromeric satellite sequences as markers [24]. However, Bonk et al. [25] used a global microarray-based approach to analyze the CpG methylation and found that mehtylation patterns of many CpG islands are different between in vivo-generated and cloned pig embryos. These apparent conflicting reports indicated different methylation status revealed by different target sequences analyzed. Therefore, it is hard to predict the global DNA methylation patterns only by evaluation of individual genes and sequences. In addition, DNA methylation mechanism relies on the catalytic activity of Dnmts. Differences in temporal patterns of transcripts could indicate possible differences in regulation and function of the Dnmts. To study the DNA methylation aspects of nuclear reprogramming, it is essential to identify DNA methylation related Dnmts gene expression profiles. In the present experiment, temporal transcript patterns of Dnmt1 and Dnmt3a gene of porcine oocytes and embryos derived from IVF and SCNT during preimplantation development were examined via real-time RT-PCR. The results revealed that Dnmt1 mRNA expression behaved in similar pattern in either IVF group or SCNT group. However, the Dnmt1 mRNA transcript level in SCNT group was observed to be significantly higher than that of IVF group at 4- and 8-cell stage. The Dnmt3a mRNA expression in SCNT group also appeared significantly higher at 16-cell and morula-stage compared with the IVF group (P < 0.05). Zhu et al. [44] also observed a significant increase in Dnmt1 mRNA expression in fetal fibroblast SCNT embryos compared with in vivo-produced embryos at the 8-cell stage. These results indicated that porcine SCNT embryos seemed to have abnormally higher Dnmts mRNA transcripts than IVF embryos do, suggesting that there existed relatively high levels of Dnmts activities during development in vitro for SCNT embryos [45]. Kwon et al. [46] also found global hypermethylated DNA at the 4-cell stage of porcine SCNT embryos compared with IVF embryos using a 5-methylcytosine (5-MeC) antibody immunostaining method. An abnormally increased expression of Dnmt1 protein has also been observed in the 8-cell stage cloned murine embryos reconstructed from cumulus cells [47]. These abnormal levels of embryonic methylation may be associated with reduced developmental potential [48]. Furthermore, it is important to realize that the results from different approaches used to evaluate methylation changes may provide different information for the very difference work on bases [41]. The immunostaining method, for example, is used for assessment of global methylation. However, the bisulfite sequencing can be used for evaluation of methylation status of individual genes that contain CpG islands.
In addition, we also interestingly found that the Dnmt1 expression in SCNT group appeared significantly higher than that of IVF group at 4-cell stage, coincident with the timing of maternal-zygotic transition (MZT). The MZT is the first major transition in which the developmental programme that is initially directed by maternally inherited proteins and transcripts is replaced by a new programme as the consequence of the embryonic genome activation for continued development [49]. However, Dnmt 1 is responsible for cytosine methylaton in mammals and influences gene silencing, the over expression of Dnmt 1 is associated with repression of gene expression. Therefore, the higher expression of Dnmt 1 during the MZT period in present study may be leading to an incomplete activation of the embryonic genome. In this study, the SCNT embryos had a similar cleavage rate but a lower further developmental rate than the IVF embryos, which may be correlated with the incomplete activation of the embryonic genome [45].
In conclusion, porcine SCNT procedures used in this study enables analysis of apoptosis and DNA methylation of cloned embryos. The porcine SCNT embryos had a lower development rate of blastocysts and a higher degree of apoptotic incidence during in vitro development as compared to IVF embryos. Despite behaved similar expression pattern in either IVF group or SCNT group, the abundance of Dnmts mRNA transcripts in SCNT embryos appeared significantly higher compared to the IVF embryos at 4-to 8-cell stage(Dnmt1) and after 16-cell stage(Dnmt3a), respectively. The low efficiency of porcine SCNT technology may be associated with either embryonic apoptosis or incomplete reprogramming of donor nuclear caused by abnormal DNA methylation.
