Abstract
The Epstein–Barr virus (EBV) nuclear antigen 1 (EBNA1) is regularly expressed in all proliferating virus-infected cells and is therefore an interesting target for immunotherapy. Alleles of the human leucocyte antigen (HLA) -A2 family are dominantly expressed in Caucasians so we sought to identify EBNA1-specific cytotoxic T-lymphocyte (CTL) responses restricted through this allele. We report on the characterization of the LQTHIFAEV (LQT) epitope. LQT-specific memory CTL responses were reactivated in three of 14 healthy EBV seropositive donors (21%) whereas responses to HLA-A2-restricted epitopes, two derived from LMP2 and one from EBNA3A, were detected in 93%, 71% and 42% of the donors, respectively. The LQT-specific CTL clones did not lyse EBV-carrying lymphoblastoid cell lines and Burkitt’s lymphoma cell lines nor EBNA1-transfected Burkitt’s lymphoma cells but specifically released interferon-γ upon stimulation with HLA-matched EBNA1-expressing cells and this response was enhanced by deletion of the Gly-Ala repeat domain that inhibits proteasomal degradation. The poor presentation of the endogenously expressed LQT epitope was not affected by inhibition of peptidases that trim antigenic peptides in the cytosol but full presentation was achieved in cells expressing a trojan antigen construct that releases the epitope directly into the endoplasmic reticulum. Hence, inefficient proteasomal processing appears to be mainly responsible for the poor presentation of this epitope.
Keywords: antigen processing, cytotoxic T lymphocyte, Epstein–Barr virus nuclear antigen 1, Epstein–Barr virus, Gly-Ala repeat
Introduction
CD8+ T lymphocytes play an important role in the control of viral infections by generating effectors that are able to recognize and kill infected cells.1,2 T-cell receptors (TCR) expressed by cytotoxic T lymphocytes (CTL) recognize virus-infected cells via interaction of the TCR with peptides derived from the processing of endogenously expressed viral proteins presented on the surface of the target cell as a complex with major histocompatibility complex (MHC) class I molecules.3 The principal enzymatic activity responsible for the generation of class I-associated peptides is that of the proteasome, a large multicatalytic protease that is essential for the degradation of intracellular proteins.4 Moreover, it has been demonstrated that other cyosolic and endoplasmic reticulum (ER) resident proteases act on products released by the proteasome.5
Although the complex network of proteases involved in antigen processing should ensure the efficient presentation of viral antigens, several viruses have evolved strategies for interfering with this pathway, allowing them to evade destruction by the immune system. One of the best examples of ubiquitin-proteasome targeting involves the Epstein–Barr virus (EBV), a γ-herpesvirus associated with several human tumours.6 This virus is widespread and establishes life-long persistent infections in the B lymphocytes in the vast majority of human adults. The EBV-infected B cells can proliferate in vitro, giving rise to lymphoblastoid cell lines (LCL) that express at least nine latency-associated viral antigens: the EBV nuclear antigens EBNA1, -2, -3A, -3B, -3C, -LP and the membrane proteins LMP1, LMP2A and LMP2B.7
The proliferation of these virus-infected cells is monitored in vivo by T lymphocytes that specifically recognize viral antigens. In particular, EBNA3A, EBNA3B and EBNA3C contain immunodominant epitopes of CTL responses over a wide range of human leucocyte antigen (HLA) backgrounds, while EBNA2, EBNA-LP, LMP1 and LMP2 are subdominant targets that are presented in the context of a limited number of HLA restrictions.8–12 Weak responses against EBNA1 have also been detected in some individuals but, so far, only a limited number of immunogenic CTL epitopes have been identified within this antigen.13–16
The poor immunogenicity of EBNA1 has been attributed to the presence of a Gly-Ala repeat (GAr) sequence, which prevents the presentation of EBNA1-derived antigenic peptides by MHC class I molecules. This GAr-mediated function has been linked to its capacity to prevent EBNA1 synthesis,17 as well as proteasomal degradation.18,19 Although the role of the GAr domain on the poor immunogenicity of EBNA1 has only partially been clarified, the characterization of CTL responses directed towards this antigen remains crucial. Because of its expression in all EBV-associated tumours, EBNA1 represents an ideal tumour-rejection target for the development of immunotherapy against EBV-associated malignancies.
To identify novel EBNA1-specific CTL target epitopes, we have tested CD8+ T-cell reactivity towards peptides derived from the EBNA1 antigen in a panel of healthy EBV-seropositive individuals. Because of the dominant expression of these alleles in White people, we focused our attention on CTL responses restricted by two different HLA alleles: A2 and A24. We report on the identification of a new EBNA1-derived CTL epitope, defined as LQTHIFAEV (LQT), presented by HLA-A2. The frequency of T cells specific for this epitope appears to be lower compared with HLA A2-restricted epitopes derived from other viral proteins and EBNA1 epitopes presented by other class I alleles. Comparison of recognition of cells expressing EBNA1 with or without the GAr domain or incubated with a Trojan antigen (TA) construct that delivers the epitope directly into the ER points to inefficient processing as the main cause for the poor recognition of this epitope.
Materials and methods
Cell lines
The .174/T2 cell line (T2) was obtained by fusion of the peptide transporter mutant .174 LCL with the CEM T-cell line. The LCL were obtained by infection of lymphocytes from HLA-typed donors with culture supernatants of the B95-8 virus-producer cell line, cultured in the presence of 0·1 μg/ml of cyclosporine A (Sandoz International GmbH, Holzkirchen, Germany). T2 cells, LCL and Burkitt cell lines were maintained in RPMI-1640 supplemented with 2 mm glutamine, 100 IU/ml penicillin, 100 μg/ml streptomycin and 10% heat-inactivated fetal calf serum (HyClone; Thermo Fisher Scientific Inc., Waltham, MA). The BJAB (B-cell lymphoma) E1 and E1ΔGA were generated upon transfection using an amaxa V solution kit (Amaxa, Cologne, Germany). Cells were maintained in selection with 0·2 mg/ml G418 (Gibco; Invitrogen, San Diego, CA). Phytohaemagglutinin (PHA)-activated blasts were obtained by stimulation of peripheral blood lymphocytes (PBL) with 1 μg/ml purified PHA (Remel Europe Ltd, Dartford, UK) for 3 days and expanded in medium supplemented with human recombinant interleukin-2 (rIL-2; Proleukin; Chiron; Novartis, Emeryville, CA) as previously described.8
Plasmids
The plasmid pFLAG-EBNA1 was kindly provided by E. Kieff (Brigham & Women's Hospital, Boston, MA); pFLAG-EBNA1dGA has been generated by swapping a BSpEI/NotI fragment from pMC-EBNA1dGA, kindly provided by Neil Blake (Institute for Cancer Studies, University of Birmingham, Birmingham, UK), to pFLAG-EBNA1 digested with the same restriction enzymes.
