Abstract
Anthocyanin content of potato tubers is a trait that is attracting increasing attention as the potential nutritional benefits of this class of compound become apparent. However, our understanding of potato tuber anthocyanin accumulation is not complete. The aim of this study was to use a potato microarray to investigate gene expression patterns associated with the accumulation of purple tuber anthocyanins. The advanced potato selections, CO97216-3P/PW and CO97227-2P/PW, developed by conventional breeding procedures, produced tubers with incomplete expression of tuber flesh pigmentation. This feature permits sampling pigmented and non-pigmented tissues from the same tubers, in essence, isolating the factors responsible for pigmentation from confounding genetic, environmental, and developmental effects. An examination of the transcriptome, coupled with metabolite data from purple pigmented sectors and from non-pigmented sectors of the same tuber, was undertaken to identify these genes whose expression correlated with elevated or altered polyphenol composition. Combined with a similar study using eight other conventional cultivars and advanced selections with different pigmentation, it was possible to produce a refined list of only 27 genes that were consistently differentially expressed in purple tuber tissues compared with white. Within this list are several new candidate genes that are likely to impact on tuber anthocyanin accumulation, including a gene encoding a novel single domain MYB transcription factor.
Keywords: Anthocyanin, antioxidant, flavonoids, potato tuber, microarray, Solanum tuberosum group Tuberosum
Introduction
Anthocyanins are responsible for the deep purple to red pigmentation of certain fruits and vegetables and play important ecophysiological roles in both plant abiotic and biotic stress resistance and as pollination attractants in flowers. In addition, anthocyanin-rich fruits and vegetables are bright and attractive to consumers and they have been documented as excellent sources of polyphenolic antioxidants (for example, Tsuda et al., 2000; Brown et al., 2007). Epidemiological and mechanistic data have been instrumental in national campaigns, designed to elevate the importance of consuming at least five portions of fruits and vegetables on a daily basis (for example, in the UK, http://www.5aday.gov/). While polyphenolic compounds such as anthocyanins are not suggested to be solely responsible for the reported health benefits of fruit and vegetable consumption, a number of reports implicate highly pigmented, anthocyanin-rich foodstuffs in suppressing or intervening in several chronic diseases (Renaud et al., 1992; Noda et al., 2000; Tsuda et al., 2000; Meiers et al., 2001; Matsumoto et al., 2003; Seeram et al., 2004; Shin et al., 2006; Butelli et al., 2008).
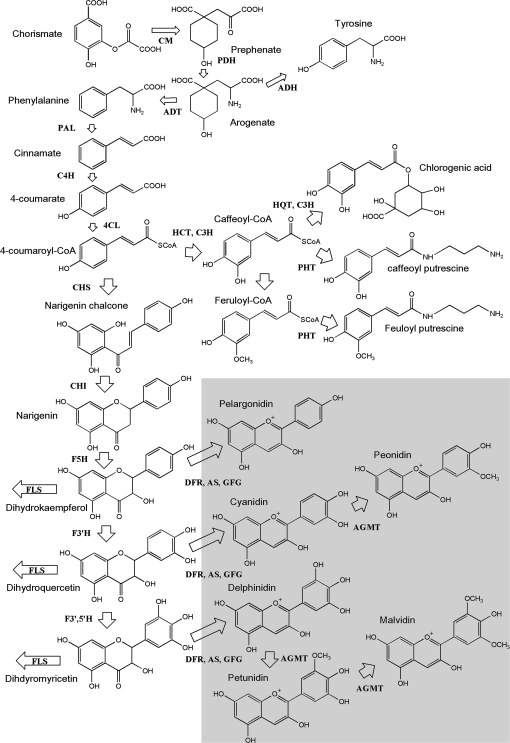
In many plants, the flavonoid biosynthetic pathway has been almost completely elucidated (Fig. 1). Genes encoding the enzymes of the pathway have been cloned from many model and crop plants (Holton and Cornisch, 1995). Considerable progress has also been made in understanding the regulation of this pathway, and transcription factors that control the expression of the structural genes have been characterized using transposon tagging approaches in model plants such as maize, Antirrhinum, and Petunia (Holton and Cornisch, 1995). More recently, similar genes have been discovered in Arabidopsis, tomato, and grape. In general, these transcription factors belong to one of two transcription factor families having sequence similarity to either the vertebrate proto-oncogene c-MYB or the vertebrate basic-Helix-Loop-Helix (bHLH) proto-oncogene c-MYC families (reviewed in Grotewald, 2006). In general, there is conservation in the structure of these transcription factors and ectopic expression of transcription factors in different plant species has become a powerful tool to activate sets of flavonoid structural genes and thus manipulate the types and amounts of flavonoids that accumulate. For example, Butelli et al. (2008) transformed tomato to obtain purple fruit pigmentation and elevated anthocyanin content by expressing transcription factor regulatory genes (Del) and MYB-related genes (Ros1) from Antirrihinum majus. Expression of these genes resulted in increased expression of several key anthocyanin pathway genes, higher antioxidant capacity, and extended life expectancy of Trp53-/-cancer knock-out mice fed with transgenic tomato.
Fig. 1.
Schematic diagram of the biosynthetic pathway for anthocyanins, caffeoylquinates, and other major phenolic derivatives in potato tuber. Enzymes abbreviated are as follows: ADH, arogenate dehydrogenase; ADT, arogenate dehydratase; AGMT, anthocyanidin-glycoside-3′-O-methyl transferase; AS, anthocyanin synthase; CHI, chalcone isomerase; CHS, chalcone synthase; C4H, cinnamate-4-hydroxylase; 4CL, 4-coumarate ligase; CM, chorismate mutase; DFR, dihydroflavonol reductase; F3H, flavonone-3-hydroxylase; F5H, ferulate 5-hydroxylase; F3′H, flavonoid-3′-hydroxylase; F3′5′H, flavonoid-3′,5′-hydroxylase; FLS, flavonol synthase; HCT, hydroxycinnamoyl-CoA:shikimate hydroxycinnamoyl transferase; HQT, hydroxycinnamoyl-CoA quinate hydroxycinnamoyl transferase; PAL, phenylalanine ammonia lyase; PDH, prephenate dehydratase; PHT, putrescine N-hydroxycinnamoyl transferase. For simplicity, only anthocyanidins and not glycosylated anthocyanins are shown. The enzymes involved in anthocyanin glycosylation (A3GT, anthocyanidin-3-O-glycosyl transferase, A5GT, anthocyanidin-5-O-glycosyl transferase, and GFG, UDP-glucose 3-O-flavonoid glucosyltransferase) are not shown due to space limitations. AGA (anthocyanidin-3′-O-glycoside-6′-O-acyl transferase) which decorates anthocyanin glycosides with hydroxycinnamoyl groups is also not represented. The steps required for anthocyanidin synthesis are highlighted in the shaded box.
While potato has not yet been widely recognized as a potent source of antioxidants for human health, recent studies suggest certain pigmented genotypes are very rich sources of polyphenols (Lewis et al., 1998a, b; Brown et al., 2003; De Jong et al., 2004; Reyes et al., 2003; Stushnoff et al., 2008). In addition, as potatoes are an important food staple with wide consumer appeal, there is enormous potential to enhance the overall nutritional status by ensuring that the consumed product is rich in desirable phytochemicals. Indeed there are recent suggestions that potato anthocyanins may protect against prostate (Reddivari et al., 2007) and breast cancers (Thompson et al., 2009).
Previous studies have addressed the biosynthesis of anthocyanin pigments in the potato tuber in both the tuber skin and flesh. In diploid potato, three classical gene loci; I, R, and P, are known to control tuber skin anthocyanin accumulation (Dodds and Long, 1955, 1956). The R locus is required for the accumulation of red pelargonidin derivatives in the tuber skin, whereas P is required for the mainly petunidin-derived purple pigments. The I locus is required for the synthesis of both red and purple skin anthocyanins and must be present in conjunction with R or P for accumulation in the skin. These three loci have been mapped in the potato genome and candidate genes have been suggested for each locus (De Jong et al., 2004). Genetic evidence based on the co-localization in the potato genetic map of R and I indicated these loci encode dihydroflavonol 4-reductase (dfr) and an R2R3MYB transcription factor designated an2, respectively. The P locus has been clearly identified, using a combination of genetic and transgenic approaches, as encoding flavonoid 3′, 5′-hydroxylase (Jung et al., 2005).
