Abstract
Embryogenesis is a critical stage during the plant life cycle in which a unicellular zygote develops into a multicellular organism. Co-ordinated gene expression is thus necessary for proper embryo development. Polycomb and Trithorax group genes are members of evolutionarily conserved machinery that maintains the correct expression patterns of key developmental regulators by repressing and activating gene transcription. TRAUCO (TRO), a gene homologous to the Trithorax group of genes that can functionally complement a BRE2P yeast mutant, has been identified in Arabidopsis thaliana. It is demonstrated that TRO is a nuclear gene product expressed during embryogenesis, and loss of TRO function leads to impaired early embryo development. Embryos that arrested at the globular stage in the tro-1 mutant allele were fully rescued by a TRO expression clone, a demonstration that the tro-1 mutation is a true loss-of-function in TRO. Our data have established that TRO is the first trithorax-group gene homologue in plants that is required for early embryogenesis.
Keywords: Arabidopsis, early embryogenesis, nuclear protein, plant embryogenesis, seed development, trithorax
Introduction
Embryo development in higher plants is a highly orchestrated process of cell division, differentiation, growth, and pattern formation (Bai et al., 2000; Willemsen and Scheres, 2004; Jenick et al., 2007). During this stage, a large number of genes must be expressed to ensure that the single-celled zygote develops into an organized multicellular structure capable of producing a viable seedling (Meinke, 1995). In Arabidopsis thaliana, the isolation of embryo-defective mutants has been a valuable approach for the identification of genes that are essential for development (Tzafrir et al., 2004; Meinke et al., 2008). A minimal set of ∼750 non-redundant Arabidopsis genes is thought to co-ordinate these developmental events in the embryo (McElver et al., 2001; Tzafrir et al., 2003). Therefore, it is of fundamental importance to understand the molecular mechanisms regulating such a wealth of genes (Jurgens, 2001; Willemsen and Scheres, 2004).
A precisely co-ordinated developmental programme is a prerequisite for the successful life cycle of multicellular organisms. Genetic studies have implicated factors that are involved in chromatin-mediated regulation in the control of a plethora of developmental processes (Reyes et al., 2002; Guyomarc'h et al., 2005; Kohler and Makarevich, 2006). Genes belonging to the Polycomb group (PcG) and trithorax group (trxG) protein complexes feature perhaps the best known evolutionarily ancient molecular machinery that prevents changes in development by maintaining transcription patterns (Ringrose and Paro, 2004). PcG and trxG genes, respectively, repress and activate transcriptional states of several loci including homeotic and cell cycle genes. Both these functional groups encode components of multimeric protein complexes that control chromatin accessibility by means of histone modifications such as methylation and acetylation (Ringrose and Paro, 2004).
Although both the PcG and trxG genes were initially described in Drosophila melanogaster and metazoans, they are found evolutionarily conserved in plants (Grossniklaus et al., 1998; Springer et al., 2002; Thakur et al., 2003). Plant PcG proteins have been found to control aspects of seed development, floral induction, and floral organogenesis (Köhler and Grossniklaus, 2002; Pien and Grossniklaus, 2007). In Arabidopsis, MEDEA (MEA) and FERTILIZATION INDEPENDENT SEED 2 (FIS2) encode one of the Polycomb group (PcG) protein homologues of Enhancer of zeste (E(Z)) and Suppressor of zeste 12 (SU(Z)12) in Drosophila, respectively (Grossniklaus et al., 1998; Luo et al., 1999). MEA and FIS2 proteins interact with FERTILIZATION INDEPENDENT ENDOSPERM (FIE), the Arabidopsis homologue of Extra Sex Combs (ESC; Spillane et al., 2000). These three proteins and a P55 homologue, MULTICOPY SUPPRESSOR OF IRA 1 (MSI1) are components of the 600 kDa Polycomb Repressive Complex 2 (PRC2; Pien and Grossniklaus, 2007). These plant PRC2 genes belong to the FERTILIZATION INDEPENDENT SEED (FIS) class, where mutations cause autonomous endosperm development in the absence of fertilization (Pien and Grossniklaus, 2007). Abnormal cell proliferation during seed development, and embryogenesis in particular, is a consistent phenotype observed in fis class mutants, suggesting an essential role for several PRC2 components. By contrast, little is known about the functions of trxG genes in Arabidopsis and other plant model systems. Mutations in an Arabidopsis homologue of a trxG gene, ATX1, cause pleiotropic developmental defects in the formation, placement, and identity of flower organs, but do not appear to play a role in plant embryogenesis (Alvarez-Venegas et al., 2003). Biochemical and genetic evidence defined ATX1 as a functional homologue of the animal TRITHORAX and the remaining plant protein complexes have yet to be identified.
In a cDNA-AFLP screen for genes specifically expressed during early embryogenesis in a conifer (Pinus radiata), a transcript-derived fragment homologous to the Drosophila ASH2 gene (Aquea and Arce-Johnson, 2008) was identified. ASH2 (absent, small, or homeotic discs 2) is a member of the trxG, discovered in a screen for late larval/early pupal lethal mutants that had imaginal disc abnormalities in Drosophila (Adamson and Shearn, 1996). Mutations in ASH2 cause homeotic transformations typical for mutations in trxG, in addition to a variety of pattern formation defects. The ASH2 genes are highly conserved among different species, including humans and yeast. ASH2L is implicated in hematopoiesis, leukemia, and has been described as a novel oncoprotein in humans (Wang et al., 2001; Lüscher-Firzlaff et al., 2008). In yeast, BRE2P is the homologue of ASH2 which belongs to a seven-member complex (Nagy et al., 2002). BRE2P is required for methylation of histone H3 at lysine residue 4, which is normally associated with transcriptional activation.
Recently, the molecular characterization of an ASH2 homologue gene from Pinus radiata (Aquea et al., 2009) was reported, but functional characterization of ASH2 is yet to be described in plant model systems. In this work, the functional analysis is reported of a trxG homologue in Arabidopsis thaliana during embryogenesis and sporophytic development. The Arabidopsis ASH2 homologue was named TRAUCO (TRO) in honour of the fertility mythology from the Chiloe island in southern Chile (Perez, 2003). TRO is a nuclear protein in agreement with the proposed role for TRO homologues in chromatin remodelling in other systems. Mutation in TRO perturbs embryo development, causing seed abortion. To our knowledge, this is the first report of a trxG gene homologue to be involved in plant embryo development in plants.