Synthetic peptides
The 9-mer and 10-mer peptide sequences were obtained using a prediction model which ranks peptides based on a predicted half-life of dissociation to HLA-A2 or HLA-A24, (Table 1). Synthetic TAs were prepared by production of synthetic peptides containing the minimal CTL epitope joined to the HIV-Tat protein transduction domain (RKKRRQRRR) using a furin-sensitive (RVKR) or furin-resistant (VRVV) linker.20,21 All peptides were synthesized by the solid-phase method using a continuous flow instrument with online ultraviolet monitoring. The stepwise syntheses were conducted by F-moc chemistry as previously described.22 Crude deprotected peptides were purified by high-performance liquid chromatography (HPLC); their purity was > 98%. Structural verification was achieved by elemental and amino acid analyses and mass spectrometry. Peptide stocks were prepared in dimethyl sulphoxide at a concentration of 10−2 m, maintained at −20°, and diluted in phosphate-buffered saline before use. The origin and restriction of the epitopes used in this study was: CLG: LMP2, A2; SVR EBNA3A, A2; LLW: LMP2, A2; YNL: EBNA1, B8; HPV: EBNA1, B35; YPL: EBNA3A, B35.
Table 1.
Predicted Epstein–Barr virus nuclear antigen 1 (EBNA1)-derived epitopes selected and tested in this study
| HLA | Sequence | Name | EBNA1 aa |
|---|---|---|---|
| A2 | FLQTHIFAEV | FLQ | 565–574 |
| SIVCYFMVFL | SIV | 557–566 | |
| A24 | VYGGSKTSL | VYG | 509–517 |
| LYNLRRGTAL | LYN | 517–526 |
Each epitope is represented by the first three amino acids of its sequence. The amino acid co-ordinates within the EBNA1 protein are also shown.
Preparation of antigen-presenting cells
T2 and T2/A24 cells (2 × 106) were cultured overnight at 26° in 1 ml of serum-free AIM-V medium. Cells were then washed, treated with mitomycin C to avoid cell proliferation, and pulsed for 3 hr with 10−5 m of the different peptides in AIM-V medium at 37°. After extensive washing, the cells were used as stimulators.23
Generation of memory CTL cultures
Monocyte-depleted PBL from EBV-seropositive subjects were plated in RPMI-1640 containing 10% fetal calf serum (HyClone) at 3 × 106 cells per well in 24-well plates, and stimulated with peptide-pulsed T2 cells at a stimulator : responder ratio of 1 : 20. Cultures were restimulated after 7 and 14 days, and the medium was supplemented from day 8 with 10 U/ml rIL-2 (Chiron). On days 14 and 21, T-cell cultures were tested by cytotoxicity assay for CTL activity.24 Single-cell cloning was achieved by limiting dilution in 96-well plates in 200 μl of 30% AIM-V medium containing 10 U/ml of human rIL-2, and 105 irradiated (3000 rads) allogeneic PHA-activated PBL as feeders.25 Growing cultures were split and tested for CTL activity by cytotoxicity assay. All EBV-specific CTL clones were expanded into 24 plates and restimulated weekly with irradiated autologous LCL in IL-2-containing medium. The EBV specificities and HLA class I restriction of the CTL preparations were investigated by testing their cytotoxic activities against a panel of EBV-positive and EBV-negative targets, including autologous LCL, allogeneic LCL and PHA-activated blasts matched through single HLA class I alleles or HLA-mismatched LCL.
Cytotoxicity assays
Cytotoxic activity was tested by standard 5-hr 51Cr-release assay, as previously described.26 Briefly, target cells were labelled with 0·1 μCi/106 cells of Na251CrO4 for 90 min at 37° and, where indicated, were pulsed for 45 min with 10−6 m of the different peptides at 37°. Cells were then washed, and 4 × 103 cells were used as targets of each CTL at different E : T ratios. The per cent specific lysis was calculated as 100 × [(c.p.m. sample − c.p.m. medium)/(c.p.m. Triton X-100 − c.p.m. medium)], where c.p.m. represents counts/min. Spontaneous release was always < 20% in all cases. None of the tested peptides affected spontaneous release.
IFN-γ ELISPOT
Enzyme-linked immunosorbent spot-forming cell assay [ELISPOT; for interferon-γ (IFN-γ)] was carried out using commercially available kits (Becton-Dickinson, Franklin Lakes, NJ) according to the manufacturer’s instructions. In brief, 96-well nitrocellulose plates were coated with 5 μg/ml of anti-IFN-γ overnight at 4°. The following day the plates were washed four times with phosphate-buffered saline and blocked for 2 hr with 10% fetal bovine serum-supplemented RPMI-1640 at 37°. The CTL were added to the wells (in triplicate) at a ratio of 3 : 1 and incubated with target cells for 24 hr at 37°. Controls were represented by cells incubated with concanavalin A (Sigma-Aldrich, St Louis, MO; 5 μg/ml) (positive control) or with the medium alone (negative control). The spots were read using an ELISPOT reader (A.EL.VIS GmbH, Hannover, Germany). Results are expressed as net number of spot-forming units (SFU)/106 cells.
Western blotting
Total cell lysates were prepared in sodium dodecyl sulphate sample buffer, fractionated in 12% sodium dodecyl sulphate –polyacrylamide gel electrophoresis gradient gels (Invitrogen, Carlsbad, CA) and transferred to polyvinylidene fluoride membranes (Millipore Corporation, Billerica, MA). The blots were then probed with anti-EBNA1 OT1X, donated by Y. Middeldorp (Vrije Universiteit, Amsterdam, the Netherlands), and β-actin (AC-15, 1 : 2000, Sigma-Aldrich, St Louis, MO), followed by the appropriate horseradish peroxidase-conjugate secondary antibodies (Zymed, San Francisco, CA) and developed by enhanced chemiluminescence.