Acknowledgements
The research was granted by National Research & Development Program of High-tech (2008AA101003) and “11th Five Years” Scientific Supporting Program of the Ministry of Agriculture of China (2008BADB2B11). We particularly thank Dr. Gary B. Anderson from the Department of Animal Science, University of California, Davis, for his comments and help with revision in English.
Footnotes
Capsule
Low efficiency of porcine SCNT technology may be associated with either embryonic apoptosis or incomplete reprogramming of donor nuclear caused by abnormal Dnmts mRNA expression.
References
- 1.Lai L, Kolber-Simonds D, Park KW, Cheong HT, Greenstein JL, Im GS, et al. Production of alpha-1, 3-galactosyltransferase knockout pigs by nuclear transfer. Science. 2002;295(5557):1089–92. doi: 10.1126/science.1068228. [DOI] [PubMed] [Google Scholar]
- 2.Rogatcheva MM, Rund LA, Swanson KS, Marron BM, Beever JE, Counter CM, et al. Creating porcine biomedical models through recombineering. Comp Funct Genomics. 2004;5(3):262–7. doi: 10.1002/cfg.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fujimura T, Takahagi Y, Shigehisa T, Nagashima H, Miyagawa S, Shirakura R, et al. Production of alpha 1, 3-galactosyltransferase gene-deficient pigs by somatic cell nuclear transfer: a novel selection method for gal alpha 1, 3-Gal antigen-deficient cells. Mol Reprod Dev. 2008;75(9):1372–8. doi: 10.1002/mrd.20890. [DOI] [PubMed] [Google Scholar]
- 4.Polejaeva IA, Chen SH, Vaught TD, Page RL, Mullins J, Ball S, et al. Cloned pigs produced by nuclear transfer from adult somatic cells. Nature. 2000;407:86–90. doi: 10.1038/35024082. [DOI] [PubMed] [Google Scholar]
- 5.Im GS, Lai L, Liu Z, Hao Y, Wax D, Bonk A, et al. In vitro development of preimplantation porcine nuclear transfer embryos cultured in different media and gas atmospheres. Theriogenology. 2004;61(6):1125–35. doi: 10.1016/j.theriogenology.2003.06.006. [DOI] [PubMed] [Google Scholar]
- 6.McElroy SL, Kim JH, Kim S, Jeong YW, Lee EG, Park SM, et al. Effects of culture conditions and nuclear transfer protocols on blastocyst formation and mRNA expression in pre-implantation porcine embryos. Theriogenology. 2008;69(4):416–25. doi: 10.1016/j.theriogenology.2007.10.010. [DOI] [PubMed] [Google Scholar]
- 7.Lai L, Prather RS. Production of cloned pigs by using somatic cells as donors. Clonging Stem Cells. 2003;5(4):233–41. doi: 10.1089/153623003772032754. [DOI] [PubMed] [Google Scholar]
- 8.Walker SC, Shin TY, Zaunbrecher GM, Romano JE, Johnson GA, Bazer FW, et al. A highly efficient method for porcine cloning by nuclear transfer using in vitro-matured oocytes. Cloning Stem Cells. 2002;4(2):105–12. doi: 10.1089/153623002320253283. [DOI] [PubMed] [Google Scholar]
- 9.Fabian D, Koppel J, Maddox-Hyttel P. Apoptotic processes during mammalian preimplantation development. Theriogenology. 2005;64(2):221–31. doi: 10.1016/j.theriogenology.2004.11.022. [DOI] [PubMed] [Google Scholar]
- 10.Betts DH, King WA. Genetic regulation of embryo death and senescence. Theriogenology. 2001;55(1):171–91. doi: 10.1016/S0093-691X(00)00453-2. [DOI] [PubMed] [Google Scholar]