Materials and methods
Plant materials and growth conditions
Both the wild type and T-DNA insertion lines were in the Arabidopsis thaliana Col-0 background. Plants were grown on soil (ED73 soil; Einheitserde, Schopfheim, Germany) in a growth room with 70% relative humidity and a day–night cycle of 16/8 h light/dark at 21/18 °C.
Yeast complementation and formamide assay
The strain of yeast (Saccharomyces cerevisiae) used in this study was Y01570. This strain was constructed from BY4741 (genotype MATa; his3D1; leu2D0; met15D0; ura3D0) by insertional mutagenesis of the gene BRE2P (YLR015w). The yeast competent cells Y01570 (a Bre2pΔ strain) were transformed with a cDNA of TRO cloned in an expression plasmid pYES-DEST52 (Invitrogen™). The empty vector was used as a control. To test for the formamide sensitivy assay, yeast complemented cells were grown to the stationary phase in liquid synthetic minimum medium (SD; Sherman, 1991) supplemented with 2% raffinose, and with 20 mg l−1 Hist, 30 mg l−1 Leu, and 20 mg l−1 Met. Then the cell density was adjusted to OD600 nm=1.0 and these density-adjusted cultures were diluted to 1/10, 1/100, 1/1000, and 1/10 000 with SD liquid medium. 5 μl of each dilution was spotted on SD solid medium with 2% galactose instead of raffinose (for the induction of the GAL1 promoter), and with or without formamide (2.5%). The plates were then incubated at 30 °C for 10 d.
Expression analyses in vivo
For in planta TRO expression analysis, a 1684 bp fragment upstream of the translation initiation codon was PCR-amplified using primers ProgwF (5′-CTGTTTAAACCTAGGTCTCTGTCATGTATAAGCTATGA-3′; PmeI/AvrII is underlined) and ProgwR (5′-GTACTAGTATGCAGCCAGTTTGTGGCACT-3′; SpecI is underlined). The PCR product was subcloned into pDONR207, and then cloned into pMDC163 (Curtis and Grossniklaus, 2003), by GATEWAY recombination cloning, according to the manufacturer's instructions (Invitrogen). The destination construct was used to generate pTRO:GUS-expressing plants by the floral dip method (Clough and Bent, 1998). Histochemical glucuronidase (GUS) activity staining was performed using 1 mg ml−1 5 bromo-4-chloro-3-indolyl-β-D-glucuronic acid in 100 mM sodium phosphate buffer (pH 7.2, 0.1% Triton X-100, and 2 mM potassium ferricyanide, and 2 mM potassium ferrocyanide) at 37 °C for 1–12 h and cleared in 70% ethanol. To visualize embryos, seeds were cleared in chloral hydrate:H2O:glycerol (8:2:1 by vol.) overnight.
mRNA in situ hybridization
Fixation, tissue embedding, and sectioning of Arabidopsis siliques, and mRNA in situ hybridization experiments were performed as described by Vielle-Calzada et al. (1999). To synthesize the TRO probe, primers 5′-ATGGAGTCTCTTCAATCAAATTC-3′ and 5′-CTGCAGTAAGTCTATCTTC-3′ spanning the region from 12 to 891 (879 bp) of the TRO sequence were used to PCR-amplify the appropriate fragment from Arabidopsis genomic DNA. PCR conditions were as follows: denaturation at 94 °C for 5 min, then 30 cycles of 30 s at 94 °C, 30 s at 55 °C, and 40 s at 72 °C, followed by 5 min at 72 °C. The PCR fragment was inserted into the pDrive Cloning Vector (Qiagen) in both sense and antisense orientations. Digoxygenin-UTP labelled riboprobes were generated by run-off transcription using T7 RNA polymerases according to the manufacturer's protocol (Roche Diagnostics). Hybridization was performed on 10 μm Paraplast Plus (Sigma-Aldrich) sections.
Protein subcellular localization
Full-length TRO (1818 bp) genomic DNA was PCR-amplified using primers TRO5′ (5′-ATGGAGTCTCTTCAATCAAATTC-3′) and TRO3′ (5′-ACTCTTCATATCCTCAGAACCAT-3′). PCR conditions were as follows: denaturation at 94 °C for 5 min, then 35 cycles of 30 s at 94 °C, 30 s at 51 °C, and 120 s at 72 °C, followed by 10 min at 72 °C. The PCR product was subcloned into pDONR207 using the BP cloning technique (Invitrogen) and then cloned into pMDC84 to generate 35S::TRO-GFP (Curtis and Grossniklaus, 2003). This construct was transfected into onion cells by particle bombardment and GFP fluorescence analysis was performed.
Genotyping tro-1 plants by PCR
DNA of the Arabidopsis T-DNA lines (SAIL_851_H01) was extracted and screened for the T-DNA insertions at the TRO locus by PCR. TRO forward and reverse primers (TRO-L1: 5′-CCATTGGAATCGTCCGGTATA-3′ and TRO-R1: 5′-CCCTGACATACACCATTTTTGAAG-3′), which flank the T-DNA insertion, were designed for PCR screening in combination with the T-DNA left border-specific primer (5′-TCATAACCAATCTCGATACAC-3′). PCR fragments were confirmed by sequencing. PCR conditions were as follows: denaturation at 94 °C for 5 min, then 30 cycles of 30 s at 94 °C, 30 s at 55 °C, and 90 s at 72 °C, followed by 5 min at 72 °C.
Segregation analysis
Seeds of tro-1 plants were surface-sterilized and plated onto Murashige and Skoog medium (MS salts, 1% sucrose, and 0.9% agar, pH 5.7) supplemented with BASTA (10 mg l−1) (AccuStandar®). After 2 d at 4 °C, the seeds were grown under 16/8 h light/dark cycles at 22 °C. The BASTA phenotype (resistant or sensitive) was scored on plates after 2 weeks.
Histological analysis
For phenotypic characterization, seeds were cleared with chloral hydrate following the protocol described by Yadegari et al. (1994). Specimens were observed using a Leica DMR microscope (Leica Microsystems) under DIC optics.
Complementation of Arabidopsis tro-1
For complementation, a construct was designed that expressed TRO under the control of its native promoter. Using 35S::TRO-GFP, the double 35S promoter was exchanged for the TRO promoter (1684 bp) using PmeI and SpecI restriction sites. The tro-1/+ heterozygous plants were transformed by the floral dip method (Clough and Bent, 1998) using Agrobacterium tumefaciens strain GV3101. Transformed plants were evaluated for their seed abortion phenotype.