Genetic and molecular studies have also addressed anthocyanin accumulation in the tuber flesh. One locus that is associated with anthocyanin accumulation in these tissues is called Pf (De Jong, 1987). Pf is tightly linked to I, whose map position co-localizes with an2. However, Pf alone is insufficient for complete tuber pigmentation implying interaction with other genes. Indeed, using genetic approaches to analyse tuber flesh pigmentation, several QTL for tuber anthocyanin content were identified on chromosomes 5, 8, and 9 (Zhang et al., 2009). Our lack of knowledge of the associated regulatory genes is consistent with the observation that constitutive over-expression of genes encoding the enzymes of flavonoid biosynthesis in potato has, in general, led to relatively modest increases in tuber flesh anthocyanins and resulted in undesirable pleiotropic effects on plant development. (Lukaszewicz et al., 2004; Aksamit-Stachurska et al., 2008, Rommens et al., 2008).
Assessment of germplasm antioxidant properties extant in the Colorado potato breeding programme revealed that genotypes with purple and red pigmented tuber flesh had substantially higher antioxidant properties than non-pigmented types (Stushnoff et al., 2008). Similar observations have been reported in other potato genotypes (Brown et al., 2003; Reyes and Cisneros-Zevallos, 2003; Lachman and Hamouz, 2005). The advanced selections, CO97216-3P/PW and CO97227-2P/PW, developed by conventional breeding procedures, produced tubers with incomplete expression of tuber flesh pigmentation. This feature permits sampling pigmented and non-pigmented tissues from the same tubers, in essence, isolating the factors responsible for pigmentation from confounding environmental and developmental effects.
In this study, an examination of the transcriptome, coupled with metabolite data from purple pigmented sectors and from non-pigmented sectors of the same tuber, was undertaken to identify genes whose expression correlated with elevated or altered polyphenol composition. Combined with a similar study using eight other conventional cultivars and advanced selections with different pigmentation, our findings expand knowledge of the associated molecular mechanisms and facilitate development of unique potato cultivars with nutritionally superior properties.
Materials and methods
Growth and sampling of plant material
Disease–free seed stock of eight cultivars and advanced selections was used to grow plants in a completely randomized design at the Colorado State University, San Luis Valley Research Center, Center, CO using commercial production practices. Tubers from five biological replicate plants, were washed and prepared for lyophilization within 24 h of harvest.
Forty to 60 grams of freshly cut radial slices (4–6 mm thick) taken only from the centres of three uniform-sized tubers were flash frozen with liquid nitrogen and freeze-dried using a ‘Virtis’ freeze drier. Freeze-dried samples were ground and passed through a 100 mesh screen to achieve uniform particle size.
Total RNA extraction from freeze-dried potato tubers
Approximately 1.5 g of freeze-dried tuber tissue was extracted with 14 ml of hot (80 °C) extraction buffer [50 mM TRIS-HCl (pH 8.0), 50 mM LiCl, 5 mM EDTA, 0.5% SDS, and 50% (v/v) phenol]. Sterile distilled water (10 ml) was added and the samples were vortexed for 2 min. The samples were placed on ice and 16 ml of chloroform:isoamyl alcohol (24:1 v/v) was added and vortexed as before. Following centrifugation at 4 °C at 14 000 g for 20 min, the upper aqueous layer was removed to a fresh, sterile 50 ml Sorval tube, containing an equal volume (16 ml) of 4 M LiCl. The samples were shaken prior to incubation overnight at –80 °C, centrifuged at 4 °C at 14 000 g for 40 min, the supernatant discarded, and the pellet resuspended in 5 ml sterile distilled water. One-tenth volume of 3 M sodium acetate (pH 5.2) and 3 vols of 100% ethanol were added and the samples were incubated at –80 °C for at least 1 h.
The precipitated RNA was pelleted by centrifugation at 4 °C at 14 000 g for 40 min, washed with 10 ml of ice-cold 70% (v/v) ethanol, and centrifuged as in the previous step. The ethanol was removed and the RNA pellet allowed to air-dry prior to resuspension in 500 μl sterile distilled water. RNA samples (100 μg) were purified and genomic DNA contamination was removed using Qiagen RNeasy columns and DNaseI according to the manufacturer's protocol (www.qiagen.com). RNA samples were quantified using a spectrophotometer and quality tested using an RNA 6000 nano chip on an Agilent 2100 Bioanalyser (www.chem.agilent.com). RNA samples were aliquotted in 20 μg (1 μg μl−1) batches and stored at –80 °C.
Microarray processing
Experimental design, array information, and complete datasets are available from ArrayExpress (accession numbers, E-TABM-787 and E-TABM-788 available at: http://www.ebi.ac.uk/microarray-as/aer/?#ae-main[0]). Briefly, two-channel POCI arrays (four in total) were used to compare white and coloured segments of four biological replicates each of line CO97216-3P/PW. A dye-swap was incorporated in a balanced design to minimize any residual dye effects. For the genotype comparison, single-channel (Cy3) POCI arrays (16 in total) allowed comparison of the four biological replicates each of CO97216-3P/PW, Purple Majesty, Rio Grande Russet, and Russet Nugget. Total RNA was labelled by indirect incorporation of fluorescent dyes following cDNA synthesis. Reverse transcription was performed using 10 μg of total RNA in a 45 μl reaction containing 50 ng μl−1 oligo d(T)18, 0.5 mM each of dATP, dCTP, and dGTP, 0.2 mM dTTP, 0.3 mM aa-dUTP, 10 mM DTT, and 400 U Superscript II (Invitrogen) in 1× reaction buffer. Primers and RNA were initially heated to 70 °C for 10 min followed by cooling on ice, and the entire reaction incubated for 16 h at 42 °C. To denature the remaining RNA, 15 μl of 1 M NaOH and 15 μl of 0.5 M EDTA (pH 8.0) were added and incubated for 10 min at 65 °C. The reaction was neutralized with 15 μl of 1 M HCl. Purification of cDNA was performed using MinElute columns as recommended (Qiagen), substituting phosphate wash buffer (4.75 mM K2HPO4, 0.25 mM KH2PO4, 84% EtOH) for PB and phosphate elution buffer (3.8 mM K2HPO4, 0.2 mM KH2PO4) for EB. Cy-dye esters were added to 10 μl of cDNA in a total volume of 13 μl, containing 150 mM sodium carbonate and 1 μl of the appropriate Cy-dye (GE Healthcare) suspended in DMSO (1/10 supplied aliquot), and incubated for 1 h at room temperature in the dark. To the labelled cDNA 750 mM hydroxylamine hydrochloride was added and incubated for a further 30 min in the dark. Labelled targets for each array were combined and diluted with 24 μl sterile water and 500 μl of PB buffer (Qiagen) prior to MiniElute purification and elution with 10 μl of elution buffer. Labelling efficiency was estimated using spectrophotometry.
Hybridization and washing were conducted according to the manufacturer's protocols (Agilent Two-Color Microarray-Based Gene Expression Analysis, version 5.5). Briefly, 20 μl labelled samples were added to 5 μl 10× blocking agent (Agilent 5188-5242) and heat denatured at 98 °C for 3 min then cooled to room temperature. 2× GE hybridization buffer HI-RPM (25 μl) was added and mixed prior to hybridization at 65 °C for 17 h at 10 rpm. Array slides were dismantled in Wash 1 buffer (Agilent, 5188-5327) and washed in Wash 1 buffer for 1 min, then Wash 2 buffer (Agilent, 5188-5327) for 1 min, and centrifuged dry. Hybridized slides were scanned using an Agilent G2505B scanner at resolution of 5 μm at 532 nm (Cy3) and 633 nm (Cy5, for two-channel analysis) wavelengths with extended dynamic range (laser settings at 100% and 10%). Microarray images were imported into Agilent Feature Extraction (v.9.5.3) software and aligned with the appropriate array grid template file (015425_D_F_20061105). Intensity data and QC metrics were extracted using the recommended FE protocol (GE1/GE2-v5_95_Feb07). Entire FE datasets for each array were loaded into GeneSpring (v.7.3) software for further analysis.