- 11.Brison DR, Schultz RM. Apoptosis during mouse blastocormation: evidence for a role for survival factors including transforming growth factor alpha. Biol Reprod. 1997;56(5):1088–96. doi: 10.1095/biolreprod56.5.1088. [DOI] [PubMed] [Google Scholar]
- 12.Hardy K, Spanos S. Growth factor expression and function in the human and mouse preimplantation embryo. J Endocrinol. 2002;172(2):221–36. doi: 10.1677/joe.0.1720221. [DOI] [PubMed] [Google Scholar]
- 13.Kamjoo M, Brison DR, Kimber SJ. Apoptosis in the preimplantation mouse embryo: effect of strain difference and in vitro culture. Mol Reprod Dev. 2002;61(1):67–77. doi: 10.1002/mrd.1132. [DOI] [PubMed] [Google Scholar]
- 14.Fahrudin M, Otoi T, Karja NW, Mori M, Murakami M, Suzuki T. Analysis of DNA fragmentation in bovine somatic nuclear transfer embryos using TUNEL. Reproduction. 2002;124(6):813–9. doi: 10.1530/rep.0.1240813. [DOI] [PubMed] [Google Scholar]
- 15.Hao Y, Lai L, Mao J, Im GS, Bonk A, Prather RS. Apoptosis and in vitro development of preimplantation porcine embryos derived in vitro or by nuclear transfer. Biol Reprod. 2003;69(2):501–7. doi: 10.1095/biolreprod.103.016170. [DOI] [PubMed] [Google Scholar]
- 16.Fabian D, Rehák P, Czikková S, Il’ková G, Baran V, Koppel J. Induced cell death of preimplantation mouse embryos cultured in vitro evaluated by comet assay. Theriogenology. 2003;60(4):691–706. doi: 10.1016/S0093-691X(03)00087-6. [DOI] [PubMed] [Google Scholar]
- 17.Dhawan A, Bajpayee M, Parmar D. Comet assay: a reliable tool for the assessment of DNA damage in different models. Cell Biol Toxicol. 2009;25(1):5–32. doi: 10.1007/s10565-008-9072-z. [DOI] [PubMed] [Google Scholar]
- 18.Wrenzycki C, Wells D, Herrmann D, Miller A, Oliver J, Tervit R, et al. Nuclear transfer protocol affects messenger RNA expression patterns in cloned bovine blastocysts. Biol Reprod. 2001;65(1):309–17. doi: 10.1095/biolreprod65.1.309. [DOI] [PubMed] [Google Scholar]
- 19.Bourc’his D, Bourhis D, Patin D, Niveleau A, Comizzoli P, Renard JP, et al. Delayed and incomplete reprogramming of chromosome methylation patterns in bovine cloned embryos. Curr Biol. 2001;11(19):1542–6. doi: 10.1016/S0960-9822(01)00480-8. [DOI] [PubMed] [Google Scholar]
- 20.Chen T, Li E. Establishment and maintenance of DNA methylation patterns in mammals. Curr Top Microbiol Immunol. 2006;301:179–201. doi: 10.1007/3-540-31390-7_6. [DOI] [PubMed] [Google Scholar]
- 21.Han YM, Kang YK, Koo DB, Lee KK. Nuclear reprogramming of cloned embryos produced in vitro. Theriogenology. 2003;59(1):33–44. doi: 10.1016/S0093-691X(02)01271-2. [DOI] [PubMed] [Google Scholar]
- 22.Dean W, Santos F, Stojkovic M, Zakhartchenko V, Walter J, Wolf E, et al. Conservation of methylation reprogramming in mammalian development: Aberrant reprogramming in cloned embryos. Proc Natl Acad Sci. 2001;98(24):13734–8. doi: 10.1073/pnas.241522698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen T, Zhang YL, Jiang Y, Liu SZ, Schatten H, Chen DY, et al. The DNA methylation events in normal and cloned rabbit embryos. FEBS Lett. 2004;578(1–2):69–72. doi: 10.1016/j.febslet.2004.10.073. [DOI] [PubMed] [Google Scholar]