Results
The Arabidopsis thaliana TRAUCO gene is a member of the conserved trithorax gene family
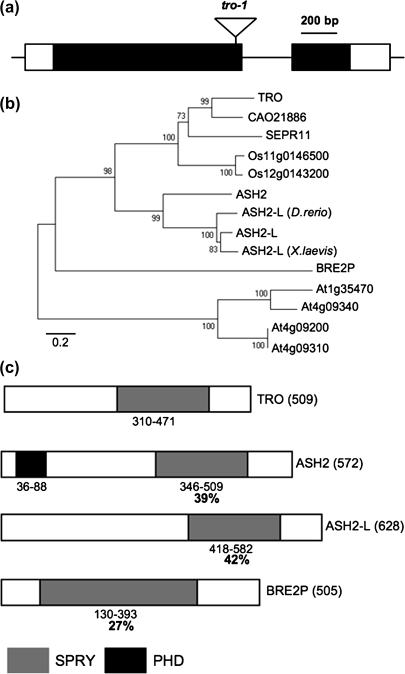
Arabidopsis thaliana TRO (At1g51450) is a single copy gene with two exons encoding a 55 kDa protein with 509 amino acids (Fig. 1a). The protein has a putative nuclear localization signal and a SPla/RYanodine receptor (SPRY) domain, involved in protein–protein interactions (Wang et al., 2002; Zhai et al., 2004). An evolutionary tree was calculated on the basis of the alignment of the complete protein sequences of ASH2 homologues, using other Arabidopsis SPRY-domain proteins as an outgroup (Fig. 1b). TRO is grouped with the other ASH2 homologous proteins and not with Arabidopsis SPRY domain-containing proteins. Figure 1c illustrates the predicted domain structure of TRO and homologous proteins in yeast, Drosophila, and human. The C-terminal portion of all four proteins harbours the SPRY domain characteristic of ASH2 homologous proteins. In this region, the predicted protein sequence of TRO is 39% identical and 55% similar to Drosophila ASH2, 42% identical and 59% similar to Human ASH2-L, and 27% identical and 41% similar to yeast BRE2P. In general, the N-termini of these proteins are poorly conserved and Drosophila ASH2 is the only protein possessing a PHD finger domain in this region. These analyses indicated that TRO is homologous to the trithorax-group proteins.
Fig. 1.
The Arabidopsis thaliana TRO is a Trithorax group homologous gene. (a) A schematic representation of the exon–intron structure of the genomic TRO gene with the location of the T-DNA insertion. (b) Phylogenetic tree analysis of TRO with TrxG genes from other organisms and with Arabidopsis SPRY-domain containing proteins. The tree was constructed by the Neighbor–Joining method with MEGA program 3.0. Branch numbers represent the percentage of bootstrap values in 1000 sampling replicates. The protein accession numbers are: TRO (NP175556), SEPR11 (ACE95183), ASH2 (AAC47328), ASH2-L (Q9UBL3), D. rerio ASH2-L (NP001103575), X. laevis ASH2-L (AAI55932), BRE2P (NP013115), At4G09200 (NP192659), At4G09310 (NP192669), and At4G09340 (NP192672). Sequences of the grape (CAO21886) and rice (Os11g0146500 and Os12g0143200) homologous genes were obtained from NCBI GeneBank. (c) Organization and comparison of TRO with homologous proteins. Locations of conserved domains with significant homologies are indicated by boxes: SPRY (light gray) and PHD (black). The percentages indicate the degree of amino acid similarity with respect to TRO for each conserved domain. Numbers below each protein indicate the initial and final amino acid of the corresponding domains.
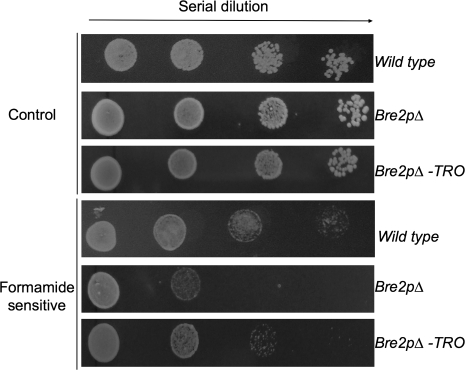
To examine the activity of TRO, the gene was expressed in the yeast strain Bre2pΔ (Nagy et al., 2002). Bre2pΔ cells lack the functional homologue of ASH2 and showed formamide-sensitive phenotypes (Nagy et al., 2002). Cells in the stationary phase cultured in liquid medium were exposed to 2.5% formamide in plates and the cell viability was determined. Based on the colony growth, it was confirmed that Bre2pΔ cells showed a clear reduction in viability in the presence of formamide (Fig. 2) confirming the published data of Nagy et al. (2002). By contrast, when the Arabidopsis TRO cDNA was expressed in Bre2pΔ yeast cells, it was observed that the transformed cells were able to grow more efficiently in formamide-containing medium (Fig. 2). These results indicated that TRO is a functional Arabidopsis homologue of the trithorax-group member BRE2P of yeast. Therefore, it is possible that, similar to BRE2P, TRO is a member of a protein complex involved in histone methylation.
Fig. 2.
Expression of TRO provides formamide tolerance to a bre2p yeast strain. Yeast cells were grown to an OD600 nm value of 1.0, and then 5 μl of 10-fold serial dilutions (left to right in each panel) were spotted onto an solid media containing 0 (control) or 2.5% formamide according to Nagy et al. (2002). Growth was recorded after 10 d of culture. Yeast cells transformed with empty vector used as control (Bre2pΔ).