Microarray data analysis
Data were normalized using default settings: for two-channel arrays, data were transformed to account for dye-swaps and data from each array were normalized using the Lowess algorithm to minimize differences in dye incorporation efficiency; for single-channel arrays, intensity values were set to a minimum of 5.0, data from each array were normalized to the 50th percentile of all measurements on the array and the signal from each probe was subsequently normalized to the median of its values from white tuber samples. Unreliable data flagged as absent in all replicate samples by the FE software were discarded. Statistical filtering of data for each experiment was performed independently using approaches and algorithms deemed suitable for the two experimental designs used: for the sectional tuber arrays, data were interpreted for analysis on the basis of flesh colour, and volcano plots were used to identify probes with significant differential expression (fold-change >2×, t test P value <0.005); for the genotype comparison, ANOVA (ANalysis Of VAriance, P value <0.05) was used with Bonferroni multiple testing correction.
Quantitative PCR using the Universal Probe Library
Reverse transcription of 10 μg of RNA was performed using Invitrogen Superscript™ II reverse transcriptase (www.invitrogen.com) using random hexamers as primer. cDNA (25 ng) was used as template for real-time PCR using the Universal Probe Library System (https://www.roche-applied-science.com/sis/rtpcr/upl/index.jsp). Reactions were performed in 25 μl containing 1× FastStart TaqMan® Probe Master (supplemented with ROX reference dye). Gene-specific primers and probe were used at a concentration of 0.2 μM and 0.1 μM, respectively. Thermal cycling conditions were: 10 min denaturation at 95 °C followed by 40 cycles (15 s at 94 °C, 60 s at 60 °C). The reactions were repeated in triplicate with independent cDNAs. Relative expression levels were calculated and the primers validated using the ΔΔCt method (Livak, 1997) using data obtained with the elongation factor-1 alpha specific primers as an internal reference control. In the case where relative efficiencies of the target and reference amplicons were not within recommended limits an alternative method for calculating relative quantification was used (Pfaffl, 2001). Universal probe library (UPL) primer and probe sequences were as follows: StEF1alpha_fwd, 5′-CTTGACGCTCTTGACCAGATT-3′, StEF1alpha rev, 5′-GAAGACGGAGGGGTTTGTCT-3′, UPL probe number 117 (5′-AGCCCAAG-3′); StAn-1 fwd, 5′-TGGTGGGCAAATATACTGGAA-3′, StAn-1 rev, 5′-CATGAAGGTAGTGTTCTTTCAGCTT-3′, UPL probe number 133 (5′-GGAGAAGG-3′); MYBTF73 fwd, 5′-CTTGGTTGGTGAGAGGTGGT-3′, MYBTF73 rev, 5′-GGCTGGTGGAATTTCTTGAGT-3′, UPL probe number 11 (5′-GCTGGAAG-3′); StLDOX fwd, 5′-GAAAGTAAGGATTTCATGGGCTATT-3′, StLDOX rev, 5′-AGGGGCTTAAGCATAATCTTCTC-3′, UPL probe number 100 (5′-CTGTGAGC-3′); StDHFR fwd, 5′-AACGCTGTGGAAAGCAGACT-3′, StDHFR rev, 5′-CAGCCTTGAATGGCTTCATC-3′, UPL probe number 86 (5′-GCAGTGGA-3′).
Phenolic extraction and analysis
Tubers from selections CO95172-3RU, CO97216-3P/PW, CO97226-2R/R, and cultivars Colorado Rose, Mountain Rose, Purple Majesty, Rio Grande Russet, and Russet Nugget were washed, sliced, frozen in liquid N2, and freeze-dried. For the sectional experiment, four replicate tubers from clone CO97216-3P/PW were selected from four different plants at the developing tuber stage. Purple and white sectors of tuber flesh were carefully excised from freeze-dried radial sections prior to grinding.
For polyphenol metabolite analysis, 600 mg of dried potato powder was extracted with 10 ml of 80% (v/v) aqueous acetone and rotated at a velocity sufficient to maintain insoluble material in suspension for 2 h in the dark at 4 °C (Thompson et al., 2009). Tubes were centrifuged (5000 g for 15 min at 4 °C) and the supernatants assayed for phenol and anthocyanin content using the Folin and Ciocalteu method and the differential colorimetric method for anthocyanins (McDougall et al., 2005). Triplicate 1 ml subsamples were dried using a Speed-Vac and resuspended in 100 μl of 5% acetonitrile for liquid chromatography–mass spectrometry (LC–MS) analysis.
Samples were analysed on an LCQ-Deca system, comprising Surveyor autosampler, pump, and photodiode array detector (PDAD) and a ThermoFinnigan ion-trap mass spectrometer. The PDAD scanned discrete channels at 280 nm, 365 nm, and 520 nm. The samples were applied to a C18 column (Synergi Hydro C18 with polar endcapping, 4.6×250 mm, Phenomenex Ltd.) and eluted using a linear gradient of 5% acetonitrile (0.1% formic acid) to 40% acetonitrile (0.1% formic acid) over 60 min at a rate of 400 μl min−1. The LCQ-Deca LC–MS was fitted with an ESI (electrospray ionization) interface and analysed the samples in positive and negative ion modes. There were two scan events; full scan analysis followed by data-dependent MS/MS of the most intense ions using collision energies (source voltage) of 45%. The capillary temp was set at 250 °C, with sheath gas at 60 psi and auxiliary gas at 15 psi.
Peaks were identified by comparing their relative retention times, PDA spectra and mass-to-charge ratio (m/z) and MS2 properties with previous reports (Fossen et al., 2003; Eichhorn and Winterhalter, 2005; Stushnoff et al., 2008) or, where available, against standard compounds (chlorogenic acid, tyrosine, and phenylalanine). Components were quantified by their m/z peak areas calculated using the software provided with the instrument and expressed as average ±standard errors. This approach is not strictly quantitative, but gives valid relative comparisons of the components between different samples or treatments. Samples were also run at 50% dilution to check that the relative amounts of the components were consistent.
Extraction and quantification of ascorbic acid
Ascorbic acid was extracted and quantified by HPLC as previously described by Tedone et al. (2004) with some modifications. Powdered lyophilized tuber material was resuspended in 10 vols of ice-cold 5% metaphosphoric acid (MPA), briefly vortexed, and placed on a blood rotator for 30 min in a cold room at 4 °C. Suspended powder was removed by centrifugation (5000 g for 10 min at 1 °C) and the supernatant stored on ice. The pellet was resuspended in a second aliquot of 5% MPA and mixing repeated. Following centrifugation, the two supernatants were combined and filtered through a 0.45 μm filter. For the analysis of reduced ascorbic acid, 20 μl of filtered supernatant was injected onto a IC-Sep ICE-Coregel 64H 300×7.8 mm column (Transgenomic Inc., CA, USA) with an isocratic mobile phase of 8 mM H2SO4 at a flow rate of 0.6 ml min−1. Ascorbic acid was detected and quantified as described by Tedone et al. (2004). Total ascorbic acid was quantified following reduction of dehydroascorbic acid by incubation of supernatant with 5 mM tris(2-carboxyethyl)phosphine hydrochloride at 4 °C for 4 h.
Extraction and quantification of glutathione
Glutathione was extracted and quantified using an enzyme recycling assay as previously described by Queval and Noctor (2007). Freeze-dried powder was extracted as described for ascorbic acid measurements with the exception that the extraction medium consisted of 0.2 M HCl. The final supernatant was diluted with 0.1 vol. 0.2 M sodium phosphate buffer, pH 5.6, and 0.8 vol. 0.2 M NaOH in order to neutralize the extract. Total glutathione was quantified in a microplate following addition of 100 μl 0.2 M sodium phosphate buffer, pH 7.5, containing 10 mM ethylenediaminetetra-acetic acid, 10 μl 10 mM NADPH, 10 μl 12 mM 5,5′-dithiobis(2-nitro-benzoic acid), and 60 μl distilled H2O to 10 μl extract. The reaction was started by the addition of 10 μl glutathione reductase (20 U ml−1 in 0.2 M sodium phosphate buffer, pH 7.5) and OD412 nm was recorded for 5 min. Glutathione quantification was achieved by reference to a standard curve constructed using authentic glutathione. Oxidized glutathione was quantified in the same way following incubation of 200 μl of neutralized extract with 1 μl 2-vinylpyridine for 30 min at room temperature to derivatize free sulphydryl groups.
Correlation matrix
Correlation coefficients were calculated to assess the relationships between gene expression levels and metabolite content and to analyse similarities in gene expression patterns using GenStat 11 (VSN International Limited, Hemel Hempstead, UK). Relationships between gene expression levels and metabolite content were displayed as a heat map. The interrelationships in gene expression levels between the genes differentially expressed in pigmented tubers was displayed graphically.