Results
Selection of HLA-A2- and HLA-A24-binding peptides derived from the EBNA1 antigen
To identify new CTL epitopes within the EBNA1 latent antigen, the amino acid sequence of the protein was analysed by a web-based algorithm that predicts peptide binding to HLA-A2, and HLA-A24 (http://www-bimas.cit.nih.gov/cgi-bin/molbio/ken_parker_comboform). This analysis yielded a list of peptide sequences containing putative binding motifs and an estimation of the half-time dissociation of the HLA–peptide complex.27 As the stability of MHC-I–peptide complexes is a crucial factor in determining CTL responsiveness,24,26,28,29 only peptide sequences with the highest scores for HLA-A2 and HLA-A24 molecules, respectively, were selected for further analysis (Table 1).
Induction of memory CTL responses by selected HLA-A2- and HLA-A24-restricted EBNA1-derived peptides
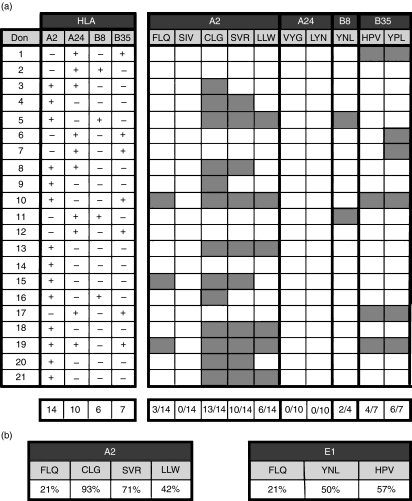
The PBL obtained from healthy HLA class I-typed EBV-seropositive donors (21 donors: 14 HLA-A2, 10 HLA-A24 and three HLA-A2, HLA-A24), were stimulated with peptides or peptide-pulsed T2 or T2/A24 cells. The specificity of the CTL cultures was tested after three stimulations using standard 51Cr-release assays against autologous PHA-blasts, pulsed or not with the relevant synthetic peptide. Only cultures that showed specific lysis (peptide-pulsed target – unpulsed target) above 20% were considered as epitope-specific. Based on this criterion, the HLA-A2-restricted 10-mer FLQ was deemed to induce peptide-specific responses in three of the 14 HLA-A2 donors tested (Fig. 1a) while no specific responses were detected against other predicted EBNA1-derived epitopes (data not shown).
Figure 1.
(a) List of healthy Epstein–Barr virus (EBV) -seropositive donors exploited and the relative human leucocyte antigen (HLA) mapping for A2, A24 and B35 molecules. Cytotoxic T-lymphocyte (CTL) memory stimulations were performed according to donor HLA-typing, and cytotoxicity activity was tested after three stimulations against autologous phytohaemagglutinin (PHA) -blasts, pulsed or not with the relevant synthetic peptide. Grey boxes indicate the presence of a peptide-specific response. The predicted EBV nuclear antigen 1 (EBNA1) -derived epitopes tested in this study are shown in bold. Other known EBV-derived CTL epitopes were used as control for EBV- and EBNA1-specific CTL responses. CLG and LLW peptides were derived from LMP2, SVR and YPL peptides from EBNA3A,and HPV and YNL peptides from EBNA1. The lowest line shows the frequency of the positive responses. (b) Comparison of FLQ responses to other HLA-A2 restricted EBV epitopes (left chart) and other EBNA1-derived epitopes (right chart).
Parallel stimulations using other known EBV-derived epitopes (Table 2) including: CLGGLLTMV (CLG) and LLWTLVVLL (LLW) from LMP2, SVRDRLARL (SVR) and YPLHEQHGM (YPL) from EBNA3A, and YNLRRGTAL (YNL) and HPVGEADYFEY (HPV) from EBNA1 were performed as controls. The presenting HLA allele of each of these peptides is indicated in Fig. 1(a). These stimulations yielded EBV-specific CTL responses in 18 of 21 donors, confirming the efficiency of the stimulation protocol. It is noteworthy that the frequency of FLQ responders (21%) was significantly lower compared with the frequency of responders to HLA-A2-restricted epitopes derived from EBNA3A (SVR) and LMP2 (CLG and LLW), which ranged between 42% and 93% of the donors. A similar observation was made when the response to FLQ was compared with that of EBNA1-derived epitopes restricted through different class I alleles (Fig. 1b).
Table 2.
Known Epstein–Barr virus-derived epitopes selected and tested in this study
| HLA | Sequence | Name | Protein |
|---|---|---|---|
| A2 | CLGGLLTMV | CLG | LMP2 |
| SVRDRLARL | SVR | EBNA3A | |
| LLWTLVVLL | LLW | LMP2 | |
| B35 | HPVGEADYFEY | HPV | EBNA1 |
| YPLHEQHGM | YPL | EBNA3A | |
| B8 | YNLRRGTAL | YNL | EBNA1 |
Each epitope is represented by the first three amino acids of its sequence. The protein of origin is also shown.
EBNA, Epstein–Barr virus nuclear antigen; HLA, human leucocyte antigen; LMP, Epstein–Barr virus membrane protein.
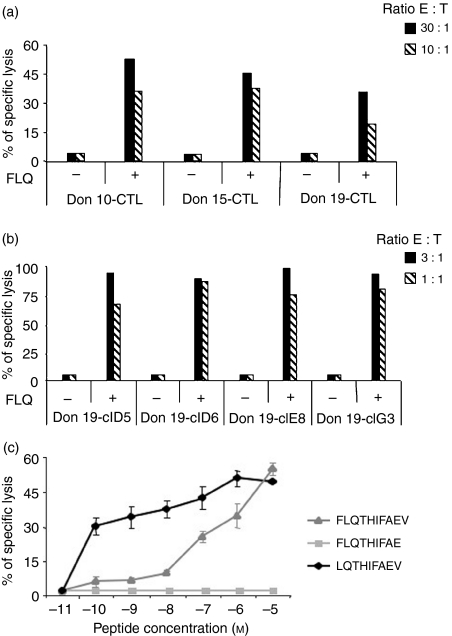
Characterization of the FLQ minimal epitope
To identify the minimal epitope seqence presented by HLA-A2, polyclonal CTL cultures derived from the three responders (Fig. 2a) were cloned by limiting dilutions and four CD8+ FLQ-specific CTL derived from donor 19 (Fig. 2b) were selected for further analysis. The PHA-blasts were pulsed with different concentrations of the FLQ peptide and used as a target of FLQ-specific CTL in cytotoxic assays. As shown in Fig. 2(c), half-maximal lysis was detected at high peptide concentrations (∼10−7 m), suggesting that, despite possessing the correct anchor residues for binding HLA-A2 molecules, FLQ 10-mer peptide may not represent the minimal epitope sequence. Two shorter peptides were therefore sysnthesized and tested for their ability to sensitize autologous PHA-blasts. As shown in Fig. 2(c), the 9-mer peptide EBNA1 565–573 (LQTHIFAEV) was able to sensitize target cells at 10−10 m concentrations, suggesting that it corresponds to the minimal epitope sequence. This was confirmed by the capacity of the LQT 9-mer to stimulate memory T-cell responses (data not shown).