TRO is expressed in vegetative and reproductive organs
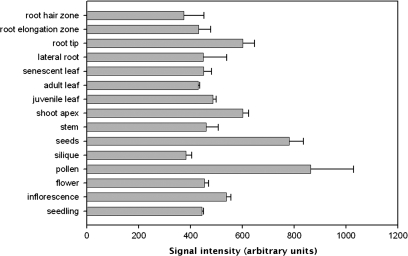
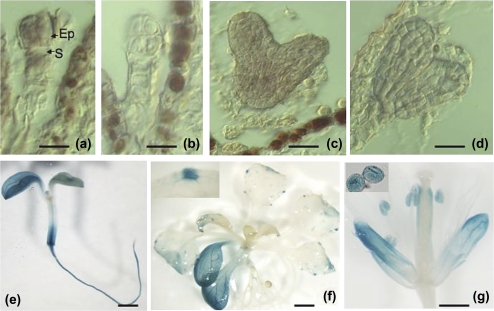
Existing data deposited in large expression databases suggested that TRO is expressed ubiquitously at different levels in all plant tissues; however, the relative transcript levels are most abundant in pollen and seeds (Fig. 3). To investigate the spatial expression pattern of TRO further, a 1684 bp portion of the TRO promoter was fused with the β-glucuronidase (GUS) reporter gene, and endogenous expression of the TRO reporter was monitored in transgenic Arabidopsis plants (pTRO:GUS). Fourteen independent transgenic lines expressing the GUS gene under the control of the TRO promoter were analysed. GUS activity was detected during embryo development (data not shown). To validate the GUS expression pattern during early embryogenesis, mRNA in situ hybridization was performed. Specific mRNA signals were detected at two stages during embryo development. Figure 4 shows that a prominent hybridization signal was observed in the embryo proper and in suspensor cells at the globular stage (Fig. 4a). Strong signals were also detected at the heart stage (Fig. 4c). No signal above background was observed in the control experiments in which the sense RNA probes were used (Fig. 4b, d). In 5-d-old seedlings, the TRO promoter was active in cotyledons and roots (Fig. 4e). In vegetative tissues of adult plants, GUS expressed under the control of the TRO promoter is associated exclusively with leaf hydathodes, structures involved in the active secretion of solutes (Fig. 4f). In reproductive tissues, GUS activity is detected only in sepals, anthers, and pollen grains (Fig. 4g). Together, these data indicate that TRO is primarily expressed in embryos, and in several tissues throughout plant development. In summary, it is confirmed that TRO is expressed in several stages of plant development, and specifically during early embryogenesis in Arabidopsis.
Fig. 3.
Gene expression of TRO in various plant tissues. Data used for the analysis were retrieved from GENEVESTIGATOR (Zimmermann et al., 2004). The values shown are means +SD.
Fig. 4.
Temporal and spatial patterns of TRO gene expression. (a–d) RNA in situ hybridization confirmed the expression of TRO during early embryogenesis. (a) Nomarski micrographs showing a strong signal in the embryo proper (EP) and suspensor (S) at the eight-cell stage. (b) Hybridization with a sense probe at the eight-cell stage. (c) Nomarski micrographs showing a strong signal at the heart stage. (d) Hybridization with a sense probe at the heart stage. (e–g) Histochemical assays of the expression pattern of GUS under the control of the TRO promoter in PTRO–GUS transgenic Arabidopsis. (e) GUS staining of a 7-d-old seedling showing TRO expression in the cotyledons, shoot base and root. (f) GUS staining of a 14-d-old seedling showing TRO expression in cotyledons and hydathodes in a rosette leaf. (g) GUS staining in adult flowers showing TRO expression in pollen grains (inset), anthers, and sepals. Scale bar: a–d, 20 μm; e–f, 1 mm; g, 0.5 mm.
TRO is a nuclear protein
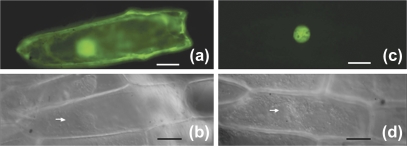
The presence of a putative N-terminal nuclear localization signal suggests that TRO is a nuclear protein. To determine the subcellular localization of TRO, we examined reporter expression of a C-terminal translational fusion of TRO with the GREEN FLUORESCENT PROTEIN (GFP), driven by the constitutive 35S promoter of Cauliflower Mosaic Virus (p35S::TRO-GFP), in onion cells by the transient transformation assay. The construct p35S::GFP served as a control. In onion cells expressing p35S::GFP, the GFP protein was localized throughout the cell, visualized as florescent signals within the cytoplasm, nucleus, and nucleolus (Fig. 5a, b). By contrast, the fluorescence of TRO-GFP colocalized exclusively within the nucleus (Fig. 5c, d). This result indicated that TRO is indeed a nuclear protein.
Fig. 5.
Subcellular localization of TRO. (a) Epifluorescence micrograph of an onion cell transiently expressing GFP (35S::GFP). (b) Nomarski micrographs showing the same cell in (a). (c) Epifluorescence micrograph of an onion cell transiently expressing GFP fused to TRO (35S::TRO-GFP). (d) Nomarski micrographs showing the same cell as (c). The white arrow highlights the nucleus. (Scale bar: 20 μm.
TRO is required for early embryogenesis
In order to determine the function of TRO in Arabidopsis development, a T-DNA insertion mutant was obtained from the Arabidopsis Biological Resource Center (ABRC) designated tro-1 for phenotypic analysis. This allele carried an insertion at the 3′-end of exon 1 (Fig. 1a). This insertion potentially interferes with the nucleotide sequence coding for the SPRY domain. The location of the T-DNA insertion was confirmed by PCR genotyping and, subsequently, by sequencing. The sequences of the PCR products demonstrated that the T-DNA left border was present at both ends of the insertion, suggesting the presence of at least two tandem full or partial T-DNA insertions. More importantly, sequence analysis established that no major deletions or chromosomal rearrangements took place during the insertional event in the TRO locus. Only a minor 24 bp deletion occurred in TRO at the insertion site (see Supplementary Fig. 1 at JXB online). Genotyping by PCR of T3 plants led to the identification of several heterozygous tro-1/+ mutant plants. However, attempts to identify homozygous tro-1 mutant plants were unsuccessful. Moreover, no homozygous mutant seedlings were obtained in the progeny of selfed heterozygous tro-1/+ plants, suggesting that embryo and/or gametophyte development are altered, and that TRO is an essential gene. The T-DNA insertion in tro-1 harbours a gene conferring resistance to BASTA. This feature facilitated segregation analysis of the mutant allele, which revealed that BASTA-resistant and BASTA-sensitive progeny segregated in a 1.7:1 ratio in selfed heterozygous plants (Table 1). This segregation ratio is significantly different from the expected 3:1 Mendelian segregation for a dominant resistant phenotype and from the expected 1:1 ratio for a fully penetrant mutation in either the male or female gametophyte. The value obtained is indicative of zygotic lethality with a possible weak gametophytic defect affecting one or both male and female gametes. To determine whether the T-DNA insertion was transmitted abnormally through the male and/or female gametophytes, reciprocal crosses with wild-type plants were performed. Transmission efficiency of the BASTA resistance marker was found to be normal through the female gametophyte, but was slightly reduced when pollen was provided from heterozygous tro-1/+ plants (Table 1). Taken together, these data suggest that the TRO is necessary for embryo development, and that TRO might also function paternally during male gametophytic or post-gametophytic development.
