Abstract
Senescence represents the last phase of petal development during which macromolecules and organelles are degraded and nutrients are recycled to developing tissues. To understand better the post-transcriptional changes regulating petal senescence, a proteomic approach was used to profile protein changes during the senescence of Petunia×hybrida ‘Mitchell Diploid’ corollas. Total soluble proteins were extracted from unpollinated petunia corollas at 0, 24, 48, and 72 h after flower opening and at 24, 48, and 72 h after pollination. Two-dimensional gel electrophoresis (2-DE) was used to identify proteins that were differentially expressed in non-senescing (unpollinated) and senescing (pollinated) corollas, and image analysis was used to determine which proteins were up- or down-regulated by the experimentally determined cut-off of 2.1-fold for P <0.05. One hundred and thirty-three differentially expressed protein spots were selected for sequencing. Liquid chromatography-tandem mass spectrometry (LC-MS/MS) was used to determine the identity of these proteins. Searching translated EST databases and the NCBI non-redundant protein database, it was possible to assign a putative identification to greater than 90% of these proteins. Many of the senescence up-regulated proteins were putatively involved in defence and stress responses or macromolecule catabolism. Some proteins, not previously characterized during flower senescence, were identified, including an orthologue of the tomato abscisic acid stress ripening protein 4 (ASR4). Gene expression patterns did not always correlate with protein expression, confirming that both proteomic and genomic approaches will be required to obtain a detailed understanding of the regulation of petal senescence.
Keywords: Abscisic acid, carbohydrates, flowers, hormones, petals, programmed cell death
Introduction
Many flowers have large, colourful petals (collectively called the corolla) that serve to attract pollinators. The longevity of flower petals varies considerably by species and is, in part, determined by the frequency of pollinator visitation (Ashman and Schoen, 1994). The maintenance of flower petals is costly in terms of metabolic energy and water loss, therefore it is advantageous for the plant to induce corolla senescence as soon as the flower has been successfully pollinated (Stead, 1992). The lifespan of most pollinated flowers is terminated by petal wilting followed by corolla abscission. During flower senescence, macromolecules are degraded and organelles are systematically dismantled so that nutrients can be remobilized to developing tissues. This process of cellular disassembly allows the plant to recover carbon, nitrogen, and phosphorus from the petals before the corolla is shed (Verlinden, 2003; Jones, 2004; Stead et al., 2006; Chapin and Jones, 2007, 2009).
Studies of both leaf and flower senescence have focused almost entirely on the identification of senescence-associated genes (reviewed in Buchanan-Wollaston et al., 2003; Jones, 2004; Stead et al., 2006; van Doorn and Woltering, 2008). While these studies have indicated that the timely initiation and progression of the senescence programme requires new gene transcription, senescence is also controlled at the post-transcriptional level (Thomas and Stoddart, 1980). Advances in mass spectrometry (MS) and 2-dimensional gel electrophoresis (2-DE) techniques and the availability of continually expanding EST and genomic databases have provided new opportunities for studying plant proteomes. Comparative proteomic analysis has been used to identify protein changes during plant development and following abiotic and biotic stresses (reviewed in Jorrin et al., 2007). Recently, plant proteomics has been used to study various developmental processes including leaf senescence, fruit ripening, and early flower development (Wilson et al., 2002; Dafny-Yelin et al., 2005; Carp and Gepstein, 2007; Faurobert et al., 2007; Hebeler et al., 2008; Desclos et al., 2009). Drought, heat, water logging, and salt-induced stress responses have also been investigated using comparative proteomic approaches (Askari et al., 2006; Horvath-Szanics et al., 2006; Huang et al., 2006; Ahsan et al., 2007; Hajheidari et al., 2007; Ito et al., 2007; Jiang et al., 2007; Veerasamy et al., 2007; Yang et al., 2007). These proteomic studies have provided a high throughput means of confirming the involvement of biochemical pathways that have been identified in genomic experiments and have provided unique insights into the role of post-translational modifications regulating and executing plant development and stress responses.
The premature senescence of floral tissues can have a detrimental impact on the quality, yield and subsequent value of agricultural and horticultural crops. It is therefore important to understand the molecular and biochemical mechanisms of senescence initiation, regulation, and execution in plants. Petunias provide an excellent model system for studies of flower senescence because they flower profusely and have large floral organs that are amenable to molecular and biochemical analysis. While the longevity of petunia corollas is under tight developmental control, petal senescence can be accelerated and synchronized by treating flowers with exogenous ethylene or by pollination (Jones et al., 2009). Changes in dry weight and petal nutrient content indicate that remobilization is occurring during petunia corolla senescence (Verlinden, 2003; Chapin and Jones, 2007, 2009; Jones, 2008).
The aim of this study was to identify proteins that increased and decreased in abundance during petunia corolla senescence. 2-DE, mass spectrometry, and bioinformatics tools were used to identify and functionally classify proteins that were differentially expressed in senescing and non-senescing petals. These studies provide insight into the dynamic changes occurring in the corolla proteome during senescence, and further support the idea that a combination of genomic and proteomic approaches will be necessary to elucidate molecular and biochemical mechanisms of senescence.
Materials and methods
Plant materials
Petunia×hybrida ‘Mitchell Diploid’ (MD) plants were used in all experiments. Seeds were sown in cell-packs on top of soil-less mix (Promix BX, Premier Horticulture, Quebec, Canada). All plants were established in the greenhouse after germination and plants were transferred to 16 cm pots after 4 weeks. Plants were fertilized at each watering with 150 mg l−1 Scott's Excel 15N-5P-15K (The Scotts Co., Marysville, OH), and a one-time treatment of Soluble Trace Element Mix (STEM, The Scotts Co., Marysville, OH) was applied 4 weeks after transferring to 16 cm pots. Growing conditions were 24/16 °C (day/night) with a 13 h photoperiod supplemented by high pressure sodium and metal halide lights.
Petunia petals (collectively called the corolla) were collected from unpollinated and pollinated flowers at 0, 24, 48, and 72 h after flower opening. Three replicates, each containing eight corollas from at least three different plants, were collected at each time point. The eight corollas in a replicate were pooled prior to protein or RNA extraction.
Protein extraction and 2-DE separation of proteins
Approximately 2.5 g corolla tissue (eight corollas) was ground in liquid nitrogen and mixed with 8000 μl homogenization buffer [100 mM HEPES-KOH (pH 7.5), 5% (v/v) glycerol, 15 mM EGTA, 5 mM EDTA, 0.5% (v/v) polyvinylpyrrolidone, 3 mM dithiothreitol, 60 μl proteinase inhibitor cocktail (Sigma-Aldrich, St Louis, Mo)]. Total soluble proteins were extracted as described by Coaker et al. (2004). The resulting pellet was resuspended in 800 μl of rehydration buffer [8 M urea, 2 M thiourea, 4% (w/v) CHAPS, 0.2% (v/v) immobilized pH gradient (IPG) buffer (pH 5–8; Bio-Rad Laboratories, Hercules, CA), 1% (v/v) Triton X-100, 0.5% (w/v) DTT] and sonicated for 2 min in a bath sonicator (Model PC5; L&R Manufacturing Co., Kearny, NJ). After shaking at 120 rpm for 1.5 h at 25 °C, the protein concentration was determined with the Amersham Biosciences 2-D quantification kit (Amersham Biosciences Corp., Piscataway, NJ).
Two hundred micrograms of total protein for each sample was used for passive rehydration of 11 cm IPG strips (pH 5–8; Bio-Rad Laboratories, Hercules, CA). Programmed isoelectric focusing (IEF) was performed with the Protean IEF Cell (Bio-Rad Laboratories, Hercules, CA) using the following preset conditions: start voltage 0 V, end voltage 8000 V, total volt-hours 35 000 V h at 20 °C. After IEF, IPG strips were immediately equilibrated in buffer 1 [6 M urea, 0.375 M TRIS (pH 8.8), 2% (w/v) SDS, 20% (v/v) glycerol, 2% (w/v) DTT] and buffer 2 (6 M uUrea, 0.375 M TRIS (pH 8.8), 2% (w/v) SDS, 20% (v/v) glycerol, 2.5% (w/v) iodoacetamide], each for 10 min. The equilibrated strips were then subjected to SDS-PAGE (12.5%) for second dimension separation. After running for 2 h under a constant 200 V, gels were stained with GelCode Blue Stain Reagent (Pierce, Rockford, IL).
Data analysis of 2-D gels
Protein profiles of pollinated corollas were compared to unpollinated corollas at the same developmental age to identify differentially expressed proteins for sequence analysis. Proteins up-regulated during senescence were defined as those where the pixel intensities associated with a spot increased in pollinated corollas compared to unpollinated corollas, or those proteins that were detected in pollinated corollas but not in unpollinated corollas. Down-regulated proteins were defined as those where the pixel intensities decreased in unpollinated corollas compared to the pollinated corollas, or proteins that were detected only in unpollinated corollas.
Three 2-D gels from each time point, representing independent replications of eight corollas each, were analysed using PDQuest v. 7.40 (Bio-Rad Laboratories, Hercules, CA, USA). For each time point the three replicate gel images for pollinated and unpollinated corollas were imported as a dataset, and a synthetic Gaussian image from one of three replicate gel images was created as a reference for all subsequent quantification and protein analyses. The quantities of protein spots on each image were normalized based on the total density in the gel image. A scaling factor was used to give a more meaningful value and the normalized quantity was multiplied by parts per million (ppm). Protein spots were visually inspected to validate all automated analyses. Average intensities and correlation coefficients between gels were obtained.
The distribution of protein intensities and residuals was inspected using the Univariate procedure of SAS (Version 9.0, SAS Institute Inc., Cary, NC, USA). Transformation of the data was necessary in order to approximate a normal distribution. Both loge [LN(value+1)] and log10 [log(value+1)] transformations of the data approximated normal distributions, and the loge (LN) transformation was subsequently applied across all quantitative data prior to statistical analysis. Analysis of variance (ANOVA) was conducted using the general linear model procedure in the SAS software. The statistical model included main effects for protein, treatment (pollinated or unpollinated), time, and replicate. All factors were considered fixed. Sufficient degrees of freedom were available to test all two-way interactions which were included in the statistical model for ANOVA. The Mean Square Error from the ANOVA was used to estimate an experiment-wide cut-off for significance, calculated as fold-change using the relationship e[t×(Sqrt(MSE/n)]/2 where t was 1.96 for P=0.05, 2.576 for P=0.01, and 3.291 for P=0.001; where n equals the number of replicates (Kerr et al., 2000).
Identification of proteins by mass spectrometry
Mass spectrometry was conducted at the Cleveland Clinic Proteomics Laboratory (Cleveland Clinic Foundation, Cleveland, OH), and trypsin digests of the proteins of interest were prepared as described by Kinter and Sherman (2000). Protein spots were excised from the SDS-PAGE gels, divided into a number of smaller pieces and washed/destained in 50% ethanol/5% acetic acid. The gel pieces were dehydrated in acetonitrile and dried in a Speed-Vac. Digestion was carried out overnight at room temperature with 5 μl of 20 ng μl−1 trypsin in 50 mM ammonium bicarbonate. The resulting peptides were extracted twice with 50% acetonitrile/5% formic acid, and the extracts were combined and evaporated to <30 μl for LC-MS analysis.
A ThermoScientific LCQ ion trap mass spectrometer (San Jose, CA) interfaced with a self-packed Phenomenex Jupiter C18 reversed-phase capillary column was used for the analysis. Two μl volumes of the trypsin digested extract were injected and peptides were eluted from the column by a 30 min binary gradient with 0.05 M acetic acid and acetonitrile as the mobile phases at a flow rate of 250 nl min−1. The microelectrospray ion source was operated at 2.5 kV. Tryptic digests were analysed using the multi-task capability of the instrument to acquire full scan mass spectra to determine peptide molecular weights and product ion spectra to determine amino acid sequences in successive instrument scans.