Results
Metabolite and transcriptome analysis of purple and white tuber sectors
The strategy for this experiment was to compare tuber gene expression profiles in purple and white sectors of tuber flesh found in clone CO97216-3P/PW in order to identify changes in gene expression that correspond with tuber flesh anthocyanin accumulation. Throughout the tuber, there are large areas of purple pigmented tissues and white sectors which both cross all tissue regions and cell types of the tuber (Fig. 2). Thus, this is excellent material for comparing gene expression profiles associated with pigment accumulation. As differentially pigmented segments were excised from the same tuber, gene expression differences due to genotype, environmental factors, and tissue type are minimized.
Fig. 2.
Variegated pigment expression in a section of a CO97216-3P/PW tuber.
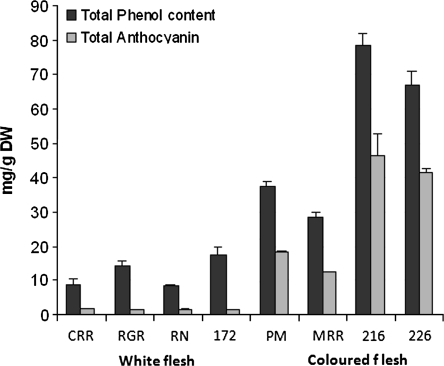
From metabolite analysis, the purple pigmentation in the tubers was due to the accumulation of anthocyanins within the pigmented sections having, on average, 6.5-fold higher total anthocyanin levels than the non-pigmented sections. The elevated levels of anthocyanins contributed to an average 2-fold increase in total phenol levels (results not shown).
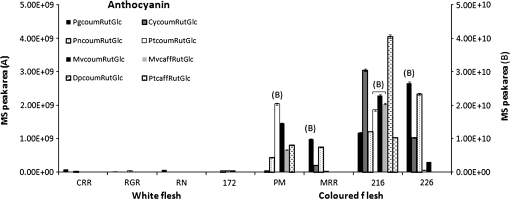
The accumulation of anthocyanins was confirmed by LC-MS analysis (Fig. 3). The pigmented sections accumulated a range of ten detectable anthocyanins, but two components, malvidin-3-p-coumaroylrutinoside-5-glucoside (trivial name negretetin) and petunidin-3-p-coumaroylrutinoside-5-glucoside (trivial name petanin, see Fig. 1; Andersen et al., 1991) which have been identified previously in purple-fleshed potatoes (Stushnoff et al., 2008), predominated. The non-pigmented section also accumulated anthocyanins, but to a much lesser degree. The ratio of accumulation of individual anthocyanins varied between 2.5-fold higher for the minor components to 20-fold higher for the dominant anthocyanins. The anthocyanin content as measured by LC-MS was, on average, 6.7-fold higher in the pigmented tissues, which fits well with the total anthocyanin measurements quantified by differential spectrophotometry. The pigmented tissues also had approximately 2.5-fold higher levels of chlorogenic acid than the non-pigmented sections, as noted previously (Lewis et al., 1998a, b; Table 1), suggesting a general increase in phenolic components. However, the levels of two other major phenolic components, feruloyl and caffeoyl putrescine, were not significantly different (Table 1). There were also no significant differences in ascorbate or glutathione levels or indeed in their oxidation state (data not shown). However, the major glycoalkaloids, solanine and chaconine, were elevated in the purple tissue over the non-pigmented tissues (Table 1).
Fig. 3.
Anthocyanin composition of coloured and uncoloured sections of tubers of CO97216-3P/PW. Values are means of triplicate injections ±standard errors. The MS peak areas for each anthocyanin [using M+H m/z values] were calculated using Xcalibur software. W1–W4 are uncoloured sections and 216P1–216P4 are separate coloured sections. Legend: PgcoumRutGlc, pelargonidin-3-p-coumaroylrutinoside-5-glucoside; CycoumRutGlc, cyanidin-3-p-coumaroylrutinoside-5-glucoside; PncoumRutGlc, peonidin-3-p-coumaroylrutinoside-5-glucoside; PtcoumRutGlc, petunidin-3-p-coumaroylrutinoside-5-glucoside; MvcoumRutGlc, malvidin-3-p-coumaroylrutinoside-5-glucoside; MvcaffRutGlc, malvidin-3-o-caffeoyl rutinoside-5-glucoside; DpcoumRutGlc, delphinidin-3-p-coumaroylrutinoside-5-glucoside; PtcaffRutGlc, petunidin-3-o-caffeoyl rutinoside-5-glucoside; MvRutGlc, malvidin 3-rutinoside-5-glucoside; PtRutGlc, petunidin 3-rutinoside-5-glucoside. Each anthocyanin is appended with the axis it relates to.
Table 1.
Relative amounts of metabolites in a sectional experiment (W denotes the white section, 216P, the purple section, data for four biological replicates are shown, and values are means of triplicate injections ±standard errors)
| Compound | White flesh |
Purple flesh |
||||||
| W1 | W2 | W4 | W5 | 216P1 | 216P2 | 216P4 | 216P5 | |
| Caffeoyl putrescine | 30.9±0.6 | 49.4±0.9 | 34.1±0.7 | 21.1±0.3 | 15.4±0.3 | 46.3±1.4 | 2.6±0.95 | 65.2±0.17 |
| Feruloyl putrescine | 23.7±0.7 | 35.4±1.1 | 28.1±0.6 | 16.6±0.3 | 9.7±0.2 | 19±0.4 | 12.2±0.4 | 41.6±1.1 |
| Tyrosine | 41.7±1.3 | 48.2±1.4 | 36.3±0.7 | 39.1±0.6 | 18.4±0.4 | 39.8±0.8 | 17.5±0.6 | 41.8±1.1 |
| Phenylalanine | 160±3.2 | 243.7±7.3 | 158±3.2 | 154.6±2.5 | 66±1.4 | 114±2.3 | 47.5±1.5 | 124.1±0.3 |
| Solanine | 307.5±8.9 | 385±11.5 | 792.4±14.0 | 643.7±12.2 | 1647.4±47.7 | 832±22.7 | 1824±36.5 | 997±29.2 |
| Chaconine | 136.8±4.3 | 160.4±5.0 | 323.7±10.1 | 288.3±9.0 | 832±26.0 | 383.1±12.0 | 792±24.7 | 486±15.2 |
| Chlorogenic acid | 25.3±0.5 | 26.3±0.5 | 32.5±0.9 | 28.8±0.9 | 79.9±2.3 | 74±1.4 | 81.2±1.4 | 69.9±0.2 |
The MS peak areas for each component were calculated using Xcalibur software.
RNA for microarray analysis was extracted from the same samples that were subjected to metabolite analysis. Microarray data were analysed using standard statistical approaches in GeneSpring software applicable to the design of each independent experiment (see Materials and methods). Using strict selection criteria (2-fold difference in expression level with a P value less than 0.005), 331 genes were up-regulated in purple sectors compared with white, whereas eight genes were down-regulated. Thus, a relatively small proportion of the 42 000 genes on the array were significantly and differentially expressed (see Supplementary Table S1 at JXB online). Contained within this gene list were many of the genes known to encode anthocyanin and flavonoid biosynthetic enzymes, as well as several genes identified from their annotation as having regulatory roles. For example, a gene with similarity to the soybean MYB73 (bf_mxflxxxx_0055g04.t3m.scf_236) was up-regulated 44-fold in the purple sectors compared with the white sectors.
Tuber anthocyanin and transcriptomic comparison of purple and white-fleshed genotypes
Relatively few genes were differentially expressed between the purple and white sectors in tubers of CO97216-3P/PW, and so it was of interest to determine whether differential expression of these genes was a general feature of high anthocyanin tubers. For this reason, metabolite and microarray analysis was carried out on tubers of eight potato cultivars, four pigmented and four non-pigmented genotypes.
The pigmented genotypes Purple Majesty (PM), Mountain Rose (MRR), CO97216-1P/P (216), and CO97226-2R/R (226) had higher anthocyanin and total phenol contents (Fig. 4) than the non-pigmented genotypes. The accumulation of anthocyanins was confirmed by LC–MS analysis. As suggested previously (Stushnoff et al., 2008), the red genotypes, MR and 226, contained mainly pelargonidin-3-p-coumaroylrutinoside-5-glucoside whereas the purple genotypes, PM and 216, contained a wider range of anthocyanins but petunidin- and malvidin-3-p-coumaroylrutinoside-5-glucoside derivatives predominated. It was notable that only 216 contained appreciable amounts of peonidin-3-p-coumaroylrutinoside-5-glucoside (Fig. 5).