Table 1.
Segregation ratio and transmission efficiency of the gene conferring BASTA resistance in tro-1
| Cross (female×male) | BASTAra | BASTAsa | Total | Rate R:Sb | P-valuec | TE (%)d |
| tro-1/+ selfed | 671 | 393 | 1064 | 1.70:1 | <0.05* | NAe |
| tro-1/+×wt | 48 | 46 | 94 | 1.04:1 | 0.6 | 104 |
| wt×tro-1/+ | 70 | 96 | 166 | 0.72:1 | <0.05* | 72 |
BASTAr, BASTA-resistant seedlings; BASTAs, BASTA-sensitive seedlings.
χ2 tests were performed to compare the BASTA-resistant and BASTA-sensitive phenotype between wt , tro-1, and reciprocal crosses.
P-value, based on an expected 2:1 or 1:1 BASTAr/BASTAs segregation ratio, respectively. * Indicate significant differences.
Transmission efficiencies (TE) = (BASTAr/BASTAs)×100%.
NA, not applicable.
Pollen of wild-type and tro-1/+ plants was analysed for viability by the staining method described by Alexander (1969). In this assay, the cytoplasm of mature viable pollen grains stains a deep purple and is surrounded by a thin green stain of the external exine layer (see Supplementary Fig. 2 at JXB online). No differences were observed in the staining pattern of pollen produced by heterozygous tro-1/+ plants, which produce both wild-type and tro-1 pollen grains. These results suggest that a mutation in TRO does not impair pollen development, rather, a tro-mediated paternal defect may occur later, possibly during pollen germination, pollen tube growth or fertilization.
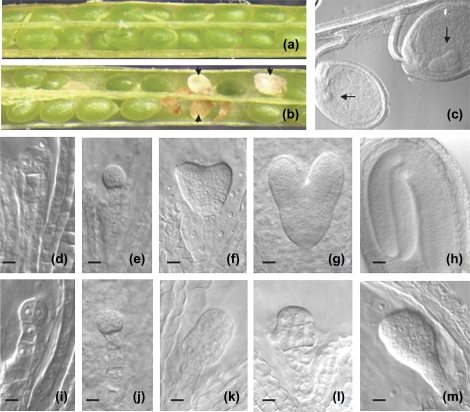
Examination of the siliques revealed that tro-1/+ mutant plants contained not only the green seeds found in wild-type siliques, but also some white and brown aborted seeds (Fig. 6a, b), a feature that indicates embryo lethality. The aborted seeds have a random distribution inside the silique (data not shown). Indeed, 22.8% of the embryos aborted in tro-1/+, suggesting that homozygous tro-1 mutants are lethal (Table 2). The aborted seeds always co-segregated with the T-DNA insertion, as observed by genotyping of plants with aborted seed (data not shown). The development of wild-type and mutant embryos was analysed using Nomarski microscopy to characterize the terminal phenotype of tro-1 embryos. To determine when embryo development was disrupted in the mutant, siliques at 7 d after self-pollination from heterozygous tro-1/+ plants were dissected and cleared by whole-mount clearing. Embryo development was arrested at the early globular stage in approximately 25% of the seeds, whereas the remaining 75% had already reached the heart stage (Fig. 6c). To follow the progression of the tro-1 mutant phenotype, normal and aborted seeds at different stages of development were isolated from tro-1/+ siliques and analysed to determine the stage at which the tro-1 homozygous embryos were arrested during embryogenesis. The development of wild-type Arabidopsis embryos progressed as described previously (Fig. 6d–h). No dramatic differences were observed in wild-type and tro-1 mutant seeds from the zygote up to the octant stage (8 cells; Fig. 6d, e, i, j). The first clear difference in development was observed at the beginning of the heart stage (Fig. 6f), when ∼25% of the seeds arrested with the embryo at the globular stage (Fig. 6k). These seeds showed alterations in the morphology of the arrested globular embryos, which developed into an amorphous agglomeration of cells (Fig. 6l). The tro-1 seeds remained at the globular stage in siliques containing wild-type seeds at the torpedo and cotyledon stages, and tro-1 embryos did not show any indication of cotyledon formation compared to sibling wild-type embryos, and eventually they aborted. No differences in early endosperm development were observed. These results provide strong evidence that TRO is an essential gene and that tro-1 mutation disrupts embryo development.
Fig. 6.
Mutant tro-1 embryos were arrested before the globular stage. (a) Wild-type silique showing a full seed set and a (b) heterozygous tro-1 silique with approximately 25% of the embryos aborted (black arrows). (c) Whole-mounted, cleared seeds from siliques of heterozygous tro-1 plants. The same silique contains a mutant embryo arrested at the early globular stage (left) compared with a normal embryo developed at the heart stage (right). (d–h) Nomarski images of wild-type embryos at the four-cell stage (d), eight-cell stage (e), early heart stage (f), heart stage (g), and torpedo (h). (i–m) Nomarski images of homozygous tro-1 embryos arrested within the same siliques as the wild-type embryos showed in (d–h). Mutant embryo with normal appearance at four-cell stage (i) and eight-cell stage (j). (k–m) Embryos with abnormal morphology compared with sibling embryos shown in (f–h), respectively. Scale bar: d–f and i–m, 20 μm; g, 25 μm; h, 40 μm.
Table 2.
Seed abortion frequency in mutant tro-1
| Allele | Normal | Aborted | Total | P-valuea |
| wt | 1085 (99.4) | 6 (0.06) | 1091 | ND |
| tro-1/+ | 859 (77.2) | 253 (22.8) | 1112 | 0.1 |
Percentage values are in parentheses. χ2 tests were performed to compare the abortion seeds between wt and tro-1.
P-value of ≥0.05 was considered consistent with the hypothesis that 25% of the seeds abort as expected for a zygotic embryonic lethal mutation.