- 24.Kang YK, Koo DB, Park JS, Choi YH, Kim HN, Chang WK, et al. Typical demethylation events in cloned pig embryos. Clues on species-specific differences in epigenetic reprogramming of a cloned donor genome. J Biol Chem. 2001;276(43):39980–4. doi: 10.1074/jbc.M106516200. [DOI] [PubMed] [Google Scholar]
- 25.Bonk AJ, Li R, Lai L, Hao Y, Liu Z, Samuel M, et al. Aberrant DNA methylation in porcine in vitro-, parthenogenetic-, and somatic cell nuclear transfer-produced blastocysts. Mol Reprod Dev. 2008;75(2):250–64. doi: 10.1002/mrd.20786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu C, Rui R, Dai J, Zhang C, Ju S, Xie B, et al. Effects of cryopreservation on the developmental competence, ultrastructure and cytoskeletal structure of porcine oocytes. Mol Reprod Dev. 2006;73(11):1454–62. doi: 10.1002/mrd.20579. [DOI] [PubMed] [Google Scholar]
- 27.Boquest AC, Grupen CG, Harrison SJ, McIlfatrick SM, Ashman RJ, d’Apice AJ, et al. Production of cloned pigs from cultured fetal fibroblast cells. Biol Reprod. 2002;66(5):1283–7. doi: 10.1095/biolreprod66.5.1283. [DOI] [PubMed] [Google Scholar]
- 28.Johnson LA, Weitze KF, Fiser P, Maxwell WM. Storage of boar semen. Anim Reprod Sci. 2000;62(1–3):143–72. doi: 10.1016/S0378-4320(00)00157-3. [DOI] [PubMed] [Google Scholar]
- 29.Nam DH, Lee SH, Kim HS, Lee GS, Jeong YW, Kim S, et al. The role of gonadotropin releasing hormone (GnRH) and its receptor in development of porcine preimplantation embryos derived from in vitro fertilization. Theriogenology. 2005;63(1):190–201. doi: 10.1016/j.theriogenology.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 30.Men H, Monson RL, Parrish JJ, Rutledge JJ. Detection of DNA damage in bovine metaphase II oocytes resulting from cryopreservation. Mol Reprod Dev. 2003;64(2):245–50. doi: 10.1002/mrd.10249. [DOI] [PubMed] [Google Scholar]
- 31.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25(4):402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 32.Ju SQ, Rui R, Lu Q, Cao AQ, Li N, Lin PF, et al. Experiment on production in vitro of porcine somatic cell cloned embryos. Scientia Agricultura Sinica. 2009;42(4):1386–93. [Google Scholar]
- 33.Dominko T, Chan A, Simerly C, Luetjens CM, Hewitson L, Martinovich C, et al. Dynamic imaging of the metaphase II spindle and maternal chromosomesin bovine oocytes: implications for enucleation efficiency verification, avoidance of parthenogenesis, and successful embryogenesis. Biol Reprod. 2000;62(1):150–4. doi: 10.1095/biolreprod62.1.150. [DOI] [PubMed] [Google Scholar]
- 34.Lee K, Hyslop JM, Nanassy L, Machaty Z. Incidence of apoptosis in parthenogenetic porcine embryos generated by using protein kinase or protein synthesis inhibitors. Anim Reprod Sci. 2009;112(3–4):261–72. doi: 10.1016/j.anireprosci.2008.04.027. [DOI] [PubMed] [Google Scholar]
- 35.Darzynkiewicz Z, Bedner E, Traganos F. Difficulties and pitfalls in analysis of apoptosis. Methods Cell Biol. 2001;63:527–46. doi: 10.1016/S0091-679X(01)63028-0. [DOI] [PubMed] [Google Scholar]
- 36.Gjørret JO, Knijn HM, Dieleman SJ, Avery B, Larsson LI, Maddox-Hyttel P. Chronology of apoptosis in bovine embryos produced in vivo and in vitro. Biol Reprod. 2003;69(4):1193–200. doi: 10.1095/biolreprod.102.013243. [DOI] [PubMed] [Google Scholar]