Fig. 4.
Total phenol and anthocyanin contents of tubers of different potato varieties. Values are triplicate assays ±standard errors. Legend: PM, Purple Majesty; MRR, Mountain Rose; CRR, Colorado Rose; RGR, Rio Grande Russet; RN, Russet Nugget; 216, CO97216_1P/P; 172, CO95172-3RU; 226, CO97226-3R/R.
Fig. 5.
Anthocyanin composition in tubers of different potato varieties. Values are means of triplicate injections ±standard errors. The MS peak areas for each anthocyanin were calculated using Xcalibur software (legend as for Fig. 3). Anthocyanins which relate to axis B are indicated on the chart.
The higher total phenol content was reflected in the levels of the major polyphenolic component of potato tubers, chlorogenic acid, which was substantially higher in the pigmented genotypes (216, 226, PM, and MRR) than the non-pigmented genotypes (Table 2). However, this trend did not apply to all polyphenolic components as illustrated by the levels of detectable hydroxycinnamic amine derivatives (data not shown). Two pigmented genotypes (216 and 226) had considerably elevated levels of glycoalkaloids compared with other genotypes (Table 2).
Table 2.
Relative amounts of metabolites in sectional experiment (abbreviations are as in the legend to Fig. 4)
| Compound | White flesh |
Coloured flesh |
||||||
| CRR | RGR | RN | 172 | PM | MRR | 216 | 226 | |
| Caffeoyl putrescine | 1±0.0 | 0.7±0.0 | 0.9±0.0 | 1.4±0.0 | 2.4±0.1 | 1.2±0.0 | 3.1±0.1 | 5.1±0.2 |
| Feruloyl putrescine | 1.1±0.0 | 0.6±0.0 | 2±0.0 | 1±0.0 | 0.4±0.0 | 0.6±0.0 | 1.1±0.0 | 3.8±0.1 |
| Tyrosine | 4.4±0.1 | 18.6±0.5 | 3.9±0.1 | 14.4±0.3 | 13±0.3 | 10.2±0.3 | 7.1±0.2 | 12.2±0.2 |
| Phenylalanine | 7.7±0.2 | 22.8±0.5 | 5.5±0.1 | 16.5±0.3 | 8.6±0.2 | 11.2±0.3 | 2.6±0.1 | 7±0.1 |
| Solanine | 11.7±3.1 | 210.7±6.6 | 87.1±2.8 | 61.2±1.4 | 230.3±5.8 | 195±3.7 | 871.5±22.0 | 91.1±29.4 |
| Chaconine | 52.1±1.6 | 164±5.2 | 38.4±1.2 | 32.5±1.0 | 112±3.8 | 138.5±3.0 | 364.7±8.6 | 443.1±8.6 |
| Chlorogenic acid | 22.1±0.7 | 38.9±1.0 | 28.3±0.8 | 32.9±0.7 | 88.3±2.7 | 79.5±1.7 | 133.3±4.3 | 114.2±2.9 |
Values are means of triplicate injections ± standard errors). The MS peak areas for each component were calculated using Xcalibur software.
As with the sectional experiment, aliquots of the same samples used for anthocyanin analysis were also subjected to microarray analysis. Two white-fleshed cultivars (Russet Nugget and Rio Grande Russet) were compared with two purple-fleshed genotypes (Purple Majesty and CO97216-1P/P). In this case, a single-channel arraying approach was adopted allowing complete transcriptional comparisons to be made at both the level of flesh colour and also as independent genotypes. Group-wise interpretation of the data comparing purple genotypes with the white-fleshed cultivars identified a set of 1817 genes that was significantly and differentially expressed (P value of less than 0.05 with strict multiple testing correction). Of these, 1307 genes were significantly up-regulated in purple-fleshed tubers, whereas 510 genes were down-regulated (see Supplementary Table 2 at JXB online). The up-regulated gene list contains many of the genes known to encode anthocyanin biosynthetic enzymes. Comparing this gene list with the gene list from the sectional experiment produced a common ‘refined’ list of only 27 genes, 24 of which were significantly up-regulated and three down-regulated (Table 3). Based on current knowledge, 14 of the up-regulated genes (as identified by microarray annotation) are implicated in anthocyanin biosynthesis or transport to the vacuole. The list contains the potato orthologue of petunia an2 (note that this gene is called an1 in potato; De Jong et al., 2004), an R2R3MYB regulator of anthocyanin production that maps to the same region of the genome as Pf and I (De Jong et al, 2004). Within this refined list, are several genes annotated as encoding the same activity, for example, four genes in the list encode glutathione S-transferase (genes 4, 8, 20, and 24). Genes 4 and 8 share 96% sequence identity and may represent different alleles of the same gene. Genes 1 and 5 share 91% sequence identity and may represent alternatively spliced transcripts of the same gene encoding caffeoyl-CoA-methyltransferase. Genes 2 and 3 encoding leucoanthocyanidin dioxygenase share 44% sequence identity and genes 14 and 15 encoding flavonone 3 β-hydroxylase are 60% identical.
Table 3.
List of common genes that are differentially expressed both in the purple versus white sector experiment and in the comparison of white and purple cultivars experiment
| Gene number | POCI Name | Annotation | Purple/white expression ratio |
| 1 | bf_mxlfxxxx_0017d09.t3m.scf_177 | Caffeoyl-CoA O-methyltransferase | 1305 |
| 2 | MICRO.15988.C1_487 | Leucoanthocyanidin dioxygenase | 1265 |
| 3 | bf_arrayxxx_0035b07.t7m.scf_757 | Leucoanthocyanidin dioxygenase | 835 |
| 4 | MICRO.16821.C1_544 | Glutathione S-transferase | 619 |
| 5 | MICRO.1634.C1_528 | Caffeoyl-CoA O-methyltransferase | 386 |
| 6 | MICRO.5054.C1_619 | Dihydroflavonol 4-reductase | 344 |
| 7 | MICRO.18000.C1_1 | Cytochrome b5 DIF-F | 198 |
| 8 | bf_mxlfxxxx_0036g04.t3m.scf_804 | Glutathione S-transferase | 217 |
| 9 | bf_mxflxxxx_0046h02.t3m.scf_308 | Cytochrome b5 | 83 |
| 10 | bf_mxflxxxx_0055g04.t3m.scf_236 | MYB transcription factor MYB73 | 44 |
| 11 | cSTB29K16TH_413 | Salicylic acid-binding protein 2 | 42 |
| 12 | MICRO.8373.C2_543 | Specific tissue protein 2 | 53 |
| 13 | MICRO.8373.C1_505 | Organ-specific protein S2 | 41 |
| 14 | MICRO.349.C3_1054 | Flavanone 3 β-hydroxylase | 27 |
| 15 | MICRO.349.C2_986 | Flavanone 3 β-hydroxylase | 31 |
| 16 | MICRO.8891.C1_1392 | Lipase, class 3 | 7 |
| 17 | MICRO.16550.C1_898 | Anthocyanin 1 | 5 |
| 18 | cSTS30I16TH_684 | Putative orcinol O-methyltransferase | 5 |
| 19 | bf_ivrootxx_0042e08.t3m.scf_406 | Cytochrome P450 | 5 |
| 20 | bf_arrayxxx_0005c08.t7m.scf_152 | Glutathione S-transferase | 4 |
| 21 | bf_lbchxxxx_0060e12.t3m.scf_471 | Phosphoprotein phosphatase | 44 |
| 22 | MICRO.4660.C1_1394 | Putative disease resistance protein | 4 |
| 23 | MICRO.12653.C1_746 | NA | 3 |
| 24 | cSTB5C21TH_466 | Glutathione S-transferase | 3 |
| 25 | MICRO.4497.C3_1021 | Epoxide Hydrolase I | 0.16 |
| 26 | BF_LBCHXXXX_0035A04_T3M.SCF_21 | NA | 0.14 |
| 27 | BF_TUBSXXXX_0060C09_T3M.SCF_519 | NA | 0.14 |
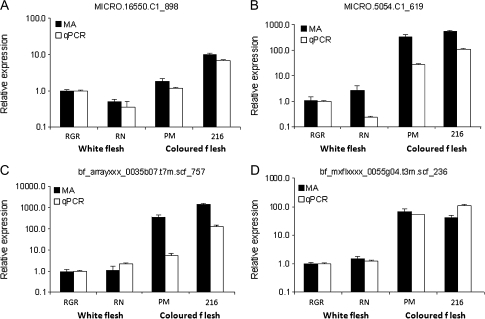
Validation of expression patterns
Independent qRT-PCR analysis was carried out for four of the genes shown to be differentially expressed in the sectional experiment, which also appear in the common gene list of 27 genes. For each, similar patterns of expression were observed whether using the microarray or qRT-PCR analysis (Fig. 6). As the qRT-PCR assay was designed to a region outwith the microarray 60-mer probe sequence, the similarity of patterns would suggest that expression differences were unlikely to be due to polymorphisms affecting one or other of the assays. Previous work has demonstrated the robustness of using the POCI microarray for the analysis of gene expression in different potato genotypes (Ducreux et al., 2008).