Complementation of the tro-1 mutant phenotype
To demonstrate further that seed lethality was caused by the interruption of the TRO gene, it was investigated whether a full-length TRO locus could rescue the seed-lethal phenotype. For this purpose, the promoter of the p35S:TRO-GFP construct was replaced with the native TRO promoter. Heterozygous BASTA-resistant tro-1/+ plants were transformed with the resulting pTRO:TRO-GFP. Five T1 plants resistant to both BASTA (marker for the tro-1) and hygromycin (marker for the TRO complementation construct) were obtained. In addition, the presence of pTRO:TRO-GFP and of the T-DNA insertion in TRO was confirmed by genotyping (data not shown). In these plants, the subcelullar localization of translated fusion protein was further evaluated by confocal microscopy (see Supplementary Fig. 3 at JXB online). As expected, the TRO protein is localized in the nucleus in vivo, suggesting that fusion protein is functional. The construct p35S::GFP-Lti6b served as a localization control and the fusion protein is localized at the cell periphery (see Supplementary Fig. 3 at JXB online, Yamada et al., 2005). The molecular complementation of a plant heterozygous for a recessive embryo-lethal mutation should result in a rescued heterozygote that exhibits 6.25% (1/16) defective seeds if a single copy of the wild-type transgene is genetically unlinked to the original locus (Stacey et al., 2002). In our results, all five lines showed a reduction in the number of defectives seeds, giving a percentage of aborted seeds that was not significantly different from 6.25% (Table 3). This value is consistent with the percentage expected described above. All the seeds aborted have the expected embryo phenotype (data not shown). These results confirm that TRO is fully responsible for the phenotypes that were recovered in the tro-1 allele, and that TRO is required for plant embryogenesis.
Table 3.
Genetic complementation of the tro-1 mutation
| Plant number | Normal | Aborted | Percentage | Expected percentage | P-valuea |
| 1 | 159 | 7 | 4.19 | 6.25 | 0.3 |
| 2 | 107 | 4 | 3.66 | 6.25 | 0.3 |
| 3 | 114 | 4 | 3.44 | 6.25 | 0.3 |
| 4 | 100 | 6 | 5.60 | 6.25 | 0.7 |
| 5 | 172 | 7 | 3.86 | 6.25 | 0.3 |
χ2 tests were performed to evaluate the genetic complementation of the tro-1 mutation.
P-value of ≥0.05 was considered consistent with the hypothesis that 6.25% of the seeds abort as expected for single copy transgene complementation of heterozygous mutant (Stacey et al., 2002).
Discussion
Arabidopsis embryo development is partly controlled by PRC2 complexes which are formed by FIS-class genes, plant proteins with homology to animal PcG (Pien and Grossniklaus, 2007). PcG, together with trxG proteins, are members of a cellular memory system that maintains the balance between proliferation and differentiation in eukaryotic cells. In contrast to plant PcG, the role of trxG during plant embryogenesis is unknown. In this work, the identification and characterization are reported of a novel plant gene named TRAUCO that is required for embryo development in Arabidopsis thaliana. Sequence analysis of the TRO protein revealed a SPRY domain in the carboxyl terminus that is characteristic of trxG members described in Drosophila ASH2 (Adamson and Shearn, 1996), human ASH2L (Wang et al., 2001), and yeast BRE2P (Nagy et al., 2002). BRE2P is member of a seven-protein complex that is required for the methylation of histone H3 and yeast lacking BRE2P (Bre2pΔ) were sensitive to formamide (Nagy et al., 2002). Heterologous expression of TRO in Bre2pΔ yeast shows that it partially restores the growth in formamide (Fig. 2). Subcellular localization analysis of TRO using onion epidermal cells and transgenic plants showed localization to the nucleus (Fig. 5; see Supplementary Fig. 3 at JXB online). Taken together, the localization and functional data suggest that TRO is a member of a nuclear protein complex in plant cells, in agreement with putative trxG function.
Molecular and genetic characterization of heterozygous tro-1/+ mutant plants allowed us to conclude that tro-1 is an embryo lethal mutation. Through genetic studies, it has been shown that the loss of TRO function caused an early arrest of embryo development before the dermatogen stage, a stage critical for pattern formation, and that tro-1 mutant embryos displayed an aberrant cell division pattern.
The transcriptional states of several loci of the genome including developmentally and cell-cycle regulated genes are repressed and activated by PcG and trxG proteins, respectively. In this way, both protein groups have an antagonistic effect during development. The Arabidopsis fis mutants are characterized by autonomous seed formation in the absence of fertilization (Luo et al., 1999; Köhler et al., 2003; Guitton et al., 2004). Development of fis seeds is delayed and seeds abort with embryos arrested at the late heart stage and possess a non-cellularized endosperm with strongly overproliferated chalazal endosperm domains (Grossniklaus et al., 1998; Köhler et al., 2003; Guitton et al., 2004). In contrast to this, tro-1 mutant embryos arrested earlier at the globular stage, and the endosperm was not affected (data not shown). In addition, mutations in FIS genes cause parent-of-origin-dependent seed abortion (Grossniklaus et al., 2004). All seeds that inherit a mutant FIS allele from the mother abort, regardless of the presence of a wild-type paternal allele. As opposed to this phenomenon, the tro-1 mutation is transmitted by both gametes, suggesting a general zygotic function.
PcG and trxG proteins form higher order complexes that contain SET domain proteins responsible for various types of lysine methylation at the N-terminal tails of the core histone proteins. Molecular analysis indicates that TRO encodes a protein containing a SPRY domain and a putative nuclear localization signal motif, suggesting that TRO is a member of a nuclear protein complex. Other than these motifs, TRO does not have extensive homology to any other sequences in the protein databases. In contrast, the Drosophila ASH2 protein possesses a SPRY domain and a putative double zinc-finger domain in the N-terminus extreme, sequences which are found in nuclear proteins that play a role in regulating chromatin (Adamson and Shearn, 1996). On the other hand, BRE2P is the homologue of ASH2 in yeast, and lacks a highly conserved PHD finger (Nagy et al., 2002). Interestingly, another component of the yeast complex contains a PHD finger (protein SAF41p), suggesting that, together, these two proteins constitute a bipartite functional homologue of ASH2 in the complex termed SET1P (Nagy et al., 2002). To date in plants, only a PRC2 complex containing MEA, FIE, and MSI1 has been biochemically characterized (Köhler et al., 2003) and no trxG complexes have been isolated. Further experiments should identify other components of the Arabidopsis trxG protein complex.
Genetic analysis in Arabidopsis indicates that, in addition to controlling seed initiation, PcG proteins control flower organ development, switch from vegetative to floral development, and vernalization (Gendall et al., 2001; Kinoshita et al., 2001; Yoshida et al., 2001). Our results show that TRO transcripts were present in specific tissues and showed higher levels of expression in sepals and anthers, suggesting that it could play a role during flower development. In our experiments, such defects were not observed, as homozygous tro-1 embryos abort and do not give rise to mature plants. Therefore, a potential role of TRO in flower development remains to be addressed. Arabidopsis Trithorax1 (ATX1) is a close homologue of trxG and is the only other member of this family that has been described previously in plants (Alvarez-Venegas et al., 2003). It contains a SET domain and additional domains characteristic of trxG proteins. In atx1 mutants, the floral homeotic genes AGAMOUS, PISTILLATA, and APETALA 3 are expressed at a lower level during flower development (Alvarez-Venegas et al., 2003). This is reflected by homeotic changes of floral organs in atx1 mutants. Additional experiments could address whether TRO and ATX1 participate in the same protein complex.