Figure 2.
(a) FLQ-specific cytotoxic T lymphocyte (CTL) cultures obtained from three different donors. CTL cultures were obtained after three consecutive stimulations and were then tested in triplicate against untreated or FLQ-pulsed autologous phytohaemagglutinin (PHA) -blasts. Results are expressed as the per cent specific lysis obtained at the indicated effector : target ratio. One representative experiment out of three is shown. (b) FLQ-specific CTL clones, obtained from a polyclonal CTL culture from donor 19, were obtained by limiting dilution. Clones were tested against untreated or FLQ-pulsed autologous PHA-blasts. The cytotoxic activity of the indicated clones is expressed as the per cent specific lysis obtained at the indicated effector : target ratio. One representative experiment out of three is shown. (c) Peptide titration. FLQ-specific CTL clones were tested against autologous PHA-blasts after preincubation with the indicated concentration of synthetic peptides from the Epstein–Barr virus nuclear antigen 1 (EBNA1) region 565–574. The minimal epitope was defined as the 9-mer LQTHIFAEV (EBNA1 566–574). Results are expressed as the per cent specific lysis obtained at an effector : target ratio of 3 : 1. Means ± SD of three independent experiments performed in triplicate are reported.
LQT epitope presentation in EBNA1-expressing cells
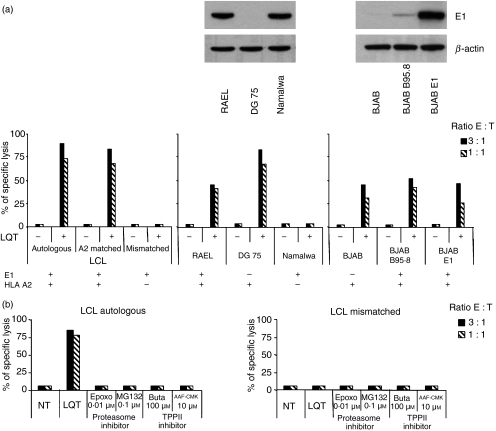
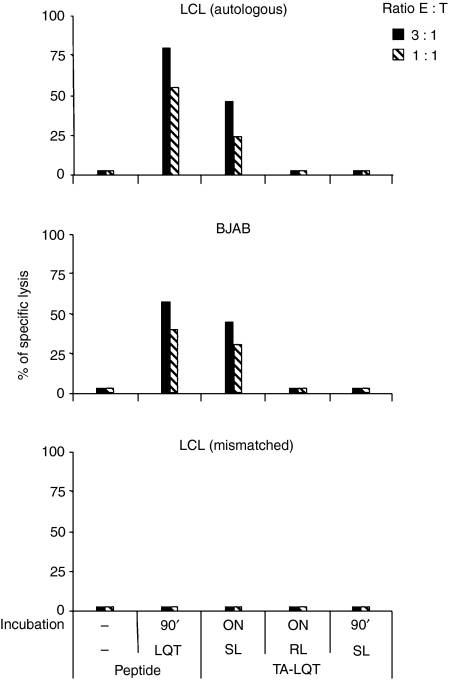
In the next set of experiments we assessed whether the endogenously produced LQT epitope is presented by HLA-A2-positive cells. To this end, the CTL clones were tested in cytotoxicity assays against a panel of LCL including the autologous LCL, and HLA-A2 single-matched or mismatched cell lines. As illustrated by the representative results obtained with clone E8, LQT-specific CTL did not lyse the autologous LCL nor allogeneic HLA-A2-matched LCL and lysis was observed only in the presence of exogenously added peptide (Fig. 3a, left panel). The failure to present the endogenous epitope to a level sufficent to trigger cytotoxic responses was further confirmed using a panel of Burkitt’s lymphoma cells, including RAEL (HLA-A2 and EBV-positive); DG75 (HLA-A2-positive and EBV-negative) and Namalwa (HLA-A2-negative and EBV-positive) (Fig. 3a middle panel) and EBV-converted or EBNA1-transfecetd sublines of the EBV-negative B-lymphoma BJAB that express different endogenous levels of EBNA1 (Fig. 3a right panel).
Figure 3.
(a) Presentation of the LQT epitope in Epstein–Barr virus (EBV) nuclear antigen 1 (EBNA1) -expressing cells. Left-hand chart: LQT-specific cytotoxic T-lymphocyte (CTL) clones were tested against autologous, human leucocyte antigen (HLA) -A2-positive single-matched and mismatched untreated or LQT-pulsed lymphocyte cell lines. Results are expressed as the per cent specific lysis obtained at an effector : target ratio of 3 : 1. One representative experiment out of three is shown. Central chart: LQT-specific CTL clones were tested against HLA-A2+, EBV+, HLA-A2+ EBV– and HLA-A2– EBV+ untreated or LQT-pulsed Burkitt’s lymphoma (BL) cell lines. Results are expressed as the per cent specific lysis obtained at an effector : target ratio of 3 : 1. One representative experiment out of three is shown. The upper panel depicts expression of the EBNA1 antigen, as detected by Western blot analysis. β-Actin was used as loading control. Right-hand chart: LQT-specific CTL clones were tested against a panel of BJAB cells including untreated or LQT-pulsed HLA-A2+ EBV– BJAB, HLA-A2+, EBV+ BJAB B95.8 and HLA-A2+ EBNA1+ BJAB E1 cells. Results are expressed as the per cent specific lysis obtained at an effector : target ratio of 3 : 1. One representative experiment out of three is shown. In the upper panel, expression of the EBNA1 antigen, as detected by Western blot analysis, is shown. β-Actin was used as loading control. (b) Effects of the proteasome and TPPII inhibitors on the LQT generation. CTL LQT clones were tested against a panel of lymphocyte cell lines (LCL) treated or not with specific proteasome and TPPII inhibitors. LCL HLA-A2 mismatched was used as a control. As a positive control the LQT peptide was added at the concentration of 10−5 m. One experiment out of three is shown. Inhibition of proteases was confirmed by in vitro assay (data not shown).