Fig. 6.
Comparison of anthocyanin biosynthesis gene expression profiles in potato tubers as determined by qPCR and microarray analysis. Cultivars analysed were Rio Grande Russet (RGR), Russet Nugget (RN), CO97216_1P/P (216), and Purple Majesty (PM). Genes examined were anthocyanin-1 (MICRO.16550.C1_898), dihydroflavonol 4-reductase (MICRO.5054.C1_619), leucoanthocyanidin dioxygenase (bf_arrayxxx_0035b07.t7m.scf_757), and MYB73-like transcription factor (bf_mxflxxxx_0055g04.t3m.scf_236).
Correlation analysis
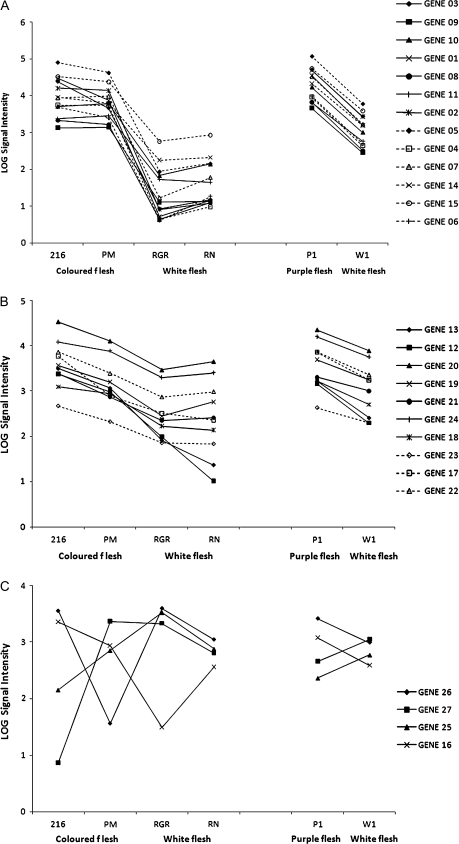
To further our understanding of the potato tuber anthocyanin metabolic map, gene–metabolite correlations were analysed. This analysis focused on the refined list of 27 genes that the microarray experiments demonstrated were associated with enhanced tuber anthocyanin content. Co-expression patterns of gene expression were also carried out by correlation analysis (Fig. 7). As all of these genes were differentially expressed in high anthocyanin tubers, a high degree of correlation would be anticipated between their expression patterns. In fact, three patterns of gene expression could be identified. In Fig. 7A, a cluster of genes that are most strongly up-regulated in pigmented tuber flesh is shown. This group of 13 includes genes encoding several pathway structural proteins but also two genes, either not annotated or not previously associated with anthocyanin biosynthesis such as the MYB73-like transcription factor (bf_mxflxxxx_0055g04.t3m.scf_236). Figure 7B shows a set of 10 genes, also expressed at higher levels in both purple-fleshed varieties and purple sectors than in non-pigmented tissues. In this group, however, the difference in expression level with pigment content is less marked. Nevertheless, this group contains genes known to be associated with tuber anthocyanin biosynthesis such as anthocyanin 1. The group of genes shown in Fig. 7C has either a negative or weak association with tuber pigment level and contains no genes currently known to be involved in anthocyanin biosynthesis.
Fig. 7.
Gene expression patterns for genes that are differentially expressed in both the comparison of pigmented and non-pigmented tubers and from the sector experiment. Gene numbers are as in Table 3.
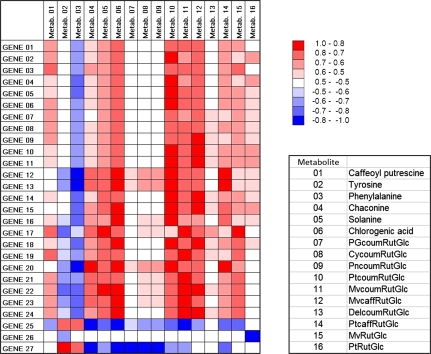
Correlation analysis of the gene expression levels of the refined gene list with the metabolite levels determined by LC-MS was also carried out (Fig. 8). Extremely high levels of correlation were observed between the expression levels of many of the genes and some of the anthocyanin metabolites. Although these positive correlations might be predicted for the known anthocyanin pathway structural and regulatory genes, the correlation with genes of unknown function (e.g. gene 9), further implicates them as having important roles in anthocyanin biosynthesis. Interestingly, negative correlations between expression levels and phenylalanine content were observed and, for some genes, there were extremely strong negative correlations with the levels of anthocyanin metabolites (e.g. gene 25; Fig. 7 MICRO.4497.C3, tentatively identified as encoding epoxide hydrolase I).
Fig. 8.
Heat map revealing correlations between gene expression levels and metabolites in anthocyanin accumulating potato tubers. Gene numbers are as in Table 3 and metabolites are shown in the key. The colour key gives the R value for the correlation calculated for the combined sectional and cultivar datasets.
Discussion
As evidence accumulates about the potential health benefits of consuming a diet rich in anthocyanins, so too does our requirement for understanding the biosynthesis of these compounds in staple foods. Several recent reports have addressed the biosynthesis of anthocyanins in potato tubers. Using a genetic approach, Zhang et al. (2009), identified several QTL for tuber anthocyanin content in a model diploid population. It was demonstrated that a gene designated as StanI (a bHLH gene homologous to petunia anthocyanin I (anI)) co-localized with a QTL for flesh colour on chromosome 9. Marker studies demonstrated that StanI was present in all 21 analysed pigmented tuber tetraploids, but was also present in 21 out of 53 white and yellow-fleshed clones, leading to the conclusion that StanI is necessary but not sufficient for anthocyanin pigment accumulation. Previous studies have also implicated a gene designated Pf in tuber anthocyanin accumulation. This gene maps to chromosome 10 and an orthologue of petunia an2, encoding an R2R3MYB transcription factor co-localizes with Pf. However, the population described in Zhang et al. (2009) was homozygous for Pf and yet 11 out of 214 progeny were unpigmented. Thus like StanI, Pf also appears to be necessary but not sufficient for tuber anthocyanin accumulation.
The complex nature of potato tuber anthocyanin accumulation was further illustrated using a transgenic approach. Rommens et al. (2008) over-expressed an R2R3MYB transcription factor, designated StMtf1 in potato. Although the map location of this gene was not determined, there are sequence similarities with petunia an2 and other potato R2R3MYB genes that map at the Pf locus such as StAn2. When expression of StMtf1 was driven by a strong constitutive promoter, Ubi7, anthocyanins accumulated throughout the potato plant in all tissues apart from the tuber flesh. Using the tuber-flesh specific GBSS promoter, anthocyanins accumulated in tuber phloem and periderm cells but were lacking in other tuber tissues, giving the tubers a mottled appearance. Thus, both genetic and transgenic studies suggest that there are additional unknown factors required for potato tuber anthocyanin accumulation.