In conclusion, the nuclear protein TRO represents a novel trxG protein that is required for early embryo development in Arabidopsis. These results reveal new biological functions for trxG proteins in plant reproduction. The molecular function of TRO as a trxG protein has not yet been demonstrated and putative downstream gene targets are not known. In addition to these investigations, discovery of the genes regulated by TRO should provide further insights into the genetic network that controls Arabidopsis embryo development.
Supplementary Material
Acknowledgments
This work was supported by a PhD fellowship from CONICYT (Gobierno de Chile) and a VRAID fellowship from the Pontificia Universidad Católica de Chile to F Aquea and P Cañon. This work was primarily financed by the Millennium Nucleus for Plant Functional Genomics (P06-009-F) and the Chilean Fruit Consortium. UG acknowledges funding from the EPIGENOME Network of Excellence. We thank J Gheyselinck for technical assistance and Michael Handford for their assistance in language support.
References
- Adamson AL, Shearn A. Molecular genetic analysis of Drosophila ash2, a member of the trithorax group required for imaginal disc pattern formation. Genetics. 1996;2:621–633. doi: 10.1093/genetics/144.2.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander MP. Differential staining of aborted and non-aborted pollen. Stain Technology. 1969;44:117–122. doi: 10.3109/10520296909063335. [DOI] [PubMed] [Google Scholar]
- Alvarez-Venegas R, Pien S, Sadder M, Witmer X, Grossniklaus U, Avramova Z. ATX-1, an Arabidopsis homolog of trithorax, activates flower homeotic genes. Current Biology. 2003;13:627–637. doi: 10.1016/s0960-9822(03)00243-4. [DOI] [PubMed] [Google Scholar]
- Aquea F, Arce-Johnson P. Identification of genes expressed during early somatic embryogenesis in Pinus radiata. Plant Physiology and Biochemistry. 2008;46:559–568. doi: 10.1016/j.plaphy.2008.02.012. [DOI] [PubMed] [Google Scholar]
- Aquea F, Matte JP, Gutiérrez F, Rico S, Lamprecht M, Sánchez C, Arce-Johnson P. Molecular characterization of a Trithorax-group homologue gene from Pinus radiata. Plant Cell Reporter. 2009;28:1531–1538. doi: 10.1007/s00299-009-0752-9. [DOI] [PubMed] [Google Scholar]
- Bai S, Chen L, Yund MA, Sung ZR. Mechanisms of plant embryo development. Current Topics in Developmental Biology. 2000;50:61–88. doi: 10.1016/s0070-2153(00)50004-0. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U. A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiology. 2003;133:462–469. doi: 10.1104/pp.103.027979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendall AR, Levy YY, Wilson A, Dean C. The VERNALIZATION 2 gene mediates the epigenetic regulation of vernalization in Arabidopsis. Cell. 2001;107:525–535. doi: 10.1016/s0092-8674(01)00573-6. [DOI] [PubMed] [Google Scholar]
- Grossniklaus U, Vielle-Calzada JP, Hoeppner MA, Gagliano WB. Maternal control of embryogenesis by MEDEA, a polycomb group gene in Arabidopsis. Science. 1998;280:446–450. doi: 10.1126/science.280.5362.446. [DOI] [PubMed] [Google Scholar]
- Grossniklaus U, Spillane C, Page DR, Köhler C. Genomic imprinting and seed development: endosperm formation with and without sex. Current Opinion in Plant Biology. 2004;4:21–27. doi: 10.1016/s1369-5266(00)00130-8. [DOI] [PubMed] [Google Scholar]
- Guitton AE, Page DR, Chambrier P, Lionnet C, Faure JE, Grossniklaus U, Berger F. Identification of new members of FERTILIZATION INDEPENDENT SEED Polycomb group pathway involved in the control of seed development in Arabidopsis thaliana. Development. 2004;131:2971–2981. doi: 10.1242/dev.01168. [DOI] [PubMed] [Google Scholar]
- Guyomarc'h S, Bertrand C, Delarue M, Zhou DX. Regulation of meristem activity by chromatin remodelling. Trends in Plant Science. 2005;10:332–338. doi: 10.1016/j.tplants.2005.05.003. [DOI] [PubMed] [Google Scholar]
- Jenik PD, Gillmor CS, Lukowitz W. Embryonic patterning in Arabidopsis thaliana. Annual Review in Cell and Developmental Biology. 2007;23:207–236. doi: 10.1146/annurev.cellbio.22.011105.102609. [DOI] [PubMed] [Google Scholar]
- Jurgens G. Apical–basal pattern formation in Arabidopsis embryogenesis. EMBO Journal. 2001;20:3609–3616. doi: 10.1093/emboj/20.14.3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T, Harada JJ, Goldberg RB, Fischer RL. Polycomb repression of flowering during early plant development. Proceedings of the National Academy of Sciences, USA. 2001;98:14156–14161. doi: 10.1073/pnas.241507798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler C, Grossniklaus U. Epigenetic inheritance of expression states in plant development: the role of Polycomb group proteins. Current Opinion in Cell Biology. 2002;14:773–779. doi: 10.1016/s0955-0674(02)00394-0. [DOI] [PubMed] [Google Scholar]
- Köhler C, Hennig L, Bouveret R, Gheyselinck J, Grossniklaus U, Gruissem W. Arabidopsis MSI1 is a component of the MEA/FIE Polycomb group complex and required for seed development. EMBO Journal. 2003;22:4804–4814. doi: 10.1093/emboj/cdg444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler C, Makarevich G. Epigenetic mechanisms governing seed development in plants. EMBO Report. 2006;7:1223–1227. doi: 10.1038/sj.embor.7400854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M, Bilodeau P, Koltunow A, Dennis ES, Peacock WJ, Chaudhury AM. Genes controlling fertilization-independent seed development in Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA. 1999;96:296–301. doi: 10.1073/pnas.96.1.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüscher-Firzlaff J, Gawlista I, Vervoorts J, et al. The human trithorax protein hASH2 functions as an oncoprotein. Cancer Research. 2008;68:749–758. doi: 10.1158/0008-5472.CAN-07-3158. [DOI] [PubMed] [Google Scholar]