All collisionally induced dissociation (CID) spectra were searched against the National Center for Biotechnology Information (NCBI) green plant protein database using the Mascot search program (Matrix Science, Boston, MA). An ion score greater than 25 was used as a cut-off for consideration, and all identifications were based on at least two peptides that could then be verified by manual inspection of the spectra. If a protein could not be identified using this method, the CID spectra were manually interpreted to determine the specific amino acid sequence of the peptides. The peptide sequence data were then subjected to either FASTA searches performed on the University of Virginia website (http://fasta.bioch.virginia.edu/fasta_www2/fasta_list2.shtml) to identify the homologous NCBI sequences or to FASTA searches of translated EST databases using the search program Sequest (ThermoScientific, San Jose, CA). The translated EST databases were created by The Ohio State University Molecular and Cellular Imaging Center (Wooster, OH) and consisted of Solanaceae ESTs translated in six reading frames. Petunia ESTs were obtained from the SOL Genomics Network (SGN) (http://sgn.cornell.edu/; Mueller et al., 2005) and Nicotiana benthamiana and Solanum tuberosum ESTs were provided by Dr Sophien Kamoun (Sainsbury Laboratory, John Innes Centre, UK). Xcorr values for each identified peptide were filtered according to the peptide charge states Xcorr ≥1.5 for +1, Xcorr ≥2.0 for +2, and Xcorr ≥2.5 for +3. To validate all identifications, the interpreted peptide sequences were manually aligned with the homologous NCBI protein sequences (see Supplementary Fig. S1 at JXB online).
Protein expression clustering and functional classification
Cluster (version 2.11) hierarchical clustering software (http://rana.lbl.gov/EisenSoftware.htm) was used to identify the main classes of protein expression (Eisen et al., 1998). Protein expression patterns were clustered as described in the software user manual. Briefly, input data were normalized by dividing the loge values by the intensity at 0 h. Weighted Pearson (centred) correlation was chosen to define the similarity matrix. A dendrogram was generated by running the average-linkage clustering algorithm. The clustering image was visualized using the TreeView software (http://rana.lbl.gov/EisenSoftware.htm).
Protein sequences were searched against the NCBI databases using the tBLASTn program to identify the most similar Arabidopsis genes. The locus identifier for this gene was then used to search the TAIR gene ontology (GO) annotation database (http://www.arabidopsis.org/tools/bulk/go/index.jsp) to identify the putative biological function of the protein of interest (Berardini et al., 2004).
Real-time PCR analysis of gene expression
Total RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA) and treated with RQ1 RNase-free DNase (Promega, Madison, WI). cDNA was synthesized from 2 μg total RNA using the Omniscript RT Kit (Qiagen, Valencia, CA). The gene sequences for selected proteins were identified from the SGN petunia EST database (Mueller et al., 2005) and gene-specific primers were designed using Primer Quest software available through IDT SciTools (http://www.idtdna.com/Scitools/Applications/Primerquest/) (Table S1). If petunia sequences corresponding to the peptides could not be identified from the EST database, degenerate primers were constructed to the most distal peptide sequences to allow for RT-PCR amplification of the largest amount of sequence from the corresponding gene. The PCR amplicon was then sequenced and Primer Quest software was used to design primers for real-time PCR. Quantitative PCR was performed in a 20 μl reaction volume using B-R SYBR Green SuperMix for iQ (Quanta BioSciences, Inc., Gaithersburg, MD). One microlitre cDNA was used as template, and all reactions were performed in triplicate. After a 3 min activation step at 95 °C, qPCR was conducted for 40 cycles of 94 °C for 10 s, 60 °C for 30 s, and 72 °C for 30 s using the iQ5 Thermocycler (Bio-Rad, Hercules, CA). The annealing temperature for some primer pairs varied and the specific temperatures are listed in Supplementary Table S1 at JXB online. Melt curves were generated to check amplification specificity. The absolute starting quantity of the target genes was normalized to that of PhACTIN1 in each cDNA sample to calculate relative target gene expression. PhACTIN1 had very consistent transcript abundance in corollas from 0 h to 72 h after pollination (ML Jones, unpublished results), and is not the same petunia actin that showed senescence-related increases in protein abundance in this study.
Results
Protein profiling during petunia corolla senescence
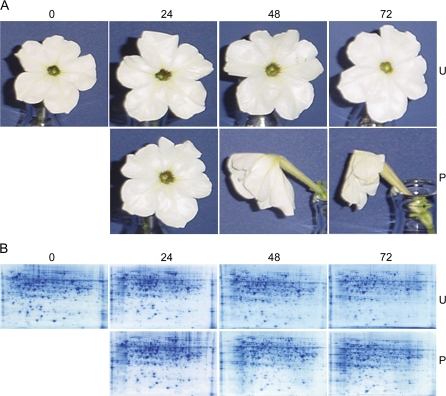
Pollination accelerated the senescence of petunia corollas, and flowers were visibly wilted by 48 h after pollination (48 P) (Fig. 1A). Corollas at 72 h after pollination (72 P) were severely wilted, but were not yet drying at the petal margins. Unpollinated petunia corollas from 24–72 h after flower opening (24 U, 48 U, and 72 U) were fully turgid, showed no senescence symptoms and were visually indistinguishable from flowers at anthesis (0 U). The 2-D separation of total soluble proteins from petunia corollas resulted in the detection of approximately 600 distinct protein spots in each gel (Fig. 1B). The molecular masses (Mr) of the proteins ranged from 15 kDa to 110 kDa (Fig. 2).
Fig. 1.
Protein changes during petunia corolla development and senescence. (A) Pollination accelerates petunia flower senescence. Petunia×hybrida ‘Mitchell Diploid’ (MD) unpollinated (U) flowers at 0, 24, 48, and 72 h and pollinated (P) flowers at 24, 48, and 72 h. (B) 2-D gel profiling of protein changes corresponding to corolla senescence. Representative 2-D gels of petunia corolla proteome variation during pollination-induced senescence. The first dimension was performed using 200 μg total soluble proteins on linear gradient IPG strips with pH 5–8. Second dimension separation was conducted on 12% SDS-PAGE gels and proteins were visualized using GelCode blue staining. Approximately 600 spots were detected on each gel. Three biological replicates at each time point were conducted.
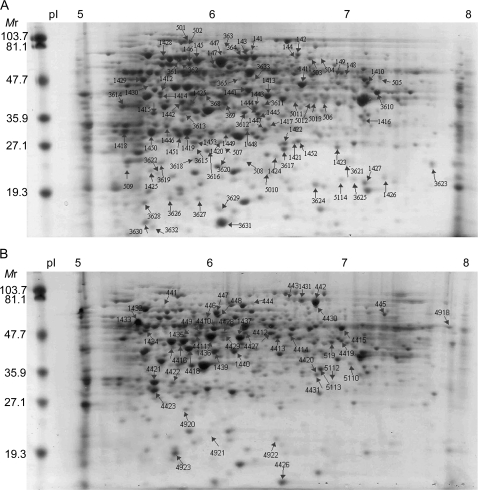
Fig. 2.
Representative 2-DE comparisons of unpollinated and pollinated gels at 72 h. (A) All of the down-regulated proteins are indictated with arrows on the 72 U gels. (B) All of the up-regulated proteins are indicated with arrows on 72 P gels. The spot numbers are the same as the sample numbers (without the dash in the middle) in Tables 2 and 3. The assigned identities of up- and down-regulated proteins and their expression data are also shown in Tables 2 and 3.
Pixel intensity data of protein spots were not normally distributed, though both natural log and log base 10 transformations were effective at creating a normal distribution of pixel intensity data. Analysis of variance (ANOVA) indicated that the main effects for differences between proteins (P <0.0001), pollination treatment (P <0.0001), and time points (P <0.0001) were all highly significant. The replication effect and all two-way interactions with replicate were non-significant (0.760 <P >1.00). The treatment by protein and time point by protein interactions were both highly significant (P <0.0001), indicating differential expression of individual proteins due to pollination and time. Based on the mean squared error associated with the ANOVA, the experimental-wide cut-off for differentially expressed spots was 2.1-fold at P <0.05, 3.4-fold at P <0.01, and 3.6-fold at P <0.001 (Kerr et al., 2000). This analysis supplemented the identification of quantitative differences between treatments (pollinated versus unpollinated) by a t test (P ≤0.05) using the PDQuest software. In addition, qualitative differences (presence/absence) were also detected.
No significant protein changes were identified at 24 h when comparing protein profiles in pollinated (24 P) and unpollinated (24 U) corollas (Table 1). By 48 h after flower opening, 72 proteins were up-regulated and 41 proteins were down-regulated in pollinated corollas by greater than 2.1-fold (P <0.05), suggesting that about 20% of the abundant proteins were differentially regulated. One-hundred-and-twelve proteins were up-regulated and 57 proteins were down-regulated at 72 h (P <0.05). This represented approximately 30% of the abundant proteins detected on the 2-D gels.
Table 1.
Profiling of differentially expressed proteins during petunia corolla senescence
| Time points and treatments |
||||||
| Differentially expressed proteinsa | 24 P versus 24 U |
48 P versus 48 U |
72 P versus 72 U |
|||
| No. | Total (%) | No. | Total (%) | No. | Total (%) | |
| Up-regulated proteinsb | 0 | 0 | 73 | 13 | 112 | 20 |
| Down-regulated proteinsc | 0 | 0 | 41 | 7 | 57 | 10 |
| Total | 0 | 0 | 114 | 20 | 169 | 30 |
Significant differences among treatments within a time point were detected by the t-test (P ≤0.05).
Proteins were defined as up-regulated if they were not detected in unpollinated petals and were detected in pollinated petals or if they increased in abundance in pollinated petals.
Proteins were defined as down-regulated if they were detected in unpollinated but not in pollinated petals or if they decreased in abundance in pollinated petals.
From the differentially expressed proteins, 133 unique spots were selected for sequencing by tandem mass spectrometry (Fig. 2A, B). Forty-six up- and 27 down-regulated spots were identified as single proteins (Tables 2, 3). Sixty-nine per cent of these proteins were identified based on peptide matches from Solanaceae sequences, with 26% of those being from petunia. A total of 47 spots included peptides that corresponded to multiple proteins (data not shown). Since the expression pattern could not unambiguously be determined for each individual protein, the data for spots with multiple protein components are not reported in this paper. Thirteen (11 up- and two down-regulated) spots could not be identified from any database (data not shown).
Table 2.