This study provides a unique and comprehensive examination of the differences in gene expression that exists between pigmented and non-pigmented tuber tissues, seeking to identify candidate genes that may encode unknown factors associated with anthocyanin biosynthesis. The approach of metabolite profiling and transcriptome co-expression analysis is a powerful method for gene discovery. Already in Arabidopsis, this type of approach has been used to assign function to novel genes involved in flavonol biosynthesis (Yonekura-Sakakibara et al., 2008). The POCI chip is the best microarray platform currently available to analyse global gene expression in potato representing 42 034 unigene sequences (described in Kloosterman et al., 2008), thereby enabling a much more complete analysis of gene expression than has hitherto been achievable. Approximately 35 000 genes are expressed in tomato (Van der Hoeven et al., 2002) and a similar number are probably expressed in potato so it is estimated that this array covers a significant proportion of the entire potato transcriptome. Our approach to obtaining a refined gene list of only 27 genes was based on novel two-stage analysis. Comparing gene expression in white and purple sectors of the same tuber gave rise to a list of 339 genes that were differentially expressed. Comparison of purple-fleshed and white genotypes identified a much larger number of differentially expressed genes (1817) even with very strict statistical filtering, presumably because other traits apart from anthocyanin content were different between the potato types. However, by comparing the gene lists only 27 genes were common to both sets. As expected, a high proportion (14/27) of these genes is known to be associated with anthocyanin biosynthesis. Further circumstantial evidence for the importance of some of these candidate genes was provided by correlation analysis. For example, genes of unknown function (genes 9 and 11) share expression correlation with anthocyanin structural genes and seem good candidates for having roles in anthocyanin biosynthesis. Moreover, the expression pattern of the MYB73-like gene (gene 10, bf_mxflxxxx_0055g04.t3m.scf_236) was also closely correlated with the expression of many of the other known genes of anthocyanin biosynthesis. There are also different degrees of correlation (both positive and negative) between the expression levels of the genes and the accumulation of metabolites determined by LC–MS. For example, the expression levels of gene 27 (annotated as unknown) and gene 25 (annotated as an epoxide hydrolase) are strongly negatively correlated with the accumulation of tuber anthocyanin metabolites. Epoxide hydrolases catalyse the conversion of epoxides to diols and there is considerable knowledge of their structure and function in many systems, including potato (Mowbray et al., 2006). Most plant epoxide hydrolase enzymes are thought to be monomers with a preference for substrates with long lipid-like substituents of the epoxide ring. Their precise biological role is unknown, however, it may be a reasonable working hypothesis that down-regulation of these genes may impact on anthocyanin accumulation. This shows the potential benefit of combining metabolite and gene expression analysis and reveals the power to implicate unsuspected gene targets as having a role in the accumulation of target metabolites. It was also of interest that the purple segments of tubers from clone CO97216-3P/PW contained elevated glycoalkaloid levels compared with the white segments. There are no obvious common metabolites in the anthocyanin and glycoalkaloid pathways and so there is no simple explanation for this relationship. A relationship between chlorogenic acid and glycoalkaloid contents in potato tubers has previously been noted (Dao and Friedman, 1994).
The appearance of the MYB73-like gene in the list of candidate genes is particularly interesting. The sequence encoded by this cDNA appears to be a single domain MYB transcription factor. Although these MYB factors lack a transcription activation domain, it is thought they exert their regulation by interacting with other transcription factors (reviewed in Jin and Martin, 1999). For example, the Arabidopsis gene CAPRICE (CPC) promotes differentiation of hair-forming cells by controlling a negative regulator, GLABRA2 (GL2), which is preferentially expressed in hairless cells (Wada et al., 2002). The N terminus of bHLH proteins interact with CPC and are responsible for GL2 expression. Very recently it has been demonstrated that CPC overexpression represses flavonoid biosynthesis in Arabidopsis by competing with the R2R3-MYB transcription factor PAP1/2, an activator of anthocyanin genes (Zhu et al., 2009). Thus there is a clear precedent for single domain MYB transcription factors such as the MYB73-like gene identified in this study, as having a role in anthocyanin biosynthesis. In contrast to the negative effect of CPC in Arabidopsis, in potato MYB73-like expression appears to have a positive effect. Based on the extremely high correlation of the MYB73-like gene expression with other anthocyanin pathway genes, it is tempting to speculate that it may encode one of the hitherto unknown co-regulators of anthocyanin biosynthesis in potato tubers. Further genetic and transgenic studies using combinations of transgenes will be required to test this hypothesis and also to investigate further the roles of other candidate genes identified in this study.
Supplementary data
Supplementary data are available at JXB online.
Supplementary Table S1. List of genes that are differentially expressed in purple and white sectors of tuber flesh found in clone CO97216-3P/PW including the normalized microarray data, probe identity and best BLAST hit.
Supplementary Table S2. List of genes that are differentially expressed in tubers from two white-fleshed cultivars (Russet Nugget and Rio Grande Russet) compared with two purple-fleshed genotypes (Purple Majesty and CO97216-1P/P).
Acknowledgments
The financial support of the Scottish Government Rural and Environment Research and Analysis Directorate and the Royal Society of Edinburgh is gratefully acknowledged. We also acknowledge additional support for this project by royalty funds collected on new potato cultivar technology by the Colorado Certified Potato Growers Association and disbursed by Colorado State University.
References
- Aksamit-Stachurska A, Korobbczak-Sosna A, Kulma A, Szopa J. Glycosyltransferase efficiently controls phenylpropanoid pathway. BMC Biotechnology. 2008;8:25,. doi: 10.1186/1472-6750-8-25. doi 0.1186/1472-6750/8/2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen OM, Opheim S, Aksnes DW, Froystein NA. Structure of petanin, an acylated anthocyanin isolated from Solanum tuberosum, using homo-and hetero-nuclear two-dimensional nuclear magnetic resonance techniques. Phytochemical Analysis. 1991;2:230–236. [Google Scholar]
- Brown CR, Wrolstadt R, Durst R, Young CP, Clevidence B. Breeding studies in potatoes containing high concentrations of anthocyanins. American Journal of Potato Research. 2003;80:241–249. [Google Scholar]
- Brown CR, Culley D, Bonierbale M, Amoros W. Anthocyanin, carotenoid content, and antioxidant values in Native South American Potato Cultivars. Horticultural Science. 2007;42:1733–1736. [Google Scholar]
- Butelli E, Titta L, Giorgio M, et al. Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors. Nature Biotechnology. 2008;26:1301–1308. doi: 10.1038/nbt.1506. [DOI] [PubMed] [Google Scholar]
- Dao L, Friedman M. Chlorophyll, chlorogenic acid, glycoalkaloid and protease inhibitor content of fresh and green potatoes. Journal of Agricultural and Food Chemistry. 1994;42:633–639. [Google Scholar]
- De Jong H. Inheritance of pigmented tuber flesh in cultivated diploid potatoes. American Potato Journal. 1987;64:337–343. [Google Scholar]
- De Jong WS, Eannetta NT, De Jong DM. Candidate gene analysis of anthocyanin pigmentation loci in the Solanaceae. Theoretical and Applied Genetics. 2004;108:423–432. doi: 10.1007/s00122-003-1455-1. [DOI] [PubMed] [Google Scholar]
- Dodds KS, Long DH. The inheritance of colour in diploid potatoes. I. Types of anthocyanidins and their genetic loci. Journal of Genetics. 1955;53:136–149. [Google Scholar]
- Dodds KS, Long DH. The inheritance of colour in diploid potatoes. II. Three-factor linkage group. Journal of Genetics. 1956;54:27–41. [Google Scholar]