- McElver J, Tzafrir I, Aux G, et al. Insertional mutagenesis of genes required for seed development in Arabidopsis thaliana. Genetics. 2001;159:1751–1763. doi: 10.1093/genetics/159.4.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinke DW. Molecular genetics of plant embryogenesis. Annual Review of Plant Physiology and Plant Molecular Biology. 1995;46:369–394. [Google Scholar]
- Meinke D, Muralla R, Sweeney C, Dickerman A. Identifying essential genes in Arabidopsis thaliana. Trends in Plant Science. 2008;13:483–491. doi: 10.1016/j.tplants.2008.06.003. [DOI] [PubMed] [Google Scholar]
- Nagy PL, Griesenbeck J, Kornberg RD, Cleary ML. A trithorax-group complex purified from Saccharomyces cerevisiae is required for methylation of histone H3. Proceedings of the National Academy of Sciences, USA. 2002;99:90–94. doi: 10.1073/pnas.221596698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez F. Mitos y leyendas de Chile. Santiago de Chile: ZIG-ZAG editorials; 2003. [Google Scholar]
- Pien S, Grossniklaus U. Polycomb group and trithorax group proteins in Arabidopsis. Biochimica et Biophysica Acta. 2007;1769:375–382. doi: 10.1016/j.bbaexp.2007.01.010. [DOI] [PubMed] [Google Scholar]
- Reyes JC, Hennig L, Gruissem W. Chromatin-remodeling and memory factors. New regulators of plant development. Plant Physiology. 2002;130:1090–1101. doi: 10.1104/pp.006791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringrose L, Paro R. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annual Review of Genetics. 2004;38:413–443. doi: 10.1146/annurev.genet.38.072902.091907. [DOI] [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. Methods in Enzymology. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- Spillane C, MacDougall C, Stock C, Köhler C, Vielle-Calzada JP, Nunes SM, Grossniklaus U, Goodrich J. Interaction of the Arabidopsis polycomb group proteins FIE and MEA mediates their common phenotypes. Current Biology. 2000;10:1535–1538. doi: 10.1016/s0960-9822(00)00839-3. [DOI] [PubMed] [Google Scholar]
- Springer NM, Danilevskaya ON, Hermon P, Helentjaris TG, Phillips RL, Kaeppler HF, Kaeppler SM. Sequence relationships, conserved domains, and expression patterns for maize homologs of the polycomb group genes E(z), esc, and E(Pc) Plant Physiology. 2002;128:1332–1345. doi: 10.1104/pp.010742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacey MG, Koh S, Becker J, Stacey G. AtOPT3, a member of the oligopeptide transporter family, is essential for embryo development in Arabidopsis. The Plant Cell. 2002;14:2799–2811. doi: 10.1105/tpc.005629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur JK, Malik MR, Bhatt V, Reddy MK, Sopory SK, Tyagi AK, Khurana JP. A POLYCOMB group gene of rice (Oryza sativa L. subspecies indica), OsiEZ1, codes for a nuclear-localized protein expressed preferentially in young seedlings and during reproductive development. Gene. 2003;314:1–13. doi: 10.1016/s0378-1119(03)00723-6. [DOI] [PubMed] [Google Scholar]
- Tzafrir I, Dickerman A, Brazhnik O, Nguyen Q, McElver J, Frye C, Patton D, Meinke D. The Arabidopsis SeedGenes project. Nucleic Acids Research. 2003;31:90–93. doi: 10.1093/nar/gkg028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzafrir I, Pena-Muralla R, Dickerman A, et al. Identification of genes required for embryo development in Arabidopsis. Plant Physiology. 2004;135:1206–1220. doi: 10.1104/pp.104.045179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vielle-Calzada JP, Thomas J, Spillane C, Coluccio A, Hoeppner MA, Grossniklaus U. Maintenance of genomic imprinting at the Arabidopsis MEDEA locus requires zygotic DDM1 activity. Genes and Development. 1999;13:2971–2982. doi: 10.1101/gad.13.22.2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Li Z, Messing EM, Wu G. Activation of Ras/Erk pathway by a novel MET-interacting protein RanBPM. Journal of Biological Chemistry. 2002;277:36216–36222. doi: 10.1074/jbc.M205111200. [DOI] [PubMed] [Google Scholar]
- Wang J, Zhou Y, Yin B, Du G, Huang X, Li G, Shen Y, Yuan J, Qiang B. ASH2L: alternative splicing and downregulation during induced megakaryocytic differentiation of multipotential leukemia cell lines. Journal of Molecular Medicine. 2001;79:399–405. doi: 10.1007/s001090100222. [DOI] [PubMed] [Google Scholar]
- Willemsen V, Scheres B. Mechanisms of pattern formation in plant embryogenesis. Annual Review of Genetics. 2004;38:587–614. doi: 10.1146/annurev.genet.38.072902.092231. [DOI] [PubMed] [Google Scholar]
- Yamada K, Fuji K, Shimada T, Nishimura M, Hara-Nishimura I. Endosomal proteases facilitate the fusion of endosomes with vacuoles at the final step of the endocytotic pathway. The Plant Journal. 2005;41:888–898. doi: 10.1111/j.1365-313X.2005.02349.x. [DOI] [PubMed] [Google Scholar]
- Yadegari R, Paiva G, Laux T, Koltunow AM, Apuya N, Zimmerman JL, Fischer RL, Harada JJ, Goldberg RB. Cell differentiation and morphogenesis are uncoupled in Arabidopsis raspberry embryos. The Plant Cell. 1994;6:1713–1729. doi: 10.1105/tpc.6.12.1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida N, Yanai Y, Chen L, Kato Y, Hiratsuka J, Miwa T, Sung ZR, Takahashi S. EMBRYONIC FLOWER2, a novel polycomb group protein homolog, mediates shoot development and flowering in Arabidopsis. The Plant Cell. 2001;13:2471–2481. doi: 10.1105/tpc.010227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai L, Dietrich A, Skurat AV, Roach PJ. Structure–function analysis of GNIP, the glycogenin-interacting protein. Archives of Biochemistry and Biophysics. 2004;421:236–242. doi: 10.1016/j.abb.2003.11.017. [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Henning L, Gruissem W. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiology. 2004;136:2621–2632. doi: 10.1104/pp.104.046367. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.