Excessive activity of the proteasome and other cytosolic peptidases such as TPPII may limit the generation of class I epitope.30–33 We therefore tested whether the presentation of LQT could be enhanced by inhibiting these but treatment with specific inhibitors did not affect the recognition of HLA-A2-positive EBNA1-expressing cells by LQT-specific CTL (Fig. 3b).
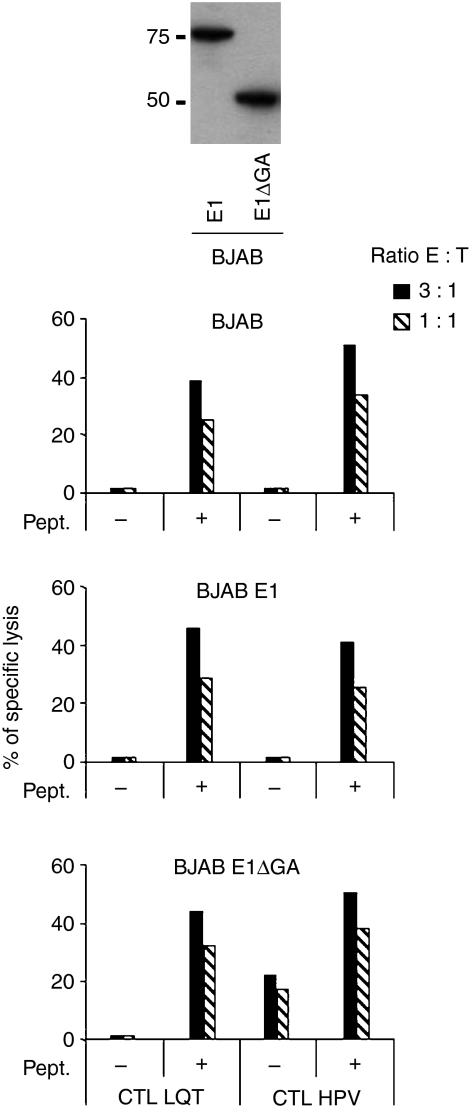
Evaluation of the effect of the Gly-Ala repeat domain on LQT-mediated lysis
The EBNA1 contains a GAr domain that inhibits proteasomal degradation.19 Removal of GAr increases proteasome-dependent processing, leading to better presentation of EBNA1-derived CTL epitopes.14,34 To assess whether the GAr is responsible for the poor presentation of the LQT epitope, stable transfected sublines of BJAB that express either EBNA1 or a GAr-deleted EBNA1 (E1ΔGA) were used as target cells in a standard 51Cr-release assay. As control, the presentation of the EBNA1 HPV epitope restricted through HLA-B35 was tested in parallel. BJAB-E1ΔGA was efficiently recognized by HPV-specific CTL, which confirmed the inhibitory effect of the GAr on antigen processing (Fig. 4). However, LQT-specific CTL were still unable to recognize the endogenously expressed epitope.
Figure 4.
Role of Gly-Ala repeat (GAr) on the presentation of the LQT epitope. LQT-specific cytotoxic T-lymphocyte (CTL) clones were tested against untreated or LQT-pulsed BJAB cells or BJAB cells stably expressing Epstein–Barr virus nuclear antigen 1 (EBNA1; E1) (central chart) or the GAr-deleted form (E1ΔGA) (lower chart). Results are expressed as the per cent specific lysis obtained at an effector : target ratio of 3 : 1. One representative experiment out of three is shown. In the upper panel, expression of EBNA1 (E1) and GAr-deleted EBNA1 (E1ΔGA) in transfected BJAB cells, as detected by Western blot analysis, is shown.
LQT-induced IFN-γ release
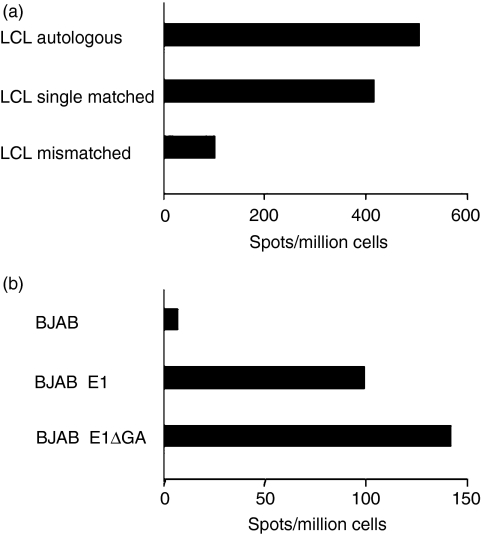
Interferon-γ production assays have been mainly used in studies documenting the presentation of EBNA1-derived MHC-I-presented epitopes.14–16 We tested therefore whether recognition of the LQT epitope could be revealed by monitoring IFN-γ release in ELISPOT. To this end, LQT-specific CTL and HLA-A2-matched LCL were seeded at an effector : target ratio of 3 : 1, and the number of LQT-specific IFN-γ-producing cells was evaluated after 24 hr. As shown in Fig. 5(a), the release of IFN-γ was specifically induced by the autologous and HLA-A2 single-matched LCL. To re-evaluate the effect of the GAr domain on the LQT presentation, the assay was repeated using BJAB, BJAB-E1 and BJAB-E1ΔGA as stimulators. As shown in Fig. 5(b), the LQT epitope was presented by both BJAB-E1 and BJAB-E1ΔGA, as assessed by IFN-γ release and presentation, was enhanced by deletion of the GAr.
Figure 5.
Presentation of the LQT epitope in Epstein–Barr virus nuclear antigen 1 (EBNA1) -expressing cells, as detected by enzyme-linked immunosorbent spot-forming cell assay (ELISPOT). (a) Autologous, HLA-A2+ single-matched and mismatched lymphocyte cell lines (LCL) were co-cultured for 24 hr with LQT-specific clones at an effector : target ratio of 3 : 1. Interferon-γ (IFN-γ) release was detected by ELISPOT. Results are expressed as net spot number/106 cells. One representative experiment out of three is shown. (b) BJAB and BJAB cells stably expressing EBNA1 (E1) or the Gly-Ala repeat (GAr) -deleted form (E1ΔGA) BJAB were cultured for 24 hr together with LQT-specific clones at an effector : target ratio of 3 : 1. IFN-γ release was detected by ELISPOT assay and results are expressed as net spot number/106 cells. One representative experiment out of three is shown.