Functional classification of petunia proteins that were up-regulated during pollination-induced corolla senescence
| Sample no.a | Putative protein assignmentb | Matched peptidesc | Matched species | Accession/EST no.d | Obser. Mr/pIe | Theor. Mr/pIf | Peptide coverageg | Mean of intensitiesh |
|||
| 48 Ui | 48 Pj | 72 U | 72 P | ||||||||
| Defence response and response to stress | |||||||||||
| 50-9 | Plasma membrane polypeptide | 16 (4,12) | Nicotiana tabacum | 2764992 | 24/5.1 | 22.9/5.0 | 62% | ud k | 1068 | ud | 470 |
| 14-23 | Manganese superoxide dismutase | 8 (4,4) | Gossypium hirsutum | 3219353 | 26/6.9 | 22.1/8.5 | 53% | 271 | 701 | 111 | 984 |
| 14-52 | Manganese superoxide dismutase | 7 (4,3) | Gossypium hirsutum | 3219353 | 28/6.6 | 22.1/8.5 | 45% | 363 | 563 | 121 | 1078 |
| 36-16 | Iron superoxide dismutase | 8 (6,2) | Solanum lycopersicum | 33413303 | 26/6.0 | 27.9/6.6 | 34% | 371 | 766 | 186 | 1106 |
| 14-51 | Superoxide dismutase; Nectarin 1 precursor | 6 (0,6) | Nicotiana plumbaginifolia | 6090829 | 30/5.8 | 24.8/7.7 | 29% | 557 | 455 | 873 | 2463 |
| 36-12 | Annexin | 17 (11,6) | Capsicum annuum | 1071660 | 37/6.3 | 35.8/5.7 | 49% | 1375 | 1345 | 579 | 1477 |
| 36-23 | Similar to NtPRp27-like protein | 11 (11,0) | Petunia×hybrida | SGN-E520812 | 24/7.7 | 29.5/7.9 | na | 156 | 108 | 235 | 898 |
| 50-4 | Auxin-responsive-like protein; DFL1 | 9 (8,1) | Arabidopsis thaliana | 15239653 | 60/6.8 | 68.9/5.5 | 16% | ud | 364 | ud | 240 |
| 36-10 | Mitochondrial formate dehydrogenase precursor | 20 (17,3) | Solanum tuberosum | 11991527 | 44/7.2 | 42.0/6.6 | 56% | 2291 | 2468 | 1173 | 2717 |
| 50-7 | Similar to abscisic stress ripening protein | 2 (2,0) | Petunia×hybrida | SGN-E524444 | 28/5.9 | 26.7/4.8 | na | ud | 202 | ud | 101 |
| 14-9 | UTP-glucose-1-phosphate uridyltransferase | 21 (13,8) | Solanum tuberosum | 21599 | 51/6.9 | 51.8/5.4 | 46% | ud | 253 | ud | 364 |
| Electron transport or energy pathways | |||||||||||
| 36-5 | Succinate dehydrogenase flavoprotein alpha subunit | 15 (10,5) | Arabidopsis thaliana | 8843734 | 49/6.3 | 69.7/5.9 | 31% | 894 | 766 | 250 | 502 |
| 14-22 | 1,4-benzoquinone reductase-like protein | 8 (4,4) | Arabidopsis thaliana | 21539481 | 27/6.5 | 21.8/6.0 | 45% | 388 | 516 | 210 | 1014 |
| 36-17 | 1,4-benzoquinone reductase-like protein | 16 (5,11) | Arabidopsis thaliana | 21539481 | 26/6.5 | 21.7/6.0 | 72% | 898 | 878 | 654 | 1322 |
| 36-30 | Similar to plastocyanin-like domain-containing protein; early nodulin-like protein 2 precursor | 3 (1,2) | Petunia×hybrida | SGN-E528550 | 16/5.6 | 29.9/9.4 | na | 171 | 582 | ud | 1067 |
| 14-25 | Photosystem II 23 kDa protein | 5 (0,5) | Solanum lycopersicum | 19317 | 22/5.7 | 27.8/8.3 | 35% | ud | 753 | ud | 576 |
| Proteolysis | |||||||||||
| 14-50 | Cysteine proteinase P21 | 8 (7,1) | Petunia×hybrida | 945081 | 30/5.6 | 39.3/5.8 | 23% | 1670 | 2150 | 1297 | 4919 |
| 36-15 | Proteasome delta-subunit | 10 (8,2) | Nicotiana tabacum | 1743356 | 27/5.9 | 25.1/5.2 | 47% | 291 | 509 | 319 | 640 |
| 14-27 | Serine carboxylase II-2 (N-term truncated) | 8 (1,7) | Oryza sativa | 55773861 | 20/7.1 | 53.4/5.5 | 18% | 444 | 431 | 36 | 1062 |
| 36-13 | Glucose acyltransferase; serine carboxypeptidase-like protein | 7 (3,4) | Solanum berthaultii | 4101703 | 35/5.8 | 53.4/5.1 | 18% | 221 | 605 | 144 | 719 |
| 36-24 | Ubiquitin-conjugating enzyme E2; ubiquitin-protein ligase | 3 (2,1) | Arabidopsis thaliana | 30693656 | 21/6.7 | 16.6/6.2 | 24% | ud | ud | ud | 927 |
| Carbohydrate metabolism | |||||||||||
| 44-7 | Beta-xylosidase 2; LeXYL2 | 13 (8,5) | Solanum lycopersicum | 37359708 | 65/6.2 | 68.9/8.0 | 20% | 3513 | 4565 | 2435 | 6521 |
| 14-13 | Beta-xylosidase 2; LeXYL1(C-terminally truncated) | 16 (8,8) | Solanum lycopersicum | 37359708 | 43/6.4 | 68.9/8.0 | 35% | ud | 925 | ud | 2601 |
| 36-3 | Beta-xylosidase 1; AtBXL1 | 7 (2,5) | Arabidopsis thaliana | 15239867 | 73/6.2 | 83.5/8.8 | 11% | ud | 404 | ud | 575 |
| 14-2 | Alpha-mannosidase | 8 (3,5) | Arabidopsis thaliana | 10177130 | 71/6.7 | 118/8.3 | 7% | 159 | 394 | 202 | 1187 |
| 14-29 | Vacuolar invertase (N or C-terminally truncated) | 14 (8,6) | Nicotiana tabacum | 29893064 | 48/5.5 | 71.2/5.4 | 19% | 896 | 1031 | 789 | 2828 |
| 36-22 | Vacuolar invertase (N-terminally truncated) | 5 (3, 2) | Nicotiana tabacum | 29893064 | 22/5.6 | 71.2/5.4 | 7% | ud | ud | ud | 455 |
| 36-21 | Vacuolar invertase precursor (N-terminally truncated) | 2 (0,2) | Nicotiana tabacum | 29893064 | 24/6.9 | 71.2/5.4 | 4% | ud | 361 | ud | 765 |
| 14-11 | Putative beta-galactosidase (C-terminally truncated) | 13 (8,5) | Solanum lycopersicum | 7939623 | 46/6.7 | 93.2/6.8 | 14% | 1668 | 2530 | 777 | 3531 |
| 50-3 | Cytosolic phosphoglucomutase | 21 (15,6) | Solanum tuberosum | 8250624 | 59/6.7 | 63.5/6.0 | 44% | 865 | 1486 | 545 | 1315 |
| Lipid metabolism | |||||||||||
| 14-6 | Lipoxygenase (N-terminally truncated) | 21 (9,12) | Nicotiana tabacum | 899344 | 62/5.9 | 97.6/5.5 | 24% | 169 | 590 | 126 | 2226 |
| 50-2 | Lipoxygenase | 8 (5,3) | Nicotiana tabacum | 899344 | 65/5.7 | 97.6/5.5 | 12% | ud | 560 | ud | 368 |
| 14-20 | Putative GDSL- motif lipase | 4 (1,3) | Vitis vinifera | 37789825 | 28/6.1 | 19.1/5.5 | 25% | 210 | 184 | 395 | 1078 |
| 36-14 | Proline-rich protein; SGNH plant-lipase-like | 8 (1,7) | Oryza sativa | 50910547 | 39/5.5 | 40.1/5.7 | 33% | 392 | 1192 | 539 | 1478 |
| Cytoskeleton organization and biogenesis | |||||||||||
| 14-30 | Actin | 20 (17,3) | Arabidopsis thaliana | 20465865 | 47/5.6 | 41.6/5.3 | 60% | 100 | 126 | 100 | 674 |
| 50-10 | Actin-depolymerizing factor 1 | 3 (3,0) | Petunia×hybrida | 14906219 | 22/6.3 | 16.0/5.8 | 19% | ud | 656 | ud | 1362 |
| 51-14 | Actin-depolymerizing factor 2 | 7 (7,0) | Petunia×hybrida | 14906210 | 21/6.9 | 16.0/5.8 | 52% | 353 | 810 | 182 | 551 |
| Nucleotide/nucleoside/nucleobase metabolism | |||||||||||
| 14-12 | Endonuclease | 8 (4,4) | Solanum tuberosum | 50657596 | 43/5.7 | 34.4/5.6 | 32% | ud | 421 | ud | 1549 |
| Amino acid metabolism | |||||||||||
| 36-32 | Methylene-tetra hydrofolate reductase; MTHFR1(C-terminally truncated) | 4 (1,3) | Arabidopsis thaliana | 30695097 | 15/5.6 | 45.6/6.2 | 10% | ud | 271 | ud | 659 |
| Nitrogen metabolism | |||||||||||
| 36-33 | Beta-Ureidopropionase; PYD3 | 3 (2,1) | Arabidopsis thaliana | 30698009 | 48/6.4 | 45.5/5.9 | 13% | ud | 164 | ud | 432 |
| Unclassified | |||||||||||
| 36-27 | Putative acid phosphatase (N-terminally truncated) | 4 (0,4) | Oryza sativa | 55296477 | 19/6.0 | 32.3/5.4 | 9% | ud | 154 | ud | 511 |
| 36-19 | Putative Kunitz-type proteinase inhibitor | 3 (1,2) | Petunia×hybrida | SGN-E536182 | 24/5.7 | 23.8/7.0 | na | 216 | 659 | 371 | 1010 |
| 36-20 | Putative Kunitz-type proteinase inhibitor | 5 (2,3) | Petunia×hybrida | SGN-E536182 | 23/6.1 | 23.8/7.0 | na | 251 | 637 | 98 | 1720 |
| 14-24 | Putative aluminium-induced protein | 6 (1,5) | Arabidopsis thaliana | 21537246 | 24/6.5 | 27.5/6.4 | 25% | 186 | 569 | 255 | 485 |
| 36-28 | Hypothetical protein | 6 (5,1) | Petunia×hybrida | SGN-E521639 | 18/5.6 | 24/9.4 | na | 368 | 751 | 289 | 960 |
| 50-11 | S-adenosyl-L-methionine:benzoic acid/salicylic acid carboxyl methyltransferase; PhBSMT2 | 21 (21,0) | Petunia×hybrida | 28629497 | 40/6.5 | 40.7/5.7 | 48% | 353 | 810 | 182 | 551 |
Sample number indicates the spot sample for in-gel digestion in the sequencing reports. These sample numbers (without dash) were also used to indicate spots on the 2-D gels in Fig. 2.
Proteins were classified based on the biological processes according to homology to genes in the TAIR Gene Ontology database.
Matched peptides indicate total number of peptides that matched to other proteins. The first and the second number in the parenthesis indicate the peptides with exact and homologous matches, respectively.
Accession/EST number indicates the sequence in the searched database identified from either NCBI non-redundant protein database or SGN petunia EST database (http://www.sgn.cornell.edu/index.pl).
Obser. Mr/pI, observed Mr/pI for each protein was calculated from the 2-D gels with Image PDQuest 7.4.0 software according to standard marker proteins.
Theor. Mr/pI, theoretical Mr/pI of the matched proteins.
Peptide coverage refers to the percentage of sequence coverage of the matched protein; no available (na) data when EST database was used.
Mean of intensities is the average of three replicated spot normalized intensities at 48 U, 48 P, 72 U, and 72 P.
U is hours after flowers are fully opened without pollination (unpollinated).
P is hours after flowers are fully opened with pollination.
ud, spot intensity was undetectable on the gel.
Table 3.