- 37.Metcalfe AD, Hunter HR, Bloor DJ, Lieberman BA, Picton HM, Leese HJ, et al. Expression of 11 members of the BCL-2 family of apoptosis regulatory molecules during human preimplantation embryo development and fragmentation. Mol Reprod Dev. 2004;68(1):35–50. doi: 10.1002/mrd.20055. [DOI] [PubMed] [Google Scholar]
- 38.Oswald J, Engemann S, Lane N, Mayer W, Olek A, Fundele R, et al. Active demethylation of the paternal genome in the mouse zygote. Curr Biol. 2000;10(8):475–8. doi: 10.1016/S0960-9822(00)00448-6. [DOI] [PubMed] [Google Scholar]
- 39.Beaujean N, Hartshorne G, Cavilla J, Taylor J, Gardner J, Wilmut I, et al. Non-conservation of mammalian preimplantation methylation dynamics. Curr Biol. 2004;14(7):R266–7. doi: 10.1016/j.cub.2004.03.019. [DOI] [PubMed] [Google Scholar]
- 40.Fulka H, Mrazek M, Tepla O, Fulka J., Jr DNA methylation pattern in human zygotes and developing embryos. Reproduction. 2004;128(6):703–8. doi: 10.1530/rep.1.00217. [DOI] [PubMed] [Google Scholar]
- 41.Fulka J, Fulka H, Slavik T, Okada K, Fulka J., Jr DNA methylation pattern in pig in vivo produced embryos. Histochem Cell Biol. 2006;126(2):213–7. doi: 10.1007/s00418-006-0153-x. [DOI] [PubMed] [Google Scholar]
- 42.Li E. Chromatin modification and epigenetic reprogramming in mammalian development. Nat Rev Genet. 2002;3(9):662–73. doi: 10.1038/nrg887. [DOI] [PubMed] [Google Scholar]
- 43.Kang YK, Koo DB, Park JS, Choi YH, Chung AS, Lee KK, et al. Aberrant methylation of donor genome in cloned bovine embryos. Nat Genet. 2001;28(2):173–7. doi: 10.1038/88903. [DOI] [PubMed] [Google Scholar]
- 44.Zhu H, Craig JA, Dyce PW, Sunnen N, Li J. Embryos derived from porcine skin-derived stem cells exhibit enhanced preimplantation development. Biol Reprod. 2004;71(6):1890–7. doi: 10.1095/biolreprod.104.032227. [DOI] [PubMed] [Google Scholar]
- 45.Suteevun T, Smith SL, Muenthaisong S, Yang X, Parnpai R, Tian XC. Anomalous mRNA levels of chromatin remodeling genes in swamp buffalo (Bubalus bubalis) cloned embryos. Theriogenology. 2006;65(9):1704–15. doi: 10.1016/j.theriogenology.2005.09.015. [DOI] [PubMed] [Google Scholar]
- 46.Kwon DJ, Park CK, Yang BK, Cheong HT. Control of nuclear remodelling and subsequent in vitro development and methylation status of porcine nuclear transfer embryos. Reproduction. 2008;135(5):649–56. doi: 10.1530/REP-06-0387. [DOI] [PubMed] [Google Scholar]
- 47.Chung YG, Ratnam S, Chaillet JR, Latham KE. Abnormal regulation of DNA methyltransferase expression in cloned mouse embryos. Biol Reprod. 2003;69(1):146–53. doi: 10.1095/biolreprod.102.014076. [DOI] [PubMed] [Google Scholar]
- 48.Santos F, Zakhartchenko V, Stojkovic M, Peters A, Jenuwein T, Wolf E, et al. Epigenetic marking correlates with developmental potential in cloned bovine preimplantation embryos. Curr Biol. 2003;13(13):1116–21. doi: 10.1016/S0960-9822(03)00419-6. [DOI] [PubMed] [Google Scholar]
- 49.Schultz RM. The molecular foundations of the maternal to zygotic transition in the preimplantation embryo. Hum Reprod Update. 2002;8(4):323–31. doi: 10.1093/humupd/8.4.323. [DOI] [PubMed] [Google Scholar]