Induction of LQT-specific lysis by a Trojan antigen construct
The presentation of MHC class I-restricted epitopes is influenced by the amount of peptide available for binding to the presenting allele as well as by the affinity of binding and stability of the complex. To assess whether the poor presentation of LQT may be ascribed to inefficient production or poor binding to the presenting allele, chimeric molecules, known as ‘Trojan antigens’ (TA),20,21,35 were designed to deliver the LQT epitope into target cells. The TA contains the LQT epitope at the N terminus and the membrane-translocating region of HIV-Tat protein at the C terminus. The two domains are linked by a furin-sensitive (SL) or furin-resistant (RL) linker.36 The membrane-translocating region carries the construct into the cytoplasm,37,38 where it is free to enter the ER, in a TAP-independent manner. Here the construct is trimmed by the furin protease, which releases the epitope that becomes available for association with the presenting class I allele. Delivery of the LQT peptide to the ER using this strategy resulted in efficient recognition of the autologous LCL and BJAB cells incubated overnight with the TA-LQT SL construct (Fig. 6) while mismatched cells and cells incubated with the construct containing the furin-resistant linker (TA-LQT RL) were not recognized. Lysis was not observed after incubation for 90 min, a time sufficient for binding of the LQT 9-mer but not for entry, confiming that presentation is dependent on ER processing of the precursor rather than on direct binding of the TA to surface HLA-A2 molecules.
Figure 6.
Presentation of the LQT epitope by Trojan antigen (TA) delivery. The LQT epitope was linked to the HIV Tat protein transduction domain (RKKRRQRRR) via a furin-sensitive (SL) or furin-resistant (RL) linker, and delivered to autologous lymphocyte cell lines (LCL), BJAB cells or mismatched LCL. Cells were incubated for the indicated time period (overnight or 90 min), and then used as a target of LQT-specific cytotoxic T-lymphocyte (CTL) clones in a standard chromium release assay. LQT-pulsed target cells were also used as control. Results are expressed as per cent specific lysis obtained at an effector : target ratio of 3 : 1. One representative experiment out of three is shown.
Discussion
As a result of its ubiquitous expression in EBV-associated malignancies, EBNA1 has long been considered an ideal target for immunotherapeutic approaches.39 The present study was aimed at exploring new potential epitopes presented by HLA-A2 and HLA-A24 molecules. An epitope prediction algorithm was used to identify four potential epitopes that were then tested for their capacity to reactivate T-cell responses in PBL from healthy EBV-positive donors. The LQT peptide corresponding to amino acids 566–574 of EBNA1 induced HLA-A2-restricted CTL responses in three out of 14 HLA-A2-positive donors tested. It should be noted that the minimal epitope was not the one predicted by the algorithm although the latter contains the correct anchor residues for binding to HLA-A2 molecules.40 This is in line with the observation that some CTL epitopes do not contain conventional anchor residues and binding to the presenting allele is instead dependent on secondary anchors. LQT-specific CTL responses were recently reported in one HLA-A206-positive donor.41 The three HLA-A2 donors included in our study were found to be HLA-A201-positive (data not shown), which suggests that the LQT epitope is likely to be presented by different HLA-A2 subtypes.
The reactivation of LQT-specific CTL in only three out of 14 of the donors tested suggests that this epitope may only account for a minor component of the EBV-specific CTL response. Indeed, in contrast to the low frequency of EBNA1-specific CTL responses, the majority of the subjects analysed in this study responded to one or more of the epitopes derived from other latent antigens. Responses to HLA-A2-restricted epitopes derived from EBNA3A (SVR) and/or LMP2 (CLG and LLW) were detected in virtually all donors and approximately half of the donors tested responded to EBNA1 epitopes restricted thorugh different class I alleles suggesting that LQT is a particularly weak epitope.
We have deliberately chosen to monitor T-cell responses based on the induction of cytolytic activity. The in vitro stimulation protocol used to reactivate memory T cells is critical for the functional outcome and in some cases results in the activation of low avidity T cells that lyse only peptide-pulsed target cells. In the case of EBNA1-specific T-cell responses, failure to lyse EBNA1-expressing target cells was frequently observed,13,34 although low levels of lysis were reported in some studies.15,16 Specific recognition of EBNA1-derived epitopes was in many cases revealed by the induction of IFN-γ release, suggesting that triggering of cytokine production may require the engagement of lower numbers of MHC–peptide complexes. This possibility is supported by the observation that removal of the GAr domain, which was shown to hamper the processing of EBNA1 by the proteasome, enhances the presentation of the LQT and HPV epitopes and, in the case of HPV, results in detectable cytotoxicity against EBNA1-expressing cells. It is noteworthy, however, that IFN-γ release may be sufficient to control EBV-induced B-cell growth even in the absence of lysis.14,42
The possibility that inefficient processing may be the main cause for the poor immunogenicity of the LQT epitope is further substantiated by the observation that delivery of the epitope to the ER results in efficient presentation that can be detected by both cytokine production and direct cytotoxicity of the target cells. Poor delivery to the ER may be the result of inefficient production of the epitope, excessive trimming of the precursor peptide by cytosolic or ER peptidases or inefficient transport as the result of poor binding to TAP. Inefficient transport seems unlikely because LQT matches the characteristics of TAP substrates43,44 and we have been unable to enhance presentation by inhibiting cytosolic peptidases such as TPPII that were shown to destroy MHC class I epitopes.
It was recently reported that EBNA1-specific CTL responses are significantly less frequent in subjects with EBV-associated nasopharyngeal carcinoma than in healthy controls, suggesting that defects in the EBNA1-specific CTL response may contribute to nasopharyngeal carcinoma pathogenesis.41 The results presented in our study suggest that strategies for enhancing the presention of weak epitopes such as LQT, for example by proteasome modulation or targeted delivery, may find useful applications in the development of immunotherapy for EBNA1-positive malignancies.
Acknowledgments
This work was supported by grants from the Associazione Italiana per la Ricerca sul Cancro (AIRC) and Fondazione Cassa di Risparmio di Ferrara, the Swedish Cancer Society, the Swedish Medical Research Council and Karolinska Institutet, Stockholm, Sweden and by the European Community Integrated Project on Infection and Cancer, INCA, Project no. LSHC-CT-2005-018704. We are grateful to A. Forster for editorial assistance, to E. Gallerani and S. Cellini for technical assistance and to Dr Alessandra Balboni for HLA typing.