Functional classification of petunia proteins that were down-regulated during pollination-induced corolla senescence
| Sample no.a | Putative protein assignmentb | Matched peptidesc | Matched species | Accession/EST No.d | Obser. Mr/pIe | Theor. Mr/pIf | Peptide coverageg | Mean of intensitiesh | |||
| 48 Ui | 48 Pj | 72 U | 72 P | ||||||||
| Defence response and response to stress | |||||||||||
| 44-12 | 3-Phosphoshikimate 1-carboxy vinyltransferase | 19 (19,0) | Petunia×hybrida | 114176 | 49/6.5 | 55.5/8.0 | 39% | 2442 | 581 | 1665 | 196 |
| 51-11 | Putative pyridoxine biosynthesis isoform B | 11 (11,0) | Nicotiana tabacum | 46399271 | 36/6.6 | 33.1/5.9 | 32% | 1100 | 465 | 671 | 411 |
| 49-24 | Copper-zinc superoxide dismutase | 5 (3,2) | Solanum lycopersicum | 13445918 | 18/6.0 | 14.8/5.3 | 45% | 3037 | 1510 | 2629 | 2511 |
| Amino acid metabolism | |||||||||||
| 14-31 | Methionine synthase | 22 (16,6) | Solanum tuberosum | 8439545 | 80/6.6 | 84.7/5.9 | 32% | 3493 | 1477 | 1322 | 230 |
| 44-1 | Methionine synthase | 2 (2, 0) | Solanum tuberosum | 8439545 | 73/5.7 | 84.7/5.9 | 5% | 576 | ud k | 322 | ud |
| 44-2 | Methionine synthase | 43 (24, 19) | Solanum tuberosum | 8439545 | 81/6.7 | 84.7/5.9 | 63% | 7428 | 3485 | 4230 | 1870 |
| 44-3 | Methionine synthase | 24 (17,7) | Solanum tuberosum | 8439545 | 80/6.6 | 84.7/5.9 | 30% | 1556 | 739 | 363 | 325 |
| 14-37 | S-adenosylmethionine synthetase | 19 (19,0) | Petunia×hybrida | 559506 | 45/6.2 | 43.8/5.5 | 73% | 2764 | 1786 | 2883 | 889 |
| 14-36 | S-adenosylmethionine synthetase | 21 (14,7) | Petunia×hybrida | 5726594 | 45/6.0 | 43.7/5.4 | 71% | 1748 | 538 | 2449 | 220 |
| 44-13 | S-adenosylmethionine synthetase | 16 (8,8) | Nicotiana tabacum | 33340517 | 47/6.5 | 43.1/5.8 | 55% | 1631 | 681 | 1520 | 888 |
| 44-14 | S-adenosylmethionine synthetase | 12 (10,2) | Nicotiana tabacum | 33340517 | 48/6.6 | 43.1/5.8 | 37% | 1859 | 676 | 1464 | 486 |
| 49-18 | Hydroxymethyltransferase | 28 (12,16) | Arabidopsis thaliana | 2244749 | 55/7.9 | 51.7/6.8 | 65% | 1968 | 593 | 476 | 299 |
| Cytoskeleton organization and biogenesis | |||||||||||
| 14-34 | Alpha tubulin | 17 (17,0) | Physcomitrella patens | 25396545 | 48/5.6 | 50.1/5.0 | 40% | 896 | 757 | 1503 | 431 |
| 49-21 | Actin-depolymerizing factor 1 | 7 (7,0) | Petunia×hybrida | 14906219 | 23/5.8 | 16.0/5.8 | 46% | 392 | ud | 308 | ud |
| 49-22 | Actin-depolymerizing factor 2 | 10 (10,0) | Petunia×hybrida | 14906210 | 22/6.4 | 16.5/5.8 | 76% | 604 | ud | 386 | ud |
| Carbohydrate metabolism | |||||||||||
| 51-13 | Xyloglucan endotransglucosylasehydrolase; XTH7 | 12 (3,9) | Solanum lycopersicum | 42795466 | 37/6.8 | 33.4/7.6 | 47% | 2910 | 1951 | 1650 | 409 |
| 44-15 | Ribulose bisphosphate carboxylase large chain precursor | 16 (15,1) | Petunia×hybrida | 132011 | 54/6.9 | 53.0/6.6 | 34% | 5642 | 4214 | 2904 | 1441 |
| 44-26 | Ribulose 1,5-bisphosphate carboxylase small chain precursor | 8 (8,0) | Petunia×hybrida | 132083 | 16/6.6 | 20.4/8.3 | 52% | 2822 | 1212 | 2384 | 799 |
| 44-21 | Caffeoyl CoA 3-O-methyltransferase | 15 (10,5) | Betula platyphylla | 57639629 | 33/5.6 | 27.8/5.3 | 66% | 2806 | 1920 | 3847 | 1187 |
| 44-22 | Caffeoyl CoA 3-O-methyltransferase | 13 (8,5) | Solanum tuberosum | 24745969 | 32/5.8 | 27.2/5.3 | 66% | 464 | ud | 499 | ud |
| 44-23 | Caffeoyl CoA 3-O-methyltransferase | 15 (8,7) | Solanum tuberosum | 24745969 | 31/5.8 | 27.2/5.3 | 77% | 2979 | 963 | 2156 | 1110 |
| Nitrogen metabolism | |||||||||||
| 44-18 | Glutamine synthetase shoot isozyme | 17(9,8) | Oryza sativa | 50912511 | 42/5.9 | 39.2/5.5 | 58% | 4066 | 2007 | 3627 | 1869 |
| Electron transport or energy pathways | |||||||||||
| 44-4 | Transketolase 1 | 10 (9,1) | Capsicum annuum | 3559814 | 77/6.4 | 80/6.2 | 13% | 1154 | 553 | 935 | 457 |
| 49-20 | Photosystem II 23 kDa protein | 14 (6,8) | Solanum lycopersicum | 19317 | 27/5.6 | 27.8/8.3 | 62% | 819 | ud | 697 | ud |
| Unclassified | |||||||||||
| 51-12 | Putative aluminium-induced protein | 9 (3,6) | Arabidopsis thaliana | 15239993 | 33/6.8 | 27.5/6.4 | 34% | 510 | ud | 382 | ud |
| 44-5 | Similar to Acyl activating enzyme 11 | 2 (2,0) | Petunia×hybrida | SGN-E523400 | 66/7.3 | 13.2/8.5 | na | 1161 | 481 | 842 | 493 |
| 14-40 | S-adenosyl-L-methionine:benzoic acid/ salicylic acid carboxyl methyltransferase; PhBSMT2 | 17 (17,0) | Petunia×hybrida | 28629497 | 40/6.2 | 40.7/5.7 | 37% | 5071 | 2018 | 2400 | 1745 |
Sample number indicates the spot sample for in-gel digestion in the sequencing reports. These sample numbers (without dash) were also used to indicate spots on the 2-D gels in Fig. 2.
Proteins were classified based on the biological processes according to homology to genes in the TAIR Gene Ontology database.
Matched Peptides indicate total number of peptides that matched to other proteins. The first and the second number in the parenthesis indicate the peptides with exact and homologous matches respectively.
Accession/EST number indicates the sequence in the searched database identified from either NCBI non-redundant protein database or SGN petunia EST database (http://www.sgn.cornell.edu/index.pl).
Obser. Mr/pI, observed Mr/pI for each protein was calculated from the 2-D gels with Image PDQuest 7.4.0 software according to standard marker proteins.
Theor. Mr/pI, theoretical Mr/pI of the matched proteins.
Peptide coverage refers to the percentage of sequence coverage of the matched protein; no available (na) data when EST database was used.
Mean of intensities is the average of three replicated spot normalized intensities at 48 U, 48 P, 72 U, and 72 P.
U is hours after flowers are fully opened without pollination (unpollinated).
P is hours after flowers are fully opened with pollination.
ud, spot intensity was undetectable on the gel.
The experimental Mr of many of the differentially regulated proteins was similar to the theoretical Mr of the proteins from the matched species (Tables 2, 3). Some protein spots had a slightly higher Mr than the theoretical Mr, and these mass differences may be due to various post-translational modifications during senescence. More striking was the number of proteins that had an experimental Mr that was much less than the theoretical Mr. A number of these proteins were truncated at the N- or C-termini, as the peptides identified by MS only aligned with a portion of the protein. These may represent proteins that have been degraded during senescence or proenzymes that have been cleaved and activated during the progression of senescence. While protease inhibitors were included in the protein extraction buffers, truncated proteins may also have resulted from in vitro protein degradation. Proteins that were identified as N- or C-terminally truncated due to MS peptide alignments and low observed Mr included the up-regulated proteins serine carboxylase II-2 (14-27), beta-xylosidase 2 (14-13), vacuolar invertase (14-29, 36-22), beta-galactosidase (14-11), lipoxygenase (14-6), methylenetetrahydrofolate reductase (36-32), cysteine protease (14-50), and acid phosphatase (36-27). Many of these truncated proteins were detected only in senescing corollas (i.e. 48 P and 72 P). None of the down-regulated proteins were identified by the peptide coverage as being N- or C-terminally truncated.
Functional classification of identified proteins based on biological processes
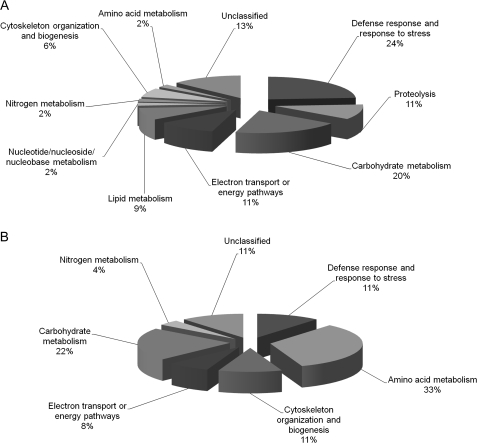
The 46 up-regulated proteins were grouped into 10 functional classifications and the 27 down-regulated proteins were grouped into seven functional classifications (Tables 2, 3). The up-regulated proteins were classified as defence and stress response (11), carbohydrate metabolism (9), electron transport or energy pathways (5), proteolysis (5), lipid metabolism (4), cytoskeleton organization and biogenesis (3), nucleotide/nucleoside/nucleobase metabolism (1), amino acid metabolism (1), nitrogen metabolism (1) or unclassified proteins (6) (Fig. 3A). The down-regulated proteins were classified as amino acid metabolism (9), carbohydrate metabolism (6), defence and stress response (3), cytoskeleton organization and biogenesis (3), electron transport or energy pathways (2), nitrogen metabolism (1), or unclassified proteins (3) (Fig. 3B).
Fig. 3.
Biological processes classification of up- (A) and down-regulated (B) proteins. Forty-six up- and 26 down-regulated proteins were assigned to putative biological process categories according to the TAIR GO annotation database (http://www.arabidopsis.org/tools/bulk/go/index.jsp).
Clustering analysis of protein expression patterns
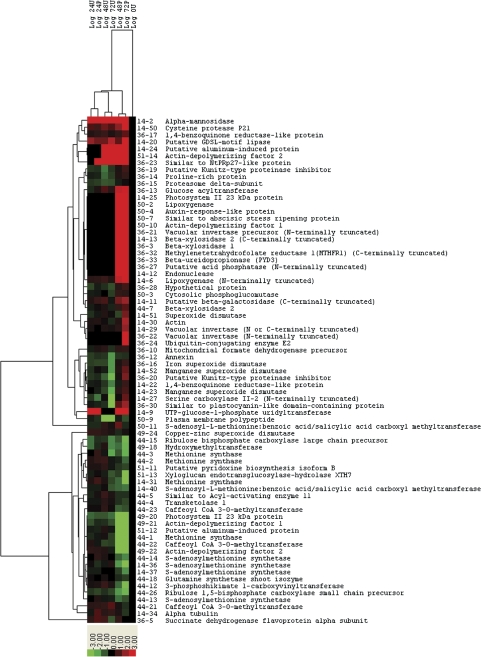
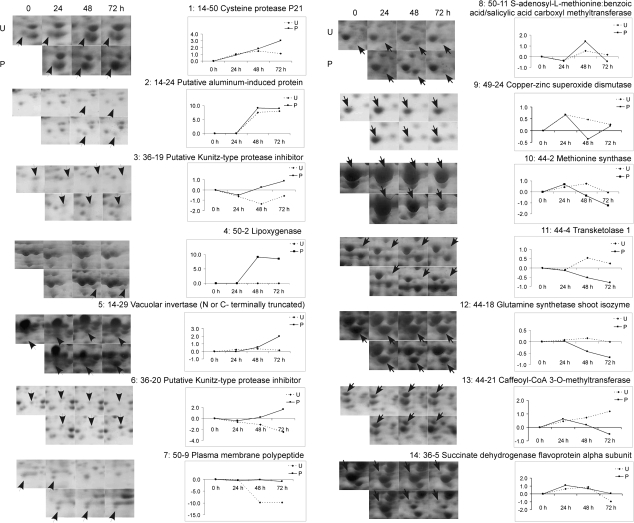
During petal senescence, certain groups of proteins will probably be co-regulated. Hierarchical clustering revealed three groups, which included 24 U and 24 P, 48 U and 72 U, and 48 P and 72 P; based on expression similarities (Fig. 4) and corresponding to developmental stages from flower opening to senescence. The two main clusters included up- and down-regulated proteins. There were eight and six subclusters of up-regulated and down-regulated proteins, respectively. The relative abundance of a representative protein from each subcluster is shown in Fig. 5. The largest up-regulated subcluster included proteins (36-13 to 44-7) with the highest abundance at 48 and 72 h after pollination. Many of these represented senescence-specific proteins that were detected only in 48 P and 72 P corollas. There were also a few proteins that were detected only at 72 h after pollination.