- Ducreux LJM, Morris WL, Prosser IM, Morris JA, Beale MH, Wright F, Shepherd T, Bryan GJ, Hedley PE, Taylor MA. Expression profiling of potato germplasm differentiated in quality traits leads to the identification of candidate flavour and texture genes. Journal of Experimental Botany. 2008;59:4219–4231. doi: 10.1093/jxb/ern264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichhorn S, Winterhalter P. Anthocyanins from pigmented potato (Solanum tuberosum L.) varieties. Food Research International. 2005;38:943–948. [Google Scholar]
- Fossen T, Ovstedal DO, Slimestad R, Andersen OM. Anthocyanins from a Norwegian potato cultivar. Food Chemistry. 2003;81:433–437. [Google Scholar]
- Grotewald E. The genetics and biochemistry of floral pigments. Annual Review of Plant Biology. 2006;57:761–780. doi: 10.1146/annurev.arplant.57.032905.105248. [DOI] [PubMed] [Google Scholar]
- Holton TA, Cornish EC. Genetics and biochemistry of anthocyanin biosynthesis. The Plant Cell. 1995;7:1071–1083. doi: 10.1105/tpc.7.7.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Martin C. Multifunctionality and diversity within the plant MYB-gene family. Plant Molecular Biology. 1999;41:577–585. doi: 10.1023/a:1006319732410. [DOI] [PubMed] [Google Scholar]
- Jung CS, Griffiths HM, De Jong DM, Cheng S, Bodis M, De Jong WS. The potato P locus codes for flavonoid 3′,5′-hydroxylase. Theoretical and Applied Genetics. 2005;110:269–275. doi: 10.1007/s00122-004-1829-z. [DOI] [PubMed] [Google Scholar]
- Kloosterman B, De Koeyer D, Griffiths R, et al. The potato transcriptome: a new look at transcriptional changes during tuber development using the POCI array. Comparative and Functional Genomics. 2008;8:329–340. doi: 10.1007/s10142-008-0083-x. [DOI] [PubMed] [Google Scholar]
- Lachman J, Hamouz K. Red and purple coloured potatoes as a significant antioxidant source in human nutrition: a review. Plant, Soil and Environment. 2005;51:477–482. [Google Scholar]
- Lewis CE, Walker JRL, Lancaster JE, Sutton KH. Determination of anthocyanins, flavonoids and phenolic acids in potatoes. I. Coloured cultivars of Solanum tuberosum L. Journal of the Science of Food and Agriculture. 1998a;77:45–57. [Google Scholar]
- Lewis CE, Walker JRL, Lancaster JE, Sutton KH. Determination of anthocyanins, flavonoids and phenolic acids in potatoes. II. Wild, tuberous Solanum species. Journal of the Science of Food and Agriculture. 1998b;77:58–63. [Google Scholar]
- Livak KJ. Foster City, CA, USA: PE Applied Biosystems; 1997. Relative quantitation of gene expression; pp. 11–15. [Google Scholar]
- Lukaszewicz M, Matysiak-Kata I, Skala J, Fecka I, Cisowski W, Szopa J. Antioxidant capacity manipulation in transgenic potato tuber by changes in phenolic compounds content. Journal of Agricultural and Food Chemistry. 2004;52:1526–1533. doi: 10.1021/jf034482k. [DOI] [PubMed] [Google Scholar]
- Matsumoto H, Nakamura Y, Tachibanaki S, Kawamura S, Hirayama M. Stimulatory effect of cyanidin 3-glycosides on the regeneration of rhodopsin. Journal of Agricultural and Food Chemistry. 2003;51:3560–3563. doi: 10.1021/jf034132y. [DOI] [PubMed] [Google Scholar]
- McDougal GJ, Fyffe S, Dobson P, Stewart D. Anthocyanins from red wine. Their stability under simulated gastrointestinal digestion. Phytochemistry. 2005;66:2540–2548. doi: 10.1016/j.phytochem.2005.09.003. [DOI] [PubMed] [Google Scholar]
- Meiers S, Kemeny M, Weyand U, Gastpar R, von Angerer E, Marko D. The anthocyanidins cyanidin and delphinidin are potent inhibitors of the epidermal growth-factor receptor. Journal of Agricultural and Food Chemistry. 2001;49:958–962. doi: 10.1021/jf0009100. [DOI] [PubMed] [Google Scholar]
- Mowbray SL, Elfstrom LT, Ahlgren KM, Andersson CE, Widersten M. X-ray structure of potato epoxide hydrolase sheds light on substrate specificity in plant enzymes. Protein Science. 2006;15:1628–1637. doi: 10.1110/ps.051792106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda Y, Kneyuki T, Igarashi K, Mora A, Packer L. Antioxidant activity of nasunin, an anthocyanin in eggplant peels. Toxicology. 2000;148:119–123. doi: 10.1016/s0300-483x(00)00202-x. [DOI] [PubMed] [Google Scholar]
- Queval G, Noctor G. A plate reader method for the measurement of NAD, NADP, glutathione, and ascorbate in tissue extracts: application to redox profiling during Arabidopsis rosette development. Analytical Biochemistry. 2007;363:58–69. doi: 10.1016/j.ab.2007.01.005. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Research. 2001;29 doi: 10.1093/nar/29.9.e45. e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddivari L, Vanamala J, Chintharlapalli S, Safe SH, Miller JC. Anthocyanin fraction from potato extracts is cytotoxic to prostate cancer cells through activation of caspase-dependent and caspase-independent pathways. Carcinogenesis. 2007;28:2227–2235. doi: 10.1093/carcin/bgm117. [DOI] [PubMed] [Google Scholar]
- Renaud S, de Lorgeril M. Wine, alcohol, platelets, and the French paradox for coronary heart disease. Lancet. 1992;339:1523–1526. doi: 10.1016/0140-6736(92)91277-f. [DOI] [PubMed] [Google Scholar]
- Reyes LF, Cisneros-Zevallos L. Wounding stress increases the phenolic content and antioxidant capacity of purple-flesh potatoes Solanum tuberosum L. Journal of Agricultural and Food Chemistry. 2003;51:5296–5300. doi: 10.1021/jf034213u. [DOI] [PubMed] [Google Scholar]
- Rommens CM, Richael CM, Yan H, Navarre DA, Ye J, Krucker M, Swords K. Engineered native pathways for high kaempferol and caffeoylquinate production in potato. Plant Biotechnology Journal. 2008;6:870–886. doi: 10.1111/j.1467-7652.2008.00362.x. [DOI] [PubMed] [Google Scholar]
- Seeram NP, Adams LS, Hardy ML, Heber D. Total cranberry extract versus its phytochemical constituents: antiproliferative and synergistic effects against human tumor cell lines. Journal of Agricultural and Food Chemistry. 2004;52:2512–2517. doi: 10.1021/jf0352778. [DOI] [PubMed] [Google Scholar]
- Shin WH, Park SJ, Kim EJ. Protective effect of anthocyanins in middle cerebral artery occlusion and reperfusion model of cerebral ischemia in rats. Life Sciences. 2006;79:130–137. doi: 10.1016/j.lfs.2005.12.033. [DOI] [PubMed] [Google Scholar]
- Stushnoff C, Holm D, Thompson MD, Jiang W, Thompson MD, Joyce NI, Wilson P. Antioxidant properties of cultivars and selections from the Colorado Potato Breeding Program. American Journal of Potato Research. 2008;85:267–276. [Google Scholar]
- Tedone L, Hancock RD, Alberino S, Haupt S, Viola R. Long-distance transport of L-ascorbic acid in potato. BMC Plant Biology. 2004;4:16. doi: 10.1186/1471-2229-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson MD, Thompson HJ, McGinley JN, Neil ES, Rush DK, Holm DG, Stushnoff C. Functional food characteristics of potato cultivars (Solanum tuberosum L.): phytochemical composition and inhibition of 1-methyl-1-nitrosourea induced breast cancer in rats. Journal of Food Composition and Analysis. 2009;22:571–576. [Google Scholar]
- Tsuda T, Horio F, Osawa T. The role of anthocyanins as an antioxidant under oxidative stress in rats. Biofactors. 2000;13:133–139. doi: 10.1002/biof.5520130122. [DOI] [PubMed] [Google Scholar]
- Van der Hoeven R, Ronning C, Giovannoni J, Martin G, Tanksley S. Deductions about the number, organization, and evolution of genes in the tomato genome based on analysis of a large expressed sequence tag collection and selective genomic sequencing. The Plant Cell. 2002;14:1441–1456. doi: 10.1105/tpc.010478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T, Kurata T, Tominaga R, Koshino-Kimura Y, Tachibana T, Goto K, Marks MD, Shimura Y, Okada K. Role of a positive regulator of hair development, CAPRICE, in Arabidopsis root epidermal cell differentiation. Development. 2002;129:5409–5419. doi: 10.1242/dev.00111. [DOI] [PubMed] [Google Scholar]
- Yonekura-Sakakibara K, Tohge T, Matsuda F, Nakabayashi R, Takayama H, Niida R, Watanabe-Takahashi A, Inoue E, Saito K. Comprehensive flavonol profiling and transcriptome coexpression analysis leading to decoding gene–metabolite correlations in Arabidopsis. The Plant Cell. 2008;20:2160–2176. doi: 10.1105/tpc.108.058040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Jung CS, De Jong WS. Genetic analysis of pigmented tuber flesh in potato. Theoretical and Applied Genetics. 2009;119:143–150. doi: 10.1007/s00122-009-1024-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu HF, Fitzsimmons K, Khandelwal A, Kranz RG. CPC, a single-repeat R3 MYB, is a negative regulator of anthocyanin biosynthesis in Arabidopsis. Molecular Plant. 2009;2:790–802. doi: 10.1093/mp/ssp030. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.