References
- 1.Schaible UE, Collins HL, Kaufmann SH. Confrontation between intracellular bacteria and the immune system. Adv Immunol. 1999;71:267–377. doi: 10.1016/s0065-2776(08)60405-8. [DOI] [PubMed] [Google Scholar]
- 2.Zinkernagel RM. Immunology taught by viruses. Science. 1996;271:173–8. doi: 10.1126/science.271.5246.173. [DOI] [PubMed] [Google Scholar]
- 3.Pamer E, Cresswell P. Mechanisms of MHC class I-restricted antigen processing. Annu Rev Immunol. 1998;16:323–58. doi: 10.1146/annurev.immunol.16.1.323. [DOI] [PubMed] [Google Scholar]
- 4.Rock KL, Goldberg AL. Degradation of cell proteins and the generation of MHC class I-presented peptides. Annu Rev Immunol. 1999;17:739–79. doi: 10.1146/annurev.immunol.17.1.739. [DOI] [PubMed] [Google Scholar]
- 5.Rock KL, York IA, Goldberg AL. Post-proteasomal antigen processing for major histocompatibility complex class I presentation. Nat Immunol. 2004;5:670–7. doi: 10.1038/ni1089. [DOI] [PubMed] [Google Scholar]
- 6.Masucci MG. Epstein–Barr virus oncogenesis and the ubiquitin–proteasome system. Oncogene. 2004;23:2107–15. doi: 10.1038/sj.onc.1207372. [DOI] [PubMed] [Google Scholar]
- 7.Kieff E, Leibowitz D. Epstein–Barr virus and its replication. In: Fields BM, Knipe DN, editors. Virology. 2nd Edn. New York: Raven press; 1990. pp. 1889–920. Vol. 2. [Google Scholar]
- 8.Gavioli R, Kurilla MG, de Campos-Lima PO, Wallace LE, Dolcetti R, Murray RJ, Rickinson AB, Masucci MG. Multiple HLA A11-restricted cytotoxic T-lymphocyte epitopes of different immunogenicities in the Epstein–Barr virus-encoded nuclear antigen 4. J Virol. 1993;67:1572–8. doi: 10.1128/jvi.67.3.1572-1578.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khanna R, Burrows SR, Kurilla MG, Jacob CA, Misko IS, Sculley TB, Kieff E, Moss DJ. Localization of Epstein–Barr virus cytotoxic T cell epitopes using recombinant vaccinia: implications for vaccine development. J Exp Med. 1992;176:169–76. doi: 10.1084/jem.176.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murray RJ, Kurilla MG, Brooks JM, Thomas WA, Rowe M, Kieff E, Rickinson AB. Identification of target antigens for the human cytotoxic T cell response to Epstein–Barr virus (EBV): implications for the immune control of EBV-positive malignancies. J Exp Med. 1992;176:157–68. doi: 10.1084/jem.176.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rickinson AB, Moss DJ. Human cytotoxic T lymphocyte responses to Epstein–Barr virus infection. Annu Rev Immunol. 1997;15:405–31. doi: 10.1146/annurev.immunol.15.1.405. [DOI] [PubMed] [Google Scholar]
- 12.Steven NM, Leese AM, Annels NE, Lee SP, Rickinson AB. Epitope focusing in the primary cytotoxic T cell response to Epstein–Barr virus and its relationship to T cell memory. J Exp Med. 1996;184:1801–13. doi: 10.1084/jem.184.5.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blake N, Lee S, Redchenko I, et al. Human CD8+ T cell responses to EBV EBNA1: HLA class I presentation of the (Gly-Ala)-containing protein requires exogenous processing. Immunity. 1997;7:791–802. doi: 10.1016/s1074-7613(00)80397-0. [DOI] [PubMed] [Google Scholar]
- 14.Lee SP, Brooks JM, Al-Jarrah H, et al. CD8 T cell recognition of endogenously expressed Epstein–Barr virus nuclear antigen 1. J Exp Med. 2004;199:1409–20. doi: 10.1084/jem.20040121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tellam J, Connolly G, Green KJ, Miles JJ, Moss DJ, Burrows SR, Khanna R. Endogenous presentation of CD8+ T cell epitopes from Epstein–Barr virus-encoded nuclear antigen 1. J Exp Med. 2004;199:1421–31. doi: 10.1084/jem.20040191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Voo K, Fu T, Wang H, Tellam J, Heslop H, Brenner M, Rooney C, Wang R. Evidence for the presentation of major histocompatibility complex class I-restricted Epstein–Barr virus nuclear antigen 1 peptides to CD8+ T lymphocytes. J Exp Med. 2004;199:459–70. doi: 10.1084/jem.20031219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yin Y, Manoury B, Fahraeus R. Self-inhibition of synthesis and antigen presentation by Epstein–Barr virus-encoded EBNA1. Science. 2003;301:1371–4. doi: 10.1126/science.1088902. [DOI] [PubMed] [Google Scholar]
- 18.Levitskaya J, Coram M, Levitsky V, Imreh S, Steigerwald-Mullen PM, Klein G, Kurilla MG, Masucci MG. Inhibition of antigen processing by the internal repeat region of the Epstein–Barr virus nuclear antigen-1. Nature (London) 1995;375:685–8. doi: 10.1038/375685a0. [DOI] [PubMed] [Google Scholar]
- 19.Levitskaya J, Sharipo A, Leonchiks A, Ciechanover A, Masucci MG. Inhibition of ubiquitin/proteasome-dependent protein degradation by the Gly-Ala repeat domain of the Epstein–Barr virus nuclear antigen 1. Proc Natl Acad Sci U S A. 1997;94:12616–21. doi: 10.1073/pnas.94.23.12616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu J, Higashimoto Y, Appella E, Celis E. Multiepitope Trojan antigen peptide vaccines for the induction of antitumor CTL and Th immune responses. J Immunol. 2004;172:4575–82. doi: 10.4049/jimmunol.172.7.4575. [DOI] [PubMed] [Google Scholar]
- 21.Lu J, Wettstein PJ, Higashimoto Y, Appella E, Celis E. TAP-independent presentation of CTL epitopes by Trojan antigens. J Immunol. 2001;166:7063–71. doi: 10.4049/jimmunol.166.12.7063. [DOI] [PubMed] [Google Scholar]
- 22.Gavioli R, Guerrini R, Masucci MG, Tomatis R, Traniello S, Marastoni M. High structural side chain specificity required at the second position of immunogenic peptides to obtain stable MHC/peptide complexes. FEBS Lett. 1998;421:95–9. doi: 10.1016/s0014-5793(97)01540-8. [DOI] [PubMed] [Google Scholar]
- 23.Reali E, Guerrini R, Marastoni M, Tomatis R, Masucci MG, Traniello S, Gavioli R. A single specific amino acid residue in peptide antigens is sufficient to activate memory cytotoxic T lymphocytes: potential role of cross-reactive peptides in memory T cell maintenance. J Immunol. 1999;162:106–13. [PubMed] [Google Scholar]