Fig. 4.
Hierarchical clustering analysis of the 73 differentially expressed proteins. Protein expression levels were presented as the Log ratio relative to the 0 h unpollinated reference. Colours ranging from green to red represent protein expression from the highest level of down-regulation to the highest level of up-regulation, respectively. Black colour indicates no change compared to 0 h. Two main clusters were formed, representing up- and down-regulated proteins during senescence. The similarities of protein expression patterns represented by the distance of tree branches are shown on the left side. The spot sample numbers with putative protein identification are indicated on the right side of the heat map.
Fig. 5.
Changes in relative protein abundance during corolla development and senescence. Protein expression patterns of each of the 14 subclusters from the hierarchical tree in Fig. 4 are shown by one representative protein. Spots that were up- or down-regulated are indicated with arrows on the 2-D gels. The solid and dotted lines indicate the protein expression changes in pollinated and unpollinated corollas from 0 h to 72 h, respectively.
Protein isoforms were identified from multiple spots
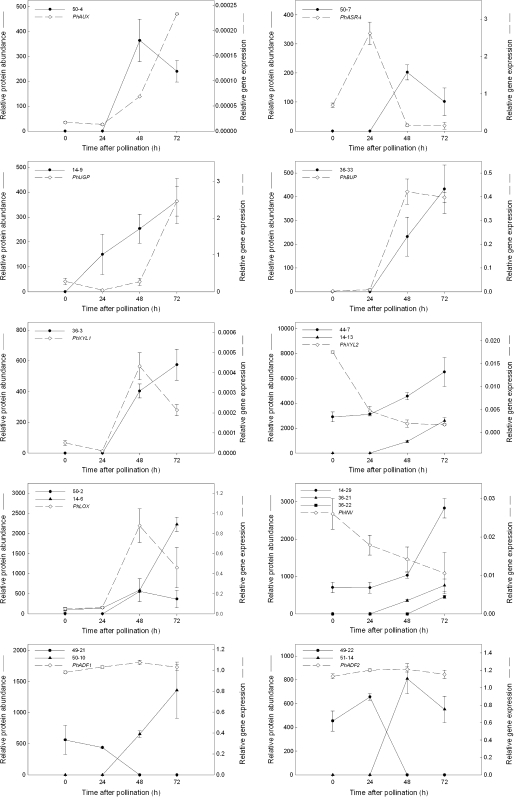
Forty-four per cent of the proteins in Tables 2 and 3 were identified in multiple spots with different pI and/ or Mr. Thus almost half of the differentially expressed proteins corresponded to various protein isoforms. Protein isoforms may be produced from different members of multigenic protein families or they may result from alternative splicing or post-translational modifications. In most instances, the multiple spots representing a protein function had the same pattern of either up- or down-regulation during the progression of corolla senescence. The proteins manganese superoxide dismutase (14-23 and 14-52), 1,4-benzoquinone reductase-like protein (14-22 and 36-17), lipoxygenase (14-6 and 50-2), Kunitz-type protease inhibitor family protein (36-19 and 36-20), vacuolar invertase (14-29, 36-21, and 36-22) and beta-xylosidase 2 (PhXYL2; 44-7 and 14-13) were all up-regulated during senescence (Table 2). The spots identified as methionine synthase (14-31, 44-1, 44-2, and 44-3), caffeoyl CoA 3-O-methyltransferase (44-21, 44-22, and 44-23), and S-adenosylmethionine synthetase (SAMS) (14-36, 14-37, 44-13, and 44-14) were all down-regulated during corolla senescence. By contrast, S-adenosyl-L-methionine:benzoic acid/salicylic acid carboxyl methyltransferase (PhBSMT2; 14-40 and 50-11), actin depolymerizing factor 1 (PhADF1; 49-21 and 50-10), actin depolymerizing factor 2 (PhADF2; 49-22 and 51-14) and photosystem II protein (14-25 and 49-20) had isoforms with opposite expression patterns (Tables 2, 3; Fig. 6).
Fig. 6.
Relative protein abundance of selected proteins and the corresponding gene expression in petunia corollas after pollination. Protein abundance of all isoforms corresponding to the protein of interest are presented as the spot volume from the replicated 2-DE gels (n=3) and gene expression was quantified using real-time PCR (n=3) as described in the methods. Error bars represent ±SD.
Transcript abundance does not always correspond with protein expression
Genes encoding 10 selected proteins were cloned by RT-PCR. These included the senescence-specific proteins, auxin-response-like protein (PhAUX; 50-4), UTP-glucose-1-phosphate uridyltransferase (PhUGP; 14-9); abscisic stress ripening protein (PhASR4; 50-7), and beta-ureidopropionase (PhBUP; 36-33); and the protein isoforms of beta-xylosidase 1 (PhXYL1; 36-3), beta-xylosidase 2 (PhXYL2; 44-7, 14-13), lipoxygenase (PhLOX; 50-2, 14-6), vacuolar invertase (PhINV; 14-29, 36-22, 36-21), actin depolymerizing factor 1 (PhADF1; 49-21, 50-10), and actin depolymerizing factor 2 (PhADF2; 49-22, 51-14). Of those proteins that were up-regulated during corolla senescence, only 60% of their corresponding genes had similar senescence-related increases in transcript abundance (Fig. 6).
Gene expression patterns for PhXYL2 (encoding proteins 44-7 and 14-13) were opposite to that observed for protein abundance, with higher transcript levels in unpollinated corollas. A large decrease in mRNA levels was detected by 24 h after pollination with little subsequent change from 48 h to 72 h. Three proteins were identified as vacuolar invertase (14-29, 36-21, and 36-22). The five peptides identified in spot 14-29 were identical to those identified in spot 36-22, suggesting that they may represent a single protein that is encoded by the same gene (PhINV). The accumulation of PhINV transcripts also had an opposite pattern to that of the protein spots. Transcript levels were highest at 0 h and decreased after pollination.
Four proteins were identified as an actin depolymerizing factor. Two of these proteins corresponded to the petunia actin depolymerizing factor PhADF1 (Genbank no. AF183903) and two corresponded to petunia actin depolymerizing factor PhADF2 (Genbank no. AF183904) (Mun et al., 2000). The PhADF1 spot 50-10 was not detectable in unpollinated corollas (0 h to 72 h) or at 24 h after pollination, but was newly detected in senescing corollas at 48 h and 72 h after pollination (Fig. 6). By contrast, the PhADF1 spot 49-21 was detected at basal levels in non-senescing corollas (0 to 72 U and 24 P) but was undetectable in 48 P and 72 P corollas. Similar patterns were observed for the PhADF2 spots (49-22 and 51-14). PhADF1 and PhADF2 transcripts were neither up- nor down-regulated by pollination, and showed very little change from 0 h to 72 h.
Discussion
Two-dimensional gel electrophoresis followed by mass spectroscopy was used successfully to identify proteins that were differentially regulated during petunia corolla senescence. While large-scale gene expression changes can be detected early in the senescence programme (24 h; ML Jones, unpublished results), protein differences were not detected until the mid-senescence stage (48 h) when corollas were starting to show visible symptoms of wilting. Proteomic techniques have been used in recent publications to investigate the post-transcriptional and post-translational regulation of senescence in leaves, but this is the first proteomic analysis of petal senescence. While a recent proteomic analysis of leaf senescence in Arabidopsis indicated that more than 50% of the soluble proteins that were visualized by 2-DE declined in abundance during the progression of senescence (Carp and Gepstein, 2007), our analysis of petals indicated that the largest percentage of differentially regulated proteins increased in abundance during senescence. At the mid-senescence stage (48 h after pollination) 64% of the differentially regulated proteins increased and at the late senescence stage (72 h after pollination) 66% of the differentially expressed proteins were up-regulated. Genomic analyses of petal senescence have also reported a greater number of senescence up-regulated compared to down-regulated genes (Breeze et al., 2004; Hoeberichts et al., 2007).
Functional classification of the differentially expressed proteins revealed that the largest number of up-regulated proteins fell into the single biological processes category of stress and defence response genes. Genomic studies of leaves and petals have also reported that a large percentage of the senescence up-regulated genes encode proteins putatively involved in plant–pathogen interactions and response to stress (primarily reactive oxygen species metabolism) (Bhalerao et al., 2003; van Doorn et al., 2003; Breeze et al., 2004; Buchanan-Wollaston et al., 2005; Hoeberichts et al., 2007; Price et al., 2008). It has been hypothesized that the pathogenesis-related and stress-response proteins may protect these tissues from pathogen attack or the accumulation of damaging reactive oxygen species (ROS) during senescence to allow for maximum degradation of cellular constituents and remobilization of nutrients (Buchanan-Wollaston et al., 2005). Some defence-related proteins were also identified as being down-regulated by pollination and may function during early petal development. Similarly, a large number of stress-related proteins were identified from rose petals in a proteomic analysis of early rose flower development from the tight bud to the fully open flower stages (Dafny-Yelin et al., 2005).
While it is well known that ethylene is the primary hormone regulating petunia flower senescence, a number of the senescence up-regulated proteins identified in this study were putative orthologues of stress response genes that are regulated by other hormones including abscisic acid (ABA) and auxin. Protein 50-4 was identified as a putative orthologue of Arabidopsis DFL1 (Dwarf in Light), a member of the auxin-responsive GH3 gene family. The IAA/AUX, SAUR, and GH3 gene family members are auxin primary response genes and DFL1 functions as an indole-3-acetic acid amido synthetase involved in auxin signal transduction (Nakazawa et al., 2001). A GH3 gene (CcGH3) in pepper is highly expressed during the late stage of fruit ripening rather than the early stage when auxin levels are highest. Promoter analysis shows that CcGH3 has both auxin and ethylene responsive elements and that CcGH3 expression is mainly responsive to ethylene during fruit ripening (Liu et al., 2005). These results suggest that CcGH3 may be regulated by ethylene or that signal crosstalk between auxin and ethylene during fruit development and ripening is involved in its regulation (Liu et al., 2005). While auxin treatment can accelerate senescence in some flowers, this response is mediated by ethylene. It is therefore possible that the auxin response-like protein (50-4) identified in our study plays a role in mediating the ethylene-induced senescence of pollinated flowers.
A senescence-specific petunia protein (50-7) was identified as having homology to tomato abscisic stress ripening protein 4 (LeASR4). The ASR gene family includes ASR1, ASR2, ASR3, and ASR4. ASR1, ASR3, and ASR4 are induced by dehydration, ABA, and cold stresses, whereas ASR2 is ABA-independent and is specifically induced by dehydration (Doczi et al., 2005). Overexpression of tomato ASR1 or lily ASR1 in transgenic tobacco or Arabidopsis enhances tolerance to drought and salt stresses (Goldgur et al., 2007). Expression analysis using promoter::GUS fusion lines show that DS2 not only responds to dehydration, but it is also expressed in pollen, flowers, and fruits, suggesting that it is regulated by both developmental cues and stress stimuli (Doczi et al., 2002, 2005). The grape ASR1 orthologue and Solanum DS2 have been hypothesized to function as transcription factors (Silhavy et al., 1995; Cakir et al., 2003).
The PhASR4 protein that was identified by 2-DE was detected only in senescing corollas at 48 h and 72 h after pollination, while PhASR4 transcript abundance was greatly increased at 24 h after pollination and decreased at 48 h and 72 h. To our knowledge, ASR genes have not previously been identified in senescing petals. This early senescence induction of PhASR4 gene expression supports a potential role in senescence signalling. The senescence of petunia corollas is accompanied by a decrease in fresh weight and an increase in ABA content (Chang et al., 2003). Changes in the relative abundance of the ASR/DS2-type protein identified in our experiment may be the result of increased petal ABA levels or petal dehydration. Whether this protein plays a role in regulating the senescence response or is a side-effect of senescence is unknown.
Up-regulated proteins in the biological classes of carbohydrate, lipid, nucleotide, amino acid, and nitrogen metabolism primarily included enzymes involved in catabolic processes that putatively function in the large-scale degradation of macromolecules and organelles that occurs during senescence to allow for nutrient remobilization. Proteins within these categories could collectively be referred to as remobilization proteins and make up 46% of the up-regulated proteins identified in this study. Most of the changes in gene expression identified using microarray and differential display techniques are also related to genes involved in remobilization, in keeping with the primary function of the senescence programme (Jones, 2004; Stead et al., 2006; van Doorn and Woltering, 2008). One major difference that can be seen by comparing the genes that were up-regulated in a recent study of wallflower petal senescence is the absence of transporter proteins in our proteomic study (Price et al., 2008). This is probably due to our protein extraction procedure, which does not adequately isolate membrane bound proteins. The proteomic techniques employed in this work are most useful for identifying highly abundant soluble proteins involved in the execution phase of senescence, while complementary genomic approaches are required to identify regulatory proteins involved in the initiation phase of senescence.
Proteins within the functional categories of amino acid, carbohydrate, and nitrogen metabolism were also observed for the down-regulated protein spots. Many of these down-regulated proteins may be anabolic enzymes that catalyse synthesis reactions that cease with the onset of senescence. It should also be noted that some of the proteins that were detected only at 0 h and 24 h after pollination may represent proteins that have changed location on the gel due to a post-translational modification that resulted in a change in the Mr and/or pI of the protein.
The next largest single functional category of up-regulated proteins after defence and stress response was carbohydrate metabolism. Microarray analyses of petal senescence in Alstroemeria and Dianthus caryophyllus (carnation) also revealed that a larger number of up-regulated genes were putatively involved in carbohydrate metabolism than were involved in lipid, nucleic acid or protein metabolism (Breeze et al., 2004; Hoeberichts et al., 2007). Many of the petunia proteins identified within this category have a putative function in cell wall degradation, while down-regulated proteins were also identified that might be involved in the synthesis of cell wall components. Cell wall degradation is a component of senescence in some flower species. Sandersonia flowers show very little cell wall degradation, while considerable degradation of the cell walls has been reported during the senescence of Hemerocallis (daylily), Ipomoea nil (morning glory), Iris, carnation, and petunia petals (Winkenbach, 1970; de Vetten et al., 1991; Panavas et al., 1998; O'Donoghue et al., 2002, 2009; van Doorn et al., 2003).
A recent study of flower development in Petunia×hybrida ‘Mitchell Diploid’ reported that the metabolism of cell wall-associated polymeric galactose was a major feature of flower opening and senescence (O'Donoghue et al., 2009). Our 2-D profiling identified a β-galactosidase protein (14-11) that was highly abundant in non-senescing corollas and which increased in abundance at 48 h and then again at 72 h after pollination. The metabolism of cell wall galactose catalysed by β-galactosidases plays a role in cell wall expansion during growth and in the loss of cell wall adhesion during senescence. β-galactosidase enzyme activity increases during flower bud opening and continues to increase during the progression of petal senescence (O'Donoghue et al., 2009). β-galactosidases are encoded by a multigene family and two β-galactosidase genes were recently cloned from petunia. PhGal1 is expressed at high levels in flower buds before flower opening and PhGAL2 is expressed at a similar level in open flowers and during petal senescence (O'Donoghue et al., 2009). In our study, the abundance of the putative β-galactosidase protein 14-11 was highly up-regulated during petal senescence. The peptide sequences from 14-11 do not match the predicted amino acid sequences of either PhGAL1 or PhGAL2, and 14-11 seems to represent a third member of the petunia β-galactosidase gene family with a role in cell wall galactose mobilization during petal senescence.
Beta-xylosidases are also involved in cell wall polysaccharide disassembly or modification and may function in cell wall degradation (Itai et al., 2003; Hayama et al., 2006). Multiple differentially regulated spots were identified as beta-xylosidases (36-3, 44-7, and 14-13). Proteins 44-7 and 14-13 were orthologues of the tomato beta-D-xylosidase LeXYL2, while 14-13 was most homologous to the Arabidopsis beta-D-xylosidase AtBXL1(Goujon et al., 2003; Itai et al., 2003). Studies in tomato have shown that the different members of the beta-D-xylosidase family are differentially regulated during fruit development. LeXYL1 mRNA levels increase at the late fruit ripening stage, while LeXYL2 mRNA levels are higher in premature fruit and decrease at the late ripening stage (Itai et al., 2003). The increased transcript abundance of PhXYL2 in unpollinated flowers at 0 h suggests that it may play a primary role in cell wall loosening and expansion during flowering opening and that the up-regulation of the C-terminally truncated form of the PhXYL2 protein (14-13) during senescence may result from targeted degradation of the protein after pollination. By contrast, PhXYL1 may catalyse cell wall degradation specifically during petunia corolla senescence.
A decrease in the total protein, RNA, and DNA content of senescing petals has been reported in many flowers including petunia (Winkenbach, 1970; Xu and Hanson, 2000; Jones, 2004; Jones et al., 2005; Langston et al., 2005). While this change could be the result of decreased protein and nucleic acid synthesis, large increases in protease and nuclease activity suggest that large-scale degradation is occurring (Stephenson and Rubinstein, 1998; Xu and Hanson, 2000; Jones et al., 2005; Langston et al., 2005; Pak and van Doorn, 2005). While the integrity of cellular membranes must be maintained during senescence to allow for maximum cellular recycling, the later stages of petal senescence are accompanied by increased activity of enzymes involved in phospholipid and fatty acid catabolism (Borochov et al., 1994). In support of this role in nutrient remobilization, a number of senescence up-regulated proteins were identified with a putative role in protein, nucleic acid or lipid degradation. These included putative proteases (14-50, 36-13, and 14-27), endonucleases (14-12), lipoxygenases (50-2 and 14-6), and lipases (14-20). These types of hydrolytic enzymes have previously been reported in petunia (Borochov et al., 1994; Xu and Hanson, 2000; Jones, 2004; Jones et al., 2005; Langston et al., 2005) and their putative roles in senescence have been discussed in a number of recent reviews (Hopkins et al., 2007; Reid and Chen, 2007; van Doorn and Woltering, 2008; Jones et al., 2009).
While senescence up-regulated proteases may be involved in large-scale protein degradation, proteins involved in the ubiquitin–proteasome pathway appear to be involved in selective protein degradation during senescence (Buchanan-Wollaston et al., 2005; Stead et al., 2006; van Doorn and Woltering, 2008). In plants, ubiquitin- and proteasome-dependent proteolysis is involved in the regulation of transcription factors and ER-associated protein degradation during development and in response to stress. It also directly impacts ethylene signalling during senescence by regulating the degradation of the EIN3/EIL transcription factors and the ethylene receptors (ETR) (Guo and Ecker, 2003; Kevany et al., 2008). Increased expression of genes putatively involved in protein ubiquitination and those encoding protein subunits of the proteasome have been identified in senescing Alstroemeria, Narcissus pseudonarcissus, carnation, morning glory, Mirabilis jalapa (four o'clocks), and petunia petals (Hunter et al., 2002; Breeze et al., 2004; Hoeberichts et al., 2007; Xu et al., 2007a, b; Yamada et al., 2007). A recent proteomic analysis of leaves also identified a proteasome beta subunit A1 that was up-regulated during nitrate starvation-induced senescence (Desclos et al., 2009).
Our proteomic analysis identified an ubiquitin-conjugating enzyme family protein (E2) (36-24) and a delta-subunit of the 20S proteasome (36-15) that were up-regulated by pollination. MS analysis revealed that a number of the up-regulated spots contained truncated proteins that had a much lower than predicted Mr. The appearance of these truncated proteins in senescing petals may be explained by the selective degradation of proteins that are no longer needed during the execution phase of senescence. The mass spectra for the peptides in band 14-29 (vacuolar invertase) support the theory that proteins are ubiquitinated during senescence. A peptide with a molecular mass of 1744 Da was detected that contained the sequence 231YSGNPVMoVPPPGIGVK246 plus an additional 114 Da, and the same peptide in its unmodified form (1630 Da) was also detected in band 14-29 (data not shown). The additional 114 Da occurred on the C-terminal fragment of the GVK residue. This mass increase can be explained by ubiquitination of the K residue, as the addition of two glycines to the K would account for the 114 Da increase. Down-regulating the expression of a RING domain E3 protein delays visual symptoms of petal wilting in petunia by 2 d, suggesting that the ubiquitin–proteasome pathway does play a regulatory role in senescence (Xu et al., 2007a).
Actin-depolymerizing factor (ADF) is one of the small actin-binding proteins that modulates actin cytoskeleton dynamics in eukaryotic cells (Mun et al., 2002; Thomas et al., 2006). In our 2-DE experiments, two proteins encoded by PhADF1 (50-10 and 49-21) and two proteins encoded by PhADF2 (50-14 and 49-22) were identified. Previous studies have shown that the petunia PhADF1 and PhADF2 genes are expressed in all tissues except pollen, with the highest transcript abundance in petals (Mun et al., 2000). In our proteomic analysis the two PhADF1 isoforms and the two PhADF2 isoforms had opposite expression patterns. MS analysis indicated that the down-regulated PhADF1 and PhADF2 isoforms were acetylated at the second amino acid of the N-terminus. We do not know if the two up-regulated proteins were also acetylated, as that same peptide was not sequenced. The down-regulated PhADFs had a higher observed Mr than the PhADF proteins that were up-regulated, supporting the theory that they may represent different stages in the post-translational modification of the ADF proteins. Although there is no evidence to indicate that acetylation regulates ADF function, the activity of the ADF protein and its interaction with actin is known to be regulated by pH, phosphorylation, and phosphoinositide (Lopez et al., 1996). The stabilization of actin depolymerization has been shown to induce PCD in plants (Thomas et al., 2006), and the post-transcriptional regulation of PhADF1 and PhADF2 may trigger or modulate corolla senescence.