- 24.Micheletti F, Bazzaro M, Canella A, Marastoni M, Traniello S, Gavioli R. The lifespan of major histocompatibility complex class I/peptide complexes determines the efficiency of cytotoxic T-lymphocyte responses. Immunology. 1999;96:411–5. doi: 10.1046/j.1365-2567.1999.00707.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Torsteinsdottir S, Masucci MG, Ehlin-Henriksson B, Brautbar C, Ben Bassat H, Klein G, Klein E. Differentiation dependent sensitivity of human B-cell derived lines to major histocompatibility complex-restricted T-cell cytotoxicity. Proc Natl Acad Sci U S A. 1986;83:5620–4. doi: 10.1073/pnas.83.15.5620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Micheletti F, Guerrini R, Formentin A, et al. Selective amino acid substitutions of a subdominant Epstein–Barr virus LMP2-derived epitope increase HLA/peptide complex stability and immunogenicity: implications for immunotherapy of Epstein–Barr virus-associated malignancies. Eur J Immunol. 1999;29:2579–89. doi: 10.1002/(SICI)1521-4141(199908)29:08<2579::AID-IMMU2579>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 27.Parker KC, Bednarek MA, Coligan JE. Scheme for ranking potential HLA-A2 binding peptides based on independent binding of individual peptide side-chains. J Immunol. 1994;152:163–75. [PubMed] [Google Scholar]
- 28.Neefjes JJ, Dierx J, Ploegh HL. The effect of anchor residue modifications on the stability of major histocompatibility complex class I–peptide interactions. Eur J Immunol. 1993;23:840–5. doi: 10.1002/eji.1830230411. [DOI] [PubMed] [Google Scholar]
- 29.Chen W, Khilko S, Fecondo J, Margulies DH, McCluskey J. Determinant selection of major histocompatibility complex class I-restricted antigenic peptides is explained by class I-peptide affinity and is strongly influenced by nondominant anchor residues. J Exp Med. 1994;180:1471–83. doi: 10.1084/jem.180.4.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luckey CJ, King GM, Marto JA, et al. Proteasomes can either generate or destroy MHC class I epitopes: evidence for nonproteasomal epitope generation in the cytosol. J Immunol. 1998;161:112–21. [PubMed] [Google Scholar]
- 31.Schwarz K, de Giuli R, Schmidtke G, et al. The selective proteasome inhibitors lactacystin and epoxomicin can be used to either up- or down-regulate antigen presentation at nontoxic doses. J Immunol. 2000;164:6147–57. doi: 10.4049/jimmunol.164.12.6147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Basler M, Groettrup M. No essential role for tripeptidyl peptidase II for the processing of LCMV-derived T cell epitopes. Eur J Immunol. 2007;37:896–904. doi: 10.1002/eji.200636372. [DOI] [PubMed] [Google Scholar]
- 33.Preta G, Marescotti D, Fortini C, Carcoforo P, Castelli C, Masucci M, Gavioli R. Inhibition of serine-peptidase activity enhances the generation of a survivin-derived HLA-A2-presented CTL epitope in colon-carcinoma cells. Scand J Immunol. 2008;68:579–88. doi: 10.1111/j.1365-3083.2008.02175.x. [DOI] [PubMed] [Google Scholar]
- 34.Blake N, Haigh T, Shaka’a G, Croom-Carter D, Rickinson A. The importance of exogenous antigen in priming the human CD8+ T cell response: lessons from the EBV nuclear antigen EBNA1. J Immunol. 2000;165:7078–87. doi: 10.4049/jimmunol.165.12.7078. [DOI] [PubMed] [Google Scholar]
- 35.Kim DT, Mitchell DJ, Brockstedt DG, Fong L, Nolan GP, Fathman CG, Engleman EG, Rothbard JB. Introduction of soluble proteins into the MHC class I pathway by conjugation to an HIV tat peptide. J Immunol. 1997;159:1666–8. [PubMed] [Google Scholar]
- 36.Nakayama K. Furin: a mammalian subtilisin/Kex2p-like endoprotease involved in processing of a wide variety of precursor proteins. Biochem J. 1997;327:625–35. doi: 10.1042/bj3270625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Derossi D, Chassaing G, Prochiantz A. Trojan peptides: the penetratin system for intracellular delivery. Trends Cell Biol. 1998;8:84–7. [PubMed] [Google Scholar]
- 38.Fawell S, Seery J, Daikh Y, Moore C, Chen LL, Pepinsky B, Barsoum J. Tat-mediated delivery of heterologous proteins into cells. Proc Natl Acad Sci U S A. 1994;91:664–8. doi: 10.1073/pnas.91.2.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Khanna R, Moss DJ, Burrows SR. Vaccine strategies against Epstein–Barr virus-associated diseases: lessons from studies on cytotoxic T-cell-mediated immune regulation. Immunol Rev. 1999;170:49–64. doi: 10.1111/j.1600-065x.1999.tb01328.x. [DOI] [PubMed] [Google Scholar]
- 40.Rammensee HG, Friede T, Stevanoviic S. MHC ligands and peptide motifs: first listing. Immunogenetics. 1995;41:178–228. doi: 10.1007/BF00172063. [DOI] [PubMed] [Google Scholar]
- 41.Fogg MH, Wirth LJ, Posner M, Wang F. Decreased EBNA-1-specific CD8+ T cells in patients with Epstein–Barr virus-associated nasopharyngeal carcinoma. Proc Natl Acad Sci U S A. 2009;106:3318–23. doi: 10.1073/pnas.0813320106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shi Y, Lutz CT. Interferon-gamma control of EBV-transformed B cells: a role for CD8+ T cells that poorly kill EBV-infected cells. Viral Immunol. 2002;15:213–25. doi: 10.1089/088282402317340350. [DOI] [PubMed] [Google Scholar]
- 43.Doytchinova I, Hemsley S, Flower DR. Transporter associated with antigen processing preselection of peptides binding to the MHC: a bioinformatic evaluation. J Immunol. 2004;173:6813–9. doi: 10.4049/jimmunol.173.11.6813. [DOI] [PubMed] [Google Scholar]
- 44.van Endert PM, Riganelli D, Greco G, Fleischhauer K, Sidney J, Sette A, Bach JF. The peptide-binding motif for the human transporter associated with antigen processing. J Exp Med. 1995;182:1883–95. doi: 10.1084/jem.182.6.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]