While most studies of senescence have focused on gene expression changes, senescence is also controlled by post-transcriptional and post-translational regulation (Thomas et al., 2003). Treating flowers with the protein synthesis inhibitor, cycloheximide, significantly delays corolla senescence and provides evidence for the role of newly translated proteins in the senescence program (Wulster et al., 1982; Lay Yee et al., 1992; van Doorn et al., 1995). In this study, ten genes corresponding to senescence up-regulated proteins were cloned to determine patterns of gene expression during petal senescence. Only six of the ten genes (60%) had senescence up-regulated increases in transcript abundance. Genes encoding PhXYL2 and PhINV showed an opposite expression pattern, with transcript abundance decreasing during senescence. Even more interestingly, the genes encoding PhADF1 and PhADF2 showed little change in relative mRNA abundance during senescence, even though protein isoforms of ADF1 and ADF2 were identified that were up- and down-regulated during petal senescence. In Arabidopsis, where extensive microarray data are available to complement proteomic analyses, a poor correlation between protein and gene expression has been reported (Jiang et al., 2007). Similar to what was observed in petals, Carp and Gepstein (2007) reported that only approximately 60% of the up-regulated proteins identified during leaf senescence had corresponding increases in mRNA abundance.
Conclusion
In this study, a proteomic analysis of petunia petals identified 46 proteins that were up-regulated and 27 proteins that were down-regulated during senescence. The proteins identified were implicated in a wide range of biological processes with the majority having a putative role in stress and defence responses or catabolism and remobilization. This is the first proteomic analysis of petal senescence and as such it provides new insights into the pathways that execute senescence and the post-transcriptional regulation of senescence. While our MS analysis did not allow for the precise identification of specific protein modifications, 37% of the senescence up-regulated proteins had higher than predicted Mr and changes in the pI that indicate potential post-translational modifications. This study provides a good starting point for further genetic analysis of the functional role of these proteins in petal senescence.
Supplementary data
Supplementary data are available at JXB online.
Supplementary Table S1 lists the primers and Tm used for real-time RT-PCR analysis.
Supplementary Fig. S1 is a flow chart outlining the methodology for mass spectroscopy and protein identification.
Supplementary Material
Acknowledgments
This work was supported by the USDA Floriculture and Nursery Research Initiative, the Ohio State University DC Kiplinger Endowment, an OARDC Research Enhancement Competitive Grant, and the Fred C Gloeckner Foundation. Salaries and research support were provided in part by State and Federal funds appropriated to the Ohio Agricultural Research and Development Center, The Ohio State University. Journal Article Number HCS 09-01.
We thank The Ohio State University Plant-Microbe Genomics Facility for use of PDQuest software, and David Mandich for excellent technical assistance. We thank Dr Sophien Kamoun (Sainsbury Laboratory, John Innes Centre, UK) for sharing the Nicotiana ESTs database and Ian Holford at the OARDC MCIC for bioinformatics assistance.
References
- Ashman TL, Schoen DJ. How long should flowers live. Nature. 1994;317:788–791. [Google Scholar]
- Ahsan N, Lee DG, Lee SH, Kang KY, Bahk JD, Choi MS, Lee IJ, Renaut J, Lee BH. A comparative proteomic analysis of tomato leaves in response to waterlogging stress. Journal of Plant Physiology. 2007;131:555–570. doi: 10.1111/j.1399-3054.2007.00980.x. [DOI] [PubMed] [Google Scholar]
- Askari H, Edqvist J, Hajheidari M, Kafi M, Salekdeh GH. Effects of salinity levels on proteome of Suaeda aegyptiaca leaves. Proteomics. 2006;6:2542–2554. doi: 10.1002/pmic.200500328. [DOI] [PubMed] [Google Scholar]
- Berardini TZ, Mundodi S, Reiser R, et al. Functional annotation of the Arabidopsis genome using controlled vocabularies. Plant Physiology. 2004;135:1–11. doi: 10.1104/pp.104.040071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalerao R, Keskitalo J, Sterky F, et al. Gene expression in autumn leaves. Plant Physiology. 2003;131:430–442. doi: 10.1104/pp.012732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breeze E, Wagstaff C, Harrison E, Bramke I, Rogers H, Stead A, Thomas B, Buchanan-Wollaston V. Gene expression patterns to define stages of post-harvest senescence in Alstroemeria petals. Plant Biotechnology Journal. 2004;2:525–525. doi: 10.1111/j.1467-7652.2004.00059.x. [DOI] [PubMed] [Google Scholar]
- Borochov A, Cho MH, Boss WF. Plasma membrane lipid metabolism of Petunia petals during senescence. Physiologia Plantarum. 1994;90:279–284. [Google Scholar]
- Buchanan-Wollaston V, Earl S, Harrison E, Mathas E, Navabpour S, Page T, Pink D. The molecular analysis of leaf senescence: a genomics approach. Plant Biotechnology Journal. 2003;1:3–22. doi: 10.1046/j.1467-7652.2003.00004.x. [DOI] [PubMed] [Google Scholar]
- Buchanan-Wollaston V, Page T, Harrison E, et al. Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. The Plant Journal. 2005;42:567–585. doi: 10.1111/j.1365-313X.2005.02399.x. [DOI] [PubMed] [Google Scholar]
- Cakir B, Agasse A, Gaillard C, Saumonneau A, Delrot S, Atanassova R. A grape ASR protein involved in sugar and abscisic acid signalling. The Plant Cell. 2003;15:2165–2180. doi: 10.1105/tpc.013854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carp M-J, Gepstein S. Genomics and proteomics of leaf senescence. In: Gan S, editor. Senescence processes in plants. Annual Plant Reviews, Vol. 26. Oxford, UK: Blackwell Publishing, Ltd.; 2007. pp. 202–230. [Google Scholar]
- Chapin L, Jones ML. Nutrient remobilization during pollination-induced corolla senescence in Petunia. Acta Horticulturae. 2007;55:181–190. [Google Scholar]
- Chapin LJ, Jones ML. Ethylene regulates phosphorus remobilization and expression of a phosphate transporter (PhPT1) during petunia corolla senescence. Journal of Experimental Botany. 2009;60:2179–2190. doi: 10.1093/jxb/erp092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H, Jones ML, Banowetz GM, Clark DG. Overproduction of cytokinins in petunia flowers transformed with PSAG12- IPT delays corolla senescence and decreases sensitivity to ethylene. Plant Physiology. 2003;132:2174–2183. doi: 10.1104/pp.103.023945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coaker GL, Willard B, Kinter M, Stockinger EJ, Francis DM. Proteomic analysis of resistance mediated by Rcm 2.0 and Rcm 5.1, two loci controlling resistance to bacterial canker of tomato. Molecular Plant–Microbe Interactions. 2004;17:1019–1028. doi: 10.1094/MPMI.2004.17.9.1019. [DOI] [PubMed] [Google Scholar]
- Dafny-Yelin M, Guterman I, Menda N, et al. Flower proteome: changes in protein spectrum during the advanced stages of rose petal development. Planta. 2005;222:37–46. doi: 10.1007/s00425-005-1512-x. [DOI] [PubMed] [Google Scholar]
- Desclos M, Etienne P, Coquet L, Jouenne T, Bonnefoy J, Segura R, Reze S, Ourry A, Avice J- C. A combined 15N tracing/proteomics study in Brassica napus reveals the chronology of proteomics events associated with N remobilization during leaf senescence induced by nitrate limitation or starvation. Proteomics. 2009;9:3580–3608. doi: 10.1002/pmic.200800984. [DOI] [PubMed] [Google Scholar]
- de Vetten NC, Huber DJ, Gross KC. Endoglycanase-catalysed degradation of hemicelluloses during development of carnation (Dianthus caryophyllus L.) petals. Plant Physiology. 1991;95:853–860. doi: 10.1104/pp.95.3.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doczi R, Csanaki C, Banfalvi Z. Expression and promoter activity of the desiccation-specific Solanum tuberosum gene, StDS2. Plant, Cell and Environment. 2002;25:1197–1203. [Google Scholar]
- Doczi R, Kondrak M, Kovacs G, Beczner F, Banfalvi Z. Conservation of the drought-inducible, DS2 genes and divergences from their ASR paralogues in solanaceous species. Plant Physiology and Biochemistry. 2005;43:269–276. doi: 10.1016/j.plaphy.2005.02.002. [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proceedings of the National Academy of Sciences, USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faurobert M, Mihr C, Bertin N, Pawlowski T, Negroni L, Sommerer N, Causse M. Major proteome variations associated with cherry tomato pericarp development and ripening. Plant Physiology. 2007;143:1327–1346. doi: 10.1104/pp.106.092817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldgur Y, Rom S, Ghirlando R, Shkolnik D, Shadrin N, Konrad Z, Bar-Zvi D. Desiccation and zinc binding induce transition of tomato abscisic acid stress ripening 1, a water stress- and salt stress-regulated plant-specific protein, from unfolded to folded state. Plant Physiology. 2007;143:617–628. doi: 10.1104/pp.106.092965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goujon T, Minic Z, Amrani AE, Lerouxel O, Aletti E, Lapierre C, Joseleau J-P, Jouanin L. AtBXL1, a novel higher plant (Arabidopsis thaliana) putative beta-xylosidase gene, is involved in secondary cell wall metabolism and plant development. The Plant Journal. 2003;33:677–690. doi: 10.1046/j.1365-313x.2003.01654.x. [DOI] [PubMed] [Google Scholar]
- Guo HW, Ecker JR. Plant responses to ethylene gas are mediated by SCF (EBF1/EBF2)-dependent proteolysis of EIN3 transcription factor. Cell. 2003;115:667–677. doi: 10.1016/s0092-8674(03)00969-3. [DOI] [PubMed] [Google Scholar]
- Hajheidari M, Eivazi A, Buchanan BB, Wong JH, Majidi I, Salekdeh GH. Proteomics uncovers a role for redox in drought tolerance in wheat. Journal of Proteome Research. 2007;6:1451–1460. doi: 10.1021/pr060570j. [DOI] [PubMed] [Google Scholar]
- Hayama H, Shimada T, Fujii H, Ito A, Kashimura Y. Ethylene-regulation of fruit softening and softening-related genes in peach. Journal of Experimental Botany. 2006;57:4071–4077. doi: 10.1093/jxb/erl178. [DOI] [PubMed] [Google Scholar]
- Hebeler R, Oeljeklaus S, Reidegeld KA, et al. Study of early leaf senescence in Arabidopsis thaliana by quantitative proteomics using reciprocal 14N/15N labeling and difference gel electrophoresis. Molecular and Cellular Proteomics. 2008;7:108–120. doi: 10.1074/mcp.M700340-MCP200. [DOI] [PubMed] [Google Scholar]
- Hoeberichts FA, van Doorn WG, Vorst O, Hall RD, van Wordragen MF. Sucrose prevents up-regulation of senescence-associated genes in carnation petals. Journal of Experimental Botany. 2007;58:2873–2885. doi: 10.1093/jxb/erm076. [DOI] [PubMed] [Google Scholar]
- Hopkins M, Taylor C, Liu Z, Ma F, McNamara L, Wang T-W, Thompson JE. Regulation and execution of molecular disassembly and catabolism during senescence. New Phytologist. 2007;175:201–214. doi: 10.1111/j.1469-8137.2007.02118.x. [DOI] [PubMed] [Google Scholar]
- Horvath-Szanics E, Szabo Z, Janaky T, Pauk J, Hajos G. Proteomics as an emergent tool for identification of stress-induced proteins in control and genetically modified wheat lines. Chromatographia. 2006;63:S143–S147. [Google Scholar]
- Huang B, Chu CH, Chen SL, Juan HF, Chen YM. A proteomics study of the mung bean epicotyl regulated by brassinosteroids under conditions of chilling stress. Cellular Molecular Biology Letters. 2006;11:264–278. doi: 10.2478/s11658-006-0021-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter DA, Steele BC, Reid MS. Identification of genes associated with perianth senescence in daffodil (Narcissus pseudonarcissus L. ‘Dutch Master’) Plant Science. 2002;163:13–21. [Google Scholar]
- Itai A, Ishihara K, Bewley JD. Characterization of expression and cloning of β-d-xylosidase and α-l-arabinofuranosidase in developing and ripening tomato (Lycopersicon esculentum Mill.) fruit. Journal of Experimental Botany. 2003;54:2615–2622. doi: 10.1093/jxb/erg291. [DOI] [PubMed] [Google Scholar]
- Ito J, Heazlewood JL, Millar AH. The plant mitochondrial proteome and the challenge of defining the posttranslational modifications responsible for signalling and stress effects on respiratory functions. Physiologia Plantarum. 2007;129:207–224. [Google Scholar]
- Jiang Y, Yang B, Harris NS, Deyholos MK. Comparative proteomic analysis of NaCl stress-responsive proteins in Arabidopsis roots. Journal of Experimental Botany. 2007;58:3591–3607. doi: 10.1093/jxb/erm207. [DOI] [PubMed] [Google Scholar]
- Jones ML. Changes in gene expression during senescence. In: Noodén L, editor. Plant cell death processes. San Diego: Elsevier Science; 2004. pp. 51–71. [Google Scholar]
- Jones ML. Ethylene signalling is required for pollination-accelerated corolla senescence in Petunias. Plant Science. 2008;175:190–196. [Google Scholar]
- Jones ML, Chaffin GS, Eason JR, Clark DG. Ethylene-sensitivity regulates proteolytic activity and cysteine protease gene expression in petunia corollas. Journal of Experimental Botany. 2005;56:2733–2744. doi: 10.1093/jxb/eri266. [DOI] [PubMed] [Google Scholar]
- Jones ML, Stead AD, Clark DG. Petunia flower senescence. In: Gerats T, Strommer J, editors. Petunia: a model system for comparative research. New York: Springer; 2009. pp. 301–324. [Google Scholar]
- Jorrin JV, Maldonado AM, Castillejo MA. Plant proteome analysis: a 2006 update. Proteomics. 2007;7:2947–2962. doi: 10.1002/pmic.200700135. [DOI] [PubMed] [Google Scholar]
- Kerr MK, Martin M, Churchill GA. Analysis of variance for gene expression microarray data. Journal of Computational Biology. 2000;7:819–837. doi: 10.1089/10665270050514954. [DOI] [PubMed] [Google Scholar]
- Kevany BM, Taylor MG, Klee HJ. Fruit-specific suppression of the ethylene receptor LeETR4 results in early-ripening tomato fruit. Plant Biotechnology Journal. 2008;6:295–300. doi: 10.1111/j.1467-7652.2007.00319.x. [DOI] [PubMed] [Google Scholar]
- Kinter M, Sherman NE. Protein sequencing and identification using tandem mass spectrometry. New York: John Wiley and Sons; 2000. [Google Scholar]
- Langston BJ, Bai S, Jones ML. Increases in DNA fragmentation and induction of a senescence-specific nuclease are delayed during corolla senescence in ethylene-insensitive (etr1-1) transgenic petunias. Journal of Experimental Botany. 2005;56:15–23. doi: 10.1093/jxb/eri002. [DOI] [PubMed] [Google Scholar]
- Lay Yee M, Stead AD, Reid MS. Flower senescence in daylily (Hemerocallis) Physiologia Plantarum. 1992;86:308–314. [Google Scholar]
- Liu KD, Kang BC, Jiang H, Moore SL, Li HX, Watkins CB, Setter TL, Jahn MM. A GH3-like gene, CcGH3, isolated from Capsicum chinense L. fruit is regulated by auxin and ethylene. Plant Molecular Biology. 2005;58:447–464. doi: 10.1007/s11103-005-6505-4. [DOI] [PubMed] [Google Scholar]
- Lopez I, Anthony RG, Maciver SK, Jiang C-J, Khan S, Weeds AG, Hussey PJ. Pollen-specific expression of maize genes encoding actin depolymerizing factor-like proteins. Proceedings of the National Academy of Sciences, USA. 1996;3:7415–7420. doi: 10.1073/pnas.93.14.7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller LA, Solow TH, Taylor N, et al. The SOL Genomics Network. A comparative resource for Solanaceae biology and beyond. Plant Physiology. 2005;138:1310–1317. doi: 10.1104/pp.105.060707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mun JH, Lee SY, Yu HJ, Jeong YM, Shin MY, Kim H, Lee I, Kim SG. Petunia actin-depolymerizing factor is mainly accumulated in vascular tissue and its gene expression is enhanced by the first intron. Gene. 2002;292:233–243. doi: 10.1016/s0378-1119(02)00646-7. [DOI] [PubMed] [Google Scholar]
- Mun JH, Yu HJ, Lee HS, Kwon YM, Lee JS, Lee I, Kim SG. Two closely related cDNAs encoding actin-depolymerizing factors of petunia are mainly expressed in vegetative tissues. Gene. 2000;257:167–176. doi: 10.1016/s0378-1119(00)00412-1. [DOI] [PubMed] [Google Scholar]
- Nakazawa M, Yabe N, Ichikawa T, Yamamoto YY, Yoshizumi T, Hasunuma K, Matsui M. DFL1, an auxin-responsive GH3 gene homologue, negatively regulates shoot cell elongation and lateral root formation, and positively regulates the light response of hypocotyl length. The Plant Journal. 2001;25:213–221. doi: 10.1046/j.1365-313x.2001.00957.x. [DOI] [PubMed] [Google Scholar]
- O'Donoghue EM, Somerfield SD, Heyes JA. Organization of cell walls in Sandersonia aurantiaca floral tissue. Journal of Experimental Botany. 2002;53:513–523. doi: 10.1093/jexbot/53.368.513. [DOI] [PubMed] [Google Scholar]
- O'Donoghue EM, Somerfield SD, Watson LM, Brummell DA, Hunter DA. Galactose metabolism in cell walls of opening and senescing petunia petals. Planta. 2009;229:709–721. doi: 10.1007/s00425-008-0862-6. [DOI] [PubMed] [Google Scholar]
- Pak C, van Doorn WG. Delay of Iris flower senescence by protease inhibitors. New Phytologist. 2005;165:473–480. doi: 10.1111/j.1469-8137.2004.01226.x. [DOI] [PubMed] [Google Scholar]
- Panavas T, Reid PD, Rubinstein B. Programmed cell death of daylily petals: activity of wall-based enzymes and effects of heat shock. Plant Physiology and Biochemistry. 1998;36:379–388. [Google Scholar]
- Price AM, Aros Orellana DF, Mohd Salleh F, Stevens R, Acock R, Buchanan-Wollaston V, Stead AD, Rogers HJ. A comparison of leaf and petal senescence in wallflower reveals common and distinct patterns of gene expression and physiology. Plant Physiology. 2008;147:1898–1912. doi: 10.1104/pp.108.120402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid MS, Chen J-C. Flower senescence. In: Gan S, editor. Senescence processes in plants. Annual Plant Reviews, Vol. 26. Oxford, UK: Blackwell Publishing, Ltd.; 2007. pp. 256–277. [Google Scholar]
- Silhavy D, Hutvagner G, Barta E, Banfalvi Z. Isolation and characterization of a water-stress-inducible cDNA clone from Solanum chacoense. Plant Molecular Biology. 1995;27:587–595. doi: 10.1007/BF00019324. [DOI] [PubMed] [Google Scholar]
- Stead AD. Pollination-induced flower senescence: a review. Plant Growth Regulation. 1992;11:13–20. [Google Scholar]
- Stead AD, van Doorn WG, Jones ML, Wagstaff C. Flower senescence: fundamental and applied aspects. In: Ainsworth C, editor. Flowering and its manipulation. Annual Plant Reviews, Vol. 20. Oxford, UK: Blackwell Publishing Ltd.; 2006. pp. 261–296. [Google Scholar]
- Stephenson P, Rubinstein B. Characterization of proteolytic activity during senescence in daylilies. Physiologia Plantarum. 1998;104:463–473. [Google Scholar]
- Thomas SG, Huang SJ, Li ST, Staiger CJ, Franklin-Tong VE. Actin depolymerization is sufficient to induce programmed cell death in self-incompatible pollen. Journal of Cell Biology. 2006;174:221–229. doi: 10.1083/jcb.200604011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas H, Ougham JH, Wagstaff C, Stead AD. Defining senescence and death. Journal of Experimental Botany. 2003;54:1127–1132. doi: 10.1093/jxb/erg133. [DOI] [PubMed] [Google Scholar]
- Thomas H, Stoddart JL. Leaf senescence. Annual Review of Plant Physiology and Plant Molecular Biology. 1980;31:83–111. [Google Scholar]
- van Doorn WG, Balk PA, van Houwelingen AM, Hoeberichts FA, Hall RD, Vorst O, van der Schoot C, van Wordragen MF. Gene expression during anthesis and senescence in Iris flowers. Plant Molecular Biology. 2003;53:845–863. doi: 10.1023/B:PLAN.0000023670.61059.1d. [DOI] [PubMed] [Google Scholar]
- van Doorn WG, Harkema H, Song JS. Water relations and senescence of cut Iris flowers: effects of cycloheximide. Postharvest Biology and Technology. 1995;5:345–351. [Google Scholar]
- van Doorn WG, Woltering EJ. Physiology and molecular biology of petal senescence. Journal of Experimental Botany. 2008;59:453–480. doi: 10.1093/jxb/erm356. [DOI] [PubMed] [Google Scholar]
- Veerasamy M, He YL, Huang BR. Leaf senescence and protein metabolism in creeping bentgrass exposed to heat stress and treated with cytokinins. Journal of the American Society for Horticultural Science. 2007;132:467–472. [Google Scholar]
- Verlinden S. Changes in mineral nutrient concentrations in petunia corollas during development and senescence. HortScience. 2003;38:71–74. [Google Scholar]
- Wilson KA, McManus MT, Gordon ME, Jordan TW. The proteomics of senescence in leaves of white clover, Trifolium repens(LProteomics. 2002;2:1114–1122. doi: 10.1002/1615-9861(200209)2:9<1114::AID-PROT1114>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Winkenbach F. Zum Stoffwechsel der aufbluhenden und welkenden Korolle der Prunkwinde Ipomoea purpurea. I. Beziehungenzwischen Gestaltwande 1, Stofftransport, Atmung und Invertaseaktivitat. Berichte Schweiz Bot Gesellschaft. 1970;80:374–390. [Google Scholar]
- Wulster G, Sacalis J, Janes H. The effect of inhibitors of protein-synthesis on ethylene induced senescence in isolated carnation petals. Journal of the American Society for Horticultural Science. 1982;107:112–115. [Google Scholar]
- Xu X, Jiang CZ, Donnelly L, Reid MS. Functional analysis of a RING domain ankyrin repeat protein that is highly expressed during flower senescence. Journal of Experimental Botany. 2007a;58:3623–3630. doi: 10.1093/jxb/erm212. [DOI] [PubMed] [Google Scholar]
- Xu XJ, Gookin T, Jiang CZ, Reid M. Genes associated with opening and senescence of Mirabilis jalapa flowers. Journal of Experimental Botany. 2007b;58:2193–2201. doi: 10.1093/jxb/erm058. [DOI] [PubMed] [Google Scholar]
- Xu Y, Hanson MR. Programmed cell death during pollination-induced petal senescence in Petunia. Plant Physiology. 2000;122:1323–1333. doi: 10.1104/pp.122.4.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T, Ichimura K, Kanekatsu M, van Doorn WG. Gene expression in opening and senescing petals of morning glory (Ipomoea nil) Planta. 2007;224:1279–1290. doi: 10.1007/s00299-006-0285-4. [DOI] [PubMed] [Google Scholar]
- Yang QS, Wang YQ, Zhang JJ, Shi WP, Qian CM, Peng XX. Identification of aluminium-responsive proteins in rice roots by a proteomic approach: cysteine synthase as a key player in Al response. Proteomics. 2007;7:737–749. doi: 10.1002/pmic.200600